Abstract
Small-molecule CCR5 inhibitors such as vicriviroc (VVC) and maraviroc (MVC) are allosteric modulators that impair HIV-1 entry by stabilizing a CCR5 conformation that the virus recognizes inefficiently. Viruses resistant to these compounds are able to bind the inhibitor-CCR5 complex while also interacting with the free coreceptor. CCR5 also interacts intracellularly with G proteins, as part of its signal transduction functions, and this process alters its conformation. Here we investigated whether the action of VVC against inhibitor-sensitive and -resistant viruses is affected by whether or not CCR5 is coupled to G proteins such as Gαi. Treating CD4+ T cells with pertussis toxin to uncouple the Gαi subunit from CCR5 increased the potency of VVC against the sensitive viruses and revealed that VVC-resistant viruses use the inhibitor-bound form of Gαi-coupled CCR5 more efficiently than they use uncoupled CCR5. Supportive evidence was obtained by expressing a signaling-deficient CCR5 mutant with an impaired ability to bind to G proteins, as well as two constitutively active mutants that activate G proteins in the absence of external stimuli. The implication of these various studies is that the association of intracellular domains of CCR5 with the signaling machinery affects the conformation of the external and transmembrane domains and how they interact with small-molecule inhibitors of HIV-1 entry.
INTRODUCTION
The sequential binding of the trimeric envelope glycoprotein (Env) complex to the CD4 receptor and the CCR5 coreceptor mediates the entry of human immunodeficiency virus type 1 (HIV-1) into host cells (1–3). The interaction between the Env gp120 subunit and CCR5 involves two structural elements: a gp120 site comprising the CD4-induced, 4-stranded bridging sheet region and the base of V3 recognizes the CCR5 N terminus (NT), while residues near the V3 tip interact with the second extracellular loop (ECL2) (4, 5). Small-molecule CCR5 inhibitors such as the licensed drug maraviroc (MVC) and the experimental compound vicriviroc (VVC) impair this interaction by a predominantly noncompetitive mechanism. They do so by binding in a hydrophobic cavity located within the transmembrane (TM) helices, thereby stabilizing a CCR5 conformation that HIV-1 recognizes inefficiently (6, 7). Viruses resistant to small-molecule CCR5 inhibitors can be generated in vitro and in vivo; the dominant route to resistance involves the acquisition of sequence changes, particularly in V3, that render gp120 capable of recognizing the inhibitor-CCR5 complex while retaining the ability to also interact with the free coreceptor (8–10).
The chemokine receptor CCR5 is a member of the seven-transmembrane G-protein-coupled receptor (GPCR) superfamily. GPCRs can acquire multiple conformations as they transit from the agonist-bound, signaling-active conformation to the more stable antagonist-bound, signaling-inactive conformation (11, 12). When CCR5 is activated by chemokine ligands, its intracellular domains can associate with four classes of heterotrimeric G proteins: Gαi, Gαs, Gαq, and Gα12/13 (13). Among these intracellular components of the signal transduction machinery, Gαi is uniquely sensitive to ADP ribosylation by pertussis toxin (PTX) (14, 15). Questions have arisen as to whether gp120 binding to CCR5 induces signals similar to those triggered by chemokines and whether such signals play any role in the entry or postentry stages of the HIV-1 replication cycle. There have been no definitive answers, not least because various studies have used multiple different cell types that differ in their activation state and therefore yield inconsistent or contradictory outcomes (reviewed by Wu and Yoder [16]). Early studies in which Gαi-mediated CCR5 signaling was inhibited with PTX or that used CCR5 mutants with an impaired ability to couple to G proteins (e.g., CCR5-R126N) demonstrated that signaling via CCR5 is dispensable for HIV-1 replication in cell lines or activated T cells (17–19). More recently, Env was found to trigger actin rearrangement through Gαi-mediated CXCR4 signaling or Gαq-mediated CCR5 signaling. These events were shown to be critical at both entry and postentry stages of HIV-1 replication, particularly in resting T cells (20–23). We have obtained evidence that CCR5 exists in multiple cell surface conformations that differ in their antigenicity, i.e., their reactivity with specific monoclonal antibodies (MAbs) (24). Some of these antigenic forms represent distinct CCR5 subsets with different functional properties and membrane localizations (24). We hypothesized that these CCR5 subsets might differ in the ability to bind various ligands, including the HIV-1 Env complex and small-molecule inhibitors (24, 25). One source of conformational diversity is whether CCR5 is or is not associated with G proteins and other signaling components (25).
In this study, we investigated the effects of uncoupling CCR5 from G proteins on the actions of VVC against VVC-sensitive and -resistant viruses in primary CD4+ T cells and the U87-CD4 cell line. To do so, we used PTX to uncouple CCR5 from Gαi in the primary T cells and expressed a signaling-deficient mutant (SDM), CCR5-R126N, in the cell line. Conversely, we also tested two constitutively active mutants (CAM), CCR5-T82P and CCR5-T82K, which activate G proteins in the absence of any external stimulus (26). Overall, we conclude that VVC-resistant viruses use the inhibitor-bound form of Gαi-coupled CCR5 more efficiently than the uncoupled CCR5 configuration. In addition, uncoupling CCR5 from Gαi increases the potency of VVC against sensitive viruses. The implication is that CCR5 association with the signaling machinery induces conformational changes that affect how small-molecule CCR5 inhibitors act against HIV-1 entry.
MATERIALS AND METHODS
Cells and plasmids.
U87-CD4 cells, contributed by HongKui Deng and Dan Littman, were obtained from the NIH AIDS Research and Reference Reagent Program (ARRRP), and 293T cells were obtained from the American Type Culture Collection (ATCC). Cell lines were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine.
Peripheral blood mononuclear cells (PBMC) were purified and stimulated as previously described (8). CD4+ T cells were purified from PBMC by use of a Dynal CD4 positive selection kit (Invitrogen, Carlsbad, CA) after 3 days of stimulation. CD4+ T cells were treated with PTX or B oligomer (Sigma-Aldrich) on the same day they were isolated.
The CCR5-WT plasmid was generated by cloning human CCR5 cDNA tagged with the C-terminal 1D4 epitope (TETSQVAPA) into pcDNA3.1 (+) as described previously (27). CCR5 mutants were generated by site-directed mutagenesis of CCR5-WT. The primers used to generate CCR5-R126N were 5′-CTCCTGACAATCGATAACTACCTGGCTGTCGTCC-3′ (forward) and 5′-GGACGACAGCCAGGTAGTTATCGATTGTCAGGAG-3′ (reverse), those for CCR5-T82P were 5′-GACCTGTTTTTCCTTCTTCCTGTCCCCTTCTGGGCTC-3′ (forward) and 5′-GAGCCCAGAAGGGGACAGGAAGAAGGAAAAACAGGTC-3′ (reverse), and those for CCR5-T82K were 5′-GACCTGTTTTTCCTTCTTAAAGTCCCCTTCTGGGCTCAC-3′ (forward) and 5′-GTGAGCCCAGAAGGGGACTTTAAAAAGGAAAAACAGGTC-3′ (reverse). The pNLluc-AM and PCI-env plasmids used for Env-pseudovirus production have been described previously (28). The pNL4-3/env chimeric clonal proviral plasmids used for the production of infectious molecular clones were constructed as described elsewhere (8, 29). The Par-4V3, Par-3FP, Res-4V3, and Res-3FP env genes in PCI-env and pNL4-3/env correspond to clones CC1/85 cl.7, CC1/85 cl.6, CC101.19 cl.7, and D1/85.16 cl.23, respectively (8, 30). The Par-4V3 (CC1/85 cl.7; GenBank accession no. AY357341) and Par-3FP (CC1/85 cl.6; GenBank accession no. AY357338) env genes were directly cloned from the VVC-sensitive patient isolate CC1/85. When CC1/85 was propagated in vitro in the presence of the CCR5 inhibitors AD101 and VVC, two inhibitor-resistant isolates were selected: CC101.19 and D1/85.16, respectively. The Res-4V3 (CC101.19 cl.7; GenBank accession no. AY357465) and Res-3FP (D1/85.16 cl.23; GenBank accession no. FJ713453) env genes were cloned from the CC101.19 and D1/85.16 isolates, respectively (8, 30). Compared to other sensitive env genes from the CC1/85 isolate, the Par-4V3 and Par-3FP env genes shared the most sequence similarity to the Res-4V3 and Res-3FP env genes, respectively; they were therefore chosen as the comparator parental viruses (28).
