Abstract
Respiratory syncytial virus (RSV) is a negative-sense single-stranded RNA virus responsible for lower respiratory tract infections. During infection, the presence of double-stranded RNA (dsRNA) activates the interferon (IFN) regulatory factor 3 (IRF3) transcription factor, an event triggering expression of immediate early, IFN-stimulated genes (ISGs). We examine the role of transcriptional elongation in control of IRF3-dependent ISG expression. RSV infection induces ISG54, ISG56, and CIG5 gene expression in an IRF3-dependent manner demonstrated by IRF3 small interfering RNA (siRNA) silencing in both A549 epithelial cells and IRF3−/− MEFs. ISG expression was mediated by the recruitment of IRF3, CDK9, polymerase II (Pol II), and phospho-Ser2 carboxy-terminal domain (CTD) Pol II to the IFN-stimulated response element (ISRE) binding sites of the IRF3-dependent ISG promoters in native chromatin. We find that RSV infection enhances the activated fraction of cyclin-dependent kinase 9 (CDK9) by promoting its association with bromodomain 4 (BRD4) and disrupting its association with the inhibitory 7SK small nuclear RNA. The requirement of CDK9 activity for ISG expression was shown by siRNA-mediated silencing of CDK9 and by a selective CDK9 inhibitor in A549 cells. In contrast, RSV-induced beta interferon (IFN-β) expression is not influenced by CDK9 inhibition. Using transcript-selective quantitative real-time reverse transcription-PCR (Q-RT-PCR) assays for the ISG54 gene, we observed that RSV induces transition from short to fully spliced mRNA transcripts and that this transition is blocked by CDK9 inhibition in both A549 and primary human small airway epithelial cells. These data indicate that transcription elongation plays a major role in RSV-induced ISG expression and is mediated by IRF3-dependent recruitment of activated CDK9. CDK9 activity may be a target for immunomodulation in RSV-induced lung disease.
INTRODUCTION
The paramyxovirus respiratory syncytial virus (RSV) is among the most important respiratory pathogens in young children worldwide (1, 2). Each year, over 33 million children under the age of 5 years are affected by this disease, leading to over 3 million hospitalizations and almost 200,000 deaths. In immunocompromised patients, institutionalized elderly, and immunologically naive children (1), RSV produces significant morbidity in the form of lower respiratory tract infection (LRTI). LRTI accounts for 120,000 hospitalizations annually in the United States and is associated with postinfectious recurrent episodic wheezing (3, 4). Because most patients with RSV-induced LRTI present at the time when viral titers are falling (5), the host signaling response to RSV infection is thought to play a significant role in disease pathogenesis. In mouse models of acute inflammation, for example, previous work has shown that inhibition of mucosal innate immune pathways reduces expression of RANTES, MIP-1α, monocyte chemoattractant protein (MCP), and TCA3 chemokines and attenuates pathology (6).
Upon inoculation, RSV replicates in the nasal mucosa, spreading from cell to cell into the lower respiratory tract through intraepithelial bridges or via free virus in respiratory secretions binding to epithelial cilia (7). RSV replicates in all mucosal epithelial cell types, where it induces necrosis, peribronchial monocytic infiltration, and submucosal edema (4, 8). Our work has indicated that RSV replication activates innate immune response pathways in airway epithelial cells (9–13) constituting a first line of pulmonary host defense. Cytoplasmic viral genomic RNA is recognized principally by retinoic acid-inducible gene I (RIG-I) and to a lesser extent by melanoma differentiation-associated gene 5 (MDA5) (14, 15). Gene silencing experiments have shown that RIG-I is indispensable for expression of type I interferons (alpha and beta interferon [IFN-α and -β, respectively]) in response to RSV (14). Additionally, the presence of double-stranded RNA (dsRNA) is recognized in endosomal compartments by membrane-bound Toll-like receptor 3 (TLR3) (14). Stimulation of these viral pattern recognition receptors (PRRs) initiates a cascade of antiviral responses that converge on activation of two major transcriptional effectors, IFN regulatory factor 3 (IRF3) and NF-κB, whose actions are responsible for expression of antiviral responses in host cells by production of type I IFN, activation of inflammasomes, and expression of proinflammatory cytokines (16, 17).
The RIG-like receptors (RLRs), RIG-I and MDA5, are primarily coupled to activation of IRF3. Upon binding to cytoplasmic dsRNA or 5′ triphosphorylated RNA, RIG-I undergoes a conformational switch coupled to inducible Lys-63-linked polyubiquitylation (18, 19). This exposes two caspase activation and recruitment domains (CARDs) in the RLR NH2 termini, which mediate signaling by binding to the CARD of the MAVS adaptor protein on the outer mitochondrial membrane (20, 21). This signaling promotes the activation of downstream TBK1/IKKε complex. This rate-limiting kinase phosphorylates the transcription factor IFN regulatory factor 3 (IRF3), leading to its dimerization and translocation to the nucleus, where it drives expression of type I IFNs (22). Another mechanism is mediated by TLR3 recognition of endosomal dsRNA, an event that induces binding of the TRIF adaptor protein, inducing TBK1/IKKε activation and subsequent serine/threonine phosphorylation of IRF3 (23).
Type I IFNs are essential in the innate mucosal host defense and are highly induced by RSV infection. Of these IFN types, IFN-β is highly induced in both human cell and mouse models of RSV disease (24–26); IFN-α1 and -4 are induced in human airway epithelial cells (27), whereas IFN-α is induced in the mouse (28). IFNs are undetectable under physiological conditions but rapidly induced after viral infection in most cell types. Virus-triggered type I IFN production amplifies the initial IRF3 signal to induce transcription of >300 IFN-stimulated genes (ISGs), whose products reduce viral replication and propagation (29, 30). In fact, treatment with IFN-α reduces RSV titers and lung inflammation in a mouse model (28). Although most individual ISG functions remain to be elucidated, collectively they function as antiviral effectors. Mice with mutations in these proteins, or key components of their downstream pathways, have increased susceptibility to viral infection (30), including that by RSV (26). ISG56 (IFIT1) and ISG54 (IFIT2) are two family members containing multiple tetratricopeptide repeats (31, 32) whose antiviral function, in part, is mediated by interacting with the eIF-3 translation initiation factor, leading to the inhibition of translational initiation and protein synthesis (33). As a result, ISGs play a central role in coordinating the innate immune response to single-stranded RNA (ssRNA) viruses.
Numerous studies have shown that ISGs are rapidly inducible immediate early genes. ISGs are initially triggered by IRF3 binding to their promoter and later amplified by the autocrine IFN-I pathway (34). Although activated IRF3 induces complex formation with the histone acetyltransferase p300/CBP (35), the rapid response of ISGs is not consistent with a gene expression mechanism involving extensive chromatin remodeling. Inducible transcription of rapidly inducible genes in open chromatin domains has recently been shown to involve regulated transcriptional elongation (36, 37). In this mechanism, immediate early genes located in open chromatin are associated with promoter-paused RNA polymerase II (Pol II). Upon recruitment of the transcriptional elongation factor P-TEFb, a complex containing the essential cyclin-dependent kinase 9 (CDK9) and the bromodomain 4 (BRD4) proteins, Ser2 of the heptad repeat of the RNA Pol II carboxy-terminal domain (CTD) is phosphorylated, licensing the polymerase to enter productive RNA processing (36, 37). Recently, we found that RelA Ser276 phosphorylation mediates RelA acetylation, BRD4/CDK9 association, and activation of a subset of downstream inflammatory genes by transcriptional elongation in RSV infection (10). Our previous findings of the regulation of transcriptional elongation on RSV-induced NF-κB-dependent gene expression prompted us to explore whether transcriptional elongation regulates IRF3-dependent ISG expression.
We therefore examined the role of transcriptional elongation in ISG expression induced by RSV or by the dsRNA analog [poly(I·C)]. We observe that RSV infection and intracellular poly(I·C) induce a cytoplasmic-nuclear translocation of IRF3 and ISG expression. Furthermore, RSV infection induces recruitment of IRF3 along with CDK9, Pol II, and phospho-Ser2 Pol II CTD to ISG promoters. Using a highly quantitative immunoprecipitation (IP)-stable isotope dilution (SID)-selected reaction monitoring (SRM)-mass spectroscopy (MS) assay, we observe that RSV infection induces an activated CDK9 complex by promoting its association with BRD4 and by disrupting its association with inhibitory 7SK small nuclear RNA (snRNA). In both human small airway epithelial cells (SAECs) and A549 cells, virus-induced ISG expression was inhibited by CDK9 small interfering RNA (siRNA)-mediated knockdown and by the use of a selective pharmacological inhibitor of CDK9, while virus-induced IFN-β expression was resistant. CDK9 inhibition blocked the expression of RSV-induced spliced ISG54 transcripts, demonstrating its requirement for transcriptional elongation. We conclude that transcription elongation plays a major role in RSV-induced IRF3-dependent ISG expression.
MATERIALS AND METHODS
Cell culture and treatment.
Human A549 pulmonary epithelial cells (American Type Culture Collection, Manassas, VA) were grown in F-12K medium (Invitrogen, Carlsbad, CA) with 10% fetal bovine serum (FBS), penicillin (100 U/ml), and streptomycin (100 μg/ml) at 37°C in a 5% CO2 incubator (27). MEFs were cultured in Eagle's minimum essential medium (Gibco) with 0.1 mM nonessential amino acids, 1.0 mM sodium pyruvate, and 10% FBS (27). Human primary small airway epithelial cells (American Type Culture Collection, Manassas, VA) were grown in small airway epithelial cell growth medium (SAGM; Lonza, Walkersville, MD). Poly(I·C) was obtained from Sigma (St. Louis, MO), and the CDK9 inhibitor II (CAN508) was purchased from Calbiochem-EMD Millipore (Billerica, MA).
For poly(I·C) electroporation, A549 cells were trypsinized, washed in phosphate-buffered saline (PBS), and pelleted by centrifugation. The cell pellet was then resuspended in 100 μl Nucleofector solution (Amaxa; Lonza) with poly(I·C) into an electroporation cuvette. The cell suspension was electroporated using a Nucleofector I device (Amaxa; Lonza). The Nucleofector Program X-001 was used.
For determinations of cell viability, A549 cells were trypsinized. The total cell numbers were counted, and cell viability was determined by using an 0.4% solution of trypan blue in PBS (Cellgro) in an automatic cell counter (Invitrogen) (38).
Virus preparation and infection.
The human RSV Long strain was grown in HEp-2 cells and prepared as described previously (39). The viral titer of purified RSV pools varied from 8 to 9 log PFU/ml, as determined by a methylcellulose plaque assay. Viral pools were aliquoted, quick-frozen on dry ice-ethanol, and stored at −70°C until they were used.
Reverse siRNA transfection.
Control, IRF3, and CDK9 siRNAs (Dharmacon, ThermoFisher Scientific, Lafayette, CO) were reverse transfected by plating trypsinized A549 cells into a dish containing 100 nM siRNA-TransIT-siQUEST complexes (Mirus Bio Corp.). Forty-eight hours later, cells were inoculated with RSV (multiplicity of infection [MOI], 1.0) or electroporated with poly(I·C) for indicated times. At indicated periods, cells were washed with phosphate-buffered saline (PBS) twice and lysed in Tri reagent (Sigma-Aldrich).
Retroviral IRF3 expression.
Full-length human IRF3 was expressed as FLAG-Strawberry fluorescent protein (Straw) fusion proteins using the pCX4-hygro expression vector (40, 41). Retrovirus stocks were produced by cotransfecting pCX4-hygro-Straw-IRF3 and amphotropic packaging plasmid pCL-10A1 into Bosc23 cells (40). A549 cells were infected with retrovirus in the presence of 8 μg/ml Polybrene and selected for hygromycin B (100 ng/ml) resistance.
Subcellular fractionation and Western immunoblot analyses.
Nuclear and cytoplasmic proteins were fractionated as previously described (42). For Western blotting, equal amounts of nuclear protein were fractionated by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were incubated with affinity-purified rabbit polyclonal antibodies to IRF3 (Santa Cruz Biotechnology). Washed membranes were then incubated with IRDye 800-labeled anti-rabbit IgG antibodies (Rockland Immunochemicals, Gilbertsville, PA), and immune complexes were quantified using the Odyssey infrared imaging system (Li-Cor Biosciences, Lincoln, NE).
Quantitative real-time reverse transcription-PCR (Q-RT-PCR).
