Abstract
Type III secretion systems (T3SSs) are central virulence mechanisms used by a variety of Gram-negative bacteria to inject effector proteins into host cells. The needle polymer is an essential part of the T3SS that provides the effector proteins a continuous channel into the host cytoplasm. It has been shown for a few T3SSs that two chaperones stabilize the needle protein within the bacterial cytosol to prevent its premature polymerization. In this study, we characterized the chaperones of the enteropathogenic Escherichia coli (EPEC) needle protein EscF. We found that Orf2 and Orf29, two poorly characterized proteins encoded within the EPEC locus of enterocyte effacement (LEE), function as the needle protein cochaperones. Our finding demonstrated that both Orf2 and Orf29 are essential for type III secretion (T3S). In addition, we found that Orf2 and Orf29 associate with the bacterial membrane and form a complex with EscF. Orf2 and Orf29 were also shown to disrupt the polymerization of EscF in vitro. Prediction of the tertiary structures of Orf2 and Orf29 showed high structural homology to chaperones of other T3SS needle proteins. Overall, our data suggest that Orf2 and Orf29 function as the chaperones of the needle protein, and therefore, they have been renamed EscE and EscG.
INTRODUCTION
Many bacterial pathogens that cause life-threatening conditions employ a type III secretion system (T3SS). The T3SS functions in the secretion and injection of bacterial virulence factors (effectors) into the cytoplasm of host cells. The effectors perform diverse biochemical activities within the host cell that are beneficial to the bacteria (1, 2). One such T3SS-expressing pathogen is enteropathogenic Escherichia coli (EPEC). This pathogen belongs to the attaching and effacing (A/E) family of pathogens, which adhere to host enterocytes and induce extensive host cell cytoskeletal rearrangements (3–5). EPEC is the main causative agent of infantile diarrhea, a major cause of death for children under the age of 5 years in developing countries (6).
The T3SS is a large protein complex composed of approximately 20 different proteins that form a syringe-like structure spanning both the inner and outer membranes of the bacteria. The T3SS apparatus is highly conserved structurally and functionally among different pathogens and shares strong homology to components of the flagellar system (7). The basal portion of the T3SS consists of several protein rings that are connected to a hollow needle consisting of a single polymerizing protein.
The needle protein of EPEC, EscF, has a few homologs, including YscF in Yersinia spp., AscF in Aeromonas hydrophila, PrgI in Salmonella enterica serovar Typhimurium pathogenicity island I (SPI-1), MxiH in Shigella flexneri, and PscF in Pseudomonas aeruginosa (8–12). This small protein (∼8 kDa) has been shown to polymerize spontaneously (10, 13). In various pathogens, chaperones have been reported to bind the needle protein monomer and prevent its premature polymerization within the bacterial cytosol prior to the assembly of the needle (14–18). The sequence identity between these chaperones is notably low. However, they possess common features, such as a small size and, often, an acidic pI (19).
The first chaperones of a T3SS needle protein to be discovered and characterized were PscE and PscG in P. aeruginosa (15). PscE and PscG were shown to form a stable, soluble complex with PscF in the cytoplasm at a 1:1:1 ratio, thus blocking premature polymerization of PscF (15, 16, 20). Subsequently, the chaperones of AscF and YscF were shown to have structures homologous to those of the Pseudomonas proteins (14, 17, 18).
Although the identities of the chaperones that recognize the T3SS needle protein are known for some pathogens, their identities in EPEC remain elusive. A recent bioinformatics study predicted that Orf2, encoded by an uncharacterized open reading frame (ORF) in the EPEC pathogenicity island, termed the locus of enterocyte effacement (LEE), shared sequence homology with SsaE in S. Typhimurium, Cv2595 in Chromobacterium violaceum, and YscE in Yersinia enterocolitica (21). Although the same bioinformatics analysis failed to find a homolog to YscG in the LEE-encoded system, the investigators speculated that Orf29 may possess a function homologous to that of YscG. This was based on a study that detected an interaction between Orf2 and Orf29 by using a yeast two-hybrid system (22).
In this study, we experimentally established Orf2 and Orf29 as the chaperones of the EPEC T3SS needle protein EscF. Our findings demonstrated that Orf2 and Orf29 are essential for type III secretion (T3S). Although predicted to be cytoplasmic, they localize to the bacterial membrane. Consistent with the suggestion that Orf2 and Orf29 serve as the needle protein chaperones, they were found to interact with EscF. Moreover, they were shown to prevent the polymerization of EscF when added in vitro. Overall, our findings support the prediction that Orf2 and Orf29 of EPEC are homologs of YscE and YscG in Yersinia spp., respectively, and we therefore propose to rename them EscE and EscG, respectively, according to the standard T3SS nomenclature. We use this terminology throughout this article.
MATERIALS AND METHODS
Bacterial strains.
Wild-type EPEC O127:H6 strain E2348/69 (streptomycin resistant [Smr]) and E. coli strain BL21(λDE3) were used in this study. Strains were grown in Luria-Bertani (LB) broth supplemented with the appropriate antibiotics at 37°C. Antibiotics were used at the following concentrations: streptomycin at 50 μg/ml, ampicillin at 100 μg/ml, kanamycin at 50 μg/ml, and chloramphenicol at 34 μg/ml.
Construction of escE and escG nonpolar mutants.
Nonpolar deletion mutants of the escE and escG genes in the Smr EPEC strain E2348/69 were generated using the sacB-based allelic exchange method (23). Briefly, for every mutant, two PCR fragments were generated using the corresponding primer pairs (Table 1), cloned into pCR2.1-TOPO (Invitrogen), verified by DNA sequencing, and then subcloned as a KpnI/NheI fragment and an NheI/SacI fragment into a KpnI/SacI-digested suicide vector, pRE112 (24). The resulting plasmids, containing the flanking regions of escE and escG, with 93% of escE and 84% of escG deleted, were transformed into E. coli SM10λpir and were then introduced into EPEC by conjugation. After sucrose selection, EPEC colonies that were resistant to sucrose and susceptible to chloramphenicol were screened for the deletion of escE or escG by PCR.
Table 1.