CCR5 transfection and Env-pseudovirus infection.
U87-CD4 cells were transfected with CCR5-expressing plasmids by use of Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. One day later, the cells were washed twice with culture medium and then seeded into 96-well plates at a density of 1 × 104 cells per well in 50 μl of medium for one more day. They were then infected in the presence or absence of VVC (50 μl) with Env-pseudoviruses, as previously described (28). Briefly, Env-pseudoviruses were incubated with magnetic beads (ViroMag R/L; Boca Scientific, Boca Raton, FL) for 15 min, added to the transfected cells, and placed on a Super Magnetic plate (Boca Scientific) for 10 min. The luciferase signal was measured at 72 h postinfection, using Bright-Glo luciferase substrate (Promega Inc., Madison, WI). There was no measurable luminescence from uninfected cells (i.e., background control). Inhibition of HIV-1 entry in the presence of VVC was calculated as 100 × [1 − (LucVVC/Luccontrol)], with the control being infection without any inhibitor.
Infection inhibition assay.
Infectious clonal virus stocks were prepared by transient transfection of 293T cells with pNL4-3/env plasmids by use of Lipofectamine 2000 (Invitrogen), as described previously (8). All stocks of infectious viruses were passed through a 0.45-μm filter and stored in aliquots at −80°C. The 50% tissue culture infective doses (TCID50) for PBMC were determined by standard methods (31).
PTX (or B oligomer)-treated or control CD4+ T cells were seeded at 1 × 105 cells per well in a 96-well plate. The CD4+ T cells, obtained from a single donor, consisted of equal numbers from each of the two stimulation conditions outlined above. VVC was diluted in culture medium (with or without 10 μM H89, as indicated) to twice the final concentration and added (50 μl) to the cells (50 μl) for 1 h at 37°C. Infection was initiated by adding 1,000 TCID50 of a clone (100 μl) for 6 h, and then the cells were washed twice and resuspended in culture medium containing the appropriate final VVC concentration. Unlike in the pseudovirus infection assays described above, magnetic beads were not used with replication-competent viruses. The production of HIV-1 p24 antigen after 7 days was quantified by enzyme-linked immunosorbent assay (ELISA) (32). Inhibition of HIV-1 replication in the presence of VVC was calculated as 100 × [1 − (p24VVC/p24control)], with the control being infection without any inhibitor.
125I-RANTES binding assay.
CCR5-transfected or control U87-CD4 cells were scraped from the surface of the culture flask, washed once with phosphate-buffered saline (PBS), and resuspended in the same buffer. The cells (800,000) were then plated in a 96-well plate and resuspended in 100 μl of binding buffer (50 mM HEPES, pH 7.4, 1 mM CaCl2, 5 mM MgCl2, 0.5% bovine serum albumin [BSA]) in the presence of increasing VVC concentrations. After incubation with VVC for 1 h at room temperature, 400 pM 125I-RANTES (PerkinElmer) was added for 1 h. The plates were then centrifuged and the cells washed twice with 200 μl of PBS. After the last wash, the cells were transferred to scintillation vials for measurement of bound 125I-RANTES, using a Packard model 5530 gamma counter (Packard Instruments, Meriden, CT). Inhibition of 125I-RANTES in the presence of VVC was calculated as 100 × [1 − (BindingVVC/Bindingcontrol)], with the control being infection without any inhibitor.
PKA assay.
CD4+ T cells (106 cells) were incubated with the protein kinase A (PKA) inhibitor H89 (Sigma-Aldrich) for 1 h, washed with PBS, and then incubated for 10 min with 500 μl of lysis buffer (20 mM morpholinepropanesulfonic acid [MOPS], 50 mM β-glycerol phosphate, 50 mM sodium fluoride, 1 mM sodium vanadate, 5 mM EGTA, 2 mM EDTA, 1% NP-40, 1 mM dithiothreitol [DTT], 1 mM benzamidine, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 10 μg/ml leupeptin and aprotinin). Cell lysates were then spun down, and the total protein content was quantified using a Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Scientific, Rockford, IL). PKA activity in 100 ng of the lysate was measured using a PKA assay kit according to the manufacturer's instructions (Enzo Lifesciences, Farmingdale, NY).
Western blotting.
U87-CD4 cells transfected with CCR5-WT, CCR5-T82P, or CCR5-T82K for 48 h were incubated with or without RANTES for 5 min or with TAK779 for 15 min. Cells were lysed in buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 2 mM EDTA, and 1% Triton X-100. The total protein content was quantified using a BCA assay kit (Thermo Scientific, Rockford, IL). The cellular proteins (10 μg in total) were separated in an 8% Tris-glycine gel (Invitrogen) for 2 h, transferred to a polyvinylidene difluoride (PVDF) membrane overnight, and probed with antibodies that recognize either phosphorylated extracellular signal-regulated kinase 1/2 (P-ERK1/2) (clone D13.14.4E) or ERK1/2 (clone 3A7) (Cell Signaling Technology, Danvers, MA), as previously described (33). The signal intensities were quantified using ImageJ (National Institutes of Health, Bethesda, MD).
Data analysis.
All titration curves were generated using Prism (Graphpad Software, San Diego, CA) and used to derive the maximum percent inhibition (MPI) and 50% effective concentrations (EC50s). P values were calculated using the one-tailed Mann-Whitney U test, and P values of ≤0.05 were considered statistically significant.
RESULTS
Uncoupling CCR5 from the Gαi subunit affects HIV-1 sensitivity to VVC.
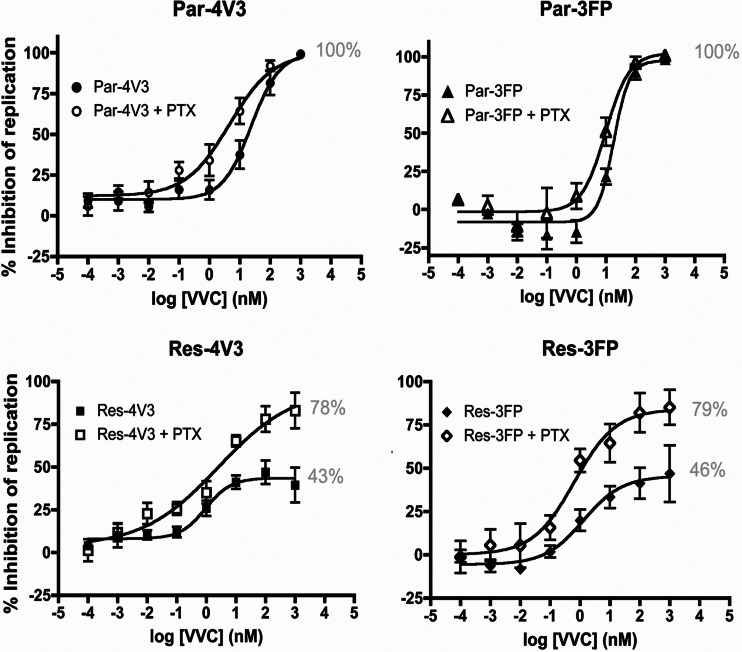
We treated primary human CD4+ T cells with PTX to uncouple the Gαi subunit from CCR5 and then infected the cells with genetically related, fully infectious, clonal, VVC-sensitive or -resistant viruses in the presence of increasing VVC concentrations. PTX treatment increased the potency of VVC against the two parental, VVC-sensitive viruses, Par-4V3 and Par-3FP, albeit only modestly; the EC50s were reduced by ∼4-fold (P = 0.019) and ∼2-fold (P = 0.028), respectively (Fig. 1A; Table 1). A more substantial effect of VVC was seen with the VVC-resistant viruses, Res-4V3 and Res-3FP. One measure of resistance is the MPI value, which reflects how efficiently a resistant virus uses the VVC-CCR5 complex for entry (34): the higher the MPI, the less efficiently the complex is used than the inhibitor-free form, and the lower the extent of residual VVC-insensitive replication (34–36). At high VVC concentrations, the MPI values for the Res-4V3 and Res-3FP VVC-resistant viruses increased from 43% and 46%, respectively, in control cells to 78% and 79%, respectively, in PTX-treated cells (P = 0.016 and P = 0.048, respectively) (Fig. 1A; Table 1). Similar inhibition trends were obtained with Par-4V3 and Res-4V3 when MVC was used instead of VVC (data not shown). The effect of PTX on VVC inhibition of virus entry was also assessed in a single-cycle assay using Tzm-bl cells. In the presence of PTX, there was a 7-fold decrease in the EC50 for VVC against Par-4V3 and an increase in the MPI against Res-4V3 (from 51% to 87%) compared to those for control cells (data not shown). These effects of PTX are consistent with the ones we observed using CD4+ T cells, suggesting that PTX treatment does modify how VVC affects HIV-1 entry, irrespective of any additional effects it may have on postentry events. Note that PTX treatment caused no overall change in the level of CCR5 expression on the CD4+ T cell surface, as judged by staining with MAb PA14 or 2D7 (data not shown).