Total RNA was extracted using acid guanidium phenol extraction (Tri reagent; Sigma). For gene expression analyses, 1 μg of RNA was reverse transcribed using SuperScript III in a 20-μl reaction mixture (27). One microliter of cDNA product was diluted 1:2, and 2 μl of diluted product was amplified in a 20-μl reaction mixture containing 10 μl of SYBR green Supermix (Bio-Rad) and 0.4 μM (each) forward and reverse gene-specific primers (Table 1). The reaction mixtures were aliquoted into a Bio-Rad 96-well clear PCR plate, and the plate was sealed with Bio-Rad Microseal B film. The plates were denatured for 90 s at 95°C and then subjected to 40 cycles of 15 s at 94°C, 60 s at 60°C, and 1 min at 72°C in an iCycler (Bio-Rad). PCR products were subjected to melting curve analysis to ensure that a single amplification product was produced. Quantification of relative changes in gene expression was done using the threshold cycle (ΔΔCT) method. In brief, the ΔCT value was calculated (normalized to glyceraldehyde-3-phosphate dehydrogenase [GAPDH]) for each sample by using the equation ΔCT = CT (target gene) − CT (GAPDH). Next, the ΔΔCT was calculated by using the equation ΔΔCT = ΔCT (experimental sample) − ΔCT (control sample). Finally, the fold differences between the experimental and control samples were calculated using the formula 2−ΔΔCT.
Table 1.
Primer sets for Q-RT-PCR
| Primer set | Sequence (5′→3′) |
|
|---|---|---|
| Forward | Reverse | |
| hIRF3 | AGAGGCTCGTGATGGTCAAG | AGGTCCACAGTATTCTCCAGG |
| hISG56 | TCAGGTCAAGGATAGTCTGGAG | AGGTTGTGTATTCCCACACTGTA |
| hISG54 | GGAGGGAGAAAACTCCTTGGA | GGCCAGTAGGTTGCACATTGT |
| hISG54-5′UTR | ACGTCAGCTGAAGGGAAACAAACA | CAGGGCTCGGTTCAGGCAGC |
| hISG54-exon1-2 | TGCAGCTGCCTGAACCGAGC | GCCGTAGGCTGCTCTCCAAGG |
| hISG54-intron1 | GACCTGGTTGCCTAACCCTC | GACAGAGGGCAAGGTGTCTC |
| hCIG5 | TGGGTGCTTACACCTGCTG | GAAGTGATAGTTGACGCTGGTT |
| hCDK9 | ATGGCAAAGCAGTACGACTCG | GCAAGGCTGTAATGGGGAAC |
| hIFN-β | ATGACCAACAAGTGTCTCCTCC | GGAATCCAAGCAAGTTGTAGCTC |
| hRN7SK | AGGACGACCATCCCCGATAG | CGTATACCCTTGACCGAAGACC |
| hGAPDH | ATGGGGAAGGTGAAGGTCG | GGGGTCATTGATGGCAACAATA |
| mIRF3 | GAGAGCCGAACGAGGTTCAG | CTTCCAGGTTGACACGTCCG |
| mISG56 | CTGAGATGTCACTTCACATGGAA | GTGCATCCCCAATGGGTTCT |
| mISG54 | AGTACAACGAGTAAGGAGTCACT | AGGCCAGTATGTTGCACATGG |
| mCIG5 | TGCTGGCTGAGAATAGCATTAGG | GCTGAGTGCTGTTCCCATCT |
| mGAPDH | AGGTCGGTGTGAACGGATTTG | TGTAGACCATGTAGTTGAGGTCA |
| RSVN | AAGGGATTTTTGCAGGATTGTTT | TCCCCACCGTAACATCACTTG |
XChIP.
Dual cross-link chromatin immunoprecipitation (XChIP) was performed as described previously (43). A549 cells (∼6 × 106 per 100-mm dish) were washed twice with PBS. Protein-protein cross-linking was first performed with disuccinimidyl glutarate (2 mM; Pierce), followed by protein-DNA cross-linking with formaldehyde. Equal amounts of sheared chromatin were immunoprecipitated overnight at 4°C with 4 μg of the indicated antibody (Ab) in ChIP dilution buffer (43). Immunoprecipitates were collected with 40 μl protein A magnetic beads (Dynal Inc.), washed, and eluted in 250 μl elution buffer for 15 min at room temperature. Samples were de-cross-linked in 0.2 M NaCl at 65°C for 2 h. The precipitated DNA was phenol-chloroform extracted, precipitated with 100% ethanol, and dried.
Q-gPCR.
Gene enrichment in XChIP was determined by quantitative real-time genomic PCR (Q-gPCR) as previously described (43, 44) using region-specific PCR primers (Table 2). Standard curves were generated using a dilution series of genomic DNA (from 1 ng to 100 ng) for each primer pair. The fold change of DNA in each immunoprecipitate was determined by normalizing the absolute amount to the input DNA reference and calculating the fold change relative to that amount in unstimulated cells.
Table 2.
Primer sets for XChIP
| Gene | Sequence (5′→3′) |
|
|---|---|---|
| Forward | Reverse | |
| hISG54-5′ ISRE | ACGTCAGCTGAAGGGAAACAAACA | CGGTTCAGGCAGCTGCACTCTT |
| hISG54-3′ UTR | TGGTTCCCCTGAACTTTACTGT | TGGGCACCAGAGGCATGATA |
| hISG56-5′ ISRE | GGTTGCAGGTCTGCAGTTTATCTGT | AGCTGTGGGTGTGTCCTTGC |
| hISG56-3′ UTR | GGCAGACTGGCAGAAGCCCA | CCTCCACACTTCAGCAAGGCCC |
| hCIG5-5′ ISRE | CCAGGCATCTGCCACAATG | CACTCAAGAGCTTCCCAGCAA |
| hCIG5-3′ UTR | GGCCACATGAGGCTGTCAAGCA | TGCCAACCCAGTGTGCCGTC |
| hCDKNIA-5′ | TCGTGGGGAAATGTGTCCAG | CTGGCCGAGTTCCAGCAG |
| mISG56-5′ ISRE | TCAGTGGAGAATGCAGTAGGGCAA | ACTGTCACACCAACTGGAAGCTCA |
Immunoprecipitation (IP).
A549 cells (4 × 106 to 6 × 106 per 100-mm dish) were washed twice with PBS. Protein-protein cross-linking was first performed with disuccinimidyl glutarate. The cross-linked cells were then collected into Eppendorf tubes and washed with PBS twice. After washing, the cells were suspended in radioimmunoprecipitation assay (RIPA) buffer with complete protease inhibitor cocktail (Sigma-Aldrich) and 0.1% Igepal CA-630 (MP Biomedicals) and incubated on ice for 30 min. After incubation, the cells were sonicated 4 times and centrifuged at 13,000 rpm for 10 min. The supernatants were collected and were quantified for protein concentrations. Equal volumes of whole-cell lysates were immunoprecipitated overnight at 4°C with 4 μg of the indicated Ab in ChIP dilution buffer (43). Immunoprecipitates were collected with 40 μl protein A magnetic beads (Dynal Inc.). The samples were dissociated with beads and were prepared for SID-SRM-MS analysis.
SID-SRM-MS.
The SID-SRM-MS assays of CDK9, BRD4, IRF3, IRF1, IRF7, STAT1, RSV N protein, and RSV NS1 were developed using a workflow described in previous publications (45, 46). The signature peptides and SRM parameters are listed in Table 3. The peptides were chemically synthesized incorporating isotopically labeled [13C615N4]arginine or [13C615N2]lysine to a 99% isotopic enrichment (Thermo Scientific).
Table 3.
SRM parameters of SRM assays of proteins for sample amount normalizationa
| Gene name | Swiss-Prot accession no. | Sequence | Q1 m/z | Q3 m/z | Ion type | CE (V) |
|---|---|---|---|---|---|---|
| NCAP | P03418 | SGLTAVIR | 408.75 | 458.308 | y4 | 17 |
| 408.75 | 559.356 | y5 | 17 | |||
| 408.75 | 672.44 | y6 | 17 | |||
| 408.75 | 729.461 | y7 | 17 | |||
| 408.75 | 816.493 | y8 | 17 | |||
| BRD4 | O60885 | DAQEFGADVR | 554.257 | 664.341 | y6 | 22 |
| 554.257 | 793.383 | y7 | 22 | |||
| 554.257 | 921.442 | y8 | 22 | |||
| CDK9 | P50750 | DPYALDLIDK | 581.803 | 603.334 | y5 | 23 |
| 581.803 | 716.418 | y6 | 23 | |||
| 581.803 | 787.455 | y7 | 23 | |||
| 581.803 | 950.519 | y8 | 23 | |||
| IRF3 | Q14653 | LVGSEVGDR | 466.2422 | 446.235 | y4 | 20 |
| 466.2422 | 575.278 | y5 | 16 | |||
| 466.2422 | 719.331 | y7 | 17 | |||
| IRF1 | P10914 | EEPEIDSPGGDIGLSLQR | 956.468 | 1,015.553 | y10 | 36 |
| 956.468 | 1,112.605 | y11 | 36 | |||
| 956.468 | 1,199.637 | y12 | 36 | |||
| 956.468 | 1,314.664 | y13 | 36 | |||
| IRF7 | Q92985 | GGGPPPEAETAER | 634.299 | 676.326 | y6 | 25 |
| 634.299 | 805.368 | y7 | 25 | |||
| 634.299 | 902.421 | y8 | 25 | |||
| 634.299 | 999.474 | y9 | 25 | |||
| 634.299 | 1,096.526 | y10 | 25 | |||
| TBK1 | Q9UHD2 | TTEENPIFVVSR | 696.362 | 817.493 | y7 | 24 |
| 696.362 | 931.535 | y8 | 24 | |||
| 696.362 | 1,060.578 | y9 | 23 | |||
| 696.362 | 1,189.621 | y10 | 23 | |||
| STAT1 | P42224 | DQQPGTFLLR | 587.814 | 649.403 | y5 | 23 |
| 587.814 | 706.424 | y6 | 23 | |||
| 587.814 | 803.477 | y7 | 23 | |||
| 587.814 | 931.535 | y8 | 23 | |||
| RIG-I | O95786 | DLENLSQIQNR | 665.341 | 745.395 | y6 | 26 |
| 665.341 | 858.479 | y7 | 26 | |||
| 665.341 | 972.522 | y8 | 26 | |||
| 665.341 | 1,101.564 | y9 | 26 | |||
| HST4 | P62805 | VFLENVIR | 495.293 | 501.314 | y4 | 20 |
| 495.293 | 630.356 | y5 | 20 | |||
| 495.293 | 743.441 | y6 | 20 | |||
| 495.293 | 890.509 | y7 | 20 | |||
| 495.293 | 989.577 | y8 | 20 | |||
| NS1 | P04544 | LQNLFDNDEVALLK | 816.435 | 901.498 | y8 | 31 |
| 816.435 | 1,016.525 | y9 | 31 | |||
| 816.435 | 1,163.594 | y10 | 31 | |||
| 816.435 | 1,276.678 | y11 | 31 | |||
| 816.435 | 901.498 | y8 | 31 |
Masses listed are for the native forms of the peptides. Abbreviations: CE, collision energy; Q, quadrupole.
The proteins immunoprecipitated with anti-CDK9 antibody were captured by protein A magnetic beads (Dynal Inc.). The proteins were trypsin digested on the beads as described previously (45, 46). Briefly, the beads were washed with PBS three times and then resuspended in 30 μl of 50 mM ammonium hydrogen carbonate (pH 7.8), and 20 μl of a 0.1-μg/μl solution of trypsin was added. The samples were mixed and trypsinized by gentle vortexing overnight at 37°C. After digestion, the supernatant was collected. The beads were washed with 50 μl of 50% acetonitrile (ACN) three times, and the supernatant was pooled and dried. The tryptic digests were then reconstituted in 30 μl of 5% formic acid-0.01% trifluoroacetic acid (TFA). An aliquot of 10 μl of diluted stable isotope-labeled standard (SIS) peptides was added to each tryptic digest. These samples were desalted with a ZipTip C18 cartridge. The peptides were eluted with 80% ACN and dried. The peptides were reconstituted in 30 μl of 5% formic acid-0.01% TFA and were directly analyzed by liquid chromatography (LC)-SRM-MS. Similarly, the proteins in the nuclear extracts from A549 cells inoculated with RSV were processed for trypsin digestion and addition of internal standard as described previously (45).
SRM assays were performed as described previously (47), with slight modification. LC-SRM-MS analysis was performed with a TSQ Vantage triple quadrupole mass spectrometer equipped with a nanospray source (Thermo Scientific, San Jose, CA). The online chromatography was performed using an Eksigent NanoLC-2D high-pressure liquid chromatography (HPLC) system (AB Sciex, Dublin, CA). An aliquot of 10 μl of each tryptic digest was injected on a C18 reverse-phase nano-HPLC column (PicoFrit; 75 μm by 10 cm; tip inside diameter [i.d.], 15 μm) at a flow rate of 500 nl/min with a 20-min 98% A, followed by a 15-min linear gradient from 2 to 30% mobile phase B (0.1% formic acid-90% acetonitrile) in mobile phase A (0.1% formic acid). The TSQ Vantage was operated in high-resolution SRM mode with Q1 and Q3 set to 0.2- and 0.7-Da full-width half-maximum (FWHM). All acquisition methods used the following parameters: 1,800-V ion spray voltage, a 275°C ion transferring tube temperature, and a collision-activated dissociation pressure at 1.5 mtorr; also, the S-lens voltage used the values in the S-lens table generated during MS calibration.
