Abstract
Health care-associated methicillin-resistant Staphylococcus aureus (HA-MRSA) forms biofilm in vitro that is dependent on the surface-located fibronectin binding proteins A and B (FnBPA, FnBPB). Here we provide new insights into the requirements for FnBP-dependent biofilm formation by MRSA. We show that expression of FnBPs is sustained at high levels throughout the growth cycle in the HA-MRSA strain BH1CC in contrast to laboratory strain SH1000, where expression could be detected only in exponential phase. We found that FnBP-mediated biofilm accumulation required Zn2+, while the removal of Zn2+ had no effect on the ability of FnBPA to mediate bacterial adherence to fibrinogen. We also investigated the role of FnBPA expressed on the surface of S. aureus in promoting biofilm formation and bacterial adhesion to fibrinogen. The minimum part of FnBPA required for ligand binding has so far been defined only with recombinant proteins. Here we found that the N1 subdomain was not required for biofilm formation or for FnBPA to promote bacterial adherence to fibrinogen. Residues at the C terminus of subdomain N3 required for FnBPA to bind to ligands using the “dock, lock, and latch” mechanism were necessary for FnBPA to promote bacterial adherence to fibrinogen. However, these residues were not necessary to form biofilm, allowing us to localize the region of FnBPA required for biofilm accumulation to residues 166 to 498. Thus, FnBPA mediates biofilm formation and bacterial adhesion to fibrinogen using two distinct mechanisms. Finally, we identified a hitherto-unrecognized thrombin cleavage site close to the boundary between subdomains N1 and N2 of FnBPA.
INTRODUCTION
Staphylococcus aureus is a commensal bacterium that is carried persistently in the anterior nares of about 20% of the human population. The organism can cause superficial skin lesions or more-serious, life-threatening invasive infections such as endocarditis, osteomyelitis, and septic arthritis (1). S. aureus is also a major cause of infections associated with indwelling medical devices such as central venous catheters, cardiovascular devices, and artificial joints (2, 3). The ability to form biofilm is crucial to success in device-related infection. Bacteria in the biofilm matrix are in a semidormant state, are impervious to host neutrophils and macrophages, and are difficult to inhibit with antibiotics (4–6). The involvement of methicillin-resistant S. aureus (MRSA) further complicates treatment of biofilm-associated infection.
Attachment to the surfaces of biomedical devices is the first stage of biofilm formation. The major autolysin Atl mediates primary attachment of S. aureus to hydrophobic plastic surfaces (7). The primary attachment stage of MRSA biofilm requires Atl autolytic activity to release DNA from some cells (8). Adhesion to surfaces that have been conditioned by host plasma proteins is promoted by S. aureus microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) (9). The accumulation phase of biofilm formation can be mediated by the icaADBC-encoded polysaccharide intercellular adhesin (PIA) (10). Alternatively, S. aureus surface proteins such as Bap, SasG, SasC, protein A, and fibronectin binding proteins (FnBPs) can promote biofilm accumulation in an ica-independent manner (11–15).
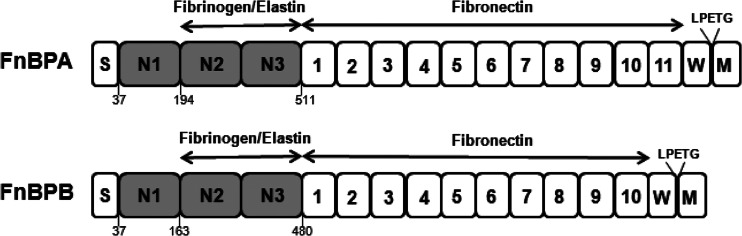
Fibronectin binding proteins A and B (FnBPA and FnBPB) (Fig. 1) promote biofilm accumulation in certain health care-associated MRSA (HA-MRSA) strains (15, 16). The FnBPs are closely related cell wall-associated proteins which promote bacterial attachment to fibrinogen, elastin, and fibronectin. The C-terminal domains of FnBPs comprise a sorting signal (LPXTG), a hydrophobic membrane-spanning domain, and positively charged residues that are required for covalent attachment of the proteins to cell wall peptidoglycan by sortase A (17, 18). A signal sequence is removed from the N terminus during secretion. The N termini of the mature proteins (A domains) comprise related amino acid sequences and are predicted to fold into three subdomains, N1, N2, and N3, similar to clumping factor A (ClfA) (19). The N2 and N3 subdomains of FnBPA and FnBPB have undergone considerable amino acid sequence divergence. At least seven different isotypes of FnBPA and FnBPB exist, and all bind to ligands with similar affinities despite considerable antigenic differences (20, 21).
Fig 1.
Schematic representation of FnBPA and FnBPB. The position of the signal sequence (S), the fibrinogen and elastin binding A domain (N1, N2, N3), and the fibronectin binding motifs (numbered) are indicated. The wall/membrane-spanning region (W and M) and LPETG motif at the C terminus are required for covalent attachment to cell wall peptidoglycan.
A hydrophobic trench located between the N2 and N3 subdomains forms the fibrinogen- and elastin-binding site (22). FnBPs recognize the same site in fibrinogen as ClfA, the extreme C terminus of the γ-chain. Ligand binding occurs through the dynamic “dock, lock, and latch” mechanism, which has been demonstrated for purified recombinant ClfA binding to fibrinogen. Docking of the peptide in the hydrophobic trench induces a redirection of the flexible C-terminal extension of subdomain N3, resulting in it covering the trench and locking the peptide in place. The remainder of the C-terminal extension interacts with subdomain N2 by β-strand complementation (latching) (23, 24). Amino acid substitutions in the recombinant FnBPA and FnBPB ligand binding trench or deletion of residues comprising the lock and latch (C-terminal extension) resulted in a reduced ability to bind to fibrinogen and elastin, suggesting that FnBPs bind to their ligands using the dock, lock, and latch mechanism (22, 25). C terminal to the A domain is the fibronectin binding region comprising 11 (FnBPA) or 10 (FnBPB) tandemly repeated motifs. The binding sites for FnBPs are the type I modules at the N terminus of fibronectin, and binding occurs by a tandem β-zipper interaction mechanism (26).
HA-MRSA strains from clonal complex 8 (CC8) and CC22 have been reported to form biofilms in vitro that require expression of FnBPs. FnBP-mediated biofilm was dependent on mildly acidic growth conditions (pH 5.5) triggered by growth in a glucose-supplemented medium (15). Biofilm accumulation occurs independently of icaADBC expression, and FnBPs are required (15, 16). Studies with HA-MRSA strain BH1CC have revealed that expression of either fnbA or fnbB from a complementing plasmid was sufficient to restore biofilm formation in a double fnbA fnbB mutant (15). The region required for biofilm formation was localized to the A domain of FnBPA, with the fibronectin binding repeats playing no role (15). Asparagine 304, a residue located in the ligand binding trench of FnBPA and crucial for fibrinogen and elastin binding, was not involved in biofilm formation (15). FnBPs enhance colonization of catheters in mouse models of MRSA foreign body infection, while absence of the ica operon has no effect, highlighting the in vivo importance of FnBP-mediated biofilm formation (16).