Fig 1.
Effect of PTX treatment on inhibition of HIV-1 replication by VVC. CD4+ T cells were treated overnight with PTX (100 ng/ml), or not, incubated for 1 h with a range of VVC concentrations, and then infected with the indicated viruses. The cells were washed 6 h later and then supplemented with medium containing the same VVC concentrations. The data represent the percent inhibition of HIV-1 replication (measured at 7 days postinfection by p24 ELISA) relative to that in the absence of VVC (0%). The average values (± standard errors of the means [SEM]) for at least five independent experiments, using CD4+ T cells from a different donor in each experiment, are shown. The numbers in gray indicate the MPI values.
Table 1.
VVC inhibition of HIV-1 replication in PTX-treated or control CD4+ T cellsa
| Virus and treatment | Avg ± SEM |
|
|---|---|---|
| MPI | EC50 | |
| Par-4V3 | 104 ± 1.7 | 53 ± 25 |
| Par-4V3 + PTX | 101 ± 3.7 | 12 ± 7.4 |
| Res-4V3 | 43 ± 7.0 | 0.81 ± 0.49 |
| Res-4V3 + PTX | 78 ± 9.0 | 1.5 ± 0.86 |
| Par-3FP | 107 ± 1.6 | 33 ± 5.0 |
| Par-3FP + PTX | 105 ± 2.7 | 16 ± 5.8 |
| Res-3FP | 46 ± 11 | 4.0 ± 2.3 |
| Res-3FP + PTX | 79 ± 10 | 0.56 ± 0.20 |
EC50 and MPI values were derived from fitted curves generated for each independent donor. Average EC50 and MPI values (± SEM) for all donors were calculated and are shown in this table. The MPI values are fitted plateau values. For the VVC-resistant viruses, the EC50 values are the VVC concentrations that inhibit replication to an extent corresponding to half the plateau values (i.e., <50% inhibition). As such, these EC50 values do not reflect the potency of VVC against infection in the same way as for VVC-sensitive viruses, which limits how the data can be interpreted. The same data set is displayed in Fig. 1.
Treating the CD4+ T cells with PTX reduced the replication of all four of the test viruses by 2- to 3-fold (data not shown). When the virus inoculum added to the PTX-treated CD4+ T cells was increased to compensate for this reduction in replication compared to that in control cells, the VVC inhibition profiles for the inhibitor-sensitive and -resistant viruses Par-4V3 and Res-4V3 were similar to those obtained previously (data not shown). This result suggests that the above changes in EC50 and MPI values were not due to PTX affecting the susceptibility of the target cells to virus infection. We also used the data set in Table 1 to perform a correlation analysis between the susceptibilities of the cells to infection by the various viruses, based on the extent of virus replication, and the EC50 or MPI values (as appropriate). These correlations were weak and did not reach statistical significance (P > 0.05).
Overall, the above-described experiments showed that PTX treatment increased the potency of VVC against the sensitive viruses (reduced EC50) while decreasing the ability of the resistant viruses to use the VVC-CCR5 complex (increased MPI). Our interpretation is that VVC-resistant viruses use the VVC-CCR5 complex less efficiently when CCR5 is uncoupled from Gαi.
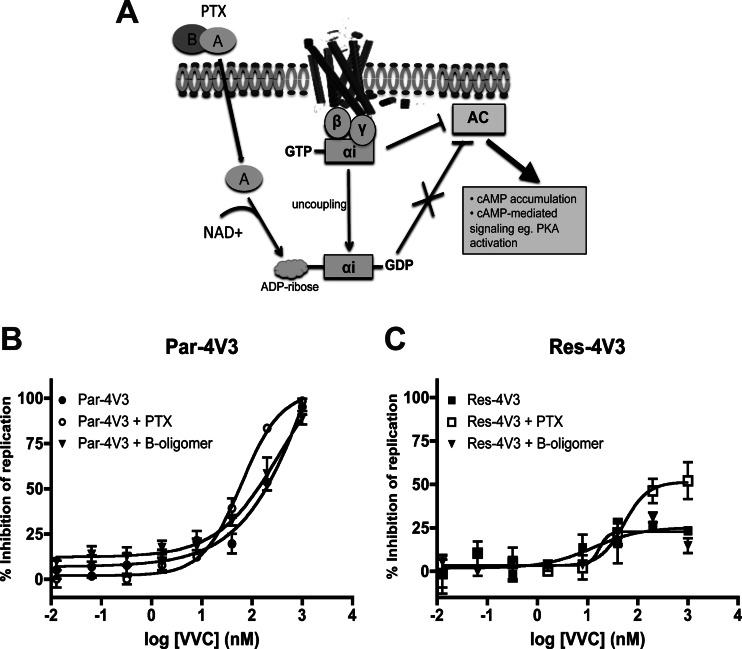
The effect of PTX on HIV-1 sensitivity to VVC is mediated by its catalytic subunit.
PTX consists of two subunits: an enzymatically active A protomer that ribosylates the Gαi subunit and prevents it from coupling to the cognate GPCR and a binding (B) oligomer that mediates the toxin's biological effects independently of Gαi (Fig. 2A) (37, 38). To determine whether the effect of PTX on VVC activity is mediated by Gαi uncoupling, we performed an experiment similar to that described for Fig. 1A, but now using the noncatalytic B oligomer. Unlike PTX, treating the cells with the B oligomer had no effect on the VVC inhibition profile for either Par-4V3 or Res-4V3 (Fig. 2B and C). Similar results were obtained using the second pair of VVC-sensitive (Par-3FP) and -resistant (Res-3FP) viruses (data not shown).
Fig 2.
Effect of the B oligomer, the noncatalytic subunit of PTX, on inhibition of HIV-1 replication by VVC. (A) Schematic representation of the mechanism of action of PTX. The PTX catalytic subunit, the A protomer, ribosylates the Gαi subunit and uncouples it from the GPCR (i.e., CCR5). As a result of Gαi uncoupling, cAMP levels increase, driving the activation of PKA and other cAMP-dependent molecules (37). (B and C) CD4+ T cells were treated overnight with 100 ng/ml of B oligomer or PTX or left untreated and then were incubated for 1 h with a range of VVC concentrations prior to infection with the VVC-sensitive virus Par-4V3 (B) or the VVC-resistant virus Res-4V3 (C), as indicated. The cells were washed 6 h later and then supplemented with medium containing the same VVC concentrations. The data represent the percent inhibition of HIV-1 replication (measured at 7 days postinfection by p24 ELISA) relative to that in the absence of VVC (0%). The average values (± SEM) for three independent experiments, using CD4+ T cells from a different donor in each experiment, are shown in panels B and C.