All SRM data were manually inspected to ensure peak detection and accurate integration. The chromatographic retention time and the relative product ion intensities of the analyte peptides were compared to those of the SIS peptides. The variation of the retention time between the analyte peptides and their SIS counterparts should be within 0.05 min, and no significant difference in the relative product ion intensities of the analyte peptides and SIS peptides were observed. The peak areas in the extract ion chromatography of the native and SIS version of each signature peptide were integrated using Xcalibur 2.1. The default values for noise percentage and baseline subtraction window were used. The ratio between the peak areas of native and SIS versions of each peptide was calculated.
Quantification of CDK9-associated 7SK snRNA.
A549 cells were either inoculated with sucrose-purified RSV (MOI, 1.0) for 0, 12, and 30 h or electroporated with 10 μg poly(I·C) for 0 and 4 h as indicated, and nuclear extracts were prepared. Nuclei were disrupted in 4 volumes of RIPA buffer (150 mM NaCl, 1 mM Na2-EDTA, 1 mM EGTA, 1% NP-40, 1% sodium deoxycholate, 20 mM Tris-HCl, pH 7.5) containing 10 U/ml RNasin (New England BioLabs) and complete protease inhibitor cocktail (Sigma-Aldrich) by sonication for 10 s with a pulse setting of 4 in a Branson Sonifier 150 (Branson Ultrasonics Corporation) (48). Sonicated nuclear extract was clarified by centrifugation (10,000 rpm, 10 min at 4°C) and immunoprecipitated for 4 h at 4°C with 4 μg of CDK9 antibody (Santa Cruz) in RIPA buffer with complete protease inhibitor cocktail and RNasin. Immunoprecipitates were collected with 40 μl protein A magnetic beads (Dynal Inc.) for 1 h at 4°C, captured on a magnetic stand, and washed 4 times with PBS. Afterwards, the magnetic beads were dissolved in 1 ml Tri reagent (Sigma-Aldrich) for total RNA extraction and processed according to the manufacturer's recommendation. The extracted RNA was dissolved in the same volume of diethylpyrocarbonate (DEPC)-treated water, and the same volume of RNA of samples was reverse transcribed using SuperScript III in a 20-μl reaction mixture (27). The cDNA products were amplified in a 20-μl reaction mixture containing 10 μl of SYBR green Supermix (Bio-Rad) and 0.4 μM (each) forward and reverse 7SK snRNA qPCR primers (Table 1). The amount of 7SK snRNA in each sample was quantified relative to a standard curve generated from 10-fold serial dilutions of human genomic DNA (1 ng to 100 ng).
Fluorescence microscopy and dynamic imaging of A549-Strawberry-IRF3 cells.
A549-Strawberry-IRF3 cells were plated on cover glasses pretreated with rat tail collagen (Roche Applied Sciences). After the indicated stimulation, the cells were fixed with 4% paraformaldehyde in PBS and counterstained with 4′,6-diamidino-2-phenylindole (DAPI). The cells were visualized with a Nikon fluorescence confocal microscope at a magnification of ×63 (10, 27).
For dynamic imaging, the cells were electroporated with poly(I·C). A549-Strawberry-IRF3 stable cells were split into a 6-well cell culture plate containing collagen-coated 25-mm round coverslips and washed twice with PBS, and 1 ml of electroporation solution was added (BTX; Harvard Apparatus) with 50 μg poly(I·C). An electroporation petri pulsar (BTX; Harvard Apparatus) was put over the top of cells, and an electric square pulse (100 V, 12 μs) was generated by an ECM830 Electro Square Porator (BTX; Harvard Apparatus). The electroporated cells were immediately placed into a confocal imaging chamber maintained at 37°C and 5% CO2. The red channel was dynamically recorded every 6 min.
Dynamic imaging data analysis.
Cell images were segmented, quantified, and tracked by a custom-written pipeline program using CellTracker (49). In each time-lapse field, cells that showed overlap with other cells, divided during the time course, or moved out of view were excluded from analysis. Images were smoothed using a median filter of size 6 and normalized using the local average and its standard deviation. Both cell and nuclear boundaries were then identified first automatically by intensity thresholding and then by manual correction. The same cells were tracked from one frame to the next frame through the time course. Where indicated, the nuclear Strawberry-IRF3 fluorescence signal was divided by the cytoplasmic Strawberry-IRF3 signal to obtain the nuclear-to-cytoplasmic (N/C) ratio.
Statistical analysis.
One-way analysis of variance (ANOVA) was performed when looking for time differences, followed by Tukey's post hoc test to determine significance. Mann-Whitney tests were used for nonparametric data. A P value of <0.05 was considered significant (10).
RESULTS
RSV infection induces cytoplasmic-nuclear IRF3 translocation.
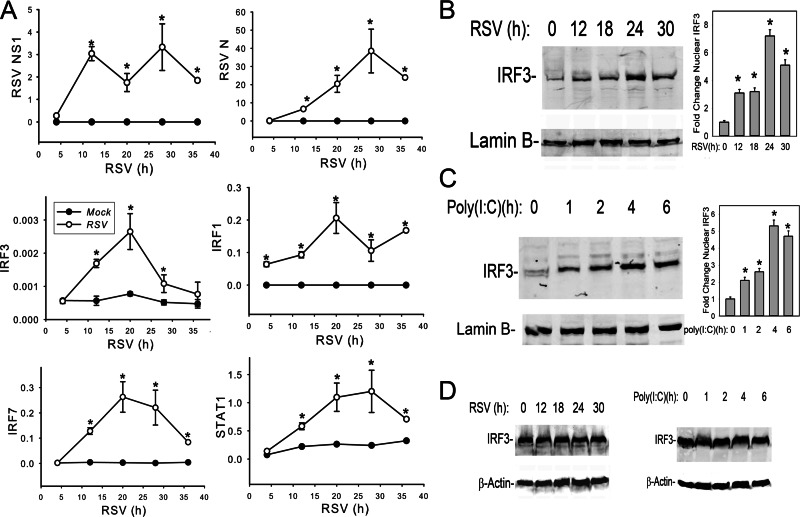
Human type II-like A549 airway epithelial cells productively replicate RSV and are a well-established model for study of the lower airway epithelial cell response to RSV infection (9, 24, 50). Previous work has shown that RSV encodes nonstructural (NS) proteins that antagonize IRF3 signaling (51). To examine the dynamics of the innate immune response relative to NS1 protein expression, we monitored the expression of viral and host proteins using a multiplexed SID-SRM-MS assay. SID-SRM-MS assays monitor precursor-product transitions of selected proteins and are superior to Western blotting in terms of quantification and analyte specificity (45, 46). The inclusion of SIS peptide standards enables accurate target protein quantification. A panel of SID-SRM-MS assays was used to determine the temporal files of nuclear innate immune response transcription factors (IRF3, IRF1, IRF7, and STAT1) and that of RSV proteins (N and NS1). Nuclear extracts from A549 cells inoculated with sucrose-purified RSV (MOI, 1.0) for 4, 12, 20, 28, and 36 h, respectively, along with mock treatments at the same time points were prepared for SID-SRM-MS analysis (Fig. 1A). We observed that RSV NS1 protein was the most rapidly expressed RSV protein, peaking within 12 h of RSV adsorption and prior to the peak of RSV N expression seen at 28 h (Fig. 1A). This expression pattern is consistent with sequential 3′-5′ expression of the RSV genome. Nuclear levels of the IRF3 and downstream IRF1 and IRF7 transcription factors significantly increased 12 h after RSV infection compared with those of mock treatments (Fig. 1A). Together, these data indicate that, despite expression of the NS1 IFN antagonist, the primary (IRF3) and secondary (IRF1/IRF7 and STAT1) signaling pathways are intact. Among the IRF family, IRF3 is considered the major transcription factor responsible for the initial ISG expression in host cells (16, 17). Here, preformed cytoplasmic IRF3 is activated by both RIG-I and TLR3 pattern receptors through the activation of the rate-limiting TBK1/IKKε kinase complex (22, 23) in response to RSV infection (14). Therefore, we explored the mechanism of how IRF3 mediates the innate immune response.
Fig 1.
RSV induces cytoplasmic-nuclear IRF3 translocation concomitantly with NS1 expression. (A) Dynamics of innate immune response in RSV-infected A549 cells. A549 cells were inoculated with sucrose-purified RSV (MOI, 1.0) for 4, 12, 20, 28, and 36 h, respectively, along with mock treatments at the same time points. Equal amounts of nuclear extract were applied for SID-SRM-MS analysis to determine the temporal profiles of nuclear IRF3, IRF1, IRF7, RSV N protein, RSV NS1, and STAT1. All of the values are presented as the ratios of native to SIS peptides. *, statistical significance compared with mock treatments (P < 0.05). (B) Kinetics of RSV-induced IRF3 nuclear translocation. A549 cells were inoculated with sucrose-purified RSV (MOI, 1.0) for 0 to 30 h as indicated. Equal amounts of nuclear extracts were resolved by 10% SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane. The membrane was incubated with anti-IRF3 (top) or anti-lamin B (bottom) Abs. Right panel, the intensity of each band of IRF3 normalized to lamin B loading in a Western blot was quantified. *, statistical significance compared with mock treatments (P < 0.05). (C) Kinetics of poly(I·C)-induced IRF3 nuclear translocation. A549 cells were electroporated with 10 μg of synthetic dsRNA analog poly(I·C) for 0 to 6 h as indicated, and nuclear extracts were processed for Western blotting. The membrane was incubated with anti-IRF3 (top) or anti-lamin B (bottom) Abs. Right panel, the normalized intensity of IRF3 was quantified. *, statistical significance compared with mock treatments (P < 0.05). (D) Abundance of cytoplasmic IRF3 in A549 cells during RSV infection. A549 cells were infected with RSV (MOI, 1.0) (left) or were electroporated with 10 μg of poly(I·C) (right) at indicated time points. Cytoplasmic extracts were processed for Western blotting, and the membrane was incubated with anti-IRF3 (top) or anti-β-actin (bottom) Abs.
To independently confirm RSV-induced cytoplasmic-nuclear IRF3 translocation, A549 cells were inoculated with RSV (MOI, 1.0) and sucrose-cushion purified nuclear extracts were assayed for IRF3 abundance by Western immunoblotting (52). We found that nuclear IRF3 was detectably increased at 12 h and reached a 7.2-fold increase at 24 h after RSV infection relative to control (Fig. 1B), qualitatively confirming the SID-SRM-MS assays. To compare the effects of the dsRNA molecular pattern on nuclear IRF3 translocation, poly(I·C) was electroporated into A549 cells, and nuclear extracts were isolated for IRF3 Western blotting. Poly(I·C) induced a more rapid nuclear IRF3 translocation than that observed for quantification by RSV. An initial increase of IRF3 was observed within 1 h, and the increase peaked at 5.2-fold 4 h after electroporation (Fig. 1C). Meanwhile, we did not observe significant changes of cytoplasmic IRF3 levels in either RSV-infected or poly(I·C)-stimulated A549 cells (Fig. 1D), suggesting that nuclear IRF3 accumulation is primarily due to nuclear translocation.
Cytoplasmic Strawberry-IRF3 fusion protein translocates into nuclei during viral infection.
To quantitate nuclear translocation, we stably expressed Strawberry (Straw) fluorescent protein-labeled IRF3 in A549 cells using retrovirus-mediated transduction. To validate that the Straw-IRF3 fusion protein translocates with kinetics similar to that of endogenous IRF3, a time course of nuclear translocation was measured in wild-type (WT) A549 and A549-Straw-IRF3 cells using anti-IRF3 Ab in a Western blot assay. We observed that Straw-IRF3 translocated into the nucleus with kinetics similar to that of endogenous IRF3 in both cell types (Fig. 2A). Similarly, inducible ISG expression and RSV replication were indistinguishable in WT A549 cells and A549-Straw-IRF3 cells (data not shown). We therefore conclude that the Strawberry-IRF3 fusion protein exhibits the same biological characteristics as does endogenous IRF3, allowing us to use dynamic imaging to more precisely quantify cellular IRF3 responses.
Fig 2.
Analysis of Strawberry-IRF3 fusion protein in RSV infection and poly(I·C) treatment. (A) Kinetics of RSV-induced nuclear translocation. A549 cells stably expressing Straw-IRF3 fusion protein were RSV inoculated (MOI, 1.0) for 0 to 24 h as indicated, and nuclear extracts were processed for Western blotting. Wild-type A549 cells were used as a control. The membrane was incubated with anti-IRF3 Ab. The high-molecular-weight band is the Straw-IRF3 fusion protein, and the lower band indicates endogenous IRF3. (B) Confocal imaging of Strawberry-IRF3 fusion protein nuclear translocation. A549-Straw-IRF3 cells were inoculated with RSV at an MOI of 1.0 for different time periods. The infected cells were fixed with 4% paraformaldehyde, stained with nuclear marker DAPI, and mounted onto slides for confocal imaging. (C) Quantification of nuclear translocation. Cytoplasmic-to-nuclear translocation (N/C) of IRF3 was analyzed using CellTracker software for multiple cells. For each time point, the percentage of N/C ratio was averaged among all the traceable cells. Data are plotted as means ± standard deviations. *, statistical significance compared with mock treatments (P < 0.05). (D) Dynamic imaging of poly(I·C)-induced IRF3 nuclear translocation in A549-Strawberry-IRF3 stable cells. A549-Straw-IRF3 cells were split into 6-well plates, electroporated with 50 μg poly(I·C), and immediately placed into a confocal imaging system maintained at 37°C and 5% CO2. The red (for Strawberry) channel was automatically recorded every 6 min for 14 h. (E) Quantification of single-cell nuclear translocation. Cytoplasmic-to-nuclear translocation of IRF3 was analyzed using CellTracker software, and individual profiles were plotted as a function of time. a, b, and c represent different images for the same time course, analyzed for traceable cells. N/C, nuclear/cytoplasmic.