This study aimed to increase our understanding of the mechanism by which FnBPs promote biofilm formation and bacterial adhesion to fibrinogen. The minimum region of the A domain of FnBPA required for ligand binding has so far only been defined with recombinant proteins. Here we investigated the role of the A domain of FnBPA expressed on the surface of S. aureus in promoting bacterial adhesion to immobilized fibrinogen and biofilm formation.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Escherichia coli was grown in Luria broth at 37°C. S. aureus was grown in tryptic soy broth (TSB) or brain heart infusion (BHI; Oxoid) broth at 37°C. Media were supplemented with glucose (1%, wt/vol), ampicillin (100 μg/ml), chloramphenicol (10 μg/ml), tetracycline (125 ng/ml), thrombin (1 U/ml), or hirudin (50 μg/ml) where appropriate. Stationary-phase cultures were grown for approximately 16 h. For exponential phase, bacteria were diluted 1:100, washed in BHI broth, and grown to an optical density at 600 nm (OD600) of 0.4. Strains used in this study are listed in Table 1.
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Characteristics | Reference or source |
|---|---|---|
| S. aureus strains | ||
| BH1CC | MRSA, SCCmec type II, MLSTa type 8, clonal complex 8 | 27 |
| BH1CC atl | atl::Cmr | 8 |
| BH1CC clfA | clfA::Tetr | 15 |
| BH1CC ΔfnbAfnbB | Deletion of both fnbA and fnbB | This study |
| SH1000 | Derivative of laboratory strain 8325-4 with repaired defect in rsbU | 28 |
| SH1000 clfA clfB fnbA fnbB | clfA clfB::Emr fnbA::Emr fnbB::Tetr | 15 |
| E. coli strains | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac | Stratagene |
| DC10B | dam+ Δdcm ΔhsdRMS endA1 recA1 | 29 |
| Plasmids | ||
| pFnBA4 | Multicopy plasmid expressing entire fnbA gene from strain 8325; Cmr | 30 |
| pFnBA4ΔLL499–511 | Plasmid expressing the fnbA gene lacking DNA encoding the C-terminal extension of the N3 subdomain (residues 499–511); Cmr | This study |
| pFnBA4ΔN138–193 | Plasmid expressing the fnbA gene lacking DNA encoding the entire N1 subdomain (residues 38–193); Cmr | This study |
| pFnBA4ΔN138–167 | Plasmid expressing the fnbA gene lacking DNA encoding N1 subdomain residues 38–167; Cmr | This study |
| pFnBB4 | Plasmid expressing entire fnbB gene; Cmr | 30 |
| pIMAY | Temperature-sensitive vector for allelic exchange in staphylococci; Cmr | 29 |
| pIMAYΔfnbAfnbB | Plasmid pIMAY carrying 600 bp of sequence upstream of fnbA and 600 bp of sequence downstream of fnbB; Cmr | This study |
| pQE30::FnBPA37–511 | Plasmid pQE30 containing codons for amino acids 37–511 of FnBPA from S. aureus 8325; Ampr | 22 |
MLST, multilocus sequence type.
Plasmid and strain construction.
Plasmids used in this study are listed in Table 1. Deletion of fnbA and fnbB to generate strain BH1CC ΔfnbAfnbB was achieved by allelic replacement using pIMAY as previously described (29). DNA fragments consisting of 600 nucleotides upstream of fnbA and 600 nucleotides downstream of fnbB were amplified by PCR using genomic DNA isolated from strain Newman as the template. The fragments were joined by PCR and cloned between EcoRI and SacI sites of pIMAY. Plasmid pIMAYΔfnbAfnbB was isolated from E. coli DC10B and transformed into electrocompetent BH1CC, and deletion of fnbA and fnbB genes was achieved by allelic replacement as previously described (29). Deletion of the fnbA and fnbB genes was confirmed by PCR. To generate plasmid pFnBA4ΔLL499–511, inverse PCR was carried out using pFnBA4 (30) as the template and 5′-phosphorylated primers 5′-AAAAATGGTCCGATTATTC-3′ and 5′-TAAACCATTATCCCAAGTTAAG-3′. Plasmid pFnBA4 was used as the template for inverse PCR to generate pFnBA4ΔN138–167 using 5′-phosphorylated primers 5′-TCAGAAAGTAAGCCACGTGTG-3′ and 5′-TGCTGCAGCTTCTTTGTC-3′, and primers 5′-GGTACAGATGTAACAAGTAAAG-3′ and 5′-TGCTGCAGCTTCTTTGTC-3′ were used to generate pFnBA4ΔN138–193. PCR products were digested with DpnI to eliminate parental DNA. Following blunt-end ligation, plasmids were transformed into electrocompetent BH1CC ΔfnbAfnbB.
Recombinant proteins and thrombin cleavage.
DNA encoding FnBPA comprising subdomains N1, N2, and N3 (FnBPA N1N2N3) was previously cloned into pQE30 (22). Recombinant FnBPA N1N2N3 (rFnBPA37-511) was expressed with an N-terminal hexahistidine tag and purified from E. coli Topp3 by Ni2+ affinity chromatography as previously described (12). Recombinant FnBPA37-511 (500 μg/ml) was incubated with bovine thrombin (10 U/ml; GE Healthcare) or thrombin and hirudin (50 μg/ml; Refludan; Pharmion) for 18 h at room temperature. Samples were analyzed by SDS-PAGE. For N-terminal sequencing, samples were transferred to polyvinylidene difluoride (PVDF), and sequencing was carried out by Alta Biosciences, Birmingham, United Kingdom.
Biofilm and accumulation assays.
S. aureus was grown for 16 h in TSB and diluted 1:200 in BHI broth or BHI broth supplemented with glucose (1%, wt/vol). For accumulation assays, wells of sterile microtiter plates (Sarstedt) were coated with a solution of fibrinogen (18 ng/ml) at 4°C for 18 h and washed with sterile phosphate-buffered saline (PBS). This low concentration of fibrinogen would not allow bacterial adhesion to be measured by crystal violet staining. Diluted bacteria (200 μl) were added to fibrinogen-coated wells or, alternatively, to wells of sterile tissue culture-treated polystyrene plates (Nunclon Δ). Plates were incubated statically at 37°C for 24 h. Wells were washed three times with PBS and dried by inversion for 30 min. Adherent cells were stained with 0.5% (wt/vol) crystal violet, and the A570 was measured. Where appropriate, diethylenetriaminepentaacetic acid (DTPA), ZnCl2 (100 μM), HCl, thrombin (1 U/ml), hirudin (50 μg/ml), fibrinogen γ-chain peptide (1 mM), or tetracycline (125 ng/ml) was added to inoculated wells at the beginning of a biofilm assay, and wells were incubated and treated as described above. The γ-chain peptide comprises the 17 C-terminal residues of the γ-chain of human fibrinogen (GEGQQHHLGGAKQAGDV) and was synthesized by Genscript (Piscataway, NJ).
