Abstract
The Gram-positive Corynebacterium glutamicum efficiently metabolizes maltose by a pathway involving maltodextrin and glucose formation by 4-α-glucanotransferase, glucose phosphorylation by glucose kinases, and maltodextrin degradation via maltodextrin phosphorylase and α-phosphoglucomutase. However, maltose uptake in C. glutamicum has not been investigated. Interestingly, the presence of maltose in the medium causes increased expression of ptsG in C. glutamicum by an unknown mechanism, although the ptsG-encoded glucose-specific EII permease of the phosphotransferase system itself is not required for maltose utilization. We identified the maltose uptake system as an ABC transporter encoded by musK (cg2708; ATPase subunit), musE (cg2705; substrate binding protein), musF (cg2704; permease), and musG (cg2703; permease) by combination of data obtained from characterization of maltose uptake and reanalyses of transcriptome data. Deletion of the mus gene cluster in C. glutamicum Δmus abolished maltose uptake and utilization. Northern blotting and reverse transcription-PCR experiments revealed that musK and musE are transcribed monocistronically, whereas musF and musG are part of an operon together with cg2701 (musI), which encodes a membrane protein of unknown function with no homologies to characterized proteins. Characterization of growth and [14C]maltose uptake in the musI insertion strain C. glutamicum IMcg2701 showed that musI encodes a novel essential component of the maltose ABC transporter of C. glutamicum. Finally, ptsG expression during cultivation on different carbon sources was analyzed in the maltose uptake-deficient strain C. glutamicum Δmus. Indeed, maltose uptake by the novel ABC transport system MusEFGK2I is required for the positive effect of maltose on ptsG expression in C. glutamicum.
INTRODUCTION
Corynebacterium glutamicum, a Gram-positive, nonsporulating bacterium, is widely employed in the large-scale production of amino acids, mainly l-glutamate and l-lysine (1, 2). The organism can be cultivated on a large variety of substrates, such as carbohydrates, alcohols, and organic acids (3, 4). Maltose serves as an excellent substrate for cultivation of and amino acid production with C. glutamicum (5, 6). Furthermore, addition of maltose to the culture broth positively affects glucose utilization by this bacterium, a unique phenomenon which can be exploited to significantly improve amino acid production (7). Based on enzyme assays and analyses of the genome sequence, a metabolic pathway for maltose utilization in C. glutamicum has been described (6) which involves maltodextrin and glucose formation catalyzed by the 4-α-glucanotransferase MalQ, maltodextrin degradation to glucose 6-phosphate via the maltodextrinphosphorylase MalP and the α-phosphoglucomutase Pgm, and phosphorylation of the glucose derived from the MalQ reaction by the glucose kinases Glk and PPgk (8, 9). Nonetheless, hitherto a maltose uptake system has not been identified in C. glutamicum.
Similar metabolic pathways for maltose utilization have already been identified in the Gram-negative bacterium Escherichia coli (reviewed in reference 10) and the archaeon Thermococcus litoralis (11), which both use well-characterized ATP-binding cassette (ABC) transporters for maltose uptake (12–18). However, in Gram-positive bacteria, maltose uptake is accomplished by various types of uptake systems, such as maltose-H+ symporters in, e.g., Bacillus licheniformis (19) and Lactobacillus sanfrancisco (20), phosphoenolpyruvate sugar phosphotransferase systems (PTS) in, e.g., Streptococcus bovis (21), Streptococcus mutans (22), and Bacillus subtilis (23), or ABC transporters in, e.g., Alicyclobacillus acidocaldarius (24). Analyses of the genome sequence of C. glutamicum indicated the presence of about 260 putative transport proteins (25, 26), the majority belonging to the categories of secondary carriers and of primary transporters (117 and 106, respectively). Furthermore, four phosphotransferase system enzyme II genes are present in C. glutamicum.
The positive effect of maltose on glucose utilization in C. glutamicum comes with the increased expression of ptsG (7, 27), which encodes the glucose-specific EII permease (EIIGlc) of the PTS (28, 29). Based on this observation, it seems reasonable to suggest that the ptsG-encoded EIIGlc additionally catalyzes maltose uptake; however, inactivation of ptsG itself or any of the three further genes for EII permeases in C. glutamicum did not affect growth with maltose as the sole carbon source (29).
In this study, we identified the maltose/maltodextrin uptake system (MUS) of C. glutamicum as an ABC transporter by combination of data obtained through characterization of maltose uptake with recently published transcriptome data. This ABC sugar uptake system is unusual, as it requires an additional, uncharacterized membrane protein for maltose transport activity. Here, we show that this uptake system is essential for growth of C. glutamicum on maltose and maltotriose as sole carbon sources. Furthermore, we investigated the transcriptional organization of the genes encoding the maltose uptake system in C. glutamicum and analyzed the substrate-dependent influence of the maltose uptake system on ptsG expression.
MATERIALS AND METHODS
Microorganisms, plasmids, oligonucleotides, and cultivation conditions.
C. glutamicum and E. coli strains and their relevant characteristics are listed in Table 1. Plasmids, their relevant characteristics and sources, and oligonucleotides used in this study are listed in Table 2 and Table S1 in the supplemental material. The CgC minimal medium used for the cultivation of C. glutamicum has been described previously (30) and contained maltose, maltotriose, glucose, and/or acetate at concentrations indicated in Results. TY broth (31) was utilized as complex medium for cultivation of C. glutamicum and E. coli. When appropriate, the media contained kanamycin (50 μg ml−1 for plasmids and 15 μg ml−1 for integration mutants and transformant selection) and/or chloramphenicol (25 μg ml−1). C. glutamicum was grown aerobically at 30°C, and E. coli at 37°C, as 50-ml cultures in 500-ml baffled Erlenmeyer flasks on a rotary shaker at 120 rpm. Growth experiments with maltotetraose or maltopentaose as the substrate were performed as 10-ml cultures in 100-ml baffled Erlenmeyer flasks. Growth of the bacteria was monitored by measuring the optical density at 600 nm (OD600). Maltose and maltotriose concentrations were analyzed by high-performance liquid chromatography (HPLC) as described previously (6).
Table 1.
Strains used in this study
| Strain | Relevant characteristic(s) | Reference/source |
|---|---|---|
| E. coli | ||
| DH5α | F− thi-1 endA1 hsdR17(r− m−) supE44 ΔlacU169 (Φ80lacZΔM15) recA1 gyrA96 relA1 | 80 |
| C. glutamicum | ||
| WT | ATCC 13032; wild-type strain | American Type Culture Collection |
| ΔramB | ramB-negative mutant of C. glutamicum WT | 41 |
| ΔramA | ramA-negative mutant of C. glutamicum WT | 42 |
| ΔsugR | sugR-negative mutant of C. glutamicum WT | 27 |
| Δmus | In-frame deletion of genes cg2708 to cg2703 of C. glutamicum WT | This work |
| IMcg2708 | C. glutamicum WT with insertion of pDrive in cg2708 | This work |
| IMcg2707 | C. glutamicum WT with insertion of pDrive in cg2707 | This work |
| IMcg2705 | C. glutamicum WT with insertion of pDrive in cg2705 | This work |
| IMcg2704 | C. glutamicum WT with insertion of pDrive in cg2704 | This work |
| IMcg2701 | C. glutamicum WT with insertion of pDrive in cg2701 | This work |
| ΔptsG | In-frame deletion of ptsG gene (cg1537) of C. glutamicum WT | This work |
Table 2.
Plasmids used in this study
| Plasmid | Relevant characteristic(s) | Reference/source |
|---|---|---|
| pK19mobsacB | Kanr, mobilizable E. coli vector for the construction of insertion and deletion mutants in C. glutamicum (oriV, sacB, lacZα) | 81 |
| pK19mobsacB-delMus | Kanr, pK19mobsacB with the deletion construct for region cg2708 to cg2703 | This work |
| pK19mobsacB-delPtsG | Kanr, pK19mobsacB with the deletion construct for cg1537 | This work |
| pXMJ19 | Expression vector, ptac, lacIq, Cmr | 36 |
| pXMJ19-musKEFG | pXMJ19 carrying region cg2708 to cg2703 | This work |
| pXMJ19-musKEFGI | pXMJ19 carrying region cg2708 to cg2701 | This work |
| pXMJ19-cg2701 | pXMJ19 carrying gene cg2701 | This work |
| pXMJ19-cg2701-strep | pXMJ19 carrying gene cg2701 with sequence for C-terminal Strep-tag | This work |
| pJET1.2 | Ampr, PCR cloning vector, PT7, eco47IR | Fermentas |
| pDrive | Ampr, Kanr, PCR cloning vector, lacZα, orif1,ori-pUC | Qiagen |
| pDrive-IMcg2708 | Ampr, Kanr, pDrive with internal fragment of cg2708 | This work |
| pDrive-IMcg2707 | Ampr, Kanr, pDrive with internal fragment of cg2707 | This work |
| pDrive-IMcg2705 | Ampr, Kanr, pDrive with internal fragment of cg2705 | This work |
| pDrive-IMcg2704 | Ampr, Kanr, pDrive with internal fragment of cg2704 | This work |
| pDrive-IMcg2701 | Ampr, Kanr, pDrive with internal fragment of cg2701 | This work |
| pDrive-RACE-cg2704 | Ampr, Kanr, pDrive carrying the PCR-amplified fragment from the cg2704 RACE assay | This work |
| pET2 | Promoter probe vector carrying the promoterless cat gene, Kmr | 82 |
| pET2-PRmusF | pET2 containing the musF promoter fragment | This work |
| pET2-PRmusF-TS | pET2 containing a truncated musF promoter fragment | This work |
| pET2-PRptsG | pET2 containing the ptsG promoter fragment | 27 |
DNA preparation, manipulation, and transformation.
Standard procedures were employed for plasmid isolation and for molecular cloning and transformation of E. coli, as well as for electrophoresis (31). Isolation of chromosomal DNA and plasmids of C. glutamicum was performed as described previously (32). Transformation of C. glutamicum was performed by electroporation as described by Tauch et al. (33). PCR experiments were performed in a Flexcycler (Analytik Jena) with Taq DNA polymerase (MBI Fermentas) or Precisior high-fidelity DNA polymerase (BioCat GmbH) with oligonucleotides obtained from Eurofins MWG Operon. All restriction enzymes, T4-DNA ligase, and shrimp alkaline phosphatase were obtained from New England BioLabs and used according to the manufacturer's instructions.