G-protein coupling to GPCRs (in this case, CCR5) involves a heterotrimeric complex containing the α, β, and γ subunits. The activation of CCR5 signaling through Gαi leads to dissociation of the heterotrimer into a GTP-bound, activated Gαi subunit and a Gβγ dimer. The activated Gαi subunit inhibits adenylyl cyclase (AC) activity, resulting in a decrease in the intracellular cyclic AMP (cAMP) concentration and a concomitant reduction in PKA activity. When PTX ribosylates the Gαi subunit, it remains locked in the GDP-bound, inactive state that is unable to inhibit AC. As a result, cAMP concentrations rise and PKA becomes more active (Fig. 2A).
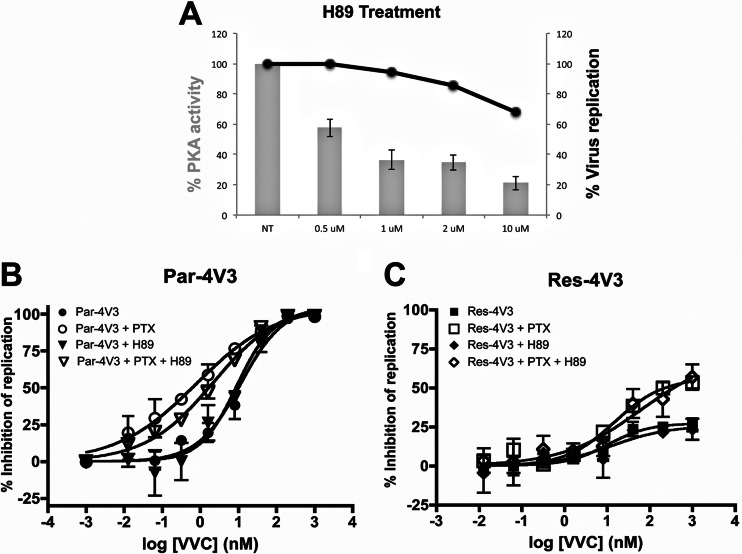
To determine whether PTX mediates its effect on VVC inhibition through cAMP-dependent PKA activation, we used the PKA inhibitor H89 (39). We first titrated H89 on CD4+ T cells to determine the optimal concentration that significantly inhibits PKA activity without compromising virus entry (Fig. 3A). At an H89 concentration of 10 μM, PKA was significantly inhibited (by 80%), but the replication of Res-4V3 and the other test viruses was only modestly reduced (by ∼35%) (Fig. 3A and data not shown). We next evaluated whether H89 altered the VVC inhibition patterns in the presence and absence of PTX. When PTX was absent, the same 10 μM H89 concentration did not affect the VVC inhibition profile for Par-4V3 or Res-4V3 in CD4+ T cells compared to the control (Fig. 3B and C). Moreover, H89 did not reverse the effect of PTX on the VVC inhibition profile for either Par-4V3 or Res-4V3 (Fig. 3B and C). Cumulatively, these results indicate that PTX most likely affects how VVC inhibits HIV-1 entry by a mechanism that is independent of downstream signaling events such as PKA activation. We propose that the effects of PTX are, instead, a direct result of conformational changes induced in CCR5 by the uncoupling of Gαi, an event driven by the PTX catalytic subunit.
Fig 3.
Effect of the PKA inhibitor H89 on inhibition of HIV-1 replication by VVC. (A) CD4+ T cells were treated for 1 h with a range of H89 concentrations and then either lysed to allow measurement of PKA activity or infected with the Res-4V3 virus for 6 h before washing. The data represent PKA activities expressed as percentages of that in the absence of inhibitor (100%). The average values (± SEM) for two independent experiments, using CD4+ T cells from a different donor in each experiment, are shown by gray bars. The infectivity of the Res-4V3 virus was measured by p24 ELISA at 7 days postinfection (black line). Data for one experiment using CD4+ T cells pooled from the same two donors used to measure PKA activity are shown. (B and C) CD4+ T cells were treated overnight with or without PTX (100 ng/ml), incubated with H89 (10 μM) and a range of VVC concentrations for 1 h, and then infected with the VVC-sensitive Par-4V3 virus (B) or the VVC-resistant Res-4V3 virus (C), as indicated. The cells were washed 6 h later and then supplemented with medium containing the same VVC concentrations. The data represent the percent inhibition of HIV-1 replication (measured at 7 days postinfection by p24 ELISA) relative to that in the absence of VVC (0%). The average values (± SEM) for three independent experiments, using CD4+ T cells from a different donor in each experiment, are shown in panels B and C.
Use of VVC-bound, signaling-deficient, or constitutively active CCR5 mutants.
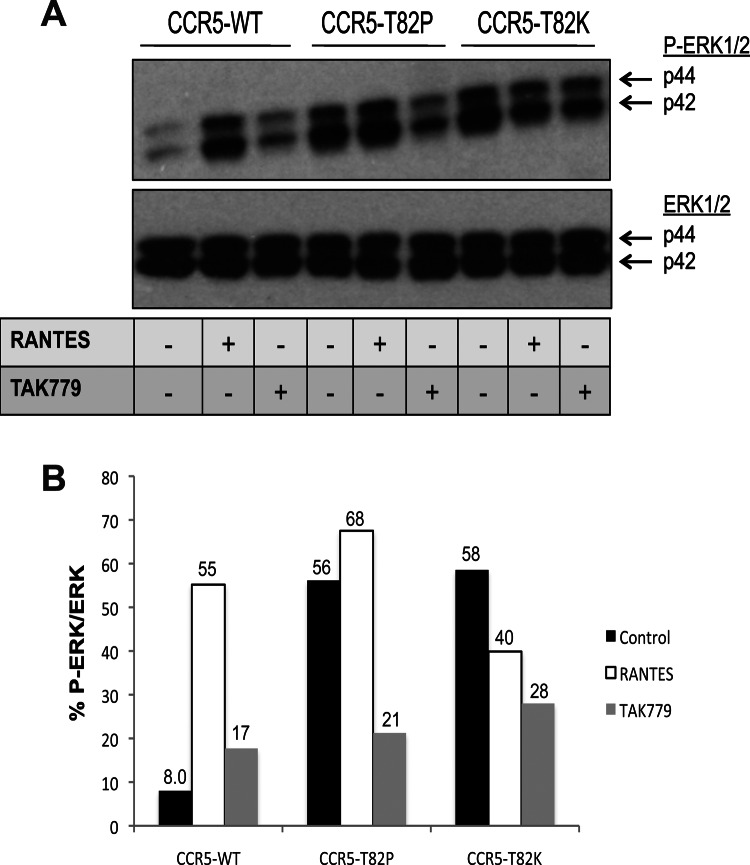
We next assessed whether the effects of PTX could be mimicked or expanded upon by the use of CCR5 mutants with different basal or chemokine-induced signaling capacities. An R126N change in the DRY motif of CCR5 and other GPCRs disables G-protein coupling and agonist-induced signal transduction (40). Conversely, several substitutions at residue Thr-82 (e.g., T82P and T82K) in the CCR5 TM2 domain cause conformational changes that make the receptor constitutively active, in that it now signals in the absence of stimulatory ligands (26). To confirm the reported properties of the CCR5-T82P and CCR5-T82K CAMs, we measured the proportion of ERK that was phosphorylated (P-ERK) as a surrogate for CCR5 activation (41, 42). The proportional phosphorylation of ERK was minimal, i.e., 8%, at baseline (i.e., in the absence of any stimulation) in cells expressing CCR5-WT, but it increased to 55% upon RANTES addition (Fig. 4). Conversely, ERK phosphorylation in cells expressing either CCR5-T82P or CCR5-T82K was high at baseline, at 56% or 58%, respectively, and comparable to the RANTES-stimulated levels seen in cells expressing CCR5-WT. RANTES treatment of CCR5-T82P- or CCR5-T82K-expressing cells did not significantly affect ERK phosphorylation. When the inverse agonist TAK779 was added to the cells, ERK1/2 phosphorylation was not affected in cells expressing CCR5-WT, but there were significant decreases in P-ERK1/2 levels in cells expressing CCR5-T82P or CCR5-T82K, from 56% to 21% and from 58% to 28%, respectively (Fig. 4). Hence, the inverse agonist TAK779 does indeed reverse the constitutive activity of the CCR5 CAMs.