A549-Straw-IRF3 cells were RSV infected for various times, fixed, and imaged by confocal microscopy. Consistent with the results of Western blotting, we observed significant nuclear translocation of Straw-IRF3 12 h after RSV infection, increasing at 24 h (Fig. 2B). To quantify this response, the static images were segmented and the fraction of Straw-IRF3 translocation into the nucleus was quantified; for comparison between cells, nuclear translocation was expressed as a nuclear/cytoplasmic (N/C) ratio due to the variable expression of the Straw-IRF3. We observed that the average fraction of IRF3 that was translocated into the nucleus increased from 6 h to 24 h; at this time point, 35% of the cytoplasmic IRF3 was translocated into the nucleus (Fig. 2C), a fraction consistent with our earlier results (14).
To more precisely measure the kinetics of rapid dsRNA-induced IRF3 translocation, we monitored live cells using confocal imaging, where we could observe the dynamic subcellular changes of Straw-IRF3 at the single-cell level (49). Here, A549-Straw-IRF3 cells were electroporated with poly(I·C) and dynamic images were recorded every 6 min (Fig. 2D). The fraction of Straw-IRF3 translocation into the nucleus was quantified. In individual nuclear translocation profiles over the first 3 h of poly(I·C) stimulation, we observed that a portion of Straw-IRF3 rapidly translocated into the nucleus (Fig. 2E). Although there is significant cell-to-cell variability, the Strawberry-IRF3 N/C ratio increased rapidly in all cells, within 12 min of poly(I·C) stimulation, peaking within 30 min. This translocation was sustained for the duration of measurement, in a manner qualitatively consistent with the Western blot analysis (Fig. 2A). Together, both experimental approaches using biochemical fractionation and confocal imaging suggested that RSV is a potent inducer of cytoplasmic-nuclear IRF3 translocation, similar to that produced by synthetic dsRNA.
Virus-induced ISG expression is IRF3 dependent.
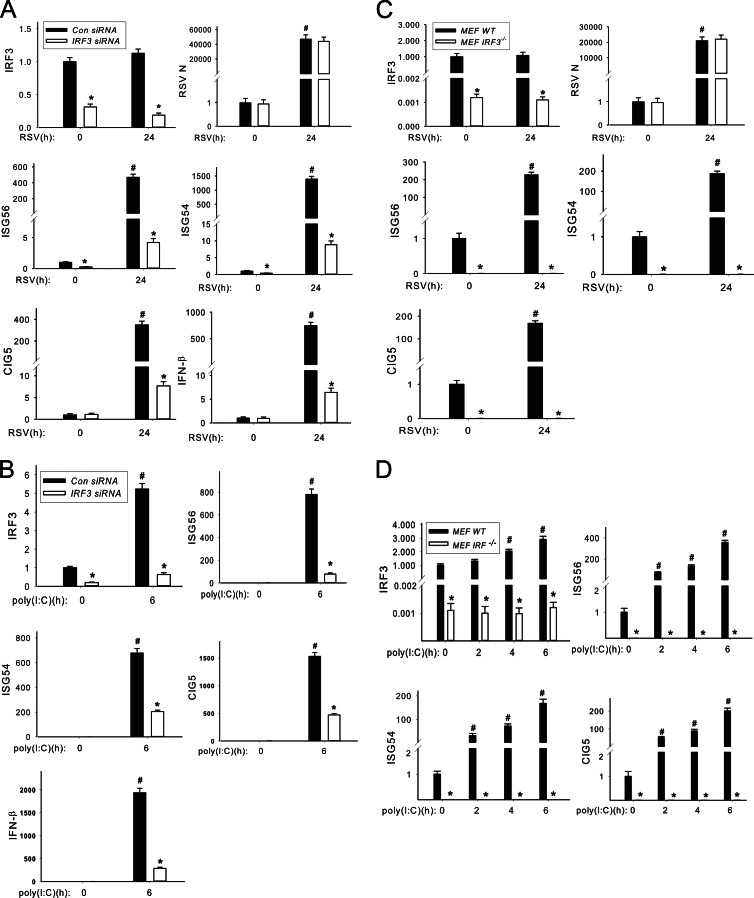
To determine whether virus-induced ISG expression is under IRF3 control, IRF3 expression was silenced using small interfering RNA (siRNA) and the ISG response was measured. For this purpose, A549 cells were transfected with duplex target (or scrambled) siRNAs specific to IRF3 48 h before RSV infection. We found that both basal and RSV-induced IRF3 mRNAs were significantly reduced by IRF3 siRNA to 31% (uninfected) and 18% (RSV infected) of that in scrambled siRNA-treated A549 cells (Fig. 3A). In scrambled siRNA-transfected A549 cells, we found that mRNA transcript levels of ISG54, ISG56, CIG5, and IFN-β genes were dramatically increased 24 h after RSV infection to 1,400-fold, 500-fold, 380-fold, and 750-fold, respectively (Fig. 3A). We further noted that RSV-induced ISG54, ISG56, CIG5, and IFN-β mRNAs were completely inhibited by IRF3 silencing (Fig. 3A), suggesting that IRF3 signaling is required to mediate their RSV-induced expression. This reduction was not due to nontarget effects on RSV replication because expression levels of RSV N mRNA were indistinguishable between scrambled and IRF3 siRNA treatments (Fig. 3A).
Fig 3.
RSV-induced ISG expression is regulated by IRF3. (A) IRF3 knockdown blocks RSV-induced ISG expression. IRF3 or control (Con) nonspecific siRNAs were reverse transfected into A549 cells (100 nM). After 48 h, the transfected cells were inoculated with or without RSV at an MOI of 1 for another 24 h and total RNA was extracted. mRNA levels of IRF3, ISG56, ISG54, CIG5, IFN-β, and RSV N protein were measured. The results were represented as the normalized fold change compared with control cells. *, statistical significance compared with scrambled siRNA treatments (P < 0.05); #, statistical significance compared with mock treatments. (B) IRF3 knockdown blocks poly(I·C)-induced ISG expression. IRF3 siRNA and control nonspecific siRNA were reverse transfected into A549 cells (100 nM). After 66 h, the transfected cells were electroporated with or without 10 μg poly(I·C) for another 6 h, and the total RNAs were extracted. Normalized mRNA levels of IRF3, ISG56, ISG54, CIG5, and IFN-β were determined and represented as fold change compared with control cells. *, statistical significance compared with scrambled siRNA treatments (P < 0.05); #, statistical significance compared with mock treatments. (C) Abrogation of RSV-induced ISG expression in MEF IRF3−/− cells. MEF IRF3−/− cells and MEF WT cells were inoculated with RSV at an MOI of 1.0 for 24 h. Normalized mRNA levels of IRF3, ISG56, ISG54, CIG5, and RSV N were determined and plotted. *, statistical significance compared with MEF WT cells (P < 0.05); #, statistical significance compared with mock treatments. (D) Abrogation of poly(I·C)-induced ISG expression in MEF IRF3−/− cells. MEF WT and MEF IRF3−/− cells were electroporated with 10 μg poly(I·C) at 0, 2, 4, and 6 h, respectively. Normalized mRNA levels of IRF3, ISG56, ISG54, and CIG5 were determined and plotted. *, statistical significance compared with MEF WT cells (P < 0.05); #, statistical significance compared with mock treatments.
We also quantified the dependence of IRF3 on poly(I·C)-induced ISG expression. In contrast to the effect seen with RSV infection, we found that that poly(I·C) treatment induced IRF3 expression by 5-fold relative to that of scrambled siRNA-treated A549 cells (Fig. 3B). In response to poly(I·C) stimulation, ISG54, ISG56, CIG5, and IFN-β mRNA transcripts were dramatically increased 6 h after electroporation of poly(I·C) in scrambled siRNA-treated A549 cells (Fig. 3B), and this expression was largely, but not completely, blocked by IRF3 siRNA (Fig. 3B). One explanation for this finding is that the incomplete inhibition may be due to inducible IRF3 expression in response to poly(I·C).
To more definitively demonstrate the absolute requirement of IRF3 for ISG expression and to circumvent the problem of its incomplete inhibition by siRNA, we examined ISG expression in IRF3-deficient mouse embryonic fibroblast cells (IRF3−/− MEFs) versus wild-type (WT) controls. For this purpose, IRF3−/− MEF and WT MEF cells were inoculated with RSV for 24 h and the abundance of IRF3, ISG54, ISG56, CIG5, and RSV N mRNAs was determined. We observed that in both uninfected and RSV-infected IRF3−/− MEFs, IRF3 mRNA is undetectable in comparison with that observed in WT MEFs (Fig. 3C). In WT MEFs, RSV infection dramatically increased ISG54, ISG56, and CIG5 gene expression by 190-, 225-, and 170-fold, respectively, relative to uninfected MEFs (Fig. 3C). Importantly, in IRF3−/− MEFs, RSV-induced ISG54, ISG56, and CIG5 mRNA expression was completely inhibited (Fig. 3C). As a control, RSV replication was measured in WT and IRF3−/− MEFs, where robust RSV N mRNA expression was observed with levels indistinguishable between the two cell types (Fig. 3C). These results are consistent with the siRNA experiments showing that RSV-induced ISG expression is under the tight control of IRF3 signaling.
We next measured the effects of poly(I·C) on ISG expression in WT and IRF3−/− MEFs after 0, 2, 4, and 6 h of transfection. In WT MEFs, poly(I·C) treatment dramatically increased the transcription levels of ISG54, ISG56, and CIG5 in a time-dependent manner, peaking at 168-, 359-, and 201-fold increases, respectively, relative to untransfected cells (Fig. 3D). In contrast, in IRF3−/− MEFs, both basal and poly(I·C)-treated mRNA levels of ISGs ISG54, ISG56, and CIG5 were also extremely low (Fig. 3D). Also, interestingly, we observed a 2.9-fold increase of IRF3 expression in poly(I·C)-stimulated WT MEFs (Fig. 3D); this finding is consistent with the induction of IRF3 expression observed in poly(I·C)-treated A549 cells. Together, these experiments suggest that IRF3 translocation is absolutely necessary for ISG54, ISG56, and CIG5 in response to RSV and poly(I·C) infection.
RSV infection induces IRF3-CDK9 recruitment to ISRE domains of ISGs.
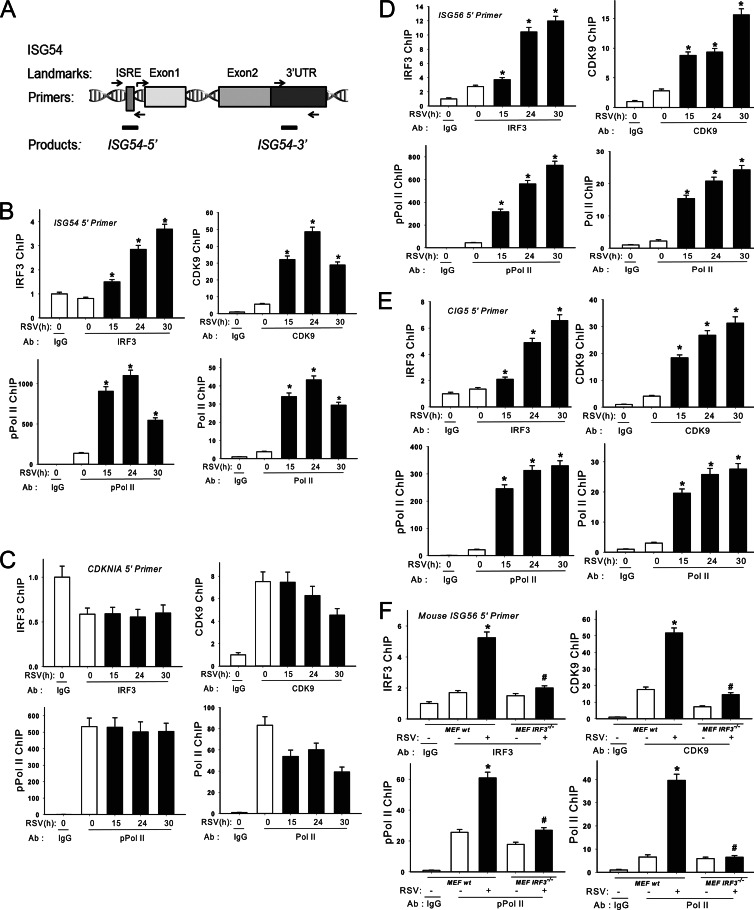
Our experimental data suggested that early RSV-induced ISG expression is absolutely IRF3 dependent. To understand the detailed mechanisms of how IRF3 regulates ISG transcription during viral infection, we examined recruitment of IRF3 to 5′ IFN-stimulated response element (ISRE) binding sites as well as the 3′ untranslated regions (UTRs) in native chromatin using a highly quantitative XChIP assay (44). For this purpose, region-specific primers were developed for quantitative genomic PCR (Q-gPCR) assays that quantify enrichment of the 5′ ISRE site or the 3′ UTR of the ISG54 gene (schematically diagrammed in Fig. 4A). A549 cells were inoculated with RSV for 0, 15, 24, and 30 h; chromatin cross-linked; and subjected to IP with Abs specific for IRF3, CDK9, RNA polymerase II (Pol II), or its CTD Ser2 phosphorylated form (phospho-Ser2 CTD Pol II). Anti-rabbit IgG was used as the negative control. We observed that IRF3 was first induced to bind the 5′ ISRE of the ISG54 promoter 15 h after RSV adsorption, and IRF3 binding continued to increase until 30 h (Fig. 4B). Strikingly, RSV also induced CDK9 binding to the 5′ ISRE site, initially detectable at 15 h, with an apparent peak at 24 h. Similar patterns of Pol II and phospho-Ser2 CTD Pol II were also observed (Fig. 4B). RSV-induced IRF3, CDK9, Pol II, and phospho-Ser2 CTD Pol II recruitment was not found at the 3′ UTR of the ISG54 gene, suggesting that the clearance of the transcriptional elongation complex is rapid (data not shown).