Construction of C. glutamicum mutant strains.
In-frame deletions of the genomic locus comprising cg2708 to cg2703 (mus genes) and of the ptsG (cg1537) gene were constructed using pK19mobsacB as described previously (34). In detail, the flanking regions of the gene/cluster were amplified using primer pairs Δ-gene/cluster_1 plus Δ-gene/cluster_2 and Δ-gene/cluster_3 plus Δ-gene/cluster_4 (see Table S1 in the supplemental material). The two flanking PCR products obtained served as the templates for a crossover PCR using the primer pair Δ-gene/cluster_1 plus Δ-gene/cluster_4. The resulting PCR product was digested with the enzymes indicated in Table S1 and cloned into pK19mobsacB cut with the same enzymes. Gene deletion with the derived pK19mobsacBΔgene/cluster plasmids was carried out as described previously (34). The deletion of the cluster comprising open reading frames (ORFs) cg2708 to cg2703 was verified by PCR using the primer pair checkΔMusFOR and checkΔMusREV, resulting in a 7,018-bp fragment for the wild type (WT) and a 1,188-bp fragment for the mus deletion mutant; ptsG deletion was verified using the primers checkΔpstgFOR and checkΔptsgREV, resulting in a 3,583-bp fragment for the WT and a 1,867-bp fragment for the deletion mutant.
Insertion mutagenesis was applied for the generation of single-gene mutants, using vector pDrive as recently described (28). For this purpose, internal fragments of the loci were amplified by PCR using the primers IM-orf-fw and IM-orf-rev (see Table S1 in the supplemental material) and cloned into vector pDrive according to the manufacturer's instructions. The resulting plasmids were isolated and used for gene disruption as described previously (35). Integration into the genome in the resulting strains was verified by PCR using gene-specific primers CONTR-orf (see Table S1) and M13-FP.
Construction of expression vectors.
For isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible overexpression, vector pXMJ19 was used (36). Genes were amplified via PCR from genomic DNA of C. glutamicum ATCC 13032 (37) using the oligonucleotide primers listed in Table S1 in the supplemental material. The resulting PCR products were introduced into the cloning vector pJET1.2 (MBI Fermentas) according to the manufacturer's instructions. Primer-attached restriction sites of the PCR products (indicated in Table S1) were used to excise the inserts, and the resulting fragments were ligated into the plasmid pXMJ19 (digested with the same enzymes) and transformed into E. coli DH5α. The resulting plasmids were isolated and the nucleotide sequences controlled by sequencing (GATC Biotech).
Cloning of the musF promoter.
The promoter probe vector pET2 was used to construct a transcriptional fusion of the musF (cg2704) promoter to the promoterless cat gene. The musF promoter fragment was amplified by PCR with the primers PRmusF-for and PRmusF-rev. The 380-bp PCR product, covering the region from 226 bp upstream to 133 bp downstream of the musF translational start codon, was digested with XbaI and BamHI and ligated into the multiple cloning site (MCS) in front of the cat gene in pET2, resulting in pET2-PmusF. Furthermore, plasmid pET2-Pmus-TS, which lacks the musF transcriptional start site, was cloned. Therefore, a shortened musF promoter fragment was generated by PCR using the primers PRmusF-TS-for and PRmusF-rev, and the 214-bp PCR product was digested with XbaI and BamHI and ligated into pET2.
Enzyme assays and protein analysis.
To determine chloramphenicol acetyltransferase (CAT) activity, C. glutamicum cells were harvested, washed twice in 0.1 M Tris-HCl, pH 7.8, and resuspended in the same buffer containing 10 mM MgCl2 and 1 mM EDTA. The specific CAT activity was determined as described by Schreiner et al. (38). Protein concentrations were determined using the Roti-Nanoquant kit (Roth) with bovine serum albumin as the standard. SDS-PAGE was performed according to Laemmli (39). Loading buffer (4×) contained 8% (wt/vol) SDS, 20% (vol/vol) glycerol, 10 mM EDTA, 100 mM Tris-HCl, pH 6.8, 2% (vol/vol) β-mercaptoethanol, and 1 mg/ml bromphenol blue. Membrane preparations and Western blot experiments for detection of the Streptavidin-tagged Cg2701 protein by using antibodies raised against the Strep-tag II (IBA GmbH) were performed as described for the uptake carrier BetP (40).
Protein purification and EMSAs.
RamA was synthesized as hexahistidyl-tagged fusion proteins and purified by Ni2+ affinity chromatography as described previously (41). The binding of purified RamA protein was tested by electrophoretic mobility shift assays (EMSAs) using DNA fragments generated by PCR and purified using the Nucleospin extract kit (Macherey-Nagel). The 380-bp fragment musF-Pr, carrying the musE-musF intergenic region, was amplified using primer pair PRmusF-for and PRmusF-rev. The 211-bp fragment ramBp3b, generated with the primer pair ramBp3b_forw and ramBp_rev, was used as a negative control for RamA binding (42). In the binding assays, 10 to 300 ng of the fragments was incubated with various amounts of RamA (0 to 7.5 μg) in 20 μl 10 mM Tris-HCl reaction buffer, pH 7.6, containing 50 mM NaCl, 1 mM dithiothreitol (DTT), 1 mM EDTA, 10% (wt/vol) glycerol, and 1 μg poly[d(I · C)] for 20 min at room temperature. Subsequently, the mixture was separated on a 2% agarose gel in 1 × TBE buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA) at 70 V and 80 mA and stained with ethidium bromide.
RNA techniques.
Isolation of total RNA from C. glutamicum cells was performed using the NucleoSpin RNAII kit (Macherey& Nagel) as described by Wolf et al. (43). For Northern (RNA) hybridization, digoxigenin (DIG)-11-dUTP-labeled gene-specific antisense RNA probes were prepared from PCR products (generated with oligonucleotides listed in Table S1 in the supplemental material) carrying the T7 promoter by in vitro transcription (1 h, 37°C) using T7 RNA polymerase (MBI Fermentas). For hybridization, total RNA of C. glutamicum was separated on an agarose gel containing 17% (vol/vol) formaldehyde and transferred to a nylon membrane using the VacuGene system from Pharmacia. RNA was cross-linked to the membrane by means of UV irradiation at 125 J · cm−2. Hybridization and detection were carried out according to the DIG application manual (Roche Applied Science). Slot blot experiments were performed as described previously (44).
The transcriptional start site of cg2704 (musG) was determined using the 5′/3′-rapid amplification of cDNA ends (RACE) kit from Roche Diagnostics according to the manufacturer's manual. First-strand cDNA was synthesized from 2 μg of total RNA using the gene-specific primer designated RACE-cg2704-SP1 in Table S1 in the supplemental material. The subsequent PCRs were performed using the primer pair RACE-cg2704-SP2/oligonucleotide anchor primer (the latter is included in the kit). The purified PCR product was ligated into plasmid pDrive (Qiagen), resulting in the recombinant plasmid pDrive-RACE-cg2703, which was sequenced. The transcriptional site was deduced from the sequences obtained.
[14C]maltose uptake studies.
Maltose uptake studies were performed essentially as described for glucose (28). In detail, C. glutamicum cells were grown to mid-exponential growth phase, harvested by centrifugation, washed twice with ice-cold CgC medium, suspended to an OD600 of 2 with CgC medium, and stored on ice until measurement. Before the transport assay, cells were incubated for 3 min at 30°C; the reaction was started by addition of 1 μM to 1 mM [14C]maltose (specific activity, 679 mCi μmol−1; Amersham, Braunschweig, Germany). Inhibitors were added at the concentrations indicated 30 s before the measurements were started. At given time intervals (15, 30, 45, 60, and 120 s), 200-μl samples were filtered through glass fiber filters (Type F; Millipore, Eschborn, Germany) and washed twice with 2.5 ml of 100 mM LiCl. Radioactivity of the samples was determined using scintillation fluid (Rotiszinth; Roth, Germany) and a scintillation counter (LS 6500; Beckmann, Krefeld, Germany). Kinetic parameters as well as standard errors were derived from nonlinear regressions according to the Michaelis-Menten equation by using Sigma Plot software.
Computational analysis.
Databank searches were carried out by using BLAST (45) and the KEGG database (46). Identification of putative rho-independent transcriptional terminators was performed using TransTermHP (47), and MFold (48) was the software used for the calculation of their ΔGo′ values (change in Gibbs free energy under standard conditions). Topology predictions of membrane proteins were performed using both TMHMM (49) and SOSUI (50). Protein sequences were analyzed using CLUSTAL W (51).
RESULTS
Kinetic parameters and transport mechanism for maltose uptake.
Uptake assays with C. glutamicum cells, cultivated in minimal medium with glucose as the sole carbon source and using various concentrations (0.5 to 200 μM) of 14C-labeled maltose, were performed for the determination of kinetic parameters. Maltose uptake in glucose-cultivated cells showed a simple saturation kinetics, with a Km of 1.0 ± 0.2 μM and a Vmax of 22.6 ± 0.8 nmol min−1 mg−1 cell dry matter (cdm) (see Fig. S1 in the supplemental material). Uptake of maltose in cells of the EIIGlc-deficient strain C. glutamicum ΔptsG cultivated on TY complex medium proceeded with a rate of 27.8 ± 1.8 nmol min−1 mg−1 cdm, which is nearly identical to that of cells of C. glutamicum WT (29.4 ± 2.0 nmol min−1 mg−1 cdm), when cells were cultivated in TY and measurements were performed at a maltose concentration of 100 μM. Based on these results, it can be ruled out that the high-affinity uptake of maltose in C. glutamicum requires the ptsG-encoded permease.
To specify the class of uptake system mediating maltose transport, we measured the effects of selective inhibitors on [14C]maltose uptake at a substrate concentration of 100 μM maltose. Simultaneous addition of valinomycin (ionophore for K+) and nigericin (ionophore for H+ and K+) to C. glutamicum cells abolishes the proton-motive force necessary to drive secondary active transporters (52, 53). However, maltose uptake was only reduced to 17.0 ± 5.0 nmol min−1 mg−1 cdm when both inhibitors were present simultaneously in the assay (not shown). Addition of vanadate, which inhibits ATP-dependent primary active transporters, to the assays inhibited maltose uptake in C. glutamicum (11.0 ± 5.0 nmol min−1 mg−1 cdm; experiments not shown). Therefore, it seems feasible that maltose uptake in C. glutamicum is mediated by a high-affinity ATP-binding cassette (ABC) transport system.