Ligand affinity blot analysis.
To extract cell wall-associated proteins, cultures of S. aureus were harvested, washed in PBS, and resuspended to OD600 of 40 in lysis buffer (50 mM Tris-HCl, 20 mM MgCl2, pH 7.5) supplemented with 30% (wt/vol) raffinose and complete protease inhibitors (40 μl/ml; Roche). Cell wall proteins were solubilized by incubation with lysostaphin (200 μg/ml; AMBI, Lawrence, NY) for 10 min at 37°C. Protoplasts were removed by centrifugation at 12,000 × g for 10 min, and the supernatant containing solubilized cell wall proteins was aspirated and boiled for 5 min in Laemmli sample buffer (Sigma).
Proteins were separated on 7.5% or 12% (wt/vol) polyacrylamide gels, transferred onto PVDF membranes (Roche), and blocked in 10% (wt/vol) skimmed-milk proteins. Human fibronectin (0.5 mg/ml in PBS) was incubated with biotin (2 mg/ml) for 20 min at room temperature. The reaction was stopped by addition of 10 mM NH4Cl. Excess biotin was removed by dialysis against PBS overnight at 4°C. Blots were probed with biotinylated fibronectin and peroxidase-conjugated streptavidin (1:5,000 dilution; Roche). Reactive bands were visualized using the LumiGLO reagent and a peroxide detection system (Cell Signaling Technology).
Bacterial adherence to fibrinogen and fibronectin.
Microtiter plates (Sarstedt) were coated with a solution of fibrinogen (2.5 μg/ml; Calbiochem) or fibronectin (2 μg/ml; Calbiochem) in PBS overnight 4°C. Wells were blocked with 5% (wt/vol) bovine serum albumin (BSA) for 2 h at 37°C. Washed bacteria were adjusted to an OD600 of 1.0 in PBS, and 100 μl was added to each well and incubated for 1.5 h at 37°C. Wells were washed with PBS, adherent cells were fixed with formaldehyde (25% vol/vol) and stained with crystal violet, and the A570 was measured.
To examine the effect of DTPA on the ability of bacteria to adhere to fibrinogen in a solid-phase binding assay, bacteria were washed and resuspended in a solution of PBS, PBS containing ZnCl2 (4.8 μM), or PBS containing ZnCl2 (4.8 μM) and DTPA (200 μM) before being added to fibrinogen-coated wells.
RESULTS
Expression of fibronectin binding proteins by S. aureus BH1CC.
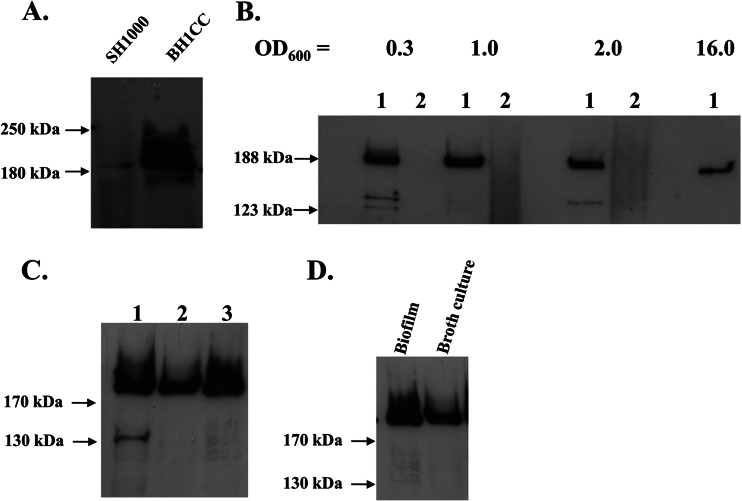
Studies with laboratory strains of S. aureus have shown that fnbA (and fnbB) is predominately expressed in the exponential phase of growth. A combination of reduced transcription in post-exponential phase (due to negative regulation by Agr) and degradation by V8 protease leads to loss of fibronectin binding in the stationary phase of growth (31, 32). Expression of FnBPs by BH1CC was examined. The sequences of the fnbA and fnbB genes of HA-MRSA strain BH1CC (CC8) were determined by DNA sequencing of PCR products amplified from genomic DNA. BH1CC expresses fibronectin binding proteins with A domain amino acid sequences identical to those of FnBPA and FnBPB from strain 8325 (isotype I) (20, 21). Ligand affinity blotting analysis of proteins solubilized from the cell wall and probed with biotinylated fibronectin revealed that FnBPs were expressed at much higher levels by BH1CC than by laboratory strain SH1000 in the exponential phase of growth (Fig. 2A). FnBPA and FnBPB have similar molecular weights and comigrate on a polyacrylamide gel. FnBP expression by BH1CC could also be detected in the late exponential and stationary phases of growth (OD600 = 1, 2, and 16) (Fig. 2B), demonstrating that intact FnBPs are present on the S. aureus BH1CC cell surface at high levels throughout growth. SH1000 expressed FnBPs detectably only in the early exponential phase of growth.
Fig 2.
Expression of FnBPA and FnBPB by BH1CC. Cell wall extracts were separated on 7.5% acrylamide gels, blotted onto PVDF membranes, and probed with biotin-labeled fibronectin. Size markers are indicated. (A) Cell wall extracts from SH1000 and BH1CC grown to exponential phase. (B) Cell wall extracts from BH1CC (lanes 1) and BH1CC ΔfnbAfnbB (lanes 2) taken at different stages of the growth phase (OD600s of 0.3, 1.0, 2.0, and 16.0). (C) Cell wall extracts from BH1CC (lane 1), BH1CC ΔfnbAfnbB(pFnBA4) (lane 2), and BH1CC ΔfnbAfnbB(pFnBB4) (lane 3) grown to stationary phase. (D) Cell wall extracts from BH1CC grown as a biofilm or in broth culture to stationary phase.
An fnbA fnbB deletion mutant of BH1CC was constructed by allelic exchange. The absence of FnBPs was confirmed by fibronectin affinity blotting analysis of proteins solubilized from the cell wall (Fig. 2B). Complementation of BH1CC ΔfnbAfnbB with plasmids pFnBA4 and pFnBB4 restored expression of FnBPA and FnBPB, respectively (Fig. 2C). BH1CC grown to stationary phase bound to fibronectin, but BH1CC ΔfnbAfnbB and stationary-phase SH1000 did not (data not shown). These data indicate that FnBPs are present at high levels on the BH1CC cell surface at all stages of growth. Cell wall extracts from BH1CC cells resuspended from biofilm growth were also examined to ensure that FnBPs were expressed under the conditions encountered during biofilm formation. Densitometric analysis of band intensity indicated that FnBPs were expressed at slightly higher levels by bacteria extracted from a biofilm (1.1-fold ± 0.4-fold) than by bacteria grown in broth culture to stationary phase (Fig. 2D).