Fig 4.
ERK phosphorylation in cells expressing CCR5-WT or CCR5 CAMs. (A) U87-CD4 cells were transfected with CCR5-WT, CCR5-T82P, or CCR5-T82K for 48 h and then incubated with or without RANTES for 5 min or with TAK779 for 15 min. The cells were lysed and their proteins analyzed by SDS-PAGE. Antibodies were used to probe for P-ERK and ERK. Both antibodies detect the p44 and p42 subunits of ERK1/2, as indicated. (B) The band intensities in panel A were quantified, and the extents of ERK phosphorylation (P-ERK1/2) relative to total ERK (ERK1/2) are expressed as percentages. Data shown are for one representative experiment out of two with similar patterns of results.
We tested the above CCR5 mutants for the ability to mediate HIV-1 entry in the presence or absence of VVC. To do so, U87-CD4 cells were transfected with CCR5-WT, the CCR5-T82P or CCR5-T82K CAM, or the CCR5-R126N SDM. The various CCR5 proteins were all expressed on the cell surface at similar levels, as detected by MAb 2D7 or PA14 (data not shown). The CCR5-transfected cells were then infected with viruses pseudotyped with the VVC-sensitive or -resistant Env protein derived from Par-4V3, Res-4V3, or Res-3FP. As Par-3FP generally behaves similarly to Par-4V3, we did not include it in this experiment.
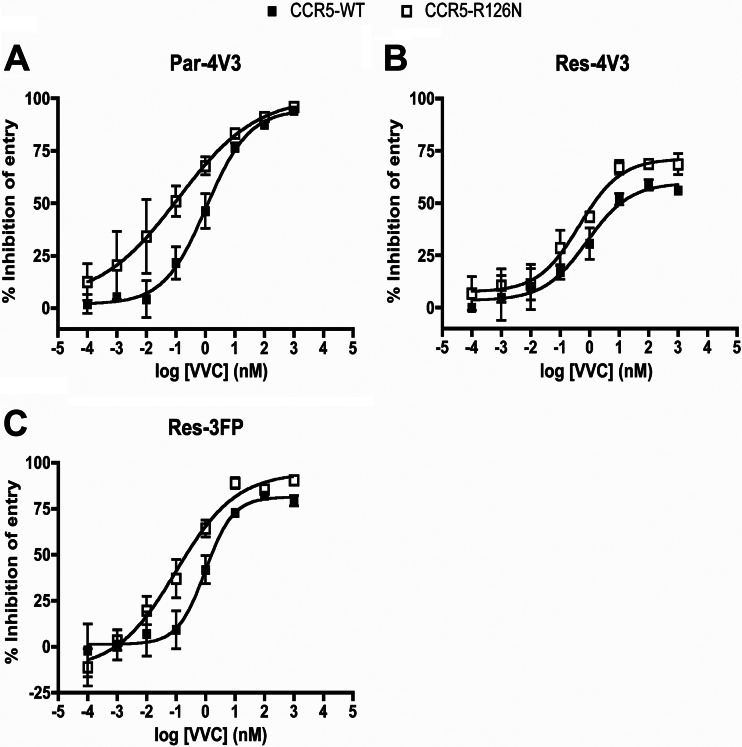
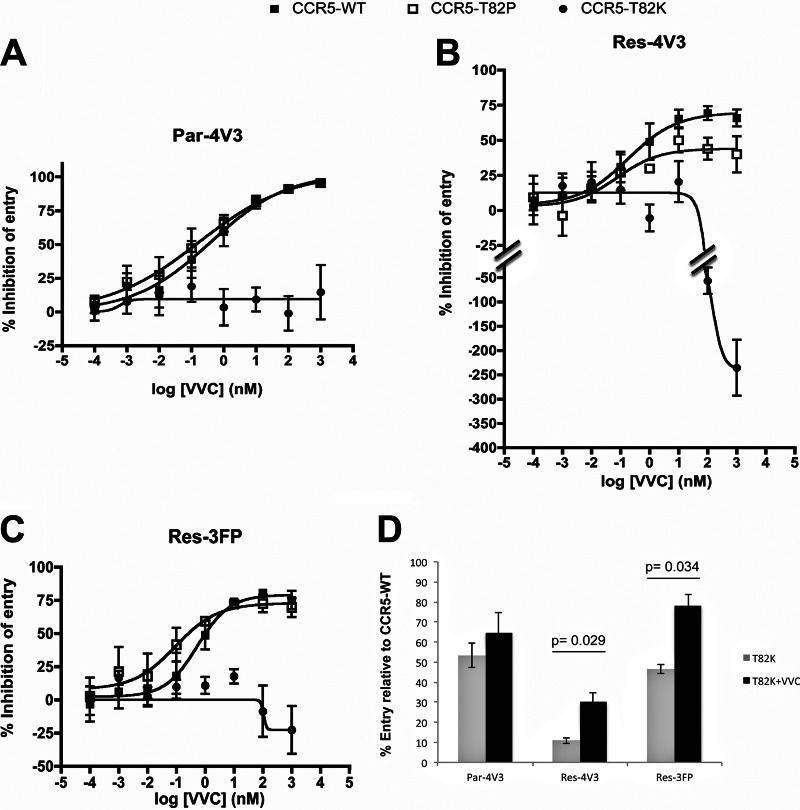
VVC inhibited the entry of Par-4V3 much more potently (∼10-fold decrease in EC50; P = 0.050) via the CCR5-R126N SDM than via CCR5-WT. Moreover, the MPIs were higher when the two resistant viruses entered via the SDM than when they entered via CCR5-WT; for Res-4V3 and Res-3FP, there were 1.5-fold and 2.7-fold (P = 0.047 and P = 0.050) decreases, respectively, in the extent of residual (i.e., VVC-insensitive) infection (Fig. 5). The VVC inhibition patterns for the two CCR5 CAMs differed from each other and from that for the CCR5-R126N SDM. VVC inhibited all three Env-pseudotyped viruses in cells expressing the CCR5-T82P CAM, although to different extents. Par-4V3 entry via CCR5-T82P was inhibited slightly more potently (2-fold decrease in EC50; P = 0.10) than that via CCR5-WT, but for both resistant viruses, the MPI values were lower when entry was via CCR5-T82P than when it was via CCR5-WT. Thus, VVC-insensitive residual entry via CCR5-T82P was increased 2-fold and 1.5-fold (P = 0.032 and P = 0.048) for Res-4V3 and Res-3FP, respectively (Fig. 6). The baseline (i.e., no VVC) entry levels of Par-4V3, Res-4V3, and Res-3FP via the CCR5-T82K CAM were reduced by approximately 50%, 90%, and 50%, respectively, compared to that via CCR5-WT (Fig. 6D). Thus, although the CCR5-T82K CAM is a coreceptor for HIV-1 entry, this function is impaired to various extents. Note that baseline entry of all viruses via the CCR5-T82P CAM or the CCR5-R126N SDM was generally comparable to entry via CCR5-WT (data not shown). In contrast to what was seen with the other CAM, CCR5-T82P, VVC inhibited none of the test viruses in cells expressing CCR5-T82K (Fig. 6). One explanation for this finding is that the T82K substitution destroys the VVC-binding site. However, Res-4V3 (and, to a lesser extent, Res-3FP) entry via CCR5-T82K was enhanced by VVC concentrations above ∼100 nM (Fig. 6B and C). Thus, when VVC was added at 1 μM, Res-4V3 entry was significantly increased (3-fold; P = 0.029), and Res-3FP entry marginally so (1.7-fold; P = 0.034), compared to when VVC was absent (Fig. 6D). VVC must therefore still bind to CCR5-T82K, at least at concentrations above ∼100 nM. The resulting VVC–CCR5-T82K complex is a more effective coreceptor for Res-4V3 and Res-3FP than the corresponding VVC-free form. Under the same conditions, entry of the Par-4V3 parental virus was not detectably affected by VVC (Fig. 6D). Par-4V3 may therefore be able to enter cells via the VVC complex of CCR5-T82K about as efficiently as it can via free CCR5-T82K (see the model in Fig. 8); alternatively, VVC may bind to CCR5-T82K with an affinity that is too low to prevent Env from interacting with this receptor. Note that similar results were obtained with CCR5-T82K when MVC was used instead of VVC (data not shown). The entry-enhancing effect of VVC via CCR5-T82K was not attributable to a VVC-mediated increase in the cell surface expression of this mutant, as judged by staining with MAb PA14 or 2D7 (data not shown). These antibodies were chosen because their binding to CCR5 is not affected by VVC or other related inhibitors (6, 24, 43). Overall, the T82K substitution must not just activate constitutive signaling but also change the conformation of CCR5 in various ways that affect both the VVC- and Env-binding sites.