Sequences of primers used in this study
| Construct and primer designation | Primer sequence (restriction site)a |
|---|---|
| escE deletion mutant | |
| ESCE-01F | GGGTACCCCACCTTAGCGCTATGCAC (KpnI) |
| ESCE-01R | GGCTAGCAGCAGAAATATCATTAACC (NheI) |
| ESCE-02F | GGCTAGCAATAATAAATAGAGGATG (NheI) |
| ESCE-02R | GGAGCTCGATATCTTTAACCCATTTTATCGATTGG (SacI) |
| escG deletion mutant | |
| ESCG-01F | GGAGCTCGGCGAAGGCTTCCGAAGG (KpnI) |
| ESCG-01R | GGCTAGCAGCAGAAATATCATTAACC (NheI) |
| ESCG-02F | GGCTAGCAGTGGTTGGGTACGAGG (NheI) |
| ESCG-02R | GGGTACCGGTGGTGTCCGCTATTGCC (SacI) |
| pEscE (pCR2.1-TOPO) | |
| EscE-F | CCGAGCTCCCGCTGAAAAATATTTAAC (SacI) |
| EscE-2HA-R | GGCTCGAGTTTATTATTAATCCTGATTCGC (XhoI) |
| pEscG (pCR2.1-TOPO) | |
| EscG-F | CCGAGCTCTGGTCTCAACCATTTCTAACGC (SacI) |
| EscG-2HA-R | GGCTCGAGAAATCCTCGTACCCAACCACTAA (XhoI) |
| pEscF (pET21a/pET28a) | |
| EscF-F1 | CCCATATGAATTTATCTGAAATTACTCAAC (NdeI) |
| EscF-R1 | GCAAGCTTATGGTTTGCCGCAGCTACAGCC (HindIII) |
| pEscFΔ5aa (pET21a) | |
| EscF-F2 | CCGCAGCATATGAATTTATCTGAAATTACTCAACAAATGG (NdeI) |
| EscF-R2 | CCGCAGGGATCCTTAAATGGTTGAGACCAGATCTTTTATCG (BamHI) |
| pHis-EscE (pET28a) | |
| His-EscE-F | CCGCAGCATATGATAACGATAACTGAGCTGG (NdeI) |
| His-EscE-R | CCGCAGGGATCCCTATTTATTATTAATCCTGATTCGC (BamHI) |
| pHis-EscG (pET28a) | |
| His-EscG-F | CCGCAGCATATGGTTAATGATATTTCTGCTAATAAGATACTGG (NdeI) |
| His-EscG-R | CCGCAGGGATCCTTAAAATCCTCGTACCCAACCAC (BamHI) |
| pHis-EscG-EscE (pET28a) | |
| His-EscG-EscE-F | GGATCCGAATTCTAAGAAGGAATGATAACGATAACTGAGC (BamHI) |
| His-EscG-EscE-R | CCCTCGAGCTATTTATTAATTAATCC (XhoI) |
| pHis-EscE-EscG (pET28a) | |
| His-EscE-EscG-F | CCGCAGGAATTCAAGGAGATATACCATGGTTAATGATATTTCTGCTAATAAGATACTGG (EcoRI) |
| His-EscE-EscG-R | CCGCAGCTCGAGTTAAAATCCTCGTACCCAACCAC (XhoI) |
Restriction sites are underlined in primer sequences.
Construction of plasmids expressing EscE, EscG, and EscF proteins.
The escE and escG genes were amplified using the primer pairs EscE-F/EscE-2HA-R and EscG-F/EscG-2HA-R (Table 1), respectively, cloned into pCR2.1-TOPO (Invitrogen), and then subcloned as SacI/XhoI fragments into SacI/XhoI-digested pTOPO-2HA (25). Each of the resulting constructs, pEscE-2HA and pEscG-2HA, expressed a fusion protein of EscE or EscG with a double hemagglutinin (HA) tag at its C terminus. A cysteine-to-serine mutation in the escG gene at position 68 was introduced using site-directed mutagenesis (Stratagene).
The escF gene was amplified using the primer pair EscF-F1/EscF-R1 (Table 1), digested with NdeI/HindIII, and cloned into NdeI/HindIII-digested pET21a or pET28a. A truncated version of EscF, missing the 5 C-terminal residues, was amplified using the primer pair EscF-F2/EscF-R2 (Table 1), digested with NdeI/BamHI, and cloned into NdeI/BamHI-digested pET21a. The resulting constructs, pEscF, pEscFΔ5aa, and pHis-EscF, expressed untagged EscF, truncated EscF, or EscF with an N-terminal His tag. In addition, the escE and escG genes were amplified using the primer pair His-EscE-F/His-EscE-R or His-EscG-F/His-EscG-R (Table 1), respectively, digested with NdeI/BamHI, and cloned into NdeI/BamHI-digested pET28a. The resulting constructs, pHis-EscE and pHis-EscG, expressed EscE or EscG fused to a His tag at its N terminus. To express His-EscG together with untagged EscE, we amplified EscE using the primer pair His-EscG-EscE-F/His-EscG-EscE-R (Table 1), digested it with BamHI/XhoI, and cloned it into BamHI/XhoI-digested pHis-EscG (pET28a). The reverse design, where His-tagged EscE was expressed with EscG, was carried out by amplifying EscG using the primer pair His-EscE-EscG-F/His-EscE-EscG-R (Table 1), digesting it with EcoRI/XhoI, and cloning into EcoRI/XhoI-digested pHis-EscE (pET28a). All constructs were verified by DNA sequencing. All the plasmids and strains used in this study are listed in Table 2.
Table 2.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| EPEC E2348/69 | Wild-type E. coli O127:H6 isolate | 39 |
| E2348/69 ΔescE | EPEC escE deletion mutant | This study |
| E2348/69ΔescG | EPEC escG deletion mutant | This study |
| Plasmids | ||
| pEscE-2HA | C-terminally tagged EscE in pTOPO-2HA | This study |
| pEscG-2HA | C-terminally tagged EscG in pTOPO-2HA | This study |
| pEscG(C68S)-2HA | C-terminally tagged EscG mutated at position 68 in pTOPO-2HA | This study |
| pHis-EscF | N-terminally His tagged EscF in pET28a | This study |
| pEscF | EscF in pET21a | This study |
| pEscFΔ5aa | Truncated EscF in pET21 (deletion of 5 aa at the C terminus) | This study |
| pHis-EscE | N-terminally His tagged EscE in pET28a | This study |
| pHis-EscG | N-terminally His tagged EscG in pET28a | This study |
| pHis-EscE-EscG | N-terminally His tagged EscE and untagged EscG in pET28a | This study |
| pHis-EscG-EscE | N-terminally His tagged EscG and untagged EscE in pET28a | This study |
| pCR2.1-TOPO | PCR cloning vector; Ampr Kanr | Invitrogen |
| pET28a(+) | Expression vector for His tagging; Kanr | Novagen |
| pET21a(+) | Expression vector; Ampr | Novagen |
| pTOPO-2HA | Expression vector for HA tagging; Kanr | 25 |
| pACYC184 | Cloning vector; Cmr Tcr | 40 |
| pRE112 | Suicide vector for allelic exchange; Cmr | 24 |
Secretion assay.
EPEC strains were grown in LB broth overnight at 37°C. The cultures were diluted 1:50 into Dulbecco's modified Eagle's medium (DMEM) that was prewarmed at 37°C in a CO2 tissue culture incubator (under 5% CO2) overnight. The cultures were grown for 6 h to an optical density at 600 nm (OD600) of 0.7 in the tissue culture incubator with the tubes standing. The cultures were centrifuged at 16,000 × g for 5 min to remove the bacteria; the supernatant was collected and was then filtered through a 0.22-μm filter (Millipore). The supernatant was then precipitated with 10% (vol/vol) trichloroacetic acid (TCA) to concentrate proteins secreted into the culture medium. The secreted and precipitated proteins were dissolved in SDS-PAGE sample buffer, and residual TCA was neutralized with saturated Tris. The volumes of buffer used to resuspend the secreted proteins were normalized relative to the A600 values of the cultures in order to ensure equal loading of the samples. The proteins were analyzed by SDS-12% PAGE and were stained with Coomassie blue.