The effect of zinc chelation on FnBP-mediated biofilm formation.
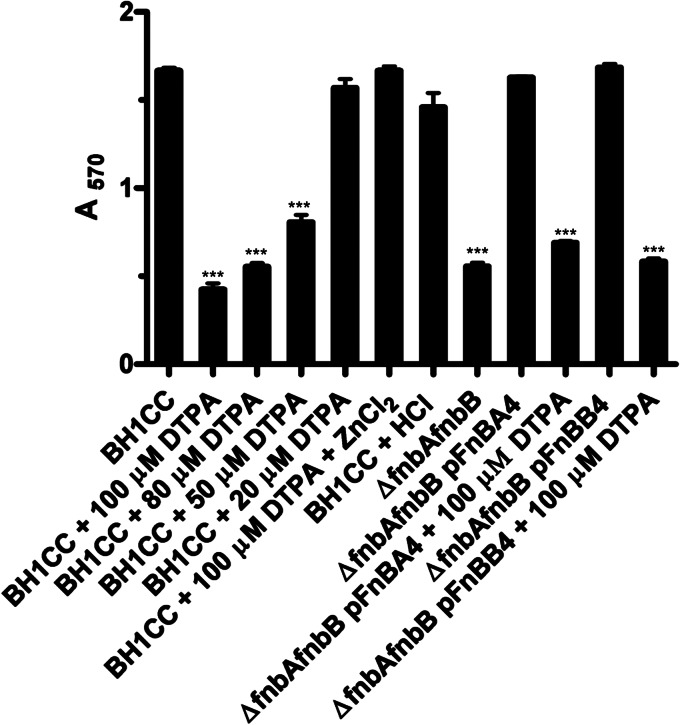
Our previous work showed that Zn2+ was required for SasG-mediated biofilm formation (12). The chelator diethylenetriaminepentaacetic acid (DTPA) also inhibited biofilm formation by Staphylococcus epidermidis RP62a and a community-associated MRSA strain (33). To determine if Zn2+ is required for FnBP-promoted biofilm formation, experiments were performed with DTPA. Biofilm formation by BH1CC was reduced in a manner that was dependent on the concentration of DTPA (Fig. 3). Biofilm formation by BH1CC ΔfnbAfnbB could be restored by expression of FnBPA or FnBPB from a plasmid, allowing tests to be performed with either FnBPA or FnBPB alone. Zn2+ chelation inhibited both FnBPA- and FnBPB-mediated biofilm formation, implying that both proteins promoted biofilm formation similarly in a Zn2+-dependent manner. In all cases biofilm was restored by the addition of ZnCl2 (Fig. 3) but not MgCl2 or CaCl2 (data not shown). These data show that Zn2+ is required for FnBP-mediated biofilm formation.
Fig 3.
Inhibition of biofilm formation by Zn2+ chelation. Bacteria were incubated with ZnCl2 (100 μM), HCl (buffer control), or the indicated concentrations of DTPA for 24 h at 37°C. Biofilm was stained with crystal violet, and the absorbance was read at 570 nm. ***, significant difference from result for BH1CC.
The effect of zinc chelation on FnBP-mediated biofilm accumulation.
Zn2+ is required for the autolytic activity of the Atl amidase domain (34). Substitution of a histidine residue involved in Zn2+ coordination at the amidase active site reduced primary attachment and biofilm formation (8). Thus, it is likely that Zn2+ chelation inhibits primary attachment. Here we investigated the role of Zn2+ in the accumulation phase of FnBP-mediated biofilm formation. We developed an assay where Atl was not required for primary attachment so the effect of Zn2+ chelation on accumulation could be examined. Microtiter plates were coated with a low concentration of fibrinogen, and biofilm was allowed to form on the surfaces of the coated wells. The coating of a surface with fibrinogen mimics the conditions that occur in vivo when surfaces of implanted devices are rapidly coated with plasma proteins. In vivo the fibrinogen-binding MSCRAMMs likely represent the major mechanism of primary attachment to immobilized fibrinogen in plasma (9).
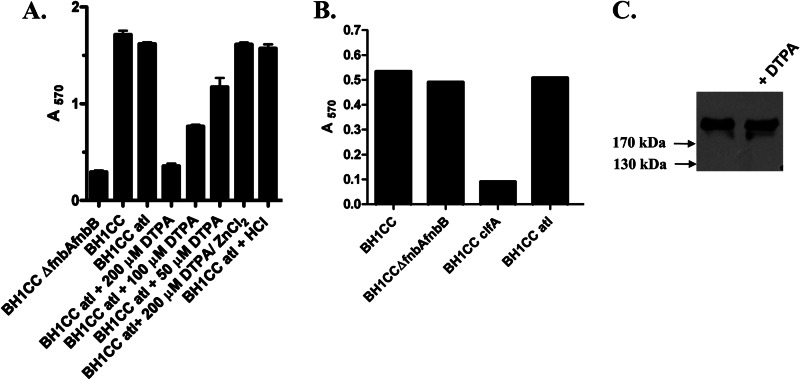
First we compared the abilities of BH1CC and an atl-deficient mutant to form biofilm on fibrinogen-coated wells. Biofilm formation abilities of both strains were very similar, demonstrating that Atl is not required for biofilm to form on a fibrinogen-coated surface (Fig. 4A). Control experiments indicated that DTPA had no effect on the ability of BH1CC atl to adhere to fibrinogen-coated plates (Fig. 4B). Thus, BH1CC atl could be used for accumulation assays to rule out effects of Zn2+ chelation on Atl activity and primary attachment. BH1CC ΔfnbAfnbB did not form biofilm on fibrinogen-coated wells (Fig. 4A) but adhered to fibrinogen-coated plates similarly to BH1CC (Fig. 4B), suggesting that the defect in biofilm formation by BH1CC ΔfnbAfnbB is due to the failure of cells to accumulate into biofilm rather than to form the initial attachment. BH1CC clfA showed reduced ability to adhere to fibrinogen (Fig. 4B), implying that ClfA contributed to adhesion to immobilized fibrinogen under these conditions.
Fig 4.
Effects of Zn2+ chelation and pH on FnBP-mediated biofilm accumulation. (A) Bacteria were incubated with ZnCl2 (100 μM), HCl (0.5 M, buffer control), or the indicated concentrations of DTPA and allowed to form biofilm on fibrinogen-coated wells (18 ng/ml) for 24 h at 37°C. Broth was supplemented with glucose unless stated otherwise. Biofilm was stained with crystal violet, and the absorbance was read at 570 nm. (B) Bacteria were grown to stationary phase, adjusted to an OD600 of 1.0, and added to wells coated with fibrinogen (1.25 μg/ml). The graph is representative of three independent experiments. (C) BH1CC atl was grown to stationary phase in BHI broth (left lane) or broth supplemented with DTPA (100 μM; right lane). Cell wall extracts were separated on 7.5% acrylamide gels, blotted onto PVDF membranes, and probed with biotin-labeled fibronectin. Size markers are indicated.