Fig 5.
HIV-1 entry via the CCR5-R126N SDM. U87-CD4 cells were transfected with CCR5-WT or the CCR5-R126N mutant for 48 h and then incubated with a range of VVC concentrations for 1 h prior to infection with the Env-pseudotyped virus Par-4V3 (A), Res-4V3 (B), or Res-3FP (C), as indicated. The data represent the percent inhibition of HIV-1 entry (measured at 3 days postinfection by using a luciferase assay) relative to that in the absence of VVC (0%). The average values (± SEM) for three independent experiments are shown in all panels.
Fig 6.
HIV-1 entry via the CCR5 CAMs. (A to C) The experimental conditions were as described in the legend to Fig. 5, except that the CCR5-T82P and CCR5-T82K mutants were compared with CCR5-WT. (D) Entry of the indicated viruses via CCR5-T82K in the presence (black bars) or absence (gray bars) of VVC (1 μM), expressed as percentages of the entry via CCR5-WT. The average values (± SEM) for four independent experiments are shown in all panels, and P values (<0.05) for differences in entry in the presence and absence of VVC are displayed in panel D. The data set in panel D is independent from those displayed in panels A to C.
Fig 8.
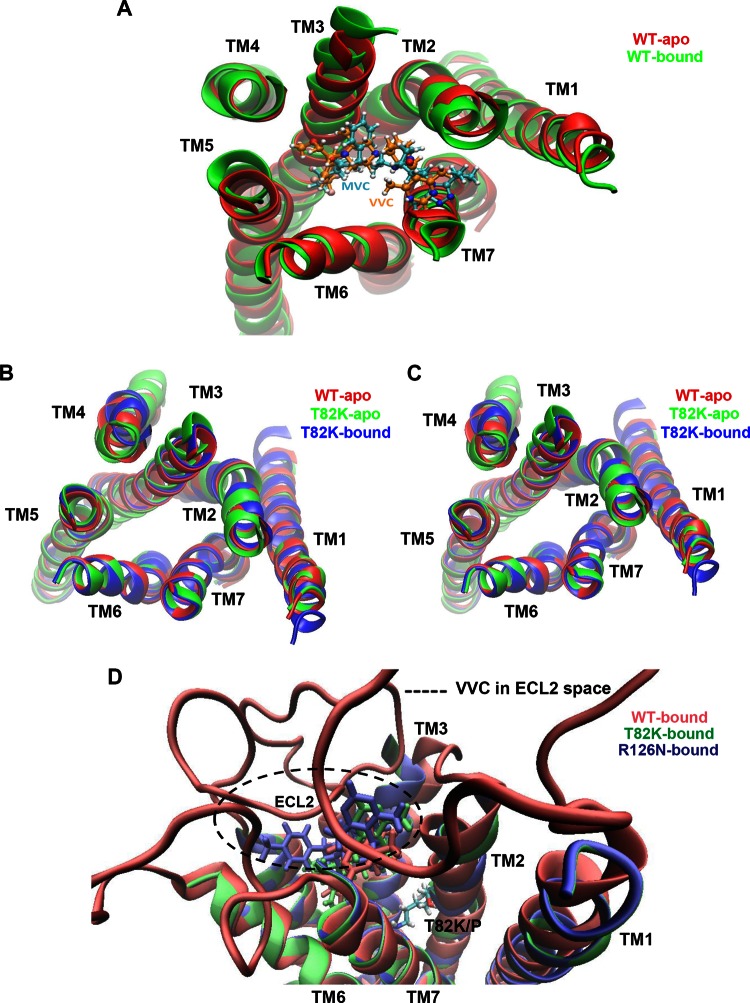
TM bundle structure and orientation of the low-energy conformational state for CCR5 variants. (A) The TM helices of the unbound CCR5-WT (WT-apo) and MVC/VVC-bound CCR5-WT (WT-bound) structures predicted to have the lowest-energy conformations by the GEnSeMBLE method are superimposed. Either MVC (cyan) or VVC (orange) is docked into the TM bundle. (B and C) The TM helices of unbound CCR5-WT (WT-apo) and the unbound and VVC-bound forms of the indicated CCR5 variant are superimposed. (D) Three VVC orientations upon docking into the TM bundles are displayed in different colors, i.e., red, green, and blue, for VVC-bound CCR5-WT (WT-bound), CCR5-T82K (T82K-bound), and CCR5-R126N (R126N-bound), respectively. The extracellular loops (ECLs) of the WT-bound VVC are shown, and the space where VVC could potentially interact with ECL2 is depicted with a dotted circle. The location of Thr (T) or Lys (K) at position 82 is indicated. In all panels, the TM region at the extracellular end is facing forward.
In summary, the overall data pattern derived using the SDM is generally consistent with the results obtained using PTX. Thus, uncoupling CCR5 from the intracellular signaling machinery, either through PTX or by mutating the DRY motif, increased the potency of VVC against the parental virus but decreased the efficiency with which the resistant viruses used the VVC-CCR5 complex. Conversely, the resistant viruses Res-4V3 and, to a lesser extent, Res-3FP use the VVC-bound form of the CCR5-T82P CAM more efficiently than the corresponding form of CCR5-WT, as indicated by the decreased MPI values (and hence the increased residual entry). The T82K substitution appears to have multiple effects, including an impairment of HIV-1 entry, but the complex between VVC (at high concentrations) and the CCR5-T82K mutant can be used for entry by VVC-resistant, and possibly also VVC-sensitive, viruses under appropriate circumstances. The latter point is explored further in the section on CCR5 modeling (see below).
Affinity of VVC for G-protein-coupled and uncoupled receptors.
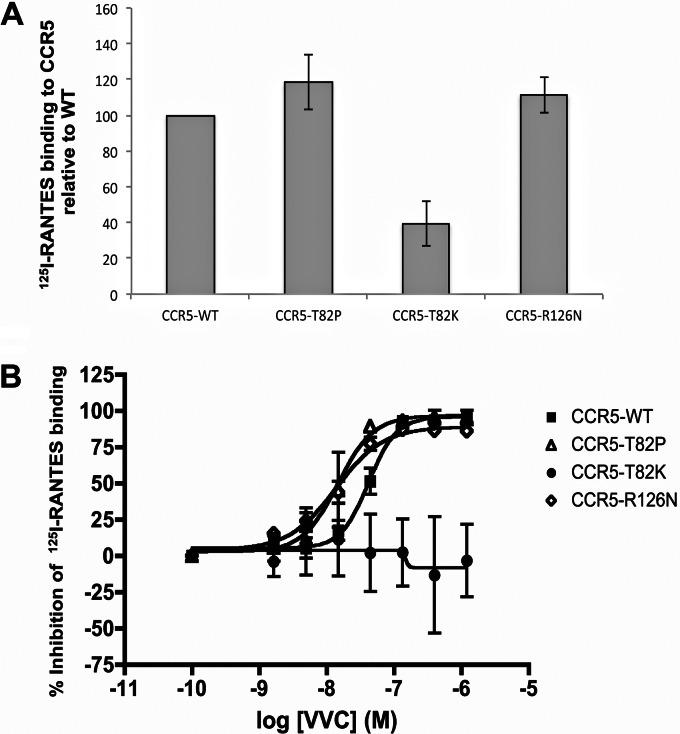
To further understand the different VVC inhibition patterns obtained for cells expressing the various CCR5 variants, we tested whether the activation state of CCR5 affects its affinity for VVC. To do so, we measured the ability of increasing VVC concentrations to compete with a saturating amount of 125I-RANTES (i.e., CCL5) for binding to CCR5-WT, the CCR5-T82P and CCR5-T82K CAMs, and the CCR5-R126N SDM, all expressed in U87-CD4 cells. Nontransfected U87-CD4 cells were used to gauge and correct for nonspecific 125I-RANTES binding. In the absence of VVC, the 125I-RANTES probe bound to CCR5-WT, CCR5-T82P, and CCR5-R126N to similar extents, but its binding to CCR5-T82K was significantly reduced, by ∼60%, relative to that to CCR5-WT (Fig. 7A). This outcome again speaks to the impact that the T82K substitution must have on the conformation of the ligand-binding regions of CCR5.