Bacterial fractionation.
Bacterial cell fractionation was based on procedures described previously (26). Briefly, EPEC from an overnight culture was subcultured 1:50 in 50 ml of DMEM for 6 h at 37°C in a CO2 tissue culture incubator. The culture was harvested, washed in phosphate-buffered saline (PBS), resuspended in 1 ml of buffer A (50 mM Tris [pH 7.5], 20% [wt/vol] sucrose, protease inhibitor cocktail [Roche Applied Science], 10 mM EDTA, and lysozyme [10 μg/ml]), and incubated for 30 min at room temperature to generate spheroplasts. MgCl2 was then added to a final concentration of 20 mM, and samples were spun for 10 min at 5,000 × g. The supernatants containing the periplasmic fractions were collected. The pellet containing the cytoplasm and the membrane fractions was resuspended in 1 ml lysis buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 3 mM MgCl2, 1 mM CaCl2, and 2 mM β-mercaptoethanol with protease inhibitors). All subsequent steps were carried out at 4°C. RNase A and DNase I (each at 10 μg/ml) were added, and the samples were sonicated (3 times, for 15 s each time; Fisher Scientific). Intact bacteria were removed by centrifugation (at 2,300 × g for 15 min), and the cleared supernatant containing cytoplasmic and membrane proteins was transferred to new tubes. To obtain the cytoplasmic fraction, supernatants were centrifuged (in a Beckman Optima TLX ultracentrifuge with a TLA-100.3 rotor) for 30 min at 100,000 × g to pellet the membranes. The supernatant containing the cytoplasmic fraction was collected; the membrane pellet was washed once with 2 ml of lysis buffer and was resuspended in 0.1 ml lysis buffer with 0.1% SDS for the collective membrane fraction. To separate the membrane fraction into inner membrane (IM) and outer membrane (OM) fractions, the washed membrane pellet was first resuspended in 0.1 ml lysis buffer with 0.5% N-lauroylsarcosine, which selectively solubilizes the IM, and was then centrifuged at 100,000 × g for 1 h. The supernatant containing the IM fraction was collected, and the OM pellet was washed in lysis buffer with 0.5% N-lauroylsarcosine. The final pellet was resuspended in 0.1 ml of lysis buffer with 0.5% N-lauroylsarcosine and 0.1% SDS. The protein contents of all samples were determined using the Coomassie Plus protein assay (Thermo Scientific) before the addition of SDS sample buffer with β-mercaptoethanol.
Purification of proteins from BL21 lysates.
E. coli BL21(λDE3) transformed with escF, escE, and escG constructs were grown in 50 ml at 37°C to the mid-exponential phase. Protein production was induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG; Sigma) for 3 h. The bacterial cultures were harvested by centrifugation at 5,000 × g for 30 min. The pellet was washed with 1 culture volume of ice-cold PBS and was pelleted by centrifugation. The washed pellet was resuspended in 1 ml lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, protease inhibitor cocktail; adjusted to pH 8.0) with 10 μg/ml lysozyme and was incubated on ice for 30 min. Bacteria were then lysed by using a probe sonicator (3 times, for 15 s each time; Fisher Scientific), and unlysed bacteria were removed by centrifugation at 3,000 × g for 15 min at 4°C. An aliquot of the cleared lysate was collected as the whole-cell extract. The cleared lysate was incubated with a Ni-nitrilotriacetic acid (NTA) agarose bead slurry (GE Healthcare) for 1 h at 4°C. The resulting mixtures were loaded onto chromatography columns (Bio-Rad) and were washed four times with 1 ml washing buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, protease inhibitor cocktail; adjusted to pH 8.0). Proteins were eluted from the beads using elution buffer (50 mM NaH2PO4, 300 mM NaCl, 300 mM imidazole, protease inhibitor cocktail; adjusted to pH 8.0). Protein purification for matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) spectroscopy was based on a previously described procedure (27).
Coimmunoprecipitation and immunoblotting.
E. coli BL21(λDE3) expressing the EPEC proteins EscF, EscE, and EscG separately or in combination with each other were subjected to a protein purification procedure excluding the elution step. Elution was performed using a modified elution buffer (elution buffer with 0.1% Tween 20, 10% glycerol, and 10 mM β-mercaptoethanol). Equal amounts of the whole-cell extracts and eluted fractions were loaded onto an SDS-PAGE gel and were transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad). Blots were blocked for 1 h in 5% (wt/vol) skim milk–PBST (0.1% Tween in PBS), incubated first with the primary antibody diluted in 5% skim milk–PBST for 1 h at room temperature and then with the secondary antibody diluted in 5% skim milk–PBST for 30 min at room temperature, and detected with ECL reagents (Bio-Rad). Primary antibodies were mouse anti-penta-His (Sigma), diluted 1/1,000; mouse anti-HA.11 (Covance, Princeton, NJ), diluted 1/1,000; rabbit anti-EscF (raised against a peptide of EPEC EscF with the sequence LNSTDLVNDPEKMLELQF; Pacific Immunology Corp.), diluted 1/500; mouse anti-DnaK (Stressgen), diluted 1/1,000; rabbit anti–maltose-binding protein (anti-MBP; New England Biolabs), diluted 1/1,000; and a rabbit antibody against the beta subunit of E. coli ATP synthase, diluted 1/15,000 (a generous gift from Gabriele Deckers-Hebestreit). Secondary antibodies were a horseradish peroxidase (HRP)-conjugated goat anti-mouse antibody (Jackson ImmunoResearch), diluted 1/5,000, and an HRP-conjugated goat anti-rabbit antibody (Sigma), diluted 1/5,000.
SEC.
The polymerization behavior of EscF in the presence and the absence of EscE and EscG was analyzed under native buffer conditions using size exclusion chromatography (SEC). First, purified His-EscF (5 mg) in the presence or absence of the His-EscG-EscE (His-EscG/E) complex (1 mg), or His-EscG/E (1 mg) alone, was sonicated in 50 mM NaH2PO4 and 300 mM NaCl buffer (pH 8.0) with protease inhibitor cocktail and was subsequently incubated at 4°C for 1 h. The samples were then analyzed by size exclusion chromatography using a Superdex 75 column (GE Healthcare) equilibrated in running buffer containing 50 mM NaH2PO4 and 300 mM NaCl (pH 8.0) at room temperature. The protein elution profile was monitored by UV detection. Three conditions were analyzed: (i) His-EscF, (ii) His-EscF in the presence of the His-EscG/E complex, and (iii) His-EscG/E. In order to assess the impact of His-EscG/E on the elution profile of His-EscF, the profile of His-EscG/E alone was subtracted from the profile of His-EscF including His-EscG/E, and the difference was then directly compared to the profile of His-EscF alone. UV signals with negative values were considered noise and were normalized to baseline.
BN-PAGE.
EscF polymerization was examined using blue native (BN)-PAGE based on the protocol described in reference 28. Briefly, protein samples were first incubated for an hour with 0.025% Coomassie brilliant blue G-250, then supplemented with 10% glycerol, and separated on a native gel (4 to 20%; Bio-Rad). After BN-PAGE at 4°C, gels were fixed (40% methanol [MeOH], 10% acetic acid), soaked in 0.5% SDS for 20 min at room temperature, and then analyzed by Western blotting.