The effect of Zn2+ chelation on biofilm formation by growth in wells that had been coated with fibrinogen was investigated. The addition of DTPA reduced biofilm formation by BH1CC atl, showing that FnBP-mediated accumulation could be inhibited by Zn2+ chelation (Fig. 4A). Biofilm could be restored by the addition of ZnCl2. The addition of DTPA had no effect on expression of or integrity of FnBPs (Fig. 4C) or on the final density of planktonic bacteria. Thus, these data show that Zn2+ is required for FnBP-mediated biofilm accumulation following primary attachment to a conditioned surface.
Localization of FnBPA residues required for S. aureus biofilm formation and adherence to fibrinogen.
The A domains of FnBPA, FnBPB, and related MSCRAMMs are predicted to fold into three subdomains called N1, N2, and N3 (Fig. 1). No function has been assigned to N1.
In order to determine if subdomain N1 is involved in biofilm formation, pFNBA4, a multicopy plasmid that expresses the entire fnbA gene was manipulated to construct deletions that removed DNA encoding the entire N1 subdomain (amino acids 38 to 193) or a large part of it (amino acids 38 to 167). We were unable to establish transformants of pFNBA4ΔN138–193 in S. aureus, but pFNBA4ΔN138–167 was established successfully in BH1CC ΔfnbAfnbB. Cell wall extracts were then probed with biotin-labeled fibronectin by ligand affinity blotting, and bacteria were tested for their ability to adhere to immobilized fibronectin to confirm that the truncated derivative of FnBPA was expressed on the cell surface and was sorted to the cell wall. Plasmid pFnBA4ΔN138–167 expressed fibronectin binding protein FnBPAΔN138–167 at the same level as the wild type (Fig. 5) and promoted bacterial adherence to fibronectin (Fig. 6A). The ability of FnBPAΔN138–167 to promote biofilm formation was also assessed. BH1CC ΔfnbAfnbB expressing FnBPAΔN138–167 formed biofilm at levels similar to those for bacteria expressing full-length FnBPA, indicating that subdomain N1 is not required for biofilm formation (Fig. 6B).
Fig 5.
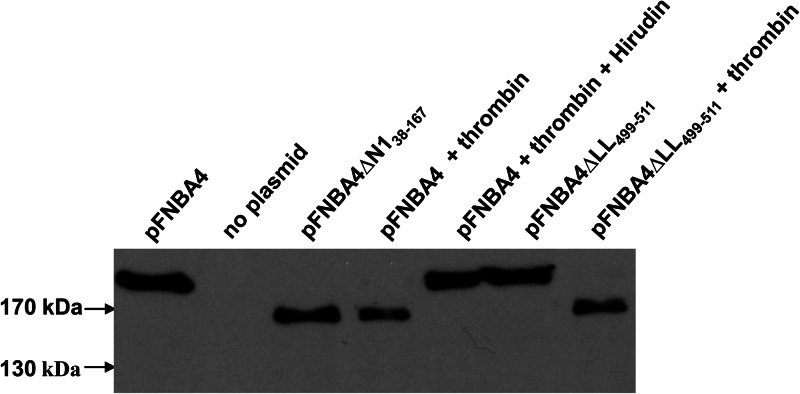
Expression of FnBPA variants. Cell wall extracts were separated on 7.5% acrylamide gels and blotted onto PVDF membranes. Membranes were probed with biotin-labeled fibronectin. Size markers are shown. Thrombin (2 U/ml) and/or hirudin (50 μg/ml) was included in growth medium where indicated.
Fig 6.
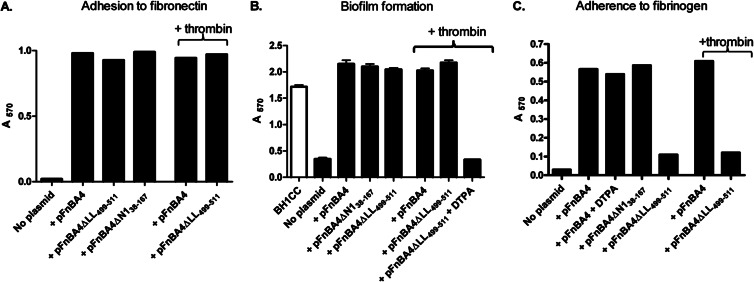
Characterization of bacteria expressing FnBPA variants. (A) Bacterial adherence to fibronectin. BH1CCΔfnbAfnbB expressing the plasmid indicated was grown to stationary phase and added to wells coated with fibronectin (2 μg/ml). Where indicated, bacteria were grown in broth supplemented with thrombin (2 U/ml). The graph is representative of three independent experiments. (B) Biofilm assays were carried out using BH1CC (open bar) or BH1CC ΔfnbAfnbB carrying the plasmid indicated (filled bars). Biofilm was allowed to form for 24 h at 37°C under static conditions in microtiter dishes. Where indicated, broth was supplemented with thrombin (2 U/ml) or DTPA (100 μM). Adherent cells were stained with crystal violet, and the absorbance was read at 570 nm. (C) Bacterial adherence to fibrinogen. SH1000 clfA clfB fnbA fnbB carrying the plasmid indicated was grown to stationary phase and added to wells coated with fibrinogen (2.5 μg/ml). Where indicated, bacteria were grown in broth supplemented with thrombin (2 U/ml) or DTPA (100 μM). The graph is representative of three independent experiments.
Studies with recombinantly expressed proteins showed that the N1 subdomain of FnBPA (and related MSCRAMMs) is not required for binding to fibrinogen (22). In order to test the ability of FnBPAΔN138–167 to mediate bacterial adherence to fibrinogen, plasmids pFnBA4 and pFnBA4ΔN138–167 were expressed in a mutant of SH1000 lacking the ability to bind to fibrinogen (SH1000 clfA clfB fnbA fnbB). Bacteria expressing FnBPA or FnBPAΔN138–167 adhered to fibrinogen with similar affinities, demonstrating that the N2N3 subdomains are sufficient to promote adhesion to fibrinogen when expressed by S. aureus and that they can fold correctly on the cell surface in the absence of subdomain N1 (Fig. 6C).
FnBPA is cleaved by thrombin.
We investigated the effect of proteolytic removal of subdomain N1 on the ability of FnBPA to promote biofilm formation and adhesion to fibrinogen. Biotin-labeled fibronectin recognized a protein of ∼165 kDa in cell wall extracts of BH1CC ΔfnbAfnbB(pFnBA4) cells that had been preincubated with thrombin. This suggested that residues at the N terminus of FnBPA had been proteolytically removed (Fig. 5). Indeed, the remaining cell wall-associated portion of the protein was of approximately the same size as FnBPAΔN138–167 (Fig. 5), suggesting that the thrombin cleavage site is located close to the junction between subdomains N1 and N2. The addition of hirudin inhibited the serine protease activity of thrombin and prevented cleavage of FnBPA (Fig. 5).