Fig 7.
Competition between VVC and 125I-RANTES for binding to CCR5 variants. U87-CD4 cells were transfected for 48 h with CCR5-WT or the mutants listed and then incubated for 1 h with 400 pM 25I-RANTES in the presence or absence of the indicated VVC concentrations. After extensive washing, the amount of cell-bound 125I-RANTES was measured using a gamma scintillation counter. (A) 125I-RANTES binding to the indicated CCR5 variants in the absence of VVC was expressed as a percentage of binding to CCR5-WT. (B) The data represent the percent inhibition of 125I-RANTES binding to each CCR5 variant relative to that in the absence of VVC (0%). The average values (± SEM) for three independent experiments are shown in all panels.
The competition curves showed that VVC inhibited 125I-RANTES binding to all the CCR5 proteins except for CCR5-T82K (Fig. 7B). The latter finding implies that RANTES can still bind to the VVC–CCR5-T82K complex, an outcome consistent with the ability of Par-4V3 to use the same complex for entry (Fig. 6A). Hence, when sufficient VVC is present to form a low-affinity complex with the CCR5-T82K mutant, the receptor may still be recognized by either HIV-1 or RANTES (see the model in Fig. 8). Other possibilities are that the affinity of VVC for CCR5-T82K is too low to block Env and RANTES binding or that RANTES and VVC bind preferentially to different subpopulations of CCR5-T82K. The EC50s for VVC inhibition of 125I-RANTES binding were modestly lower for both CCR5-T82P and CCR5-R126N, by 2.8- and 2.4-fold (P = 0.050 and P = 0.028), respectively, than for CCR5-WT (Fig. 7).
Overall, the VVC affinities of the various CCR5 variants did not correlate with their differential usage by the VVC-resistant viruses. Thus, VVC bound to both the CCR5-T82P CAM and the CCR5-R126N SDM with slightly higher affinities than to CCR5-WT, but the corresponding MPIs for the resistant viruses were modestly decreased and increased, respectively, again compared to CCR5-WT. The EC50s for VVC inhibition of the VVC-sensitive virus were somewhat lower for entry via CCR5-R126N and CCR5-T82P than for that via CCR5-WT, an outcome that is consistent with the modest increase in VVC affinity for the two mutants.
Structural modeling of the CCR5 variants.
The three-dimensional (3D) structures of the WT, T82K, and R126N CCR5 variants were generated using a method that predicts the ensemble of low-energy conformational states a GPCR can adopt (44). The GEnSeMBLE technique involves sampling all reasonable packings of the 7-helix bundle (we examined ∼13 trillion different helix bundle packings) and then selecting an ensemble of 100 low-energy packings that are likely to play a role in binding various ligands and in the GPCR activation process. The resulting set of CCR5-WT conformations, ranked from lowest to highest energy, is referred to as wt1 to wt100.
The wt1 conformation is the lowest-energy form (i.e., the most stable) that CCR5-WT can adopt. We refer to this conformation as WT-apo (Fig. 8A). We docked MVC and VVC into the 10 most structurally diverse conformations from among the 20 with the lowest energies (i.e., wt1 to wt20) and then ranked the resulting docked structures according to their energies. The outcome was that both MVC and VVC stabilized the same conformation, wt7, which became the most stable. This conformation is designated WT-bound (Fig. 8A). Comparing the WT-apo and WT-bound structures reveals that upon inhibitor binding, TM2 tilts slightly toward the center of the TM bundle, while TM3 tilts away from the center; the net effect is to reduce the distance between the intracellular ends of TM3 and TM6 (see overlay in Fig. 8A). We next introduced the T82K and R126N substitutions into all 100 conformations (wt1 to wt100) to assess their effects on CCR5 structure. The resulting conformations were then reranked and the most stable selected. Note that, for simplicity, we retained the same, “wt”-based nomenclature for the two CCR5 variants. For the CCR5-T82K mutant, conformation wt6 became the most stable (shown in Fig. 8B as T82K-apo). When unbound CCR5-T82K was compared to unbound CCR5-WT, most of the helical changes were located in TM2, TM3, and TM4. When MVC or VVC was docked into the CCR5-T82K CAM, the wt31 conformation became the lowest-energy form in both cases (shown in Fig. 8B as T82K-bound). Note that both inhibitor-bound CCR5-T82K variants acquired conformations similar to that of unbound CCR5-WT. In fact, except for TM1 and TM4, the positions of the T82K-bound and WT-apo TM helices seem to be identical. In particular, TM3 and TM2 completely overlap for T82K-bound and WT-apo but are oriented differently in T82-apo (see overlay in Fig. 8B).
The analogous modeling study on the CCR5-R126N SDM revealed that this variant can adopt multiple conformations with comparable low-energy states, which implies that it has a certain degree of structural flexibility. The lowest-energy conformation for CCR5-R126N is wt37 (shown in Fig. 8C as R126N-apo), although a few other structurally similar conformations with similarly low energies can also be adopted. As with T82K-apo, most of the helical changes in R126N-apo are located in TM2, TM3, and TM4. The binding of VVC or MVC stabilized the wt41 conformation of CCR5-R126N (shown in Fig. 8C as R126N-bound). As seen with CCR5-T82K, the inhibitor-bound form of CCR5-R126N adopted a conformation similar to that of unbound CCR5-WT.
The docking studies predict that MVC and VVC share a binding site on CCR5-WT and bind in similar orientations. The binding of MVC within the TM bundle involves interactions with W86 (TM2), Y108 (TM3), Y251 (TM6), and E283 (TM7). VVC also interacts with W86 (TM2), Y251 (TM6), and E283 (TM7), but unlike MVC, it is not predicted to interact directly with Y108 (TM3) (Fig. 8A). The different outcomes for Y108 may reflect a weakness in the side chain placement method (SCREAM) used to predict how ligands are orientated within the TM bundle. In fact, the position of the C-β atom of Y108 suggests that it is indeed located close enough to VVC for an interaction to be possible. In previous mutagenesis-based studies, the residues identified as interacting with VVC and MVC were similar to those predicted by our models, although there were subtle differences in the extent and nature of the interactions of the two inhibitors with individual residues (45–47). Overall, MVC and VVC are predicted to have a common binding site within the TM bundle and to contact similar residues, albeit with perhaps some differences in their modes of binding. The modeling study predicts that VVC acquires a different orientation within each of the three CCR5 variants (Fig. 8D). The VVC-bound conformations of CCR5-R126N and CCR5-T82K are very similar, although the orientations of VVC differ slightly between the two variants. The introduction of a positively charged bulky residue when Thr-82 is replaced by Lys changes how the drug is oriented, specifically by slightly shifting its position toward TM4 compared to where it is located in CCR5-WT. For CCR5-R126N, the VVC location shifts again, but closer to TM5 and upward toward ECL2; the smaller upward shift enables VVC to interact with the middle section of the ECL2 loop, which is not the case with the other two CCR5 variants. The location of VVC in CCR5-R126N is most likely affected by the overall conformational change that the Arg-to-Asn substitution confers on the CCR5 structure rather than by any more direct impact on the inhibitor-binding site. That supposition is consistent with the location of the R126N substitution in the DRY motif at the intracellular interface of TM3, a considerable distance from where VVC binds.