Computational analysis.
Three-dimensional (3D) protein structures of EscE and EscG were predicted by I-TASSER (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) (29, 30). Structural alignment, superposition, and visualization of 3D protein structures were performed using UCSF Chimera software, version 1.6.1 (http://www.cgl.ucsf.edu/chimera) (31).
RESULTS
EscE and EscG are essential for T3S.
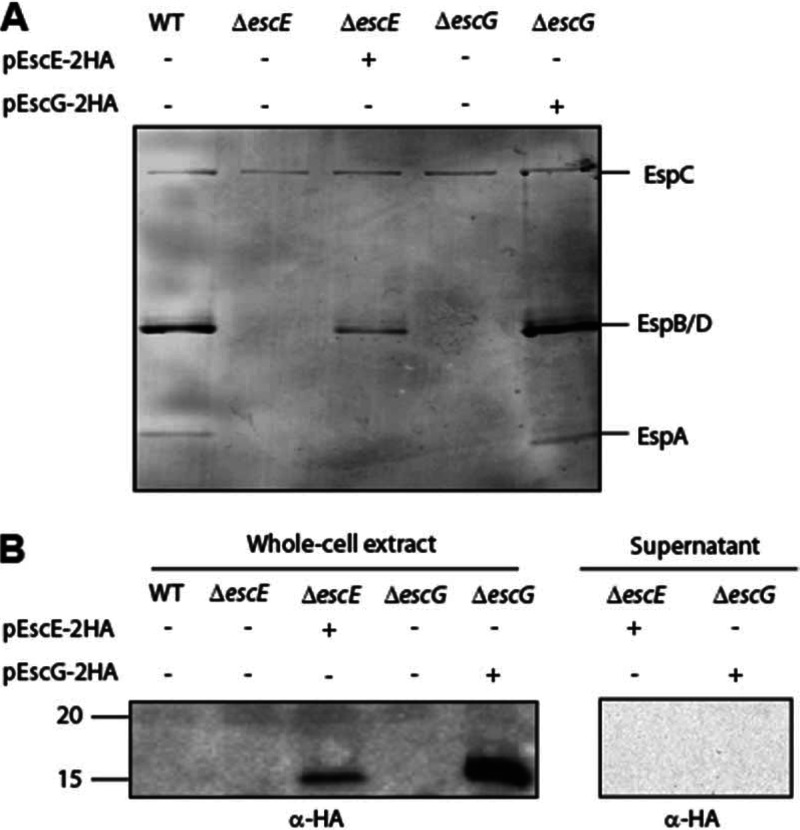
All the components of the T3S apparatus of EPEC found so far are known to be encoded within the LEE. While many of the T3SS components of EPEC are well studied, there are still a few poorly characterized LEE-encoded proteins. These include the Orf2 (EscE) and Orf29 (EscG) proteins. A BLAST search revealed that EscE and EscG are highly conserved among the pathogenicity islands of A/E pathogens (Fig. 1). To determine if EscE and EscG are essential for T3S, as expected for chaperones of a structural T3S component, we analyzed the T3S in EPEC escE- and escG-null strains (ΔescE and ΔescG strains) by SDS-PAGE and Coomassie blue staining. The null strains did not secrete type III translocators (EspA, EspB, and EspD), indicating that EscE and EscG are both essential for T3S (Fig. 2A). Similar results were shown for the related murine A/E pathogen Citrobacter rodentium (25). The T3S phenotype of the escE and escG mutants was similar of that of the escF mutant. Complementation of the mutant strain with full-length escE or escG in trans restored the secretion of the translocators, thus confirming that the deletion of escE and escG is nonpolar.
Fig 1.
Sequence alignment of proteins that have been suggested to function as the chaperones of the type III needle protein in attaching and effacing pathogens. (A) BLAST (http://blast.ncbi.nlm.nih.gov) and multiple sequence alignment (using CLUSTAL W) of EscE (Orf2) from EPEC with T3S proteins of the related A/E pathogens enterohemorrhagic E. coli (EHEC; O157:H7) and Citrobacter rodentium. NCBI protein identification numbers are as follows: for EPEC Orf2, AAC38365.1; for EHEC Orf2, NP_290287.1; and for Citrobacter rodentium (CR) Orf2, YP_003366521. Asterisks indicate positions that have a single, fully conserved residue. Colons indicate conservation of strongly similar properties between groups. Periods indicate conservation of weakly similar properties between groups. (B) BLAST and multiple sequence alignment of EscG (Orf29) from EPEC with T3S proteins of enterohemorrhagic E. coli and C. rodentium. Protein identification numbers are as follows: for EPEC Orf29, AAC38399.1; for EHEC Orf29, NP_290251.1; and for Citrobacter rodentium Orf29, YP_003366485.1.
Fig 2.
EscE and EscG are essential for T3S. (A) Protein secretion profiles of wild-type (WT) EPEC, the ΔescE mutant, the ΔescE mutant complemented with pescE, the ΔescG mutant, and the ΔescG mutant complemented with pescG. Secreted proteins were concentrated from supernatants of bacterial cultures grown in DMEM and were analyzed by Coomassie staining of an SDS-12% PAGE gel. The locations of the translocators EspA, EspB, and EspD are indicated on the right. Also indicated is the location of EPEC EspC, which is not secreted via the LEE-encoded T3SS. (B) Whole-cell extracts of EPEC strains expressing EscE-2HA or EscG-2HA grown under T3S-inducing conditions were examined for expression and secretion of EscE-2HA and EscG-2HA. Samples were analyzed by SDS-16% PAGE and immunoblotting using an anti-HA antibody. Molecular size markers (in kilodaltons) are indicated on the left.
To determine whether EscE and EscG are secreted components of the T3SS, we assessed the ability of the EPEC ΔescE or ΔescG mutant complemented with HA-tagged EscE or EscG, respectively, to secrete the tagged protein when grown under T3S-inducing conditions. Whole-cell extracts and supernatants were analyzed by immunoblotting with a mouse anti-HA antibody. Neither EscE nor EscG was detected in the secreted samples of the complemented strains, although the tagged proteins were expressed in the whole-cell extracts (Fig. 2B). These results demonstrate that EscE and EscG are nonsecreted components of the T3SS in EPEC.
Subcellular localization of EscE and EscG.