Factors affecting maltose uptake in C. glutamicum.
To identify putative parameters affecting maltose uptake properties in C. glutamicum, [14C]maltose uptake in cells cultivated in minimal medium with different carbon sources was analyzed. Rates for [14C]maltose uptake were slightly lower in cells cultivated with maltose than in cells cultivated with glucose as the sole carbon source (17.0 ± 6.4 and 22.7 ± 3.4 nmol min−1 mg−1 cdm, respectively). Reduced [14C]maltose uptake rates were observed for cells cultivated on lactate (13.3 ± 1.5 nmol min−1 mg−1 cdm), on fructose (10.7 ± 1.7 nmol min−1 mg−1 cdm), or on acetate (10.7 ± 1.0 nmol min−1 mg−1 cdm). These differences of the observed uptake rates might be due to carbon source-dependent transcriptional regulation of the genes encoding the maltose uptake system in C. glutamicum.
Three transcriptional regulators, RamA, RamB, and SugR, have been shown to be master regulators for the adjustment of the central metabolism of C. glutamicum in response to the utilization of the gluconeogenic substrate acetate (reviewed in reference 54). We therefore analyzed [14C]maltose uptake and maltose utilization by the RamA-deficient strain C. glutamicum ΔramA, the RamB-deficient strain C. glutamicum ΔramB, and the SugR-deficient strain C. glutamicum ΔsugR. As both uptake of (9.1 ± 0.3 nmol min−1 mg−1 cdm) and growth on (growth rate of 0.27 ± 0.01 h−1) maltose were significantly reduced in C. glutamicum ΔramA compared to C. glutamicum WT (uptake rate, 30.3 ± 2.4 nmol min−1 mg−1 cdm; growth rate, 0.37 ± 0.03 h−1), we concluded that RamA probably acts as an activator of genes for maltose utilization/uptake. Maltose uptake (10.8 ± 0.4 nmol min−1 mg−1 cdm) and growth on maltose (0.27 ± 0.02 h−1) were also found to be slowed in C. glutamicum ΔsugR. This indicates that SugR also acts as an activator of genes for maltose utilization, which is rather unexpected, as this DeoR-type transcriptional regulator has been described as a global repressor of genes required for carbohydrate utilization in C. glutamicum (reviewed in reference 54). Growth of the RamB-deficient strain C. glutamicum ΔramB in minimal medium with maltose (0.34 ± 0.02 h−1) as well as maltose uptake (18.9 ± 1.5 nmol min−1 mg−1 cdm for cells cultivated with glucose) were only slightly slower than those of the parental strain C. glutamicum WT. These data suggest that RamB also is involved in the transcriptional regulation of the genes encoding the maltose uptake system.
Analyses of transcriptome data lead to identification of the maltose uptake system.
Several RNA microarray studies of the transcriptomes of C. glutamicum WT, C. glutamicum ΔramA, C. glutamicum ΔramB, and C. glutamicum ΔsugR, as well as a chromatin immunoprecipitation (ChIP)-to-chip analysis of potential SugR-binding sites, were recently published (27, 55, 56, 57). Based on the results from the characterization of maltose uptake described above, the data from the RNA microarrays were analyzed to identify candidate genes for the maltose uptake system. The following criteria were used for the analyses. Genes encoding the maltose uptake system of C. glutamicum should (i) encode membrane proteins, preferentially components of ABC transporters, (ii) be expressed in cultivations with glucose and repressed in cultivations with acetate, (iii) be induced by RamA and therefore repressed in the ramA deletion mutant C. glutamicum ΔramA when cultivated on glucose, (iv) be repressed in C. glutamicum ΔsugR, and (v) be slightly affected in the ramB deletion mutant C. glutamicum ΔramB. We reanalyzed the above-mentioned transcriptome data using our set of criteria and identified two clusters of genes, cg2181 to cg2184 and cg2703 to cg2708, which indeed encode putative ABC transporters and which were repressed in C. glutamicum WT during cultivation with acetate, and they were also repressed in C. glutamicum ΔramA when cultivated with glucose. As the cluster cg2181 to cg2184 is annotated as a peptide uptake system (25), we focused on the investigation of the cluster cg2703 to cg2708, annotated as a putative ABC-type sugar transporter. In detail, cg2703 and cg2704 probably encode the transmembrane domains (TMDs), cg2705 the substrate binding domain, cg2708 the nucleotide binding domains (NBDs), and cg2707 a hypothetical protein.
The cluster of genes from cg2703 to cg2708 was deleted, and the resulting mutant strain, C. glutamicum Δmus, was tested for both growth with and uptake of maltose. As depicted in Fig. 1A and B, both utilization and uptake of maltose were abolished in C. glutamicum Δmus. At least partial complementation of this phenotype was achieved by the introduction of plasmid pXMJ19-musKEFG, which carries the complete gene cluster from cg2703 to cg2708. Introduction of the empty vector pXMJ19 into C. glutamicum Δmus did not restore growth on or uptake of maltose (Fig. 1B). From these experiments, we conclude that the gene cluster comprising cg2703 to cg2708 indeed encodes the single, high-affinity maltose uptake system of C. glutamicum; therefore, we named the genes for the components of this ABC transporter musK (cg2708), musE (cg2705), musF (cg2704), and musG (cg2703).
Fig 1.
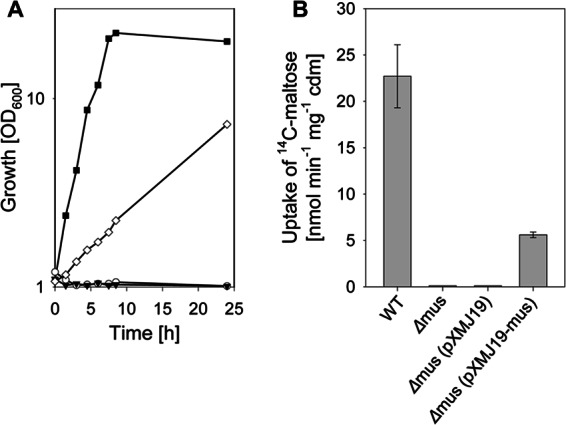
Growth (A) and rates of [14C]maltose uptake (B) of C. glutamicum WT (filled squares), C. glutamicum Δmus (open circles), C. glutamicum Δmus(pXMJ19) (filled triangles), and C. glutamicum Δmus(pXMJ19-musKEFG) (open diamonds) in CgC minimal medium initially containing 2% (wt/vol) maltose. One representative growth curve of at least three independent cultivations is shown in panel A; the results of each of the cultivations were comparable. For uptake measurements, the strains were cultivated in CgC minimal medium with 2% (wt/vol) glucose. The data represent mean values and standard deviations of three independent measurements from three independent cultivations.
Maltodextrin uptake by the musKEFG-encoded ABC transport system.
To analyze the substrate specificity of the ABC transporter encoded by the musKEFG genes, [14C]maltose uptake assays were performed with unlabeled competitors in 100-fold excess. As depicted in Fig. 2A, addition of 50 mM unlabeled maltose to the uptake assay completely quenched the uptake of label. No effects on [14C]maltose uptake were observed by addition of glucose or the disaccharides trehalose and isomaltose. Addition of the maltodextrins maltotriose and maltotetraose completely quenched the uptake of [14C]maltose; furthermore, maltopentaose reduced the [14C]maltose uptake rate from 19.5 ± 1.5 nmol min−1 mg−1 cdm to 6.8 ± 0.3 nmol min−1 mg−1 cdm. Addition of maltodextrins consisting of more than five glucose moieties, such as maltohexaose and maltoheptaose, did not affect the uptake of labeled maltose. Addition of the pseudo-oligosaccharide acarbose, which in E. coli is taken up via the maltose ABC transporter and inhibits maltose metabolism (58), had no effect on the uptake of labeled maltose in C. glutamicum. Taken together, these data indicate that in addition to maltose, short maltodextrins probably are also substrates of the musKEFG-encoded ABC transport system.
Fig 2.
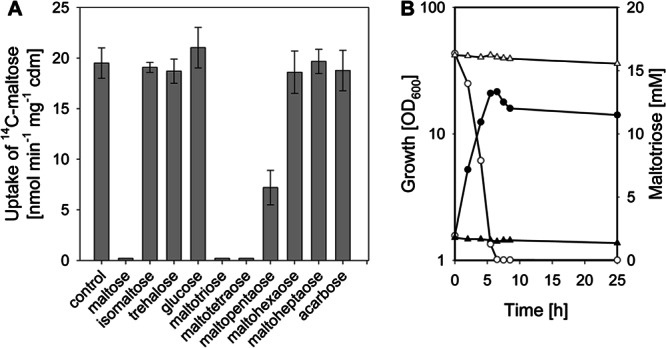
Effect of unlabeled competitors on [14C]maltose uptake rates in C. glutamicum (A) and growth (filled symbols) and substrate consumption (open symbols) of C. glutamicum WT (circles) and C. glutamicum Δmus in CgC minimal medium with 1% (wt/vol) maltotriose as the sole carbon source (B). One representative growth curve of at least three independent cultivations is shown; the results of each of the cultivations were comparable. The uptake measurements were started by simultaneous addition of 50 μM [14C]maltose and 5 mM competitor. As a control, [14C]maltose uptake was measured without competitor. Cells for the experiments were cultivated in CgC minimal medium with 1% (wt/vol) glucose. The data represent mean values and standard deviations of three independent measurements from two independent cultivations.
However, the assay utilized here does not discriminate between competition for transport or only binding. To analyze if short maltodextrins are indeed substrates of the musKEFG-encoded ABC transport system, growth experiments with C. glutamicum WT and C. glutamicum Δmus in minimal medium with maltotriose as the sole carbon source were performed. As shown in Fig. 2B, C. glutamicum utilized maltotriose as a carbon source, grew at a rate of 0.38 ± 0.02 h−1, and reached a final optical density of 14.0 ± 0.5 after 24 h of cultivation. Neither growth nor maltotriose utilization was observed for C. glutamicum Δmus (Fig. 2B). Maltotetraose was efficiently used as the sole source of carbon and energy by C. glutamicum; in growth experiments with 0.5% (wt/vol) maltotetraose, a growth rate of 0.34 ± 0.02 h−1 and a final optical density of 10.5 ± 0.5 were measured. No growth on maltotetraose was observed for C. glutamicum Δmus. In growth experiments with 0.5% (wt/vol) maltopentaose as the sole substrate, no growth of C. glutamicum was detected. From these results, we conclude that in addition to maltose, the short maltodextrins maltotriose and maltotetraose are taken up in C. glutamicum via the musKEFG-encoded ABC transport system.