Fig 4.
XChIP analysis on ISRE binding sites of ISGs during RSV infection. (A) ISG54 ChIP primers for quantitative genomic PCR (Q-gPCR). The locations of sequences of human ISRE binding sites of ISG54 promoter and primers for Q-gPCR. (B) Elongation complex binding to the ISG54 5′ ISRE site during RSV infection. A549 cells were inoculated with or without RSV (MOI of 1). The corresponding chromatin was immunoprecipitated with anti-rabbit IRF3, CDK9, Pol II, and phospho-Ser2 CTD Pol II Abs. IgG was the negative control. Q-gPCR was performed using the ISG54 5′ ISRE primer set, and the fold change was calculated compared with IgG control. *, statistical significance compared with mock treatments (P < 0.05). (C) Elongation complex binding to CDK9-independent CDKNIA gene. XChIP analysis performed on the negative-control gene CDKNIA. Note the absence of inducible IRF3, CDK9, and phospho-Ser2 Pol II recruitment. (D) Elongation complex binding to ISG56. The experiment was performed as described for panel B. (E) Elongation complex binding to the CIG5 gene. The experiment was performed as described for panel B. (F) Elongation complex binding to ISG56 5′ ISRE site in RSV-infected MEF WT and MEF IRF3−/− cells. MEF WT and MEF IRF3−/− cells were inoculated with or without RSV (MOI of 1). Chromatin was immunoprecipitated with anti-rabbit IRF3, CDK9, Pol II, and phospho-Ser2 CTD Pol II Abs. Q-gPCR was performed using the mouse ISG56 5′ ISRE primer set, and the fold change was calculated compared with IgG control. *, statistical significance compared with mock treatments (P < 0.05); #, statistical significance compared with MEF WT cells.
To understand whether RSV infection indiscriminately increases P-TEFb association with active genes, we also examined its recruitment to the p53 target gene CDKNIA in native chromatin using the XChIP assay. CDKNIA expression is P-TEFb independent, being readily activated in the absence of CDK9 activity (53). We observed that RSV infection does not induce IRF3, CDK9, Pol II, or phospho-Ser2 CTD Pol II recruitment to the CDKNIA promoter (Fig. 4C). These data indicate that P-TEFb recruitment is restricted to a subset of the A549 transcriptome.
We also examined the RSV induction of IRF3, CDK9, Pol II, and phospho-Ser2 CTD Pol II recruitment to 5′ ISRE sites of the ISG56 and CIG5 genes. We observed temporal patterns of IRF3, CDK9, Pol II, and pSer2 CTD Pol II recruitment on both genes similar to that seen on the ISG54 gene (Fig. 4D and E). Similarly, these proteins were not recruited on the 3′ UTR (data not shown), suggesting that RSV induces accumulation of IRF3, the transcriptional elongation complex, and its substrate selectively on the 5′ end of ISGs.
CDK9 recruitment is IRF3 dependent.
The recruitment of CDK9 to target genes can be mediated by association with sequence-specific transcription factor complexes or via BRD4 binding to acetylated histone H4. To determine whether RSV-induced CDK9 and phospho-Ser2 CTD Pol II recruitment is IRF3 dependent, IRF3−/− MEFs were inoculated with RSV for 24 h and processed for the XChIP assay. The experimental results show that although IRF3, CDK9, and phospho-Ser2 CTD Pol II were recruited to the 5′ ISRE site of the mouse ISG56 gene after RSV infection in WT MEFs, recruitment of the same proteins (IRF3, CDK9, and phospho-Ser2 CTD Pol II) was significantly disrupted in IRF3−/− MEFs (Fig. 4F). These results suggest that RSV-inducible CDK9, Pol II, and phospho-Ser2 CTD Pol II recruitment to the ISGs promoter is under the control of IRF3.
Intracellular poly(I·C) induces IRF3-CDK9 recruitment to ISREs of ISGs.
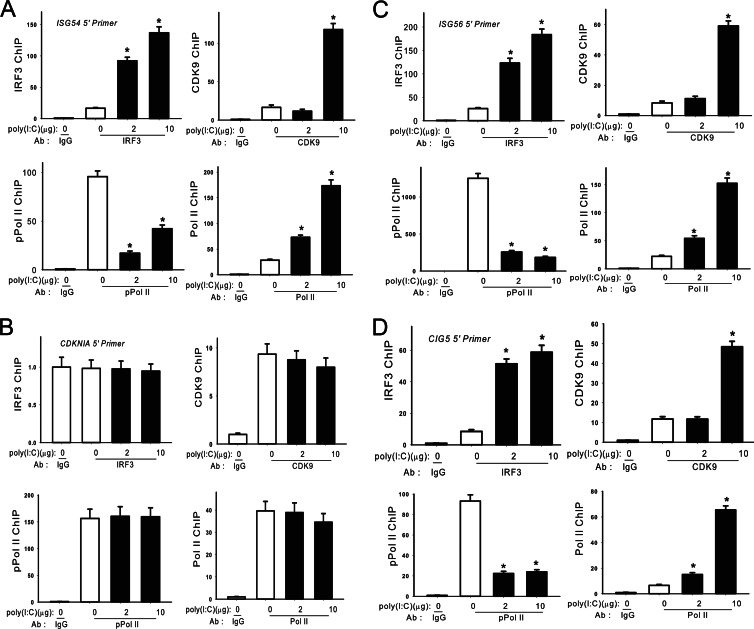
We next examined the extent of IRF3, CDK9, Pol II, and pSer2 CTD Pol II recruitment to the 5′ ISRE and 3′ UTR of ISGs in response to poly(I·C) stimulation. Here, we observed that IRF3, CDK9, and Pol II are also recruited to the 5′ ISRE binding sites while phospho-Ser2 CTD Pol II was apparently depleted (Fig. 5A, C, and D). Also, examination of the P-TEFb-independent CDKNIA gene again showed that poly(I·C) did not induce IRF3, CDK9, Pol II, or phospho-Ser2 CTD Pol II recruitment to its promoter (Fig. 5B).
Fig 5.
XChIP analysis on ISRE binding sites of ISGs in poly(I·C)-treated A549 cells. (A) Elongation complex binding to the ISG54 gene. A549 cells were electroporated with or without poly(I·C) for 4 h. Chromatin was immunoprecipitated with anti-rabbit IRF3, CDK9, Pol II, and phospho-Ser2 CTD Pol II Abs. Q-gPCR was performed using the ISG54 5′ ISRE primer set, and the fold change was calculated compared with IgG control. *, statistical significance compared with mock treatments (P < 0.05). (B) Elongation complex binding to CDKNIA. The experiment was performed as described for panel A. (C) Elongation complex binding to ISG56 ISRE. The experiment was performed as described for panel A. (D) Elongation complex binding to CIG5. The experiment was performed as described for panel A.
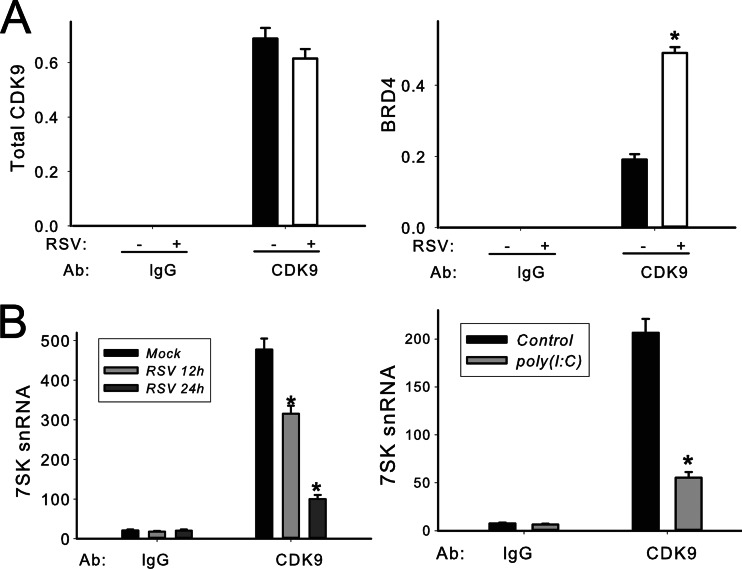
RSV infection enhances CDK9 association with its positive regulator BRD4.
BRD4 is considered a positive regulator of CDK9 and stimulates RNA polymerase II-dependent transcription (10, 36). To measure CDK9 complex formation, we adapted the SID-SRM-MS assay for CDK9 immunoprecipitates. A549 cells were inoculated with RSV for 0 and 30 h. The CDK9 complex was first enriched by IP with anti-rabbit CDK9 Ab, and the complex was analyzed for the presence of CDK9 and BRD4 proteins by SID-SRM-MS, normalized by the input protein concentration. Controls represent samples immunoprecipitated with IgG. Compared to control IgG IPs, where the CDK9 abundance was completely undetectable, CDK9 was significantly enriched in the samples immunoprecipitated with anti-CDK9 Ab (Fig. 6A, left). For more accurate comparison of the fraction of CDK9 associated with BRD4 in the control and infected lysates, the BRD4 signal was normalized to the total CDK9 in each sample (Fig. 6A, right). We detected strong BRD4 signals in the samples immunoprecipitated with anti-CDK9 Ab while BRD4 signals were completely undetectable in control samples. Importantly, the ratio of BRD4-associated CDK9 was increased 2.6-fold in the RSV-infected samples compared to mock treatment (Fig. 6A, right). We therefore conclude that RSV infection significantly increased CDK9 association with its positive regulator, BRD4.
Fig 6.
Viral infection induces the formation of active CDK9 complex. (A) Immunoprecipitation of CDK9 and SID-SRM assay in RSV-infected A549 cells. A549 cells were inoculated with RSV for 0 and 30 h (MOI of 1.0). Whole-cell lysates were prepared and immunoprecipitated with anti-rabbit CDK9 Ab and subjected to SID-SRM analysis of CDK9 and BRD4 protein abundance. The results of the SID-SRM assay were normalized by the input protein concentrations of the whole-cell lysates. The left panel of the figure shows total CDK9 detected in each sample, and the right panel shows the relative ratio of BRD4-associated CDK9 among samples. *, statistical significance compared with mock treatments (P < 0.05). (B) Quantification of CDK9-associated 7SK small nuclear RNA in A549 cells. A549 cells were either inoculated with sucrose-purified RSV (MOI, 1.0) for 0, 12, and 30 h (left panel) or electroporated with 10 μg poly(I·C) for 0 and 4 h (right panel). CDK9-associated 7SK snRNA was quantified relative to genomic DNA standards. *, statistical significance compared with mock treatments (P < 0.05).
RSV infection and intracellular poly(I·C) disrupt 7SK snRNA association with CDK9.
It has been previously shown that 7SK snRNA binds to and inhibits the kinase activity of the CDK9/cyclin T1 complex (48). To examine whether viral infection regulates 7SK snRNA-CKD9 association, A549 cells were either inoculated with RSV or stimulated with poly(I·C) and nuclear extracts were prepared. The nuclear extracts were immunoprecipitated with anti-CDK9 Ab, and CDK9-associated 7SK snRNA was quantified by Q-RT-PCR. 7SK snRNA was detected in the CDK9 complex in control cells, whereas its abundance was reduced in a time-dependent manner in response to RSV infection (Fig. 6B). Similar findings were observed in response to poly(I·C) treatment (Fig. 6B). These data indicate that RSV and dsRNA stimulation disrupts CDK9 association with its negative regulator, 7SK snRNA. Taken together with the increase in BRD4 association, we conclude that both RSV and poly(I·C) increase the activated fraction of P-TEFb.