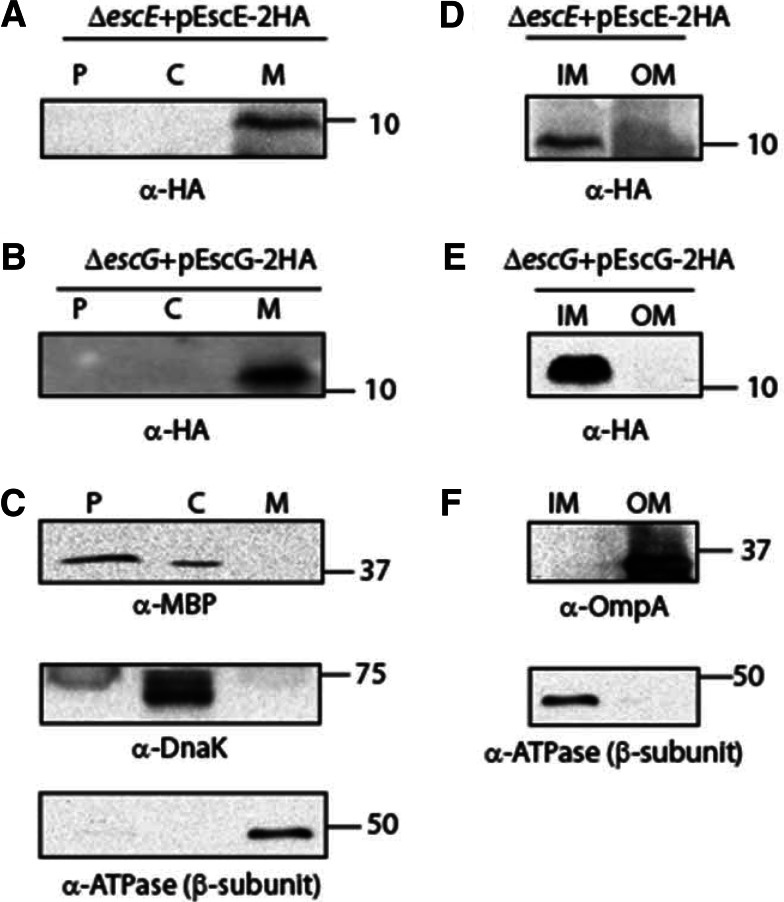
The subcellular localization of HA-tagged EscE or HA-tagged EscG expressed in the ΔescE or ΔescG strain, respectively, grown under T3S inducing conditions was examined in whole-cell extracts that were fractionated into periplasmic, cytoplasmic, and membrane fractions. EscE and EscG were detected using an antibody against HA. We found that both EscE and EscG were enriched in the membrane fraction (Fig. 3A and B, respectively). Western blots were probed with cytoplasmic, periplasmic, and membrane markers to confirm proper fractionation (Fig. 3C). In order to determine whether EscE and EscG were localized in a specific membrane fraction, i.e., in the outer or the inner membrane, a detergent-selective solubilization assay was applied to further fractionate the membranes. Both EscE and EscG were found to be localized mainly to the IM fraction (Fig. 3D and E). Western blots were probed with IM and OM markers, which confirmed proper fractionation (Fig. 3F). These results suggested that although EscE and EscG are predicted to be cytosolic according to the SubcellPredict program (32), they are found predominantly in the IM fraction. A possible explanation of the partitioning of EscG into the inner membrane was the presence of a predicted lipidation motif (LxGC) at the C terminus of the protein. To investigate this possibility, we mutated the cysteine at position 68 to a serine residue and examined the effect of this mutation on the subcellular localization of the protein. We found that EscG mutated at position 68 showed localization to the inner membrane similar to that of the wild-type protein (see Fig. S1 in the supplemental material), suggesting that the localization of EscG to the membrane was not dependent on the predicted lipidation motif.
Fig 3.
Subcellular localization of EscE and EscG. (A) The EPEC ΔescE strain expressing EscE-2HA was grown under T3S-inducing conditions and was fractionated into periplasmic (P), cytoplasmic (C), and membrane (M) fractions. Two micrograms of protein from each fraction was loaded onto an SDS-16% PAGE gel, transferred to a nitrocellulose membrane, and blotted with an anti-HA antibody to detect EscE-2HA. (B) Fractionation of the EPEC ΔescG strain expressing EscG-2HA by use of conditions similar to those described for panel A. (C) To confirm correct fractionation, the Western blots were also probed with antibodies against MBP (periplasmic marker), DnaK (cytoplasmic marker), and the beta subunit of ATPase (membrane marker). (D and E) The membrane fraction was further separated into the inner membrane (IM) and outer membrane (OM) by using selective membrane solubilization detergents in order to examine the exact locations of EscE-2HA (D) and EscG-2HA (E). (F) To confirm correct fractionation, the Western blots were also probed with antibodies against OmpA (OM marker) and the beta subunit of ATPase (IM marker).
EscF-EscE-EscG complex formation.
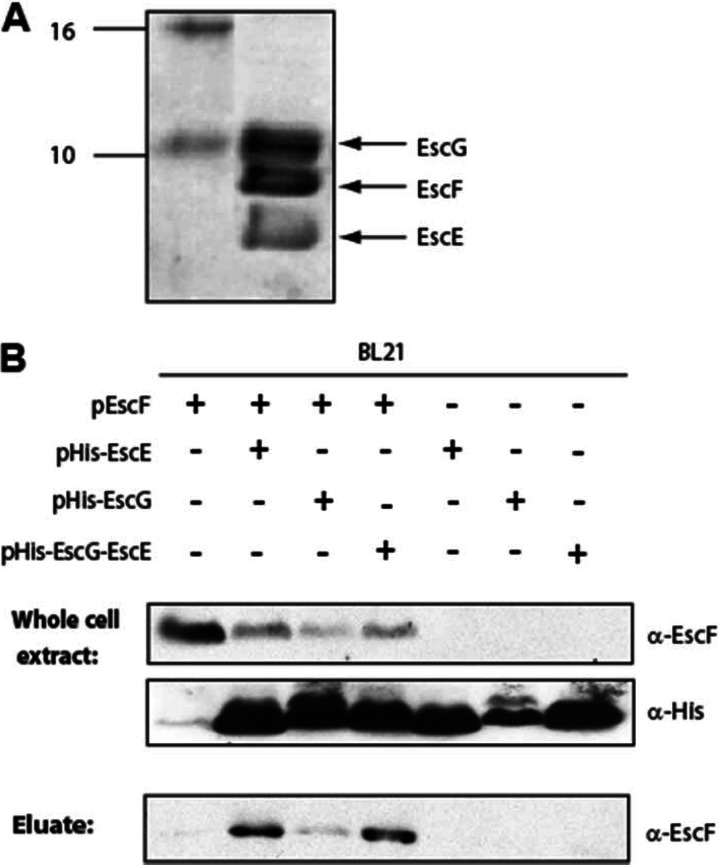
If EscE and EscG are indeed functional homologs of chaperones from other pathogens, then they may be expected to form a complex with the needle protein EscF. To test this hypothesis, we coexpressed His-EscE, EscG, and EscFΔ5aa in E. coli BL21(DE3), and we purified His-EscE using nickel affinity chromatography. Analysis of the eluted fraction by SDS-PAGE and Coomassie staining revealed three protein bands (Fig. 4A). The sizes of these bands matched the predicted sizes of EscE, EscG, and EscF, and their identities were further supported by MALDI-TOF analysis (data not shown). To confirm the formation of the EscE-EscG-EscF complex and to further characterize the interactions within the complex, we examined the abilities of EscE and EscG to interact directly with EscF. For this purpose, E. coli BL21(DE3) expressing EscF was compared to E. coli BL21(DE3) coexpressing EscF with either pHis-EscE, pHis-EscG, or pHis-EscG-EscE. Analysis of whole-cell extracts confirmed the correct expression of the proteins of interest (Fig. 4B). His-tagged proteins were extracted and purified using nickel affinity chromatography. Immunoblotting of the eluted fractions with an anti-EscF antibody showed that the EscF protein coelutes with His-EscE and, to a lesser extent, with His-EscG (Fig. 4B). When His-EscG was coexpressed with untagged EscE (pHis-EscG-EscE) and EscF and was purified using nickel affinity chromatography, EscF coeluted with EscG to a greater extent. These results suggested that both EscE and EscG can bind directly to EscF. However, binding of EscG to the needle protein EscF was enhanced in the presence of EscE.