Transcriptional organization of the musKEFG genes.
At first glance, in C. glutamicum the genes encoding the maltose/maltodextrin uptake system seem to be organized as an operon (Fig. 3A), which may comprise the two additional open reading frames cg2707 and cg2701. The two open reading frames cg2707 and cg2701 are annotated as genes for hypothetical proteins (25). In accordance with this, initial analyses of the genome sequence revealed the presence of a rho-independent transcriptional terminator upstream (centered 143 bp upstream of the musK ATG start codon; ΔGo′, −21.9 kcal mol−1; TT1) (Fig. 3A) and of a transcriptional terminator downstream of the mus genes (centered 1,058 bp downstream of the TAA stop codon of cg2701, the ORF directly following musG; ΔGo′, −13.6 kcal mol−1; TT2). To further investigate the transcriptional organization, we employed Northern blot analyses with specific RNA probes raised against musK, cg2707, musE, musG, and cg2701. As shown in Fig. 3B, with the probes specific for musK and musE, respectively, signals were detected which correspond to fragment sizes of about 1,800 nucleotides. No signal was detected with the cg2707-specific probe (data not shown). These data show that musK and musE are transcribed monocistronically, and that there is no single transcript for all mus genes, as the whole cluster (musK to musG) comprises about 6,000 nucleotides (Fig. 3A). With the probes for musF and cg2701, transcripts of about 2,500 bases were detected (Fig. 3B). From these data it can be concluded that musF, musG, and cg2701 form an operon, as these genes comprise 2,427 bp. The organization of the musF-musG-cg2701 operon was confirmed by reverse transcription-PCR (RT-PCR) analyses, which clearly showed musF and musG as well as musG and cg2701 being coexpressed (data not shown). Furthermore, employing 5′-RACE with total RNA of maltose-grown C. glutamicum WT cells, a transcriptional start site of the musF-musG-cg2701 operon was determined (results not shown). In three independent experiments, the transcriptional start site of musF (TSmusF) was found to be a cytosine residue within the musE-musF intergenic region, located 64 bp upstream of the musF ATG start codon and 122 bp downstream of the musE TAA stop codon (see Fig. S2 in the supplemental material). Upstream of TSmusF we found a −10 CATCCT motif that only slightly matches the −10 consensus motif (TANANT) determined recently for corynebacteria (59); however, it is similar to the −10 regions of aceE (TATCCT [38]) and metE (CGTCCT [60]).
Fig 3.
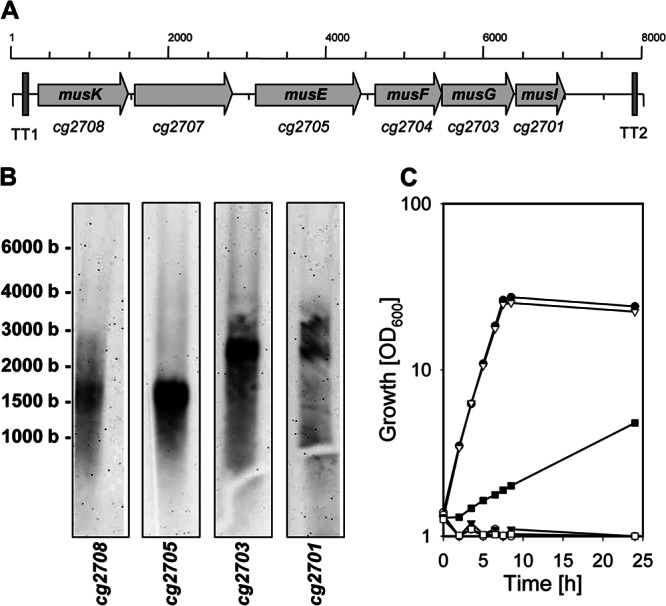
Genetic map of the cg2708 to cg2701 locus in C. glutamicum (A), Northern blot analysis of RNA samples from C. glutamicum cells grown in CgC minimal medium containing maltose as a carbon source (B), and growth of C. glutamicum WT (filled circles), C. glutamicum IMcg2708 (filled squares), C. glutamicum IMcg2707 (open triangles), C. glutamicum IMcg2705 (filled triangles), C. glutamicum IMcg2703 (open circles), and C. glutamicum IMcg2701 (open squares) in CgC minimal medium with 2% (wt/vol) maltose (C). Panel A shows the arrangements of the genes as arrows and the predicted transcriptional terminators (TT1 and TT2). For the Northern blot analyses, RNA was electrophoresed and probed with RNA probe specific for cg2708 (musK), cg2705 (musE), cg2703 (musG), or cg2701 (musI); the size of RNA fragments, determined with the RiboRuler high-range RNA ladder (Fermentas), is shown on the left. One representative growth curve of at least three independent cultivations is shown in panel C; the results of each of the cultivations were comparable.
As the data presented here showed that the mus genes are not organized as a single operon, the functions of these genes as components of the C. glutamicum ABC transporter for maltose and maltodextrins were open to question. To investigate the individual functions of these genes, we constructed the single-gene disruption mutants of musK, cg2707, musE, musF, and cg2701, named C. glutamicum IMmusK, C. glutamicum IMcg2707, C. glutamicum IMmusE, C. glutamicum IMmusF, and C. glutamicum IMcg2701, respectively. As depicted in Fig. 3C, inactivation of both musF and musE resulted in the loss of growth with maltose as the sole carbon source. In conformity with the above-mentioned results, uptake of 14C-labeled maltose was also abolished in both C. glutamicum IMmusE and C. glutamicum IMmusF (results not shown). As both utilization of maltose as the sole carbon source (Fig. 3C) and uptake of [14C]maltose were slowed down at least 5-fold in C. glutamicum IMmusK (2.5 ± 0.5 nmol min−1 mg−1 cdw; growth rate, 0.07 ± 0.5 h−1) compared to the parental strain C. glutamicum WT (22.7 ± 3.4 nmol min−1 mg−1 cdm; growth rate, 0.37 ± 0.01 h−1), we conclude that an alternative, unidentified ATPase can partially replace MusK. Inactivation of the ORF cg2707, which encodes a hypothetical soluble protein, did not affect maltose utilization (growth rate, 0.36 ± 0.02 h−1) (Fig. 3C) and uptake of maltose (maltose uptake rate for C. glutamicum IMcg2707 of 27.5 ± 2.8 nmol min−1 mg−1 cdm). However, inactivation of the second putative gene, cg2701, severely impaired maltose metabolism of C. glutamicum, as growth on maltose (Fig. 3C) and maltose uptake were abolished in C. glutamicum IMcg2701. Taken together, these results show that although the genes musK, musE, musF, and musG are not part of a single operon, their gene products encode components of the ABC transporter for maltose uptake in C. glutamicum.
The cg2701-encoded membrane protein MusI is an essential, novel component of the maltose ABC transporter of C. glutamicum.
To rule out the possibility that the effect on maltose utilization in C. glutamicum IMcg2701 is due to polar effects on musG, we carried out complementation studies with the plasmid pXMJ19-cg2701. As shown in Fig. 4A, ectopic expression of cg2701 in C. glutamicum IMcg2701 by the plasmid pXMJ19-cg2701 restored growth with maltose as the sole carbon source, while no growth was observed for the strain carrying the empty plasmid pXMJ19. In accordance with these results, uptake of 14C-labeled maltose was observed for C. glutamicum IMcg2701(pXMJ19-cg2701) (maltose uptake rate, 16.7 ± 2.3 nmol min−1 mg−1 cdm; results not shown), while no uptake of label was detected for C. glutamicum IMcg2701 (pXMJ19). In addition, the introduction of the plasmid pXM19-cg2701-strep for expression of cg2701 as a C-terminally tagged protein also restored growth with maltose as the sole carbon source (Fig. 4A) and uptake of maltose (17.1 ± 1.9 nmol min−1 mg−1 cdm) in C. glutamicum IMcg2701. The tagged protein Cg2701-Strep was detected in Western blotting experiments with antibodies binding to the N-terminal tag exclusively in the membrane fraction of C. glutamicum IMcg2701(pXM19-cg2701-strep) cells (Fig. 4B).
Fig 4.

Growth of C. glutamicum WT (open circles), C. glutamicum IMcg2701 (open triangles), C. glutamicum IMcg2701(pXMJ19-cg2701) (filled triangles), C. glutamicum IMcg2701(pXMJ19-cg2701-strep) (open squares), and C. glutamicum IMcg2701(pXMJ19) (filled circles) in CgC minimal medium with 2% (wt/vol) maltose (A), analysis of the cellular localization of MusI-Strep (B), and topology model of MusI (C). For localization of MusI-Strep, C. glutamicum IMcg2701(pXMJ19-cg2701-strep) cells were harvested, washed, and disrupted. Cytosolic and membrane fractions separated by ultracentrifugation were then electrophoresed on a 12% SDS-polyacrylamide gel and transferred to a membrane. MusI-Strep was detected using anti-Strep antibody. A PAGE ruler prestained protein ladder (MBI Fermentas) was used as the marker. The topology model of the membrane-spanning regions of MusI is based on topology modeling. Membrane-spanning sequences were determined based on consensus sequences from two topology prediction algorithms. One representative growth curve from at least three independent cultivations is shown. The results of each of the cultivations were comparable.