Next, a recombinant form of the FnBPA A domain (rFnBPA37–511) was expressed in E. coli with an N-terminal hexahistidine tag. When incubated with thrombin, rFnBPA37–511 was cleaved to generate a fragment of ca. 38 kDa (see Fig. S1 in the supplemental material). A band corresponding to the N-terminal region of the protein was not apparent, and the N-terminal portion could not be detected with anti-His antibody in a Western blot (data not shown), suggesting that the N1 subdomain undergoes extensive degradation. Cleavage by thrombin could be inhibited by addition of hirudin. The 38-kDa product generated by incubating rFnBPA37–511 with thrombin was analyzed by N-terminal sequencing to identify the proximal cleavage site. The five N-terminal residues were identified as 166TASES170, indicating that cleavage by thrombin occurs between residues R165 and T166 of FnBPA, close to the junction between subdomains N1 and N2.
Addition of thrombin to broth did not affect the ability of bacteria expressing FnBPA to form biofilm (Fig. 6B). Similarly, bacterial adhesion to immobilized fibrinogen and fibronectin was not reduced following thrombin cleavage (Fig. 6A and C). These data indicate that residues 37 to 165 of the N1 subdomain are not required for biofilm formation or fibrinogen binding.
Removal of the C-terminal extension of the FnBPA A domain.
FnBPA and FnBPB are thought to bind to fibrinogen and elastin by the dock, lock, and latch mechanism (22, 23). The C-terminal extension of subdomain N3 comprises residues involved in interactions with the bound fibrinogen γ-chain peptide (lock) and those involved in β-strand complementation (latch) to seal the ligand into the trench located between N2 and N3. The X-ray structure of the recombinant A domain of ClfB indicated that the protein dimerized during crystallization, with the C-terminal extension of one protein binding with the latching trench of its partner (35). It seemed possible that a similar interaction between FnBPA domains on neighboring cells could be responsible for cell-cell association during biofilm accumulation.
We investigated if the lock and latch peptide of FnBPA was involved in biofilm accumulation. BH1CC ΔfnbAfnbB was complemented with a plasmid expressing the entire fnbA gene (pFnBA4) or a variant lacking DNA encoding the C-terminal extension of N3 (lock and latch; pFnBA4ΔLL499–511). Bacteria expressing FnBPAΔLL499–511 formed biofilm at levels similar to those by bacteria expressing full-length FnBPA, demonstrating that the C-terminal extension is not required for biofilm formation (Fig. 6B).
Previous studies have shown that recombinantly expressed FnBPA and FnBPB A domain proteins lacking the C-terminal extension are unable to bind to fibrinogen or elastin (22, 25). When plasmids pFnBA4 and pFnBA4ΔLL499–511 were expressed in SH1000 clfA clfB fnbA fnbB, levels of fibronectin binding by both strains were very similar (Fig. 6A) but FnBPA lacking the C-terminal extension did not promote adherence to fibrinogen (Fig. 6C). The integrity of the protein was confirmed by fibronectin affinity blotting of cell wall extracts (Fig. 5). These data confirm the importance of the C-terminal extension in fibrinogen binding by FnBPA and show that the dock, lock, and latch binding mechanism applies to protein attached to the cell surface. Thus, it appears that biofilm is formed by a mechanism distinct from fibrinogen binding. In support of this, the addition of a fibrinogen γ-chain peptide that inhibits FnBPA binding to fibrinogen by the dock, lock, and latch mechanism (22) did not inhibit biofilm formation (data not shown). In addition, Zn2+ chelation did not reduce the ability of FnBPA to promote adherence to fibrinogen (Fig. 6C) but did inhibit biofilm formation (Fig. 3), again indicating that distinct mechanisms are involved in FnBP-mediated biofilm formation and adherence to fibrinogen.
In order to determine if residues 166 to 498 were sufficient for FnBPA-mediated biofilm, BH1CC expressing FnBPAΔLL499–511 was incubated with thrombin to remove residues 38 to 165 (Fig. 5). Biofilm formation was not affected, suggesting that residues 166 to 498 are sufficient for FnBPA-mediated biofilm accumulation (Fig. 6B).
DISCUSSION
Fibronectin binding proteins A and B promote the accumulation phase of biofilm formation in HA-MRSA from CC8 and CC22 (15, 16). This study revealed that FnBPs were expressed at much higher levels in the FnBP biofilm-forming MRSA isolate BH1CC (CC8) than in laboratory strain SH1000. Expression was sustained throughout the growth cycle by BH1CC in contrast to SH1000, where FnBPs could be detected only in the exponential phase. Thus, the data presented here challenge the widespread perception that some surface proteins such as FnBPA and FnBPB are expressed only during the exponential phase of growth (36). High-level expression sustained throughout the growth cycle likely contributes to the ability of FnBPs to mediate biofilm accumulation.
In addition we have identified the conditions required for FnBP-mediated accumulation. We coated wells with a low concentration of purified fibrinogen (a major component of plasma) so primary attachment could occur independently of Atl. Coating microtiter plates with plasma has previously been shown to promote biofilm formation by clinical isolates of S. aureus (37) and likely mimics in vivo conditions, where MSCRAMMs promote primary attachment to plasma-coated biomaterials. Loss of FnBPs did not reduce adherence to fibrinogen but loss of ClfA did, implying that ClfA is more important than FnBPs in promoting adhesion of BH1CC to immobilized fibrinogen.
Attachment to wells coated with fibrinogen was a useful tool for dissecting factors involved in biofilm accumulation while bypassing the role of Atl in primary attachment to unconditioned surfaces. We show that FnBP-mediated accumulation requires a physiological concentration of Zn2+. Removal of Zn2+ with the chelator DTPA inhibited biofilm accumulation but did not affect FnBP expression or integrity on the surface of S. aureus. FnBPA- and FnBPB-mediated biofilm formation was inhibited to similar extents, implying that both proteins promote biofilm formation by a similar Zn2+-dependent mechanism.
Subdomain N1 of the A domain is highly conserved among different FnBP isotypes, despite sequence variation in the N2N3 subdomains (21). While subdomains N2N3 of ClfA, ClfB, and SdrG have been crystallized both in the apo form and in the ligand-bound form, the structure of N1 is unknown. In the case of ClfB, subdomain N1 is predicted to be elongated and is composed mainly of a β-sheet structure (38). Studies with recombinantly expressed N2N3 subdomains of FnBPA and FnBPB and related MSCRAMMs showed that N1 is not required for ligand binding (22, 38, 39). Here we show that subdomains N2N3 of FnBPA, when expressed on the surface of S. aureus, are sufficient to promote bacterial adhesion to immobilized fibrinogen.