Fig 4.
EscE-EscF-EscG complex formation. (A) Purification of His-EscE by nickel affinity chromatography coeluted two extra proteins, corresponding to the sizes of the coexpressed proteins EscF and EscG. (B) Whole-cell extracts of E. coli BL21(λDE3) transformed with pEscF, either alone (pET21) or together with a vector expressing pHis-EscE (pET28), pHis-EscG (pET28), or pHis-EscG-EscE (pET28), were subjected to nickel affinity chromatography. Whole-cell extracts and eluted fractions were analyzed by Western blotting with probing for EscF (using anti-EscF) and EscE or EscG (using anti-His).
The EscE-EscG complex inhibits EscF self-polymerization in vitro.
The role of the chaperones of the needle protein is to prevent premature polymerization of the needle protein within the bacterial cytoplasm. To investigate whether EscE-EscG can trap EscF in a monomeric state within a ternary complex, we mixed purified EscF with the purified His-EscG-EscE complex. To enhance the monomeric concentration of EscF within the purified fraction, His-EscF was sonicated shortly before the addition of the His-EscG-EscE complex, as described in reference 33. Subsequently, the samples were incubated for an hour at 4°C. Then the proteins were separated according to their sizes by using size exclusion chromatography (SEC). The protein elution profile was monitored by UV detection and was recorded over time. The elution profile of purified EscF revealed five major protein peaks, suggesting different oligomeric states of EscF (Fig. 5A). To assess the effect of the EscG-EscE complex on EscF polymerization, we compared the elution profile of EscF alone with the elution profile of EscF that had been preincubated with the purified EscG-EscE complex (after subtraction of the elution profile of the EscG-EscE complex alone). This revealed a clear shift in the elution profile of EscF from large molecular complexes to smaller complexes (Fig. 5A). The areas of the major elution peaks revealed that about 25% of the total protein measured for purified EscF eluted as large molecular complexes (peak I), compared to only 1% when the EscG-EscE complex was present. A corresponding increase in the proportions of protein species of smaller sizes (peaks IV and V) was observed—from 18% of the total protein for EscF alone to 36% in the presence of the EscG-EscE complex (Fig. 5A).
Fig 5.
EscE and EscG alter the polymerization of EscF. (A) Five milligrams of purified His-EscF was incubated in the presence (red) and absence (black) of the His-EscG-EscE (EscG/E) complex for an hour and was then analyzed under nondenaturing conditions using SEC. The elution profile of EscF in the presence of the EscG/E complex is shown after the subtraction of the elution profile of the EscG/E complex alone. (B) Samples (7 μg) of purified EscF were mixed with 0, 2, 4, or 8 μg of purified EscG/E complex. The samples were analyzed on a BN-PAGE gel and were immunoblotted with anti-EscF in order to examine the effect of EscG/E on the polymeric state of EscF. Molecular size markers (in kilodaltons) are indicated on the left. EscF complexes of different sizes are marked with asterisks.
To better visualize the size distribution of various EscF oligomeric states and to further confirm the effect of the EscG-EscE complex on EscF oligomerization, BN-PAGE was employed as a complementary approach to SEC. Purified EscF samples were mixed with increasing concentrations of the purified His-EscG-EscE complex. The samples were first incubated for an hour at 4°C, then separated using BN-PAGE, and subsequently analyzed by immunoblotting using an antibody against EscF. Purified EscF (∼9 kDa, including the His tag) demonstrated the clear behavior of a self-polymerizing protein, with a dominant complex around 60 to 70 kDa (Fig. 5B). Additionally, very large molecular complexes were found close to the 1,236-kDa marker. Upon the addition of the His-EscG-EscE complex, the polymerization of EscF was affected: the very large molecular mass complex at around 1,236 kDa disappeared completely, and a decrease in the level of EscF complexes at around 66 kDa was observed, with simultaneous increases in the levels of two lower-order oligomeric species (Fig. 5B). This effect of the EscG-EscE proteins on EscF was concentration dependent. It is possible that longer incubation times are required to completely solubilize the polymerization of the needle protein. It is also likely that a portion of the purified EscF formed a nonreversible polymer state, since the protein was expressed and purified in the absence of its chaperones. Collectively, these experiments indicated that the EscG-EscE complex is capable of inhibiting the polymerization of the needle protein EscF into filaments in vitro, a conclusion that is coherent with our observations using SEC.
EscE and EscG align structurally with the known structures of the chaperones of the needle protein complex.
Since the primary sequences of many chaperones of the needle protein are not highly conserved among T3SSs or flagellar export systems, we examined whether EscE and EscG align structurally with known tertiary structures of needle protein chaperones from P. aeruginosa (14, 15, 17). The tertiary structures of EscE and EscG were predicted using the I-TASSER structural prediction software (29, 30). Both predicted structures revealed predominantly an α-helical fold similar to known structures of chaperones from other pathogens (14, 15, 17). The predicted EscE and EscG structures were aligned and superimposed on the known structures of the P. aeruginosa heterotrimeric core complex PscE-PscF-PscG (Fig. 6). The resulting structural model clearly revealed high structural homology of EscE and EscG with the chaperones of the needle protein of P. aeruginosa. This further supports the notion that EscG is indeed a homolog of PscG and YscG even though it does not show primary sequence homology to PscG or YscG (21). The structure of the P. aeruginosa PscE-PscF-PscG complex does not comprise the full-length needle protein PscF, but only an N-terminally truncated version of PscF. To better understand the intermolecular communication of the chaperones and the needle protein within the complex, we further superimposed the slightly more N-terminally extended structure of the needle protein YscF from Yersinia pestis onto our structural model. This clearly revealed a close proximity between the N-terminal domain of YscF and the EscE or PscE chaperone and thus further provided a possible explanation for the interaction between EscE and EscF observed in this study.
Fig 6.
Structural alignment of EscE and EscG with the heterotrimeric core complex PscE-PscF-PscG from P. aeruginosa. The superposition of structurally aligned EscE (red) and EscG (blue) on the P. aeruginosa needle regulatory complex (PscE [amino acids {aa} 16 to 70], PscG [aa 1 to 115], and PscF [aa 54 to 84]) (gray) (Protein Data Bank [PDB] code 2UWJ) demonstrates structural homology between EscE and PscE and between EscG and PscG. The 3D structural models of EscE and EscG were predicted by I-TASSER. The superposition of the homologous Yersinia pestis needle protein YscF (green) (aa 50 to 87) (PDB 2P58) on PscF reveals the close proximity of the Yersinia needle protein to the cochaperone EscE. The structural alignments were performed using UCSF Chimera software, version 1.6.1.