The finding that cg2701 is required for maltose uptake in C. glutamicum made us question the above-described results of the complementation studies with C. glutamicum Δmus using plasmid pXMJ19-mus, as this plasmid only harbors the genes musK to musG and did not lead to full complementation. However, the plasmid-encoded expression of all genes of the mus cluster (musK, musE, musF, and musG), together with cg2701 by plasmid pXMJ19-musEFGKI in C. glutamicum Δmus, did not further improve growth on maltose, as identical growth rates for C. glutamicum Δmus(pXMJ19-musKEFGI) and C. glutamicum Δmus(pXMJ19-musEFGK) were observed (0.26 ± 0.02 h−1 and 0.27 ± 0.03 h−1, respectively). These data seem to be contradictory to the above-described transcriptional organization of musF, musG, and cg2701 as an operon, since no transcript of cg2701 should be present in C. glutamicum Δmus(pXMJ19-musKEFG) cells. As cg2701 is essential for maltose uptake, the aforementioned strain should not grow on maltose. However, in RNA slot blot experiments using a cg2701-specific probe, transcripts of cg2701were detected in both C. glutamicum Δmus and C. glutamicum Δmus(pXMJ19-musKEFG) (data not shown). Indeed, stronger signals for cg2701 expression were detected for the C. glutamicum WT and for C. glutamicum Δmus(pXMJ19-musKEFGI), which carries the plasmid comprising the complete mus gene cluster. These data show that although the musFG-cg2701 operon is disrupted, weak transcription of cg2701 from an unknown promoter takes place which is sufficient to support growth of C. glutamicum Δmus(pXMJ19-musKEFG) on maltose.
As described before, in addition to maltose, maltotriose and maltotetraose also are taken up via the musKEFG-encoded ABC transporter. No growth of C. glutamicum IMcg2701 in minimal medium with maltotriose or maltotetraose as the sole source of carbon and energy was observed. The cg2701-encoded membrane protein therefore is also necessary for the uptake of short maltodextrins.
Taking these findings together, it can be concluded that cg2701 indeed encodes an essential component of the maltose/maltodextrin uptake system of C. glutamicum; therefore, we named the gene musI. Analyses of the MusI amino acid sequence and the predicted protein topology using both SOSUI and TMHMM revealed that MusI probably possesses an extracellular N terminus and 5 transmembrane helices (Fig. 4C); however, the cytoplasmic and periplasmic regions of MusI share no similarities with characterized proteins to date. Analyses of the deduced amino acid sequences and predicted protein topologies of the gene products of musF and musG showed that each of the two membrane proteins possesses six transmembrane domains and contains the so-called EAA sequence motifs required for interaction with the NBDs in the last cytoplasmic loop, as is common for TMDs of prokaryotic ABC importers (17, 61, 62). As TMDs of prokaryotic ABC importers contain up to 11 transmembrane helices (63), we assume that the additional protein MusI provides the additional transmembrane segments for one or both TMDs of the C. glutamicum maltose uptake system.
Effects of the transcriptional regulators RamA and SugR on musK, musE, and musFGI transcription.
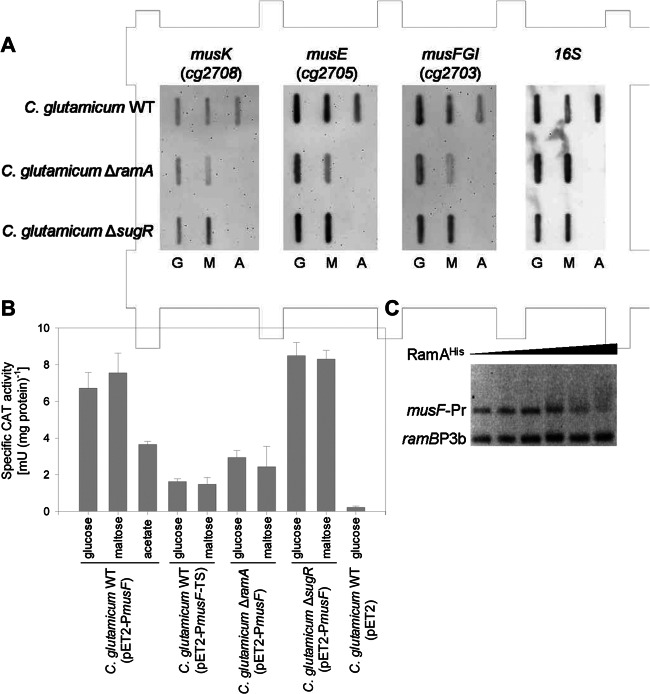
Characterization of growth and maltose uptake properties of the RamA-deficient strain C. glutamicum ΔramA and the SugR-deficient strain C. glutamicum ΔsugR pointed toward a strong role of the two regulators in the transcriptional control of the mus genes, as both maltose utilization and uptake were severely impaired in both mutant strains. The transcriptome data of the ramA and the sugR deletion mutants, reanalyzed here for the identification of the maltose uptake system, were derived from growth experiments on complex medium and/or minimal medium with glucose or acetate as the carbon source (55, 57). Hence, these transcriptome analyses provide no insights on transcriptional regulation of the mus genes toward the utilization of maltose as the sole carbon source. Therefore, we performed slot blot experiments with RNA probes raised against musK, musE, and musF and RNA samples derived from cultivations of C. glutamicum WT, C. glutamicum ΔramA, and C. glutamicum ΔsugR on glucose, maltose, or acetate. As shown in Fig. 5A, compared to the signals obtained for cultivations of C. glutamicum WT on maltose or glucose, only minor amounts of musE and musF transcripts were detected in C. glutamicum WT cells cultivated on acetate. No obvious changes in the amounts of detected musK transcripts were observed in the slot blot experiments with RNA samples from cultivations on different carbon sources (Fig. 5A). Analyses of the expression of musK, musE, and musF in the RamA-deficient strain C. glutamicum ΔramA showed that fewer transcripts of musE and musF were present in the mutant strain than in C. glutamicum WT; however, no altered expression was observed for musK. This effect of the lack of the transcriptional activator RamA on the expression of musE and musF was even more apparent in C. glutamicum ΔramA cells from cultivations with maltose than in cells from cultivations on glucose. These results correspond to the finding of putative RamA binding sites upstream of musE (ACCCCG; 19 bp upstream of the annotated TTG start codon) and of musF (CGGGGA and AGGGGA, 69 and 64 bp upstream of TSmusF, respectively; the musF promoter region is depicted in Fig. S2 in the supplemental material). EMSAs with different amounts of purified hexahistidyl-tagged RamA fusion protein (RamAHis) showed at least weak binding of RamAHis to the musF promoter region (Fig. 5C). Even at high concentrations of RamAHis, no retardation was observed with the control fragment ramBp3b, which possesses no RamA binding site (64).
Fig 5.
(A) Representative RNA hybridization experiments with RNA isolated from C. glutamicum WT, C. glutamicum ΔramA, and C. glutamicum ΔsugR cultivated in minimal medium with 2% (wt/vol) of the indicated carbon sources. The RNA levels of musK, musE, musFGI, and 16S were monitored with DIG-labeled antisense RNA probes. The results of one representative experiment from a series of three independent experiments is shown. (B) Expression of a musF′-′cat fusion in C. glutamicum WT(pET2-PmusF), C. glutamicum ΔramA(pET2-PmusF), and C. glutamicum ΔsugR(pET2-PmusF). As controls, CAT activity was analyzed in C. glutamicum WT(pET2-PmusF-TS), which lacks the musF transcriptional start site, and C. glutamicum WT(pET2), which carries the promoterless cat gene. The reporter gene activity was determined in cell extracts from cultivations in CgC minimal medium with 1% (wt/vol) of each of the indicated carbon sources. The values represent averages and standard deviations from at least three independent experiments. (C) Representative EMSA using hexahistidyl-tagged RamA protein (0 to 7.5 μg) with the 380-bp musF-Pr fragment as a probe (20 ng) and the 211-bp ramBp3b fragment as a negative control (10 ng).
To test for transcriptional regulation of the musFGI operon by RamA in vivo, a transcriptional fusion between the musF promoter region and the promoterless CAT gene was constructed in the promoter probe vector pET2 (plasmid pET2-PmusF) and transformed into C. glutamicum WT and the ramA deletion mutant C. glutamicum ΔramA. As controls, the empty pET2 plasmid and the plasmid pET2-PmusF-TS, which carries the truncated musF promoter region lacking TSmusF, were transformed into C. glutamicum WT. CAT activities were determined in the plasmid-carrying strains during exponential growth in minimal medium with 1% (wt/vol) glucose, 1% (wt/vol) maltose, or 1% (wt/vol) acetate. CAT activity was highest in cells of C. glutamicum WT(pET2-PmusF) cultivated on maltose (7.6 ± 1.1 mU [mg protein]−1), similarly high in cells cultivated on glucose (6.7 ± 0.9 mU [mg protein]−1), and significantly reduced in cells cultivated on acetate (3.6 ± 0.2 mU [mg protein]−1) (Fig. 5B). Independent of the carbon source, the musF promoter activities were 60 to 80% lower in C. glutamicum ΔramA (2.9 ± 0.4 and 2.4 ± 1.1 mU [mg protein]−1 for cells cultivated on glucose and maltose, respectively). Removal of TSmusF from the musF promoter region led to even lower CAT activities, with cell extracts of C. glutamicum(pET2-PmusF-TS) showing activities between 1.5 ± 0.3 mU and 1.6 ± 0.1 mU (mg protein)−1 (Fig. 5B). In accordance with the published microarray data (57) as well as the maltose uptake rates measured here for the RamA-deficient strain C. glutamicum ΔramA, we conclude from these data that RamA acts as a transcriptional activator for the carbon source-dependent transcription of musFGI at the musF promoter identified here.
Also in C. glutamicum ΔsugR, maltose uptake rates as well as the growth rates on maltose were shown to be significantly reduced compared to those of C. glutamicum WT. This led to the assumption that SugR, the repressor of ptsG, ptsS, and ldhA (27, 55, 56), acts as an activator of the genes for the maltose uptake system. However, microarray data of C. glutamicum ΔsugR versus C. glutamicum WT (27, 56) did not reveal significant changes in the expression of the mus genes in the sugR deletion mutant strain. Also in slot blot experiments, the intensity of the signals obtained with the probes for musK, musE, and musF for RNA samples from C. glutamicum ΔsugR and C. glutamicum WT were similar (Fig. 5A). CAT activity in cell extracts of C. glutamicum ΔsugR(pET2-PmusF) was even slightly increased, to 8.5 ± 0.7 and 8.3 ± 0.3 mU (mg protein)−1, in cells cultivated on glucose and maltose, respectively (Fig. 5B). As we did not observe sequence motifs of possible SugR binding sites in the intergenic regions of the mus genes and no SugR binding sites within the mus gene cluster were identified in recently published ChIP-to-chip experiments (55), it has to be assumed that SugR is not involved in the transcriptional control of musK, musE, and musFGI. The severe effects observed on maltose uptake and metabolization in the SugR-deficient strain C. glutamicum ΔsugR, however, indicate that SugR affects expression of MusK2EFGI, the ABC transporter for maltose, by an unknown posttranscriptional mechanism.