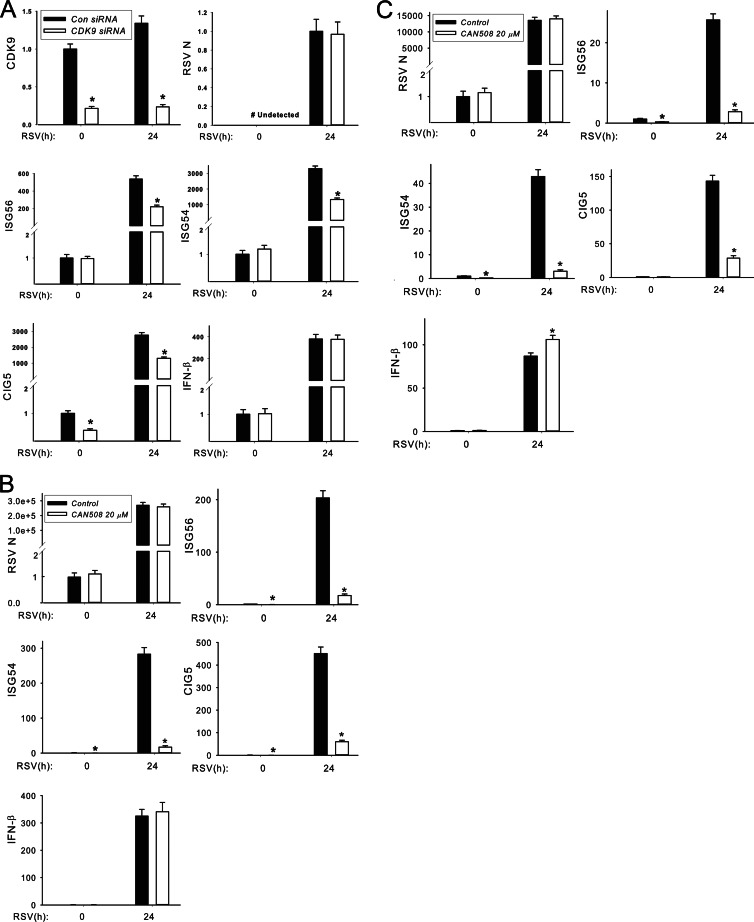
CDK9 activity is required for RSV-induced ISG expression. Our results suggest that IRF3 recruits activated CDK9 to the 5′ ISRE binding sites of ISGs during RSV infection (Fig. 4 and 5). To further examine the functional role of CDK9 in RSV-induced ISG expression, we used two approaches: (i) modulating CDK9 abundance using siRNA silencing and (ii) inhibiting CDK9 kinase activity using a CDK9-selective inhibitor.
The levels of CDK9 were inhibited by siRNA transfection in A549 cells, where CDK9 mRNA abundance was significantly reduced relative to scrambled controls in both control and RSV-infected cells (Fig. 7A). Under these conditions, 95% cell viability was detected using a trypan blue exclusion assay (not shown). Here, RSV-induced ISG54, ISG56, and CIG5 expression were all significantly inhibited (∼50%) in CDK9-silenced cells, whereas no effect on RSV replication was observed (Fig. 7A). Surprisingly, we noted that IFN-β expression was also highly induced and not inhibited by CDK9 silencing, suggesting that IFN-β transcription is CDK9 independent.
Fig 7.
CDK9 activity is required for RSV-induced ISG expression. (A) Effect of CDK9 siRNA silencing on RSV-induced ISG transcription in A549 cells. Control and CDK9 siRNAs were transfected into A549 cells, and 48 h later, cells were inoculated with or without RSV (MOI, 1.0) for another 24 h. The total RNAs were collected for gene expression analysis using Q-RT-PCR, and the mRNA levels of CDK9, ISG56, ISG54, CIG5, IFN-β, and RSV N protein were determined. The results are presented as normalized fold change compared to scrambled siRNA-transfected cells. *, statistical significance compared with scrambled siRNA treatments (P < 0.05). (B) Effect of CAN508 on RSV-induced ISG transcription in A549 cells. A 20 μM concentration of CAN508 was used to pretreat A549 cells, and 1 h later, cells were inoculated with or without RSV (MOI, 1.0) for another 24 h. The normalized mRNA levels of ISG56, ISG54, CIG5, IFN-β, and RSV N protein were determined. (C) Effect of CAN508 on RSV-induced ISG transcription in SAECs. Primary human SAECs were treated with 20 mM CAN508. The experiment was performed as described for panel B.
Despite multiple attempts at optimization, including timing, repeated dosing, and variation of transfection methods, we were never completely successful in inhibiting CDK9 expression by siRNA. To further confirm CDK9's role, we also evaluated the effect of the specific CDK9 kinase inhibitor CAN508 in RSV infection. CAN508 has a 10-fold-lower 50% inhibitory concentration (IC50) for CDK9 than for CDK1 to CDK7 and therefore is CDK9 selective (54). Various doses of CAN508 were tested for effects on cellular viability. At 20 μM CAN508, A549 cells were 95% of viable versus 97% in vehicle-treated cells (not significant). At this concentration, CAN508 potently inhibited RSV-induced ISG54, ISG56, and CIG5 expression (Fig. 7B).
We also examined the effect of CAN508 in human small airway epithelial cells (SAECs). At this 20 μM dose, SAECs were equally as viable as were vehicle-treated cells (94% versus 96% viability, respectively [not significant]). We also observed that CAN508 potently inhibited RSV-induced ISG54, ISG56, and CIG5 expression (Fig. 7C). Consistent with our findings in CDK9-silenced A549 cells (Fig. 7A), RSV-induced IFN-β expression was not inhibited by CAN508 either in A549 cells or in SAECs (Fig. 7B and C). Moreover, CAN508 has no effect on RSV replication in both A549 cells and SAECs (Fig. 7B and C). Together, the two experimental results suggested that CDK9 expression and activity are required for RSV induction of a subset of ISGs in a manner independent of effects on cellular viability or RSV replication.
Transcription elongation in RSV-induced ISG expression.
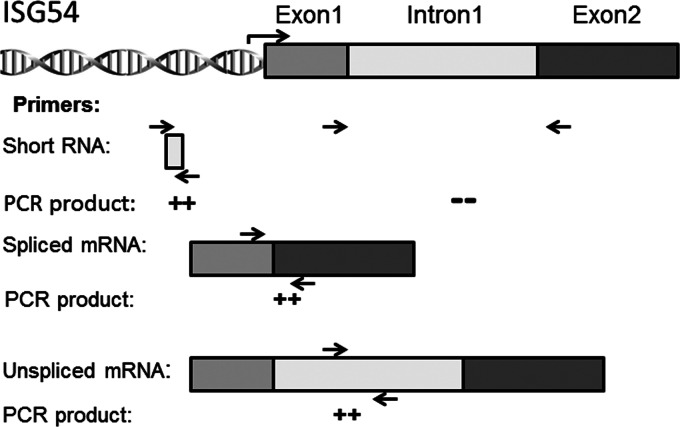
Rapid ISG expression is essential for a protective mucosal response to invading pathogens, enabling host survival (29, 30). Our findings of IRF3-dependent CDK9 recruitment and inducible formation of a complex with phospho-Ser2 CTD Pol II suggest that a component of ISG expression is under transcriptional elongation control. This network of ISGs would be located in open chromatin domains bound by hypophosphorylated RNA Pol II. For promoters bound by the hypophosphorylated form of Pol II, nonproductive cycles produce short, prematurely terminated (“abortive”) transcripts of ∼30 to 50 nucleotides (nt) in length (55). Upon inducible Ser2 phosphorylation in the CTD heptad repeat, RNA Pol II enters transcriptional elongation mode, producing full-length, fully spliced mRNAs. To quantify the process of transcriptional elongation, primers specific for the 5′ UTR of the ISG54 gene were designed to measure total RNA (which will detect both short transcripts, spliced RNA, and pre-mRNA), and separately, primers spanning exons 1 to 2 of the ISG54 gene to detect fully spliced mRNA and primers spanning intron 1 to detect pre-mRNA isoforms were also designed (schematically diagrammed in Fig. 8). The absolute amount of each transcript population was quantified and expressed using standard curves generated by the same primers to amplify known amounts of genomic DNA. The population of short transcripts was calculated as the difference between the total transcripts and the measured pools of spliced and pre-mRNA.
Fig 8.
Schematic diagram of human ISG54 RNA transcripts. The annealing regions of primer pairs are shown. The 5′ UTR primer pair amplifies the 5′ UTR of human ISG54 RNA, quantifying short, spliced, and pre-mRNA pools. The primer pair to exons 1 and 2 quantifies fully spliced mRNA, whereas the intron 1 primer pair quantifies full-length (unspliced) pre-mRNA.
RSV-induced total, spliced, and pre-mRNA ISG54 transcripts were quantified in primary human SAECs and A549 cells in the presence or absence of CAN508 (Table 4). In uninfected SAECs, the level of total transcripts was quite low, with most detected transcripts being fully spliced (93%). The portion of pre-mRNA in both uninfected and infected SAECs is a tiny fraction of the ISG54 transcript population, representing 1 to 3% of the total transcripts (Table 4). In response to CAN508 treatment alone, the abundance of total transcripts increased approximately 6-fold, representing an accumulation of short transcripts (93% of total), while the amount of spliced transcripts was reduced to ∼25% of that detected in untreated SAECs. In response to RSV infection, total ISG54 transcripts increased 25-fold (from 16.8 ng to 426 ng); this fraction represents predominately spliced transcripts (∼91% of the total transcripts). In contrast, in CAN508-treated SAECs, RSV induced only a 1.7-fold increase in total transcripts (from 99 ng to 168 ng) over CAN508 treatment alone; only 13% of the total transcripts were fully spliced, with the remainder (86%) being short transcripts (Table 4). Again, the fraction of pre-mRNA was negligible, representing less than 0.2% of the total transcript abundance.
Table 4.
Quantification of total, spliced, and pre-mRNA ISG54 transcripts
| Cell type | Treatment | Transcript, ng, ± SD |
|||
|---|---|---|---|---|---|
| Total | Spliced (% of total) | Pre-mRNA (% of total) | Short (estimated) (% of total) | ||
| SAECs | Mock, vehicle | 16.8 ± 2.13 | 15.6 ± 1.5 (93) | 0.44 ± 0.064 (2.6) | 0.75 (5) |
| Mock, CAN508 | 99 ± 10.2 | 4.8 ± 0.68 (5) | 0.113 ± 0.026 (0.1) | 94.1 (95) | |
| RSV, vehicle | 426 ± 29.7 | 389.4 ± 27.6 (91) | 3.8 ± 0.286 (0.9) | 33 (8) | |
| RSV, CAN508 | 168.3 ± 14.5 | 22.3 ± 3.7 (13) | 0.217 ± 0.042 (0.1) | 145 (86) | |
| A549 | Mock, vehicle | 33.2 ± 4.65 | 27.6 ± 4.6 (83) | 0.82 ± 0.1 (2.5) | 4.8 (14) |
| Mock, CAN508 | 175 ± 18.4 | 8.83 ± 2.1 (5) | 0.14 ± 0.03 (0.01) | 166 (95) | |
| RSV, vehicle | 1,034 ± 58.9 | 923.5 ± 64.3 (89) | 6.3 ± 0.53 (0.6) | 104 (10) | |
| RSV, CAN508 | 357 ± 25.4 | 59.6 ± 16.9 (17) | 0.54 ± 0.16 (0.2) | 296 (83) | |
Similar results were observed in A549 cells. CAN508 treatment induced an accumulation of total transcripts that were largely short transcripts (only 5% of the 175 ng of total transcripts were fully spliced; 95% were short). In vehicle-treated A549 cells, RSV induced a 31-fold increase in total ISG54 transcripts; greater than 90% of these were fully spliced (Table 4). In response to RSV infection in the presence of CAN508, only a 2-fold induction of total transcripts was observed over that of CAN508 treatment alone, and these transcripts were predominately short transcripts (only ∼17% were fully spliced, and 83% were short; Table 4). We interpret these data to indicate that transcriptional elongation plays a major role in basal and RSV-inducible ISG expression.