DISCUSSION
The needle protein of the T3SSs of many Gram-negative pathogens is an essential structural component of the secretion apparatus that is required for the injection of effector proteins into host cells (34). The strong tendency of the needle protein to polymerize enables this small protein to form a long, helical needle filament (8, 35, 36). However, this polymerization propensity has to be blocked within the bacterial cells before the protein is secreted. Such an activity has been described for the class V chaperones, which include Yersinia YscE/YscG, Pseudomonas PscE/PscG, and Aeromonas AscE/AscG (14–17). While YscE/YscG, PscE/PscG, and AscE/AscG (14, 16–18) revealed high structural similarity, their sequence identity was relatively low, making it difficult to predict the identities of their homologs in other pathogens. In this study, we aimed to test the bioinformatics prediction made by Pallen et al., which suggested that Orf2 and Orf29 are the cochaperones of the needle protein EscF from the A/E pathogens (21). Overall, our results provide experimental evidence that Orf2 and Orf29 share many common functional and structural features with the class V chaperones within T3SSs. Based on our results, we renamed Orf2 and Orf29 EscE and EscG, respectively.
In the structure of the ternary complex comprising the P. aeruginosa needle protein and its chaperones, PscG appears to be the main binding partner of PscF, while PscE binds PscF only peripherally (16, 20). A similar organization was observed for the Yersinia needle protein regulatory complex (17) and the A. hydrophila AscE/AscF/AscG complex (16, 20). Based on these observations, it was suggested that PscE, YscE, or AscE stabilizes the fold of PscG, YscG, or AscG, while the latter is the primary chaperone of PscF, YscF, or AscF, respectively. Interestingly, we found in this study that both EscE and EscG bind EscF directly (Fig. 4). Moreover, EscF coeluted to a higher extent with His-EscE than with His-EscG. The ability of His-EscG to coelute with EscF was significantly higher when His-EscG was coexpressed with untagged EscE (pHis-EscG-EscE), suggesting that EscE contributes to the intermolecular stability of the complex. Furthermore, our structural model of an EscE-EscG-needle protein complex revealed close proximity of the N terminus of the needle protein to EscE, supporting direct interaction between the two proteins. Another possibility is that EscE binds EscF prior to EscG in order to allow the needle protein to adopt the conformation required for binding to EscG. Upon the formation of the ternary complex, the initial conformation of the needle protein may change, and only its final conformation within the heterotrimeric complex can be observed in the crystal structure. However, it is necessary to determine the structure of a ternary complex comprising the full-length needle protein in order to answer this question unambiguously. Obtaining this intermolecular structural data will also provide insight into the higher stability of EscG when coexpressed with EscE, which was observed by others and in this study (15, 18, 20).
To provide a direct indication that EscE and EscG function as the cochaperones of EscF, we examined the effects of these proteins on the polymerization of EscF in vitro by using gel filtration and BN-PAGE. Analysis of purified proteins revealed that the EscE/EscG complex prevents EscF polymerization or dissociates EscF oligomers in vitro (Fig. 5), as shown previously for the chaperones of flagellin (33) and the needle protein of the T3SS of P. aeruginosa (15). The alteration to smaller EscF complexes was found to be dependent on the concentration of EscE-EscG that was added to EscF. On the basis of these results, we concluded not only that EscE and EscG are “bodyguards” that prevent the polymerization of EscF within the bacterial cells but also that the polymerization of EscF is reversible to some degree. This pointed us to the needle polymer as a potential site for prevention or therapeutics for pathogenic E. coli. Vaccination against the needle monomers has been shown to provide mice with resistance against Y. pestis (37), demonstrating that the needle is a viable target for drug design. Based on a recent study that has shown that coiled-coil peptides can be used to inhibit the polymerization of another polymerizing protein of EPEC, EspA, and reduce the cytotoxicity of the pathogen (38), we speculate that external addition of purified chaperones to the bacterial cells might disrupt EscF polymerization and prevent the colonization of the host cell. Identification of the sequences of EscE and EscG that bind EscF might allow the design of specific antivirulence peptides that will bind to the needle protein and disrupt the formation of functional T3SS needles.
In conclusion, a better understanding of the E. coli T3SS offers valuable insights into the virulence mechanisms of EPEC and other T3SS-expressing pathogens. We believe that this study improves our understanding of T3SS assembly and provides a possible target for drug design.
Supplementary Material
ACKNOWLEDGMENTS
We thank DeLu (Tyler) Yin (University of British Columbia) for help with the gel filtration experiments.
Work in the laboratory of B.B.F. is funded by the Canadian Institutes of Health Research (CIHR). B.B.F. is the University of British Columbia Peter Wall Distinguished Professor. N.S.-M. is supported by postdoctoral fellowships from the Michael Smith Foundation for Health Research (MSFHR), the Natural Sciences and Engineering Research Council of Canada (NSERC), the Rothschild Foundation, and the Mitacs Elevate program. Work in the laboratory of N.C.J.S. is funded by the CIHR, the Howard Hughes Medical Institute International Scholar Program, the MSFHR, the Canada Foundation for Innovation, and the British Columbia Knowledge Development Fund.
Footnotes
Published ahead of print 22 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00118-13.