The S. aureus zinc metalloprotease aureolysin was previously shown to cleave ClfB close to the boundary between subdomains N1 and N2, removing N1 from the bacterial surface during growth (40). Here we show that thrombin, a host serine protease, cleaves within FnBPA to remove subdomain N1 from the protein expressed on the surface of S. aureus. The thrombin cleavage site in FnBPA (AQPRTA) differs from a typical thrombin cleavage site (LVPRGS), but the Pro and Arg residues after which thrombin cleaves are conserved. Proteolytic removal of the N1 subdomain did not affect fibrinogen binding or biofilm formation.
It is possible that thrombin-mediated cleavage of FnBPs on the surface of S. aureus occurs in vivo. Thrombin is formed by the proteolytic activation of prothrombin and functions to convert fibrinogen to fibrin, promoting the clotting of plasma. S. aureus activates thrombin by secreting coagulases, proteins which bind to and activate prothrombin. It is possible that the serine protease activity of the coagulase-prothrombin complex cleaves FnBPs.
It appears that S. aureus employs various mechanisms to remove subdomain N1 from MSCRAMMs located on the bacterial cell surface. This can occur through the expression of a bacterial protease (aureolysin for ClfB) or the action of a host protease (thrombin for FnBPA). In the case of ClfB, removal of N1 eliminates ClfB-mediated adherence to fibrinogen (40). In contrast, proteolytic removal of the N1 subdomain of FnBPA did not affect fibrinogen binding or biofilm formation. Thus, the biological significance of thrombin-mediated removal of N1 is unclear. Future studies addressing the function of subdomain N1 will determine if there are any consequences of removing N1 from the surface of the bacterium during in vivo growth.
Previous studies have shown that the C termini of the N3 subdomains of ClfA, ClfB, FnBPA, and FnBPB are important for ligand binding by the dock, lock, and latch mechanism (22, 23, 25, 35). During dock, lock, and latch, the C terminus of N3, which is unbound in the apo form, undergoes a conformational change and forms an extra β-strand in subdomain N2, locking the peptide ligand into the binding trench. Here we report the first example of an MSCRAMM lacking a C-terminal extension being expressed on the surface of S. aureus and showing a reduced ability to promote adhesion to fibrinogen. This is a validation of the importance of the C-terminal extension of FnBPA during binding by the dock, lock, and latch mechanism, something that has previously been tested only using E. coli-expressed recombinant subdomains. Because the C terminus of the A domain is linked to the cell surface via a flexible stalk, the bulk of the A domain must reorient itself. The variant lacking the C-terminal extension, however, may be oriented differently because there is nothing to anchor it in the form indicated for the full-length protein. However, the C-terminal extension is not required for biofilm accumulation. Thus, it is clear that integrity of regions of the FnBPA A domain required for fibrinogen binding and conformational changes involved in ligand binding are not necessary for biofilm to form. In agreement with this, a previous study showed that residue N304, located in the ligand binding trench between subdomains N2 and N3, could be replaced without altering biofilm formation (15).
In summary, we have localized the region required for biofilm formation by FnBPA to residues 166 to 498 of the A domain. We have shown that the accumulation stage requires high-level expression of FnBPs to be sustained throughout the growth cycle. Future studies will focus on elucidating the mechanism by which FnBPs mediate biofilm formation. It is possible that specific homophilic interactions between FnBP N2N3 subdomains occur on neighboring bacteria. Alternatively, FnBPs may bind a receptor on the surface of adjacent bacteria, or there may be a bridging molecule which binds to the FnBP A domain and links it to a surface-exposed receptor.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by a Science Foundation Ireland Programme Investigator grant (to T.J.F.).
We thank F. Burke for assistance with plasmid construction and D. Muldowney for assistance with protein purification.
Footnotes
Published ahead of print 5 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02128-12.
REFERENCES
- 1. Lowy FD. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520–532 [DOI] [PubMed] [Google Scholar]
- 2. O'Gara JP, Humphreys H. 2001. Staphylococcus epidermidis biofilms: importance and implications. J. Med. Microbiol. 50:582–587 [DOI] [PubMed] [Google Scholar]
- 3. Zimmerli W, Trampuz A, Ochsner PE. 2004. Prosthetic-joint infections. N. Engl. J. Med. 351:1645–1654 [DOI] [PubMed] [Google Scholar]
- 4. Stewart PS, Costerton JW. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135–138 [DOI] [PubMed] [Google Scholar]
- 5. Otto M. 2006. Bacterial evasion of antimicrobial peptides by biofilm formation. Curr. Top. Microbiol. Immunol. 306:251–258 [DOI] [PubMed] [Google Scholar]
- 6. Vuong C, Voyich JM, Fischer ER, Braughton KR, Whitney AR, DeLeo FR, Otto M. 2004. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell. Microbiol. 6:269–275 [DOI] [PubMed] [Google Scholar]
- 7. Biswas R, Voggu L, Simon UK, Hentschel P, Thumm G, Gotz F. 2006. Activity of the major staphylococcal autolysin Atl. FEMS Microbiol. Lett. 259:260–268 [DOI] [PubMed] [Google Scholar]
- 8. Houston P, Rowe SE, Pozzi C, Waters EM, O'Gara JP. 2011. Essential role for the major autolysin in the fibronectin-binding protein-mediated Staphylococcus aureus biofilm phenotype. Infect. Immun. 79:1153–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vaudaux PE, Francois P, Proctor RA, McDevitt D, Foster TJ, Albrecht RM, Lew DP, Wabers H, Cooper SL. 1995. Use of adhesion-defective mutants of Staphylococcus aureus to define the role of specific plasma proteins in promoting bacterial adhesion to canine arteriovenous shunts. Infect. Immun. 63:585–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Heilmann C, Schweitzer O, Gerke C, Vanittanakom N, Mack D, Gotz F. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20:1083–1091 [DOI] [PubMed] [Google Scholar]