Maltose uptake by MusK2EFGI is necessary for the positive effect of maltose on ptsG expression.
In C. glutamicum, expression of ptsG is enhanced by the presence of the non-PTS substrate maltose (7, 27), which was shown here to be exclusively taken up via the ABC transporter MusK2EFGI. To distinguish whether presence of maltose in the culture broth or intermediates formed in the course of maltose metabolization give rise to this positive effect of maltose on ptsG expression, we analyzed the activity of the ptsG promoter using reporter gene assays with the promoter-probe plasmid pET2-PRptsG (27) in C. glutamicum WT and C. glutamicum Δmus cells cultivated with different carbon sources. Plasmid pET2-PRptsG contains a fusion between the ptsG promoter region and the promoterless cat gene (19). As depicted in Fig. 6, the positive effect of maltose addition on ptsG expression in C. glutamicum WT(pET2-PRptsG) led to an increase of the specific CAT activity from 0.40 ± 0.06 U (mg protein)−1 measured in extracts of glucose-grown cells to 0.53 ± 0.7 U (mg protein)−1 for extracts from cells cultivated with maltose. As previously described by Engels and Wendisch (27), the presence of acetate in the culture broth leads to significantly reduced ptsG promoter activities of 0.18 ± 0.06 and 0.19 ± 0.05 U (mg protein)−1 in C. glutamicum WT(pET2-PRptsG) cells cultivated with acetate and glucose plus acetate, respectively. In the maltose uptake-deficient strain C. glutamicum Δmus(pET2-PRptsG), ptsG promoter activities were nearly identical to the activities measured for C. glutamicum WT(pET2-PRptsG). Promoter activities of 0.44 ± 0.03, 0.17 ± 0.05, and 0.19 ± 0.05 U (mg protein)−1 were measured for C. glutamicum Δmus(pET2-PRptsG) cells cultivated with glucose, acetate, or glucose plus acetate, respectively. These data show that in both strains, expression of ptsG is repressed by the presence of acetate. As expected for C. glutamicum WT(pET2-PRptsG), maltose addition to the culture broth containing glucose plus acetate significantly increased ptsG promoter activity to 0.54 ± 0.06 U (mg protein)−1, which corresponds to the increased activity measured in cells cultivated with maltose as the sole carbon source. However, for C. glutamicum Δmus(pET2-PRptsG), no positive effect on the ptsG promoter activity was detected when maltose was added to the culture broth already containing glucose plus acetate. The specific CAT activity of 0.20 ± 0.05 U (mg protein)−1 for C. glutamicum Δmus(pET2-PRptsG) cultivated with glucose plus acetate plus maltose is about the same as the activities determined in cells cultivated with acetate or acetate plus glucose. This result clearly shows that uptake of maltose by MusK2EFGI in C. glutamicum cells is a prerequisite for the positive effect of maltose on ptsG expression.
Fig 6.
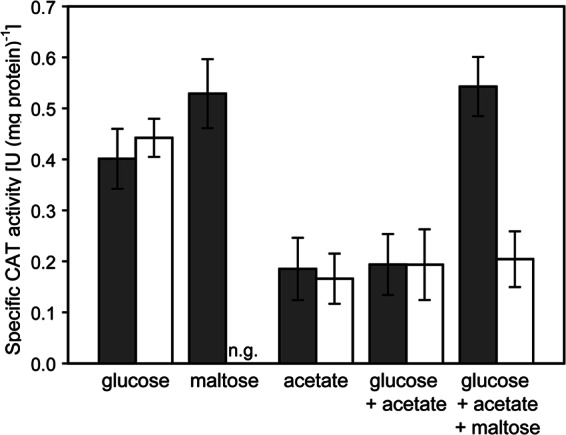
Expression of a ptsG′-′cat fusion in C. glutamicum WT(pET2-ptsG) (dark bars) and C. glutamicum Δmus(pET2-ptsG) (white bars). The reporter gene activity was determined in cell extracts from cultivations in CgC minimal medium with 1% (wt/vol) of each of the indicated carbon sources. The values represent averages and standard deviations from at least three independent experiments.
DISCUSSION
The positive effect of maltose on expression of the ptsG-encoded EIIGlc of the PTS depends on maltose uptake by the MusK2EFGI transporter identified here. It seems reasonable to suggest that intermediates of maltose metabolization trigger the positive effect on ptsG expression. In C. glutamicum, transcription of ptsG is controlled by the global regulators RamA, RamB, GlxR, and SugR (27, 57, 65), which all coordinate the adaptation of the central metabolism of C. glutamicum toward the utilization of carbon sources requiring gluconeogenesis, such as acetate (54). Repression of ptsG in the presence of acetate is mainly caused by SugR (27). Deletion of sugR indeed abolishes the negative effects of acetate addition on ptsG expression and glucose utilization (27, 66). However, the positive effect of maltose on ptsG expression is still present in the sugR deletion mutant (27). Therefore, it is unlikely that the effect of maltose addition on ptsG expression is brought about by increased intracellular concentrations of one of the negative effectors of SugR, namely, fructose-6-phosphate and/or fructose-1-phosphate (27, 56).
The cAMP receptor protein (CRP) homologue of C. glutamicum, GlxR, acts both as a transcriptional repressor (for gltA, aceB, sdhCAB [67–69]) and transcriptional activator (for pstSCAB, narKGHJI, ctaC, and atpB [70–72]) and binds in a cyclic AMP (cAMP)-dependent manner at its consensus motif, 5′-TGTGA-N6-TCACA-3′, in the promoter regions of several genes (65, 68, 72). Indeed, two GlxR binding sites are situated close to the ptsG transcriptional start site (65, 72), indicating GlxR-dependent control of ptsG expression. As neither effects of maltose addition on cAMP levels nor the role of GlxR in the transcriptional control of ptsG have been studied so far, assumptions that the positive effect of maltose addition on ptsG expression relies on changes in the cAMP levels, which might lead to GlxR-mediated activation of ptsG transcription, are hypothetical.
In reporter gene assays using the reporter plasmid pET2-PRptsG in the RamA-deficient C. glutamicum ΔramA strain, CAT activity in cells cultivated on glucose was reduced about 60%, to 0.18 ± 0.02 U (mg protein)−1, compared to activity in C. glutamicum WT (0.40 ± 0.06 U [mg protein]−1). These data show that RamA acts as the activator of ptsG transcription. Moreover, the positive effect of maltose on ptsG transcription was absent from C. glutamicum ΔramA, and CAT activity in extracts of C. glutamicum ΔramA(pET2-PRptsG) cultivated on maltose (0.17 ± 0.02 U [mg protein]−1) was identical to the activity of cells grown on glucose. These results lead to the assumption that RamA mediates the positive effect of maltose on ptsG transcription. However, in addition to ptsG expression, expression of musFGI, and probably of musE, was shown here to be activated by RamA. As a result, maltose uptake by MusK2EFGI is reduced in C. glutamicum ΔramA. It is therefore complex to discriminate between the indirect involvement of RamA as an activator of musFGI expression influencing the uptake of maltose required for the effect on ptsG and the direct participation of RamA as a mediator of the positive effect of maltose on ptsG expression. Although in C. glutamicum ΔsugR the maltose uptake rate was reduced, similar to that in C. glutamicum ΔramA, and the maltose effect on ptsG expression was still present in C. glutamicum ΔsugR, it seems reasonable to suggest that RamA is directly involved in the positive effect on ptsG. Taking these results together, an intermediate of maltose metabolism probably triggers the positive effect on ptsG expression, which most is likely mediated via RamA. As RamA effector molecules have not been identified before (57), its role in the maltose effect on ptsG expression and, therefore, the underlying mechanism remain elusive.
Maltose uptake in C. glutamicum is brought about by the ABC transporter system MusEFGK2I, which is unusual, as it requires the additional membrane protein MusI to be functional. In general, ABC importers are well understood on the mechanistic level and share a modular organization, which comprises an NBD dimer, a substrate binding protein (SBP), and two TMDs, which form the translocation pore (73–75). The domains of ABC importers exist in several transporters as single proteins or are arranged in other transporters in various protein fusions (76). This domain organization of ABC transporters is used for their classification (75, 76). The additional membrane protein MusI of the C. glutamicum maltose importer might be an essential accessory protein or a novel variant of the organization of an ABC transporter's TMDs into proteins and genes. Possible homologues of MusI are the hypothetical proteins BL0145 of Bifidobacterium longum NCC2705, SP_1677 of Streptococcus pneumoniae TIGR4, and SAG0038 of Streptococcus agalactiae 2603. Although the amino acid sequences of the MusI homologues share only low similarities (see Fig. S3 in the supplemental material), each of the three hypothetical proteins consists of about 210 amino acids and contains five transmembrane stretches and the DUF (domain of unknown function) 624 motif, and it is located in the vicinity of genes for ABC importers (see Fig. S4), as is also the case for MusI. In B. longum, the ORF bl0145 is directly following bl0143 and bl0144, which encode TMDs of the maltose ABC importer (77). In S. agalactiae 2603, the ORF sag0038 is separated by one ORF (sag0037), encoding a further hypothetical protein, from the ORFs sag0034, sag0035, and sag0036, which encode the SBP and the TMDs of the NanT ABC importer for sialic acid (78). Three ORFs (sp1680, sp1679, and sp1678) are situated between the genes satA, satB, and satC for the sialic acid ABC importer (79) and the ORF sp1677, which encodes the MusI-like protein (see Fig. S4). The relevance of these MusI-like proteins for substrate uptake by the aforementioned ABC importers has not been studied. As is the case for MusF and MusG of C. glutamicum, based on comparisons of the amino acid sequences of the TMDs, none of the general features necessary for the functionality of ABC transporters are lacking in the B. longum maltose importer or the sialic acid importers of S. agalactiae and S. pneumoniae. Even though no function can be assigned to MusI or the MusI-like proteins of other ABC importers so far, the occurrence of genes for MusI-like proteins in the vicinity of several genes for ABC importers indicates that MusEFGK2I of C. glutamicum is not the sole ABC transport system requiring an additional membrane protein for its functionality.
Supplementary Material
ACKNOWLEDGMENTS
We thank Eva Glees and Ute Meyer for excellent technical assistance.
Work in the laboratories of the authors was funded in part by grant 0315589F from BMBF in the CRP “Corynebacterium: improving flexibility and fitness for industrial production.”