REFERENCES
- 1. Cornelis GR, Van Gijsegem F. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735–774 [DOI] [PubMed] [Google Scholar]
- 2. Hueck CJ. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bhavsar AP, Guttman JA, Finlay BB. 2007. Manipulation of host-cell pathways by bacterial pathogens. Nature 449:827–834 [DOI] [PubMed] [Google Scholar]
- 4. Dean P, Kenny B. 2009. The effector repertoire of enteropathogenic E. coli: ganging up on the host cell. Curr. Opin. Microbiol. 12:101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sal-Man N, Biemans-Oldehinkel E, Finlay BB. 2009. Structural microengineers: pathogenic Escherichia coli redesigns the actin cytoskeleton in host cells. Structure 17:15–19 [DOI] [PubMed] [Google Scholar]
- 6. Black RE, Cousens S, Johnson HL, Lawn JE, Rudan I, Bassani DG, Jha P, Campbell H, Walker CF, Cibulskis R, Eisele T, Liu L, Mathers C. 2010. Global, regional, and national causes of child mortality in 2008: a systematic analysis. Lancet 375:1969–1987 [DOI] [PubMed] [Google Scholar]
- 7. Blocker A, Komoriya K, Aizawa S. 2003. Type III secretion systems and bacterial flagella: insights into their function from structural similarities. Proc. Natl. Acad. Sci. U. S. A. 100:3027–3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blocker A, Jouihri N, Larquet E, Gounon P, Ebel F, Parsot C, Sansonetti P, Allaoui A. 2001. Structure and composition of the Shigella flexneri “needle complex”, a part of its type III secreton. Mol. Microbiol. 39:652–663 [DOI] [PubMed] [Google Scholar]
- 9. Chacon MR, Soler L, Groisman EA, Guarro J, Figueras MJ. 2004. Type III secretion system genes in clinical Aeromonas isolates. J. Clin. Microbiol. 42:1285–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hoiczyk E, Blobel G. 2001. Polymerization of a single protein of the pathogen Yersinia enterocolitica into needles punctures eukaryotic cells. Proc. Natl. Acad. Sci. U. S. A. 98:4669–4674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kimbrough TG, Miller SI. 2000. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc. Natl. Acad. Sci. U. S. A. 97:11008–11013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pastor A, Chabert J, Louwagie M, Garin J, Attree I. 2005. PscF is a major component of the Pseudomonas aeruginosa type III secretion needle. FEMS Microbiol. Lett. 253:95–101 [DOI] [PubMed] [Google Scholar]
- 13. Poyraz O, Schmidt H, Seidel K, Delissen F, Ader C, Tenenboim H, Goosmann C, Laube B, Thunemann AF, Zychlinsky A, Baldus M, Lange A, Griesinger C, Kolbe M. 2010. Protein refolding is required for assembly of the type three secretion needle. Nat. Struct. Mol. Biol. 17:788–792 [DOI] [PubMed] [Google Scholar]
- 14. Chatterjee C, Kumar S, Chakraborty S, Tan YW, Leung KY, Sivaraman J, Mok YK. 2011. Crystal structure of the heteromolecular chaperone, AscE-AscG, from the type III secretion system in Aeromonas hydrophila. PLoS One 6:e19208 doi:10.1371/journal.pone.0019208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Quinaud M, Chabert J, Faudry E, Neumann E, Lemaire D, Pastor A, Elsen S, Dessen A, Attree I. 2005. The PscE-PscF-PscG complex controls type III secretion needle biogenesis in Pseudomonas aeruginosa. J. Biol. Chem. 280:36293–36300 [DOI] [PubMed] [Google Scholar]
- 16. Quinaud M, Ple S, Job V, Contreras-Martel C, Simorre JP, Attree I, Dessen A. 2007. Structure of the heterotrimeric complex that regulates type III secretion needle formation. Proc. Natl. Acad. Sci. U. S. A. 104:7803–7808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sun P, Tropea JE, Austin BP, Cherry S, Waugh DS. 2008. Structural characterization of the Yersinia pestis type III secretion system needle protein YscF in complex with its heterodimeric chaperone YscE/YscG. J. Mol. Biol. 377:819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tan YW, Yu HB, Leung KY, Sivaraman J, Mok YK. 2008. Structure of AscE and induced burial regions in AscE and AscG upon formation of the chaperone needle-subunit complex of type III secretion system in Aeromonas hydrophila. Protein Sci. 17:1748–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Page AL, Parsot C. 2002. Chaperones of the type III secretion pathway: jacks of all trades. Mol. Microbiol. 46:1–11 [DOI] [PubMed] [Google Scholar]
- 20. Ple S, Job V, Dessen A, Attree I. 2010. Cochaperone interactions in export of the type III needle component PscF of Pseudomonas aeruginosa. J. Bacteriol. 192:3801–3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pallen MJ, Beatson SA, Bailey CM. 2005. Bioinformatics, genomics and evolution of non-flagellar type-III secretion systems: a Darwinian perspective. FEMS Microbiol. Rev. 29:201–229 [DOI] [PubMed] [Google Scholar]
- 22. Creasey EA, Delahay RM, Daniell SJ, Frankel G. 2003. Yeast two-hybrid system survey of interactions between LEE-encoded proteins of enteropathogenic Escherichia coli. Microbiology 149:2093–2106 [DOI] [PubMed] [Google Scholar]
- 23. Kaniga K, Delor I, Cornelis GR. 1991. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109:137–141 [DOI] [PubMed] [Google Scholar]
- 24. Edwards RA, Keller LH, Schifferli DM. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207:149–157 [DOI] [PubMed] [Google Scholar]
- 25. Deng W, Puente JL, Gruenheid S, Li Y, Vallance BA, Vazquez A, Barba J, Ibarra JA, O'Donnell P, Metalnikov P, Ashman K, Lee S, Goode D, Pawson T, Finlay BB. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. U. S. A. 101:3597–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ilan O, Bloch Y, Frankel G, Ullrich H, Geider K, Rosenshine I. 1999. Protein tyrosine kinases in bacterial pathogens are associated with virulence and production of exopolysaccharide. EMBO J. 18:3241–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yu AC, Worrall LJ, Strynadka NC. 2012. Structural insight into the bacterial mucinase StcE essential to adhesion and immune evasion during enterohemorrhagic E. coli infection. Structure 20:707–717 [DOI] [PubMed] [Google Scholar]
- 28. Scholz R, Suter M, Weimann T, Polge C, Konarev PV, Thali RF, Tuerk RD, Viollet B, Wallimann T, Schlattner U, Neumann D. 2009. Homo-oligomerization and activation of AMP-activated protein kinase are mediated by the kinase domain αG-helix. J. Biol. Chem. 284:27425–27437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5:725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40 doi:10.1186/1471-2105-9-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 32. Niu B, Jin YH, Feng KY, Lu WC, Cai YD, Li GZ. 2008. Using AdaBoost for the prediction of subcellular location of prokaryotic and eukaryotic proteins. Mol. Divers. 12:41–45 [DOI] [PubMed] [Google Scholar]
- 33. Auvray F, Thomas J, Fraser GM, Hughes C. 2001. Flagellin polymerisation control by a cytosolic export chaperone. J. Mol. Biol. 308:221–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Loquet A, Sgourakis NG, Gupta R, Giller K, Riedel D, Goosmann C, Griesinger C, Kolbe M, Baker D, Becker S, Lange A. 2012. Atomic model of the type III secretion system needle. Nature 486:276–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cordes FS, Komoriya K, Larquet E, Yang S, Egelman EH, Blocker A, Lea SM. 2003. Helical structure of the needle of the type III secretion system of Shigella flexneri. J. Biol. Chem. 278:17103–17107 [DOI] [PubMed] [Google Scholar]
- 36. Fujii T, Cheung M, Blanco A, Kato T, Blocker AJ, Namba K. 2012. Structure of a type III secretion needle at 7-Å resolution provides insights into its assembly and signaling mechanisms. Proc. Natl. Acad. Sci. U. S. A. 109:4461–4466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Matson JS, Durick KA, Bradley DS, Nilles ML. 2005. Immunization of mice with YscF provides protection from Yersinia pestis infections. BMC Microbiol. 5:38 doi:10.1186/1471-2180-5-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Larzabal M, Mercado EC, Vilte DA, Salazar-Gonzalez H, Cataldi A, Navarro-Garcia F. 2010. Designed coiled-coil peptides inhibit the type three secretion system of enteropathogenic Escherichia coli. PLoS One 5:e9046 doi:10.1371/journal.pone.0009046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Levine MM, Bergquist EJ, Nalin DR, Waterman DH, Hornick RB, Young CR, Sotman S. 1978. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet i:1119–1122 [DOI] [PubMed] [Google Scholar]
- 40. Rose RE. 1988. The nucleotide sequence of pACYC184. Nucleic Acids Res. 16:355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.