DISCUSSION
RSV is an etiological agent of severe LRTI in pediatric and immunocompromised hosts. RSV productively replicates in the airway epithelium, producing epithelial necrosis, mucosal mononuclear inflammation, and mucous plugging that produce obstructive airway disease (4, 8). In epithelial cells, dsRNA molecular patterns produced by replicating RSV are sensed by cytoplasmic RIG-I and endosomal TLR3, triggering a highly conserved signaling cascade converging on the IRF3 and NF-κB transcription factors. Our work, and that of others, has indicated that the innate IRF3 pathways are protective through the induction of type I mucosal IFNs, one of whose actions is to induce a Th1 lymphocyte response (28). In the absence of type I IFN, enhanced eosinophilia, inflammation, and increased viral titers are observed in mouse models of disease (26, 28). In contrast, activation of the NF-κB arm is linked to disease pathogenesis, since inhibition of mucosal NF-κB signaling abrogates severity of disease (6). Therefore, a thorough understanding of detailed signaling pathways of RSV-induced innate immune response will be of paramount importance in understanding the pathogenesis of and preventive procedures and therapeutic interventions for RSV infection. In this study, we examined the mechanism for RSV induction of IRF3-dependent ISGs. The ISGs are highly inducible immediate early genes that coordinate the antiviral response, reducing intracellular viral replication, viral infectivity, and cellular necrosis (29, 56). Here, we demonstrate that RSV induces cytoplasmic-to-nuclear IRF3 translocation despite the presence of the RSV IRF antagonist NS1, along with activation of the CDK9 complex through transition from an inactive 7SK snRNA complex into the activated BRD4 complex. IRF3-mediated CDK9 recruitment to the ISG 5′ ISRE mediates phospho-Ser2 CTD Pol II formation, an event necessary for the inducible switch from unspliced to spliced ISG RNA. Together, these data indicate that inducible ISG expression is controlled at the level of transcriptional elongation.
The innate immune system is an ancient, highly conserved mode of defense against pathogens and the sole method of protection for invertebrates and plants. A critical aspect of the innate immune response is the rapid activation of RNA Pol II-dependent gene expression programs to generate effectors that restrict pathogen spread (57, 58). Transcription of protein-encoding genes by RNA Pol II is a highly regulated process involving preinitiation (assembly of basal transcription factors and coactivator recruitment), initiation, elongation, and termination phases (37). For innate genes located within closed chromatin, preinitiation complex formation is an essential first step in inducible expression. In this process, histone acetyltransferases, such as p300/CBP, are recruited to destabilize repressive histone, resulting in p300/CBP dissociation and binding of general transcription factors, including TFIID (58). This preinitiation complex assembly has been shown to be an important mediator of virus-induced IFN-β expression, where the IRF3-p300/CBP complex triggers enhanceosome formation (35, 59, 60). Our findings that IFN-β expression is resistant to CDK9 inhibition are consistent with a distinct transcriptional mechanism controlling IFN-β expression in contrast to other immediate early ISGs.
In contrast, for immediate early genes located within open chromatin, Pol II is preengaged in a hypophosphorylated state, producing short ∼30- to 50-nt transcripts (55). Promoter proximal pausing is reversed by activated P-TEFb, a multiprotein complex containing CDK9, BRD4, and cyclin T1 or T2 subunits (61). CDK9 forms the catalytic core of P-TEFb; upon phosphorylation of Ser2, the RNA Pol II CTD heptad repeats, and negative elongation factors (DSIF and NELF), phospho-Ser2 CTD RNA Pol II is released from the pause site, producing full-length transcripts that are appropriately spliced (62, 63). Our work here extends the mechanisms of IRF3 activation to include a role in recruiting activated CDK9-BRD4 complex to ISG promoters, where CDK9 activity is required for switching from unspliced to spliced mRNA formation in a subset of highly inducible ISGs.
In the present study, we have observed for the first time that RSV infection induces the formation of an activated IRF3-CDK9 complex at ISG promoters (Fig. 4 and 5). We further infer that CDK9 is activated based on several observations. First, CDK9 exists either as an inactive complex with 7SK snRNA and HEXIM (48, 63) or as an activated complex with BRD4 (10, 61), a chromatin adaptor that preferentially binds to Ac-Lys 5 histone H4 (64). Our IP-SID-SRM-MS observations that RSV induces CDK9 to form a complex with BRD4 suggest that CDK9 is shifting into an activated complex (Fig. 6A) (61, 64, 65). Second, we found that both RSV infection and intracellular poly(I·C) significantly disrupt 7SK snRNA association with CDK9 (Fig. 6B), suggesting that viral patterns reduce the population of inactive CDK9 complexes. Third, CDK9-IRF3 recruitment to the 5′ ISRE is associated with increased formation of phospho-Ser2 CTD Pol II. Fourth, the CDK9 kinase inhibitor CAN508 reduces basal and RSV-inducible transcriptional elongation of a subset of IRF3-dependent ISGs (but not that of IFN-β). Recent work has shown that P-TEFb may be associated with distinct transcriptional elongation complexes with distinct target specificities (66), and sequence-specific transcription factors, including NF-κB (67–69), Myc (70), and STAT3 (71). Our finding that RSV-inducible CDK9 binding is disrupted in the IRF3−/− MEFs suggests that IRF3 is primarily responsible for activated CDK9 recruitment to ISREs. Despite multiple attempts, we have not been able to convincingly demonstrate that IRF3 forms a stable protein complex with CDK9. Further work will be required to understand how IRF3 activation promotes CDK9 recruitment to ISREs.
In examining IRF3, CDK9, Pol II, and phospho-Ser2 CTD Pol II recruitment in response to poly(I·C) stimulation, we have made the observation that IRF3, CDK9, and Pol II are also recruited to ISRE binding sites of ISG promoters while phospho-Ser2 CTD Pol II is not (Fig. 5A, C, and D). This behavior of phospho-Ser2 CTD Pol II clearance is strikingly different from what we found in RSV infection, where increased phospho-Ser2 CTD Pol II binding was seen. Previously, we observed that dsRNA induces initial clearance of phospho-Ser2 CTD Pol II on NF-κB-dependent promoters followed by its later reassociation (10). We are unsure how to explain this difference in behavior between poly(I·C) and RSV. One possibility could be that phospho-Ser2 CTD Pol II rapidly dissociates from the ISG after entering elongation phase and because poly(I·C) is such a potent stimulus of ISG expression that the available pool of RNA Pol II is depleted.
The findings from this study (and previous work from our group) suggest that translational elongation is an important pathway in mediating immediate early gene expression in the innate immune response (10, 69, 71). We previously provided evidence that CDK9 is important for the activation of a subset of rapidly inducible NF-κB-dependent cytokine genes (69). This work was extended in studies examining the effect of blocking CDK9 activity and found decreased ICAM-1 expression in human endothelial cells and reduced leukocyte recruitment (72). Our present study further demonstrates that CDK9-initiated transcription elongation also plays a major role in the IRF3-induced ISG response.
We note that others have shown that transcriptional elongation is important in TLR signaling activated by lipopolysaccharide (57, 58, 64). This TLR4 signaling is classically described in two stages: a rapid protein synthesis-independent induction of immediate early genes (termed the primary response), followed by the subsequent protein synthesis-dependent induction of secondary response genes (58, 59). Here, primary response genes are located in an open chromatin domain and are activated by inducible transcriptional elongation mediated by BRD4 binding to acetylated H4 K5/8/12 residues (64, 65). Our findings suggest that transcriptional elongation also mediates RIG-I-inducible rapid ISG expression and that ISG54, ISG56, and CIG5 may be immediate early genes, whereas IFN-β may be a secondary response gene whose expression may require chromatin remodeling as a part of its enhanceosome formation. Finally, STAT3 represents another important effector of the interleukin-6 (IL-6) superfamily of cytokines. We observed that IL-6 induces the formation of a nuclear STAT3/CDK9 complex essential for acute-phase γ-fibrinogen synthesis (71). Together, these studies suggest that transcriptional elongation mediated by CDK9 is a shared regulatory pathway for inflammation-mediated activation of immediate early genes.
One striking finding of the present study is that RSV-induced IFN-β expression was resistant to CAN508 in both A549 cells and SAECs (Fig. 7B and C). Although both RSV- and poly(I·C)-induced IFN-β expression were inhibited by IRF3 silencing, IFN-β expression was unresponsive to CDK9 siRNA and CAN508 treatment. These data indicate that not all ISGs require CDK9 for transcriptional induction (Fig. 3A and B). In our evaluation of the effect of a distinct CDK inhibitor, flavopiridol, on IFN-β expression we found that this compound effectively blocked IFN-β expression (data not shown). Flavopiridol is a synthetic flavonoid with potent broad-spectrum CDK-inhibitory activity (72, 73), in contrast to the narrow CDK9 selectivity of CAN508. This observation suggests that IFN-β expression may be dependent on other CDK isoforms. Recently, it has been shown that CDK8 phosphorylates the STAT1 transcription factor to regulate the interferon response (74). These intriguing findings suggest that a distinct CDK, CDK8, may be involved in RSV- or poly(I·C)-induced IFN-β expression. More work will be required to understand the complex role of various CDK isoforms on ISG expression in the innate response.
In conclusion, we demonstrate that transcriptional elongation plays a major role in RSV-induced immediate early gene expression in a subset of IRF3-dependent ISGs. The presence of dsRNA molecular patterns induces cytoplasmic-nuclear IRF3 translocation and simultaneously activates CDK9 through dissociation with 7SK snRNA and BRD4 binding. Subsequently the activated CDK9 complex induces phospho-Ser2 CTD RNA Pol II on 5′ ISRE sites, producing RNA Pol II clearance and expression of fully spliced ISG mRNAs. CDK9 modulation might be a mechanism for modulating the innate response in viral infection.
ACKNOWLEDGMENTS
We thank Leoncio Vergara and the UTMB Optical Imaging Core for assistance with the dynamic fluorescence imaging studies.
This work was supported by R01 GM086885, 2P01AI062885, a Clinical and Translational Sciences Award (UL1TR000071), NIH-NHLBI-HHSN268201000037C from the NHLBI Proteomics Center for Airway Inflammation (A.R.B.), and R01 AI081977 (M.N.T.).