Footnotes
Published ahead of print 29 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01629-12.
REFERENCES
- 1. Shimizu H, Hirasawa T. 2007. Production of glutamate and glutamate-related amino acids: molecular mechanisms analysis and metabolic engineering, p 1–38 In Wendisch VF. (ed), Amino acid biosynthesis–pathways, regulation and metabolic engineering. Springer, Heidelberg, Germany [Google Scholar]
- 2. Wittmann C, Becker J. 2007. The L-lysine story: from metabolic pathways to industrial production, p 39–70 In Wendisch VF. (ed), Amino acid biosynthesis–pathways, regulation and metabolic engineering. Springer, Heidelberg, Germany [Google Scholar]
- 3. Arndt A, Eikmanns BJ. 2008. Regulation of carbon metabolism in Corynebacterium glutamicum, p 155–182 In Burkovski A. (ed), Corynebacteria: genomics and molecular biology. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 4. Blombach B, Seibold GM. 2010. Carbohydrate metabolism in Corynebacterium glutamicum and applications for the metabolic engineering of L-lysine production strains. Appl. Microbiol. Biotechnol. 86:1313–1322 [DOI] [PubMed] [Google Scholar]
- 5. Seibold G, Auchter M, Berens S, Kalinowski J, Eikmanns BJ. 2006. Utilization of soluble starch by a recombinant Corynebacterium glutamicum strain: growth and lysine production. J. Biotechnol. 124:381–391 [DOI] [PubMed] [Google Scholar]
- 6. Seibold GM, Wurst M, Eikmanns BJ. 2009. Roles of maltodextrin and glycogen phosphorylases in maltose utilization and glycogen metabolism in Corynebacterium glutamicum. Microbiology 155:347–358 [DOI] [PubMed] [Google Scholar]
- 7. Krause FS, Henrich A, Blombach B, Krämer R, Eikmanns BJ, Seibold GM. 2010. Increased glucose utilization in Corynebacterium glutamicum by use of maltose, and its application for the improvement of L-valine productivity. Appl. Environ. Microbiol. 76:370–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Park SY, Kim HK, Yoo SK, Oh TK, Lee JK. 2000. Characterization of glk, a gene coding for glucose kinase of Corynebacterium glutamicum. FEMS Microbiol. Lett. 188:209–215 [DOI] [PubMed] [Google Scholar]
- 9. Lindner SN, Knebel S, Pallerla SR, Schoberth SM, Wendisch VF. 2010. Cg2091 encodes a polyphosphate/ATP-dependent glucokinase of Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 87:703–713 [DOI] [PubMed] [Google Scholar]
- 10. Boos W, Shuman H. 1998. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol. Mol. Biol. Rev. 62:204–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xavier KB, Martins LO, Peist R, Kossmann M, Boos W, Santos H. 1996. High-affinity maltose/trehalose transport system in the hyperthermophilic archaeon Thermococcus litoralis. J. Bacteriol. 178:4773–4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Diederichs K, Diez J, Greller G, Müller C, Breed J, Schnell C, Vonrhein C, Boos W, Welte W. 2000. Crystal structure of MalK, the ATPase subunit of the trehalose/maltose ABC transporter of the archaeon Thermococcus litoralis. EMBO J. 19:5951–5961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Diez J, Diederichs K, Greller G, Horlacher R, Boos W, Welte W. 2001. The crystal structure of a liganded trehalose/maltose-binding protein from the hyperthermophilic archaeon Thermococcus litoralis at 1.85 A. J. Mol. Biol. 305:905–915 [DOI] [PubMed] [Google Scholar]
- 14. Orelle C, Ayvaz T, Everly RM, Klug CS, Davidson AL. 2008. Both maltose-binding protein and ATP are required for nucleotide-binding domain closure in the intact maltose ABC transporter. Proc. Natl. Acad. Sci. U. S. A. 105:12837–12842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Oldham ML, Khare D, Quiocho FA, Davidson AL, Chen J. 2007. Crystal structure of a catalytic intermediate of the maltose transporter. Nature 450:515–521 [DOI] [PubMed] [Google Scholar]
- 16. Daus ML, Berendt S, Wuttge S, Schneider E. 2007. Maltose binding protein (MalE) interacts with periplasmic loops P2 and P1 respectively of the MalFG subunits of the maltose ATP binding cassette transporter (MalFGK(2)) from Escherichia coli/Salmonella during the transport cycle. Mol. Microbiol. 66:1107–1122 [DOI] [PubMed] [Google Scholar]
- 17. Daus ML, Grote M, Müller P, Doebber M, Herrmann A, Steinhoff HJ, Dassa E, Schneider E. 2007. ATP-driven MalK dimer closure and reopening and conformational changes of the “EAA” motifs are crucial for function of the maltose ATP-binding cassette transporter (MalFGK2). J. Biol. Chem. 282:22387–22396 [DOI] [PubMed] [Google Scholar]
- 18. Grote M, Polyhach Y, Jeschke G, Steinhoff HJ, Schneider E, Bordignon E. 2009. Transmembrane signaling in the maltose ABC transporter MalFGK2-E: periplasmic MalF-P2 loop communicates substrate availability to the ATP-bound MalK dimer. J. Biol. Chem. 284:17521–17526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tangney M, Buchanan CJ, Priest FG, Mitchell WJ. 1992. Maltose uptake and its regulation in Bacillus subtilis. FEMS Microbiol. Lett. 76:191–196 [DOI] [PubMed] [Google Scholar]
- 20. Neubauer H, Glaasker E, Hammes WP, Poolman B, Konings WN. 1994. Mechanism of maltose uptake and glucose excretion in Lactobacillus sanfrancisco. J. Bacteriol. 176:3007–3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martin SA, Russell JB. 1987. Transport and phosphorylation of disaccharides by the ruminal bacterium Streptococcus bovis. Appl. Environ. Microbiol. 53:2388–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Webb AJ, Homer KA, Hosie AH. 2007. A phosphoenolpyruvate-dependent phosphotransferase system is the principal maltose transporter in Streptococcus mutans. J. Bacteriol. 189:3322–3327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schönert S, Seitz S, Krafft H, Feuerbaum EA, Andernach I, Witz G, Dahl MK. 2006. Maltose and maltodextrin utilization by Bacillus subtilis. J. Bacteriol. 188:3911–3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hülsmann A, Lurz R, Scheffel F, Schneider E. 2000. Maltose and maltodextrin transport in the thermoacidophilic gram-positive bacterium Alicyclobacillus acidocaldarius is mediated by a high-affinity transport system that includes a maltose binding protein tolerant to low pH. J. Bacteriol. 182:6292–6301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kalinowski J, Bathe B, Bartels D, Bischoff N, Bott M, Burkovski A, Dusch N, Eggeling L, Eikmanns BJ, Gaigalat L, Goesmann A, Hartmann M, Huthmacher K, Krämer R, Linke B, McHardy AC, Meyer F, Mockel B, Pfefferle W, Pühler A, Rey DA, Rückert C, Rupp O, Sahm H, Wendisch VF, Wiegrabe I, Tauch A. 2003. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L-aspartate-derived amino acids and vitamins. J. Biotechnol. 104:5–25 [DOI] [PubMed] [Google Scholar]
- 26. Winnen B, Felce J, Saier MH., Jr 2005. Genomic analyses of transporter proteins in Corynebacterium glutamicum and Corynebacterium efficiens, p 149–186 In Eggeling L, Bott M. (ed), Handbook of Corynebacterium glutamicum. CRC Press, Boca Raton, FL [Google Scholar]
- 27. Engels V, Wendisch VF. 2007. The DeoR-type regulator SugR represses expression of ptsG in Corynebacterium glutamicum. J. Bacteriol. 189:2955–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lindner SN, Seibold GM, Henrich A, Krämer R, Wendisch VF. 2011. Phosphotransferase system-independent glucose utilization in Corynebacterium glutamicum by inositol permeases and glucokinases. Appl. Environ. Microbiol. 77:3571–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moon MW, Kim HJ, Oh TK, Shin CS, Lee JS, Kim SJ, Lee JK. 2005. Analyses of enzyme II gene mutants for sugar transport and heterologous expression of fructokinase gene in Corynebacterium glutamicum ATCC 13032. FEMS Microbiol. Lett. 244:259–266 [DOI] [PubMed] [Google Scholar]
- 30. Eikmanns BJ, Metzger M, Reinscheid D, Kircher M, Sahm H. 1991. Amplification of three threonine biosynthesis genes in Corynebacterium glutamicum and its influence on carbon flux in different strains. Appl. Microbiol. Biotechnol. 34:617–622 [DOI] [PubMed] [Google Scholar]
- 31. Sambrook J, Russel DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 32. Eikmanns BJ, Thum-Schmitz N, Eggeling L, Lüdtke KU, Sahm H. 1994. Nucleotide sequence, expression and transcriptional analysis of the Corynebacterium glutamicum gltA gene encoding citrate synthase. Microbiology 140:1817–1828 [DOI] [PubMed] [Google Scholar]
- 33. Tauch A, Kirchner O, Loffler B, Gotker S, Puhler A, Kalinowski J. 2002. Efficient electrotransformation of Corynebacterium diphtheriae with a mini-replicon derived from the Corynebacterium glutamicum plasmid pGA1. Curr. Microbiol. 45:362–367 [DOI] [PubMed] [Google Scholar]
- 34. Niebisch A, Bott M. 2001. Molecular analysis of the cytochrome bc1-aa3 branch of the Corynebacterium glutamicum respiratory chain containing an unusual diheme cytochrome c1. Arch. Microbiol. 175:282–294 [DOI] [PubMed] [Google Scholar]
- 35. Jolkver E, Emer D, Ballan S, Krämer R, Eikmanns BJ, Marin K. 2009. Identification and characterization of a bacterial transport system for the uptake of pyruvate, propionate, and acetate in Corynebacterium glutamicum. J. Bacteriol. 191:940–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jakoby M, Ngouoto-Nkili CE, Burkovski A. 1999. Construction and application of new Corynebacterium glutamicum vectors. Biotechnol. Techniques 13:437–441 [Google Scholar]