- 11. Cucarella C, Solano C, Valle J, Amorena B, Lasa I, Penades JR. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183:2888–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Geoghegan JA, Corrigan RM, Gruszka DT, Speziale P, O'Gara JP, Potts JR, Foster TJ. 2010. Role of surface protein SasG in biofilm formation by Staphylococcus aureus. J. Bacteriol. 192:5663–5673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Merino N, Toledo-Arana A, Vergara-Irigaray M, Valle J, Solano C, Calvo E, Lopez JA, Foster TJ, Penades JR, Lasa I. 2009. Protein A-mediated multicellular behavior in Staphylococcus aureus. J. Bacteriol. 191:832–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schroeder K, Jularic M, Horsburgh SM, Hirschhausen N, Neumann C, Bertling A, Schulte A, Foster S, Kehrel BE, Peters G, Heilmann C. 2009. Molecular characterization of a novel Staphylococcus aureus surface protein (SasC) involved in cell aggregation and biofilm accumulation. PLoS One 4:e7567 doi:10.1371/journal.pone.0007567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. O'Neill E, Pozzi C, Houston P, Humphreys H, Robinson DA, Loughman A, Foster TJ, O'Gara JP. 2008. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J. Bacteriol. 190:3835–3850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vergara-Irigaray M, Valle J, Merino N, Latasa C, Garcia B, Ruiz de Los Mozos I, Solano C, Toledo-Arana A, Penades JR, Lasa I. 2009. Relevant role of fibronectin-binding proteins in Staphylococcus aureus biofilm-associated foreign-body infections. Infect. Immun. 77:3978–3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Signas C, Raucci G, Jonsson K, Lindgren PE, Anantharamaiah GM, Hook M, Lindberg M. 1989. Nucleotide sequence of the gene for a fibronectin-binding protein from Staphylococcus aureus: use of this peptide sequence in the synthesis of biologically active peptides. Proc. Natl. Acad. Sci. U. S. A. 86:699–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jonsson K, Signas C, Muller HP, Lindberg M. 1991. Two different genes encode fibronectin binding proteins in Staphylococcus aureus. The complete nucleotide sequence and characterization of the second gene. Eur. J. Biochem. 202:1041–1048 [DOI] [PubMed] [Google Scholar]
- 19. Deivanayagam CC, Wann ER, Chen W, Carson M, Rajashankar KR, Hook M, Narayana SV. 2002. A novel variant of the immunoglobulin fold in surface adhesins of Staphylococcus aureus: crystal structure of the fibrinogen-binding MSCRAMM, clumping factor A. EMBO J. 21:6660–6672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Burke FM, McCormack N, Rindi S, Speziale P, Foster TJ. 2010. Fibronectin-binding protein B variation in Staphylococcus aureus. BMC Microbiol. 10:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Loughman A, Sweeney T, Keane FM, Pietrocola G, Speziale P, Foster TJ. 2008. Sequence diversity in the A domain of Staphylococcus aureus fibronectin-binding protein A. BMC Microbiol. 8:74 doi:10.1186/1471-2180-8-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Keane FM, Loughman A, Valtulina V, Brennan M, Speziale P, Foster TJ. 2007. Fibrinogen and elastin bind to the same region within the A domain of fibronectin binding protein A, an MSCRAMM of Staphylococcus aureus. Mol. Microbiol. 63:711–723 [DOI] [PubMed] [Google Scholar]
- 23. Ponnuraj K, Bowden MG, Davis S, Gurusiddappa S, Moore D, Choe D, Xu Y, Hook M, Narayana SV. 2003. A “dock, lock, and latch” structural model for a staphylococcal adhesin binding to fibrinogen. Cell 115:217–228 [DOI] [PubMed] [Google Scholar]
- 24. Ganesh VK, Rivera JJ, Smeds E, Ko YP, Bowden MG, Wann ER, Gurusiddappa S, Fitzgerald JR, Hook M. 2008. A structural model of the Staphylococcus aureus ClfA-fibrinogen interaction opens new avenues for the design of anti-staphylococcal therapeutics. PLoS Pathog. 4:e1000226 doi:10.1371/journal.ppat.1000226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Burke FM, Di Poto A, Speziale P, Foster TJ. 2011. The A domain of fibronectin-binding protein B of Staphylococcus aureus contains a novel fibronectin binding site. FEBS J. 278:2359–2371 [DOI] [PubMed] [Google Scholar]
- 26. Schwarz-Linek U, Werner JM, Pickford AR, Gurusiddappa S, Kim JH, Pilka ES, Briggs JA, Gough TS, Hook M, Campbell ID, Potts JR. 2003. Pathogenic bacteria attach to human fibronectin through a tandem beta-zipper. Nature 423:177–181 [DOI] [PubMed] [Google Scholar]
- 27. O'Neill E, Pozzi C, Houston P, Smyth D, Humphreys H, Robinson DA, O'Gara JP. 2007. Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J. Clin. Microbiol. 45:1379–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Horsburgh MJ, Aish JL, White IJ, Shaw L, Lithgow JK, Foster SJ. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Monk IR, Shah IM, Xu M, Tan MW, Foster TJ. 2012. Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. mBio 3(2):e00277–11 doi:10.1128/mBio.00277-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Greene C, McDevitt D, Francois P, Vaudaux PE, Lew DP, Foster TJ. 1995. Adhesion properties of mutants of Staphylococcus aureus defective in fibronectin-binding proteins and studies on the expression of fnb genes. Mol. Microbiol. 17:1143–1152 [DOI] [PubMed] [Google Scholar]
- 31. Saravia-Otten P, Muller HP, Arvidson S. 1997. Transcription of Staphylococcus aureus fibronectin binding protein genes is negatively regulated by agr and an agr-independent mechanism. J. Bacteriol. 179:5259–5263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McGavin MJ, Zahradka C, Rice K, Scott JE. 1997. Modification of the Staphylococcus aureus fibronectin binding phenotype by V8 protease. Infect. Immun. 65:2621–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Conrady DG, Brescia CC, Horii K, Weiss AA, Hassett DJ, Herr AB. 2008. A zinc-dependent adhesion module is responsible for intercellular adhesion in staphylococcal biofilms. Proc. Natl. Acad. Sci. U. S. A. 105:19456–19461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zoll S, Patzold B, Schlag M, Gotz F, Kalbacher H, Stehle T. 2010. Structural basis of cell wall cleavage by a staphylococcal autolysin. PLoS Pathog. 6:e1000807 doi:10.1371/journal.ppat.1000807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ganesh VK, Barbu EM, Deivanayagam CC, Le B, Anderson AS, Matsuka YV, Lin SL, Foster TJ, Narayana SV, Hook M. 2011. Structural and biochemical characterization of Staphylococcus aureus clumping factor B:ligand interactions. J. Biol. Chem. 286:25963–25972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Novick RP. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429–1449 [DOI] [PubMed] [Google Scholar]
- 37. Beenken KE, Blevins JS, Smeltzer MS. 2003. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 71:4206–4211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Perkins S, Walsh EJ, Deivanayagam CC, Narayana SV, Foster TJ, Hook M. 2001. Structural organization of the fibrinogen-binding region of the clumping factor B MSCRAMM of Staphylococcus aureus. J. Biol. Chem. 276:44721–44728 [DOI] [PubMed] [Google Scholar]
- 39. Geoghegan JA, Ganesh VK, Smeds E, Liang X, Hook M, Foster TJ. 2010. Molecular characterization of the interaction of staphylococcal microbial surface components recognizing adhesive matrix molecules (MSCRAMM) ClfA and Fbl with fibrinogen. J. Biol. Chem. 285:6208–6216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McAleese FM, Walsh EJ, Sieprawska M, Potempa J, Foster TJ. 2001. Loss of clumping factor B fibrinogen binding activity by Staphylococcus aureus involves cessation of transcription, shedding and cleavage by metalloprotease. J. Biol. Chem. 276:29969–29978 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.