Footnotes
Published ahead of print 17 April 2013
REFERENCES
- 1. Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O'Brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita C, Simoes EA, Rudan I, Weber MW, Campbell H. 2010. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 375:1545–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hall CB, Weinberg GA, Iwane MK, Blumkin AK, Edwards KM, Staat MA, Auinger P, Griffin MR, Poehling KA, Erdman D, Grijalva CG, Zhu Y, Szilagyi P. 2009. The burden of respiratory syncytial virus infection in young children. N. Engl. J. Med. 360:588–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mohapatra SS, Boyapalle S. 2008. Epidemiologic, experimental, and clinical links between respiratory syncytial virus infection and asthma. Clin. Microbiol. Rev. 21:495–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shay DK, Holman RC, Roosevelt GE, Clarke MJ, Anderson LJ. 2001. Bronchiolitis-associated mortality and estimates of respiratory syncytial virus-associated deaths among US children, 1979–1997. J. Infect. Dis. 183:16–22 [DOI] [PubMed] [Google Scholar]
- 5. Zorc JJ, Hall CB. 2010. Bronchiolitis: recent evidence on diagnosis and management. Pediatrics 125:342–349 [DOI] [PubMed] [Google Scholar]
- 6. Haeberle HA, Casola A, Gatalica Z, Petronella S, Dieterich HJ, Ernst PB, Brasier AR, Garofalo RP. 2004. IkappaB kinase is a critical regulator of chemokine expression and lung inflammation in respiratory syncytial virus infection. J. Virol. 78:2232–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hall CB, Douglas RG, Jr, Schnabel KC, Geiman JM. 1981. Infectivity of respiratory syncytial virus by various routes of inoculation. Infect. Immun. 33:779–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aherne W, Bird T, Court SD, Gardner PS, McQuillin J. 1970. Pathological changes in virus infections of the lower respiratory tract in children. J. Clin. Pathol. 23:7–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Garofalo R, Sabry M, Jamaluddin M, Yu RK, Casola A, Ogra PL, Brasier AR. 1996. Transcriptional activation of the interleukin-8 gene by respiratory syncytial virus infection in alveolar epithelial cells: nuclear translocation of the RelA transcription factor as a mechanism producing airway mucosal inflammation. J. Virol. 70:8773–8781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brasier AR, Tian B, Jamaluddin M, Kalita MK, Garofalo RP, Lu M. 2011. RelA Ser276 phosphorylation-coupled Lys310 acetylation controls transcriptional elongation of inflammatory cytokines in respiratory syncytial virus infection. J. Virol. 85:11752–11769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tian B, Zhang Y, Luxon BA, Garofalo RP, Casola A, Sinha M, Brasier AR. 2002. Identification of NF-kappaB-dependent gene networks in respiratory syncytial virus-infected cells. J. Virol. 76:6800–6814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang Y, Luxon BA, Casola A, Garofalo RP, Jamaluddin M, Brasier AR. 2001. Expression of respiratory syncytial virus-induced chemokine gene networks in lower airway epithelial cells revealed by cDNA microarrays. J. Virol. 75:9044–9058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Casola A, Garofalo RP, Jamaluddin M, Vlahopoulos S, Brasier AR. 2000. Requirement of a novel upstream response element in respiratory syncytial virus-induced IL-8 gene expression. J. Immunol. 164:5944–5951 [DOI] [PubMed] [Google Scholar]
- 14. Liu P, Jamaluddin M, Li K, Garofalo RP, Casola A, Brasier AR. 2007. Retinoic acid-inducible gene I mediates early antiviral response and Toll-like receptor 3 expression in respiratory syncytial virus-infected airway epithelial cells. J. Virol. 81:1401–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bibeau-Poirier A, Servant MJ. 2008. Roles of ubiquitination in pattern-recognition receptors and type I interferon receptor signaling. Cytokine 43:359–367 [DOI] [PubMed] [Google Scholar]
- 16. Loo YM, Gale M., Jr 2011. Immune signaling by RIG-I-like receptors. Immunity 34:680–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thompson MR, Kaminski JJ, Kurt-Jones EA, Fitzgerald KA. 2011. Pattern recognition receptors and the innate immune response to viral infection. Viruses 3:920–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yoneyama M, Fujita T. 2008. Structural mechanism of RNA recognition by the RIG-I-like receptors. Immunity 29:178–181 [DOI] [PubMed] [Google Scholar]
- 19. Yount JS, Gitlin L, Moran TM, Lopez CB. 2008. MDA5 participates in the detection of paramyxovirus infection and is essential for the early activation of dendritic cells in response to Sendai virus defective interfering particles. J. Immunol. 180:4910–4918 [DOI] [PubMed] [Google Scholar]
- 20. Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981–988 [DOI] [PubMed] [Google Scholar]
- 21. Seth RB, Sun L, Ea CK, Chen ZJ. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF-3. Cell 122:669–682 [DOI] [PubMed] [Google Scholar]
- 22. Maelfait J, Beyaert R. 2012. Emerging role of ubiquitination in antiviral RIG-I signaling. Microbiol. Mol. Biol. Rev. 76:33–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. O'Neill LA, Bowie AG. 2010. Sensing and signaling in antiviral innate immunity. Curr. Biol. 20:R328–R333 doi: 10.1016/j.cub.2010.01.044 [DOI] [PubMed] [Google Scholar]
- 24. Jamaluddin M, Wang S, Garofalo RP, Elliott T, Casola A, Baron S, Brasier AR. 2001. IFN-beta mediates coordinate expression of antigen-processing genes in RSV-infected pulmonary epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 280:L248–L257 [DOI] [PubMed] [Google Scholar]
- 25. Garofalo R, Mei F, Espejo R, Ye G, Haeberle H, Baron S, Ogra PL, Reyes VE. 1996. Respiratory syncytial virus infection of human respiratory epithelial cells up-regulates class I MHC expression through the induction of IFN-beta and IL-1 alpha. J. Immunol. 157:2506–2513 [PubMed] [Google Scholar]
- 26. Durbin JE, Johnson TR, Durbin RK, Mertz SE, Morotti RA, Peebles RS, Graham BS. 2002. The role of IFN in respiratory syncytial virus pathogenesis. J. Immunol. 168:2944–2952 [DOI] [PubMed] [Google Scholar]
- 27. Liu P, Lu M, Tian B, Li K, Garofalo RP, Prusak D, Wood TG, Brasier AR. 2009. Expression of an IKKgamma splice variant determines IRF3 and canonical NF-kappaB pathway utilization in ssRNA virus infection. PLoS One 4:e8079 doi: 10.1371/journal.pone.0008079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guerrero-Plata A, Baron S, Poast JS, Adegboyega PA, Casola A, Garofalo RP. 2005. Activity and regulation of alpha interferon in respiratory syncytial virus and human metapneumovirus experimental infections. J. Virol. 79:10190–10199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schoggins JW, Rice CM. 2011. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 1:519–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sadler AJ, Williams BR. 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8:559–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wacher C, Muller M, Hofer MJ, Getts DR, Zabaras R, Ousman SS, Terenzi F, Sen GC, King NJ, Campbell IL. 2007. Coordinated regulation and widespread cellular expression of interferon-stimulated genes (ISG) ISG-49, ISG-54, and ISG-56 in the central nervous system after infection with distinct viruses. J. Virol. 81:860–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Levy D, Larner A, Chaudhuri A, Babiss LE, Darnell JE., Jr 1986. Interferon-stimulated transcription: isolation of an inducible gene and identification of its regulatory region. Proc. Natl. Acad. Sci. U. S. A. 83:8929–8933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Terenzi F, Hui DJ, Merrick WC, Sen GC. 2006. Distinct induction patterns and functions of two closely related interferon-inducible human genes, ISG54 and ISG56. J. Biol. Chem. 281:34064–34071 [DOI] [PubMed] [Google Scholar]
- 34. Peters KL, Smith HL, Stark GR, Sen GC. 2002. IRF-3-dependent, NF-kappaB and JNK-independent activation of the 561 and IFN-beta genes in response to double-stranded RNA. Proc. Natl. Acad. Sci. U. S. A. 99:6322–6327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Weaver BK, Kumar KP, Reich NC. 1998. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol. Cell. Biol. 18:1359–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhou Q, Li T, Price DH. 2012. RNA polymerase II elongation control. Annu. Rev. Biochem. 81:119–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chiba K, Yamamoto J, Yamaguchi Y, Handa H. 2010. Promoter-proximal pausing and its release: molecular mechanisms and physiological functions. Exp. Cell Res. 316:2723–2730 [DOI] [PubMed] [Google Scholar]
- 38. Ray S, Ju Sun XH, Finnerty CC, Herndon DN, Brasier AR. 10 January 2013. The IL-6 trans-signaling-STAT3 pathway mediates ECM and cellular proliferation in fibroblasts from hypertrophic scar. J. Invest. Dermatol. [Epub ahead of print.] doi:10.1038/jid.2012.499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ueba O. 1978. Respiratory syncytial virus. I. Concentration and purification of the infectious virus. Acta Med. Okayama 32:265–272 [PubMed] [Google Scholar]
- 40. Akagi T, Sasai K, Hanafusa H. 2003. Refractory nature of normal human diploid fibroblasts with respect to oncogene-mediated transformation. Proc. Natl. Acad. Sci. U. S. A. 100:13567–13572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Naviaux RK, Costanzi E, Haas M, Verma IM. 1996. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J. Virol. 70:5701–5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tian B, Nowak DE, Jamaluddin M, Wang S, Brasier AR. 2005. Identification of direct genomic targets downstream of the nuclear factor-kappaB transcription factor mediating tumor necrosis factor signaling. J. Biol. Chem. 280:17435–17448 [DOI] [PubMed] [Google Scholar]
- 43. Nowak DE, Tian B, Brasier AR. 2005. Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation. Biotechniques 39:715–725 [DOI] [PubMed] [Google Scholar]
- 44. Tian B, Yang J, Brasier AR. 2012. Two-step cross-linking for analysis of protein-chromatin interactions. Methods Mol. Biol. 809:105–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhao Y, Tian B, Edeh CB, Brasier AR. 15 February 2013. Quantitation of the dynamic profiles of the innate immune response using multiplex selected reaction monitoring-mass spectrometry. Mol. Cell. Proteomics [Epub ahead of print.] doi:10.1074/mcp.M112.023465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhao Y, Brasier AR. 11 February 2013. Applications of selected reaction monitoring (SRM)-mass spectrometry (MS) for quantitative measurement of signaling pathways. Methods [Epub ahead of print.] doi:10.1016/j.ymeth.2013.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhao Y, Widen SG, Jamaluddin M, Tian B, Wood TG, Edeh CB, Brasier AR. 2011. Quantification of activated NF-kappaB/RelA complexes using ssDNA aptamer affinity-stable isotope dilution-selected reaction monitoring-mass spectrometry. Mol. Cell. Proteomics 10(6):M111.008771 doi: 10.1074/mcp.M111.008771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang Z, Zhu Q, Luo K, Zhou Q. 2001. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414:317–322 [DOI] [PubMed] [Google Scholar]
- 49. Kalita MK, Sargsyan K, Tian B, Paulucci-Holthauzen A, Najm HN, Debusschere BJ, Brasier AR. 2011. Sources of cell-to-cell variability in canonical nuclear factor-kappaB (NF-kappaB) signaling pathway inferred from single cell dynamic images. J. Biol. Chem. 286:37741–37757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Foster KA, Oster CG, Mayer MM, Avery ML, Audus KL. 1998. Characterization of the A549 cell line as a type II pulmonary epithelial cell model for drug metabolism. Exp. Cell Res. 243:359–366 [DOI] [PubMed] [Google Scholar]
- 51. Spann KM, Tran KC, Collins PL. 2005. Effects of nonstructural proteins NS1 and NS2 of human respiratory syncytial virus on interferon regulatory factor 3, NF-kappaB, and proinflammatory cytokines. J. Virol. 79:5353–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Forbus J, Spratt H, Wiktorowicz J, Wu Z, Boldogh I, Denner L, Kurosky A, Brasier RC, Luxon B, Brasier AR. 2006. Functional analysis of the nuclear proteome of human A549 alveolar epithelial cells by HPLC-high resolution 2-D gel electrophoresis. Proteomics 6:2656–2672 [DOI] [PubMed] [Google Scholar]
- 53. Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Espinosa JM. 2006. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 20:601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Krystof V, Cankar P, Frysová I, Slouka J, Kontopidis G, Dzubák P, Hajdúch M, Srovnal J, de Azevedo WF, Jr, Orság M, Paprskárová M, Rolcík J, Látr A, Fischer PM, Strnad M. 2006. 4-Arylazo-3, 5-diamino-1H-pyrazole CDK inhibitors: SAR study, crystal structure in complex with CDK2, selectivity, and cellular effects. J. Med. Chem. 49:6500–6509 [DOI] [PubMed] [Google Scholar]
- 55. Sims RJ, III, Belotserkovskaya R, Reinberg D. 2004. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 18:2437–2468 [DOI] [PubMed] [Google Scholar]
- 56. Yan N, Chen ZJ. 2012. Intrinsic antiviral immunity. Nat. Immunol. 13:214–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Medzhitov RT, Horng T. 2009. Transcriptional control of the inflammatory response. Nat. Rev. Immunol. 9:692–703 [DOI] [PubMed] [Google Scholar]
- 58. Smale ST. 2012. Transcriptional regulation in the innate immune system. Curr. Opin. Immunol. 24:51–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shiama N. 1997. The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Cell Biol. 7:230–236 [DOI] [PubMed] [Google Scholar]
- 60. Parekh BS, Maniatis T. 1999. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-beta promoter. Mol. Cell 3:125–129 [DOI] [PubMed] [Google Scholar]
- 61. Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. 2005. The bromodomain protein BRD4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 19:523–534 [DOI] [PubMed] [Google Scholar]
- 62. Nechaev S, Adelman K. 2011. Pol II waiting in the starting gates: regulating the transition from transcription initiation into productive elongation. Biochim. Biophys. Acta 1809:34–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Peterlin BM, Price DH. 2006. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23:297–305 [DOI] [PubMed] [Google Scholar]
- 64. Hargreaves DC, Horng T, Medzhitov R. 2009. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 138:129–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhao R, Nakamura T, Fu Y, Lazar Z, Spector DL. 2011. Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nat. Cell Biol. 13:1295–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Luo Z, Lin C, Shilatifard A. 2012. The super elongation complex (SEC) family in transcriptional control. Nat. Rev. Mol. Cell Biol. 13:543–547 [DOI] [PubMed] [Google Scholar]
- 67. Huang B, Yang XD, Zhou MM, Ozato K, Chen LF. 2009. BRD4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol. Cell. Biol. 29:1375–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. 2001. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 8:327–337 [DOI] [PubMed] [Google Scholar]
- 69. Nowak DE, Tian B, Jamaluddin M, Boldogh I, Vergara LA, Choudhary S, Brasier AR. 2008. RelA Ser276 phosphorylation is required for activation of a subset of NF-kappaB-dependent genes by recruiting cyclin-dependent kinase 9/cyclin T1 complexes. Mol. Cell. Biol. 28:3623–3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. 2010. c-Myc regulates transcriptional pause release. Cell 141:432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hou T, Ray S, Brasier AR. 2007. The functional role of an interleukin 6-inducible CDK9.STAT3 complex in human gamma-fibrinogen gene expression. J. Biol. Chem. 282:37091–37102 [DOI] [PubMed] [Google Scholar]
- 72. Schmerwitz UK, Sass G, Khandoga AG, Joore J, Mayer BA, Berberich N, Totzke F, Krombach F, Tiegs G, Zahler S, Vollmar AM, Fürst R. 2011. Flavopiridol protects against inflammation by attenuating leukocyte-endothelial interaction via inhibition of cyclin-dependent kinase 9. Arterioscler. Thromb. Vasc. Biol. 31:280–288 [DOI] [PubMed] [Google Scholar]
- 73. Shapiro GI. 2004. Preclinical and clinical development of the cyclin-dependent kinase inhibitor flavopiridol. Clin. Cancer Res. 10(12 part 2):4270s–4275s [DOI] [PubMed] [Google Scholar]
- 74. Bancerek J, Poss ZC, Steinparzer I, Sedlyarov V, Pfaffenwimmer T, Mikulic I, Dölken L, Strobl B, Müller M, Taatjes DJ, Kovarik P. 2013. CDK8 kinase phosphorylates transcription factor STAT1 to selectively regulate the interferon response. Immunity 38:250–262 [DOI] [PMC free article] [PubMed] [Google Scholar]