- 37. Eikmanns BJ, Rittmann D, Sahm H. 1995. Cloning, sequence analysis, expression, and inactivation of the Corynebacterium glutamicum icd gene encoding isocitrate dehydrogenase and biochemical characterization of the enzyme. J. Bacteriol. 177:774–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schreiner ME, Fiur D, Holatko J, Patek M, Eikmanns BJ. 2005. E1 enzyme of the pyruvate dehydrogenase complex in Corynebacterium glutamicum: molecular analysis of the gene and phylogenetic aspects. J. Bacteriol. 187:6005–6018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 40. Rübenhagen R, Rönsch H, Jung H, Krämer R, Morbach S. 2000. Osmosensor and osmoregulator properties of the betaine carrier BetP from Corynebacterium glutamicum in proteoliposomes. J. Biol. Chem. 275:735–741 [DOI] [PubMed] [Google Scholar]
- 41. Gerstmeir R, Cramer A, Dangel P, Schaffer S, Eikmanns BJ. 2004. RamB, a novel transcriptional regulator of genes involved in acetate metabolism of Corynebacterium glutamicum. J. Bacteriol. 186:2798–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cramer A, Auchter M, Frunzke J, Bott M, Eikmanns BJ. 2007. RamB, the transcriptional regulator of acetate metabolism in Corynebacterium glutamicum, is subject to regulation by RamA and RamB. J. Bacteriol. 189:1145–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wolf A, Kramer R, Morbach S. 2003. Three pathways for trehalose metabolism in Corynebacterium glutamicum ATCC13032 and their significance in response to osmotic stress. Mol. Microbiol. 49:1119–1134 [DOI] [PubMed] [Google Scholar]
- 44. Möker N, Brocker M, Schaffer S, Krämer R, Morbach S, Bott M. 2004. Deletion of the genes encoding the MtrA-MtrB two-component system of Corynebacterium glutamicum has a strong influence on cell morphology, antibiotics susceptibility and expression of genes involved in osmoprotection. Mol. Microbiol. 54:420–438 [DOI] [PubMed] [Google Scholar]
- 45. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 46. Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36:D480–D484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kingsford CL, Ayanbule K, Salzberg SL. 2007. Rapid, accurate, computational discovery of Rho-independent transcription terminators illuminates their relationship to DNA uptake. Genome Biol. 8:R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridisation prediction. Nucleic Acids Res. 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 50. Hirokawa T, Boon-Chieng S, Mitaku S. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378–379 [DOI] [PubMed] [Google Scholar]
- 51. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Farwick M, Siewe RM, Kramer R. 1995. Glycine betaine uptake after hyperosmotic shift in Corynebacterium glutamicum. J. Bacteriol. 177:4690–4695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Krämer R, Lambert C. 1990. Uptake of glutamate in Corynebacterium glutamicum. 2. Evidence for a primary active transport system. Eur. J. Biochem. 194:937–944 [DOI] [PubMed] [Google Scholar]
- 54. Schröder J, Tauch A. 2010. Transcriptional regulation of gene expression in Corynebacterium glutamicum: the role of global, master and local regulators in the modular and hierarchical gene regulatory network. FEMS Microbiol. Rev. 34:685–737 [DOI] [PubMed] [Google Scholar]
- 55. Engels V, Lindner SN, Wendisch VF. 2008. The global repressor SugR controls expression of genes of glycolysis and of the L-lactate dehydrogenase LdhA in Corynebacterium glutamicum. J. Bacteriol. 190:8033–8044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gaigalat L, Schluter JP, Hartmann M, Mormann S, Tauch A, Puhler A, Kalinowski J. 2007. The DeoR-type transcriptional regulator SugR acts as a repressor for genes encoding the phosphoenolpyruvate:sugar phosphotransferase system (PTS) in Corynebacterium glutamicum. BMC Mol. Biol. 8:104 doi:10.1186/1471-2199-8-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Auchter M, Cramer A, Hüser A, Rückert C, Emer D, Schwarz P, Arndt A, Lange C, Kalinowski J, Wendisch VF, Eikmanns BJ. 2011. RamA and RamB are global transcriptional regulators in Corynebacterium glutamicum and control genes for enzymes of the central metabolism. J. Biotechnol. 154:126–139 [DOI] [PubMed] [Google Scholar]
- 58. Brunkhorst C, Andersen C, Schneider E. 1999. Acarbose, a pseudooligosaccharide, is transported but not metabolized by the maltose-maltodextrin system of Escherichia coli. J. Bacteriol. 181:2612–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Patek M, Nesvera J. 2011. Sigma factors and promoters in Corynebacterium glutamicum. J. Biotechnol. 154:101–113 [DOI] [PubMed] [Google Scholar]
- 60. Suda M, Teramoto H, Imamiya T, Inui M, Yukawa H. 2008. Transcriptional regulation of Corynebacterium glutamicum methionine biosynthesis genes in response to methionine supplementation under oxygen deprivation. Appl. Microbiol. Biotechnol. 81:505–513 [DOI] [PubMed] [Google Scholar]
- 61. Hunke S, Mourez M, Jehanno M, Dassa E, Schneider E. 2000. ATP modulates subunit-subunit interactions in an ATP-binding cassette transporter (MalFGK2) determined by site-directed chemical cross-linking. J. Biol. Chem. 275:15526–15534 [DOI] [PubMed] [Google Scholar]
- 62. Mourez M, Hofnung M, Dassa E. 1997. Subunit interactions in ABC transporters: a conserved sequence in hydrophobic membrane proteins of periplasmic permeases defines an important site of interaction with the ATPase subunits. EMBO J. 16:3066–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Biemans-Oldehinkel E, Doeven MK, Poolman B. 2006. ABC transporter architecture and regulatory roles of accessory domains. FEBS Lett. 580:1023–1035 [DOI] [PubMed] [Google Scholar]
- 64. Cramer A, Eikmanns BJ. 2007. RamA, the transcriptional regulator of acetate metabolism in Corynebacterium glutamicum, is subject to negative autoregulation. J. Mol. Microbiol. Biotechnol. 12:51–59 [DOI] [PubMed] [Google Scholar]
- 65. Kohl TA, Baumbach J, Jungwirth B, Pühler A, Tauch A. 2008. The GlxR regulon of the amino acid producer Corynebacterium glutamicum: in silico and in vitro detection of DNA binding sites of a global transcription regulator. J. Biotechnol. 135:340–350 [DOI] [PubMed] [Google Scholar]
- 66. Blombach B, Arndt A, Auchter M, Eikmanns BJ. 2009. L-valine production during growth of pyruvate dehydrogenase complex-deficient Corynebacterium glutamicum in the presence of ethanol or by inactivation of the transcriptional regulator SugR. Appl. Environ. Microbiol. 75:1197–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bussmann M, Emer D, Hasenbein S, Degraf S, Eikmanns BJ, Bott M. 2009. Transcriptional control of the succinate dehydrogenase operon sdhCAB of Corynebacterium glutamicum by the cAMP-dependent regulator GlxR and the LuxR-type regulator RamA. J. Biotechnol. 143:173–182 [DOI] [PubMed] [Google Scholar]
- 68. Kim HJ, Kim TH, Kim Y, Lee HS. 2004. Identification and characterization of glxR, a gene involved in regulation of glyoxylate bypass in Corynebacterium glutamicum. J. Bacteriol. 186:3453–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. van Ooyen J, Emer D, Bussmann M, Bott M, Eikmanns BJ, Eggeling L. 2010. Citrate synthase in Corynebacterium glutamicum is encoded by two gltA transcripts which are controlled by RamA, RamB, and GlxR. J. Biotechnol. 154:14–18 [DOI] [PubMed] [Google Scholar]
- 70. Nishimura T, Teramoto H, Toyoda K, Inui M, Yukawa H. 2011. Regulation of the nitrate reductase operon narKGHJI by the cAMP-dependent regulator GlxR in Corynebacterium glutamicum. Microbiology 157:21–28 [DOI] [PubMed] [Google Scholar]
- 71. Panhorst M, Sorger-Herrmann U, Wendisch VF. 2011. The pstSCAB operon for phosphate uptake is regulated by the global regulator GlxR in Corynebacterium glutamicum. J. Biotechnol. 154:149–155 [DOI] [PubMed] [Google Scholar]
- 72. Toyoda K, Teramoto H, Inui M, Yukawa H. 2011. Genome-wide identification of in vivo binding sites of GlxR, a cyclic AMP receptor protein-type regulator in Corynebacterium glutamicum. J. Bacteriol. 193:4123–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hollenstein K, Dawson RJ, Locher KP. 2007. Structure and mechanism of ABC transporter proteins. Curr. Opin. Struct. Biol. 17:412–418 [DOI] [PubMed] [Google Scholar]
- 74. Oldham ML, Davidson AL, Chen J. 2008. Structural insights into ABC transporter mechanism. Curr. Opin. Struct. Biol. 18:726–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Eitinger T, Rodionov DA, Grote M, Schneider E. 2011. Canonical and ECF-type ATP-binding cassette importers in prokaryotes: diversity in modular organization and cellular functions. FEMS Microbiol. Rev. 35:3–67 [DOI] [PubMed] [Google Scholar]
- 76. Davidson AL, Dassa E, Orelle C, Chen J. 2008. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 72:317–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Parche S, Amon J, Jankovic I, Rezzonico E, Beleut M, Barutcu H, Schendel I, Eddy MP, Burkovski A, Arigoni F, Titgemeyer F. 2007. Sugar transport systems of Bifidobacterium longum NCC2705. J. Mol. Microbiol. Biotechnol. 12:9–19 [DOI] [PubMed] [Google Scholar]
- 78. Pezzicoli A, Ruggiero P, Amerighi F, Telford JL, Soriani M. 2012. Exogenous sialic acid transport contributes to group B streptococcus infection of mucosal surfaces. J. Infect. Dis. 206:924–931 [DOI] [PubMed] [Google Scholar]
- 79. Marion C, Aten AE, Woodiga SA, King SJ. 2011. Identification of an ATPase, MsmK, which energizes multiple carbohydrate ABC transporters in Streptococcus pneumoniae. Infect. Immun. 79:4193–4200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580 [DOI] [PubMed] [Google Scholar]
- 81. Schäfer A, Tauch A, WJäger Kalinowski J, Thierbach G, Puhler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73 [DOI] [PubMed] [Google Scholar]
- 82. Vasicova P, Abrhamova Z, Nesvera J, Patek M, Sahm H, Eikmanns BJ. 1998. Integrative and autonomously replicating vectors for analysis of promoters in. Corynebacterium glutamicum. Biotechnol. Techniques 12:743–746 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.