The predicted structures of the VVC complexes with CCR5-WT, CCR5-T82K, and CCR5-R126N indicate that the drug binds to CCR5-T82K and CCR5-R126N more weakly and more strongly, respectively, than to CCR5-WT, as approximated by the binding enthalpy (ΔH). Note that the relative affinity estimates for VVC for CCR5-R126N and CCR5-WT, derived from the 125I-RANTES competition assay (Fig. 7), are consistent with the predictions for MVC, with CCR5-R126N having the higher affinity.
Overall, 3D modeling of the CCR5 structure shows that VVC still binds to CCR5-T82K, but in doing so it changes the mutant's conformation to one that is similar to unbound CCR5-WT. This outcome explains why Par-4V3 could still use the inhibitor-bound form of CCR5-T82K for entry and why the VVC-resistant viruses used this complex more efficiently than unbound CCR5-T82K. Although the inhibitor-bound form of CCR5-R126N also has a conformation similar to that of unbound CCR5-WT, the VVC-sensitive viruses did not use this complex, and the VVC-resistant viruses did so only inefficiently. The modeling study suggests that when VVC is bound to CCR5-R126N, the inhibitor is oriented in a way that allows it to interact with ECL2, which is an important element of the Env-binding site. A plausible outcome is a change in the conformation of ECL2 that is sufficient to impede HIV-1 entry either substantially or completely, depending on the test virus.
DISCUSSION
In this study, we assessed how CCR5 conformational/functional variants influence the sensitivity of HIV-1 to small-molecule CCR5 inhibitors, exemplified by VVC. In particular, we investigated whether VVC's action against inhibitor-sensitive and -resistant viruses is affected by the state of CCR5 coupling to G proteins. One finding is that uncoupling CCR5 from the Gαi subunit, either by treating cells with PTX or by expressing the CCR5-R126N SDM, increased the potency of VVC against a VVC-sensitive virus but decreased the efficiency with which resistant viruses entered cells via the VVC-CCR5 complex. Our results also indicate that the observed effects of Gαi subunit uncoupling on sensitivity to VVC are unlikely to be mediated by downstream signaling events, such as PKA activation, but rather by conformational changes in CCR5. When we assessed the VVC sensitivity of viruses entering cells via two constitutively active CCR5 variants, we observed that VVC-resistant viruses used the inhibitor-bound form of the CCR5-T82P CAM more efficiently than the corresponding VVC complex of CCR5-WT. Entry via the CCR5-T82K CAM was impaired compared to that via CCR5-WT, but the resistant viruses used this variant more efficiently when a VVC concentration high enough to allow formation of the inhibitor-CCR5 complex was present.
Differences in the affinities of the CCR5 variants for VVC do not explain the variations in how efficiently the VVC-resistant viruses used the various inhibitor-CCR5 complexes. Thus, based on its ability to inhibit 125I-RANTES binding, we infer that VVC has slightly higher affinities for both the CCR5-T82P CAM and the CCR5-R126N SDM than for CCR5-WT. However, the resistant viruses entered via the VVC-bound forms of these two variants with higher and lower efficiencies, respectively, than that for the VVC complex with CCR5-WT. Having said that, the EC50s for VVC against the sensitive virus were modestly lower when entry was via CCR5-R126N and CCR5-T82P than when it was via CCR5-WT. For these three CCR5 variants, the EC50s for the sensitive virus, but not the MPI values for the resistant viruses, rank similarly to the corresponding VVC affinities (i.e., to the EC50s for VVC inhibition of 125I-RANTES binding).
The T82K substitution adversely affected HIV-1 entry, but infection by the VVC-resistant viruses via the CCR5-T82K CAM was enhanced by high VVC concentrations. Hence, VVC and CCR5-T82K must form a complex that the resistant viruses use more efficiently than the corresponding inhibitor-free configuration. Conversely, VVC did not prevent inhibitor-sensitive viruses from entering via CCR5-T82K; the extents of entry were similar whether high VVC concentrations were present or not. The modeling data suggest that when the CCR5-T82K variant binds VVC, it acquires a conformation similar to that of unbound CCR5-WT. The VVC-sensitive virus can use that CCR5 conformation(s) for entry with the same efficiency as it can enter via unbound CCR5-T82K, while the VVC-resistant viruses actually use it more efficiently than the inhibitor-free form, leading to enhanced entry. Although the predicted TM bundle structures are similar for unbound CCR5-WT and inhibitor-bound CCR5-T82K, the loop orientations for these receptors might differ, particularly when VVC is bound. Such an effect might explain the observed submaximal entry for Par-4V3 via CCR5-T82K in the presence of VVC relative to that via CCR5-WT. The virology and modeling studies strongly suggest that VVC and MVC behave very similarly in how they interact with CCR5-WT and its variants. Although we cannot exclude the possibility of subtle, molecule-specific differences, none was evident when MVC or VVC was docked into CCR5, as both compounds stabilized the same TM bundle conformations of all three CCR5 variants. However, there must be inhibitor-specific conformational changes in the more flexible extracellular loop structures, given the variation in binding of ECL-specific MAbs to CCR5 complexes with different small-molecule inhibitors (43). Having said that, the MAb-binding profiles of MVC-CCR5 and VVC-CCR5 complexes were very similar to one another, although both were different from CCR5 complexes with other small-molecule inhibitors, such as aplaviroc (APL) (45). These inhibitor-specific conformational changes in the ECLs are, however, still sufficient to influence the cross-resistance profiles of MVC- versus VVC-selected viruses (8–10, 43).
The effect the T82K substitution has on VVC sensitivity does not imply that residue 82 is necessarily part of the inhibitor-binding site, just as it is unlikely to be involved directly in HIV-1 binding. Instead, our modeling shows that introducing a positively charged lysine at this position within TM2 causes a substantial structural rearrangement within the body of CCR5. Note that an Ile-Met change at position 189 in TM5 indirectly compromises the antiviral activity of a different CCR5 inhibitor, SCH-C, again without this residue being part of the inhibitor-binding site (48). Another probable long-range effect on the CCR5 structure involves the T82A substitution, which partially impairs the inhibitory activity of TAK779 but not that of other small molecules, including AD101 and SCH-C (7, 27). Overall, the two changes at residue 82 that create the CCR5-T82K and CCR5-T82P CAMs have different effects on VVC and MVC binding and HIV-1 entry, presumably because they alter the overall geometry of CCR5 in different ways that are not yet understood at the molecular level.
The conserved TXP motif is present in TM2 of all GPCRs of the angiotensin, opioid, and chemokine families (49, 50). This sequence, residues 82 to 84, plays a crucial role in chemokine signaling via CCR5. Depending on the type of substitution made at residue Thr-82, the effect can range from strong impairment of CCR5 activation by chemokines to constitutive activation in their absence (26, 49, 50). Using ERK phosphorylation as a surrogate for CCR5 activation, our results suggest that the CCR5-T82P and CCR5-T82K variants have higher constitutive activity than that of CCR5-WT. Earlier work has also shown that both the CCR5-T82P and CCR5-T82K CAMs bind higher basal levels of GTPγS than that with CCR5-WT (i.e., they are constitutively coupled to G proteins), but also that their overall properties are different (26). Thus, CCR5-T82P can reach an active (i.e., G protein coupled) state in the absence of chemokines but undergoes agonist-mediated activation in calcium flux and chemotaxis assays. In contrast, CCR5-T82K is more strongly activated constitutively but has a dramatically impaired responsiveness to natural agonists that is associated with reduced binding affinities (26). The data from our chemokine binding assay are consistent with this earlier report: 125I-RANTES binding to CCR5-T82K, but not CCR5-T82P, was severely compromised. Overall, the replacement of Thr-82 with Pro or Lys creates two constitutively active CCR5 variants that have distinct conformational and functional properties and are recognized differently by HIV-1 and small-molecule inhibitors. This point was further confirmed in a recent study in which both CCR5-T82K and CCR5-T82P were reported to be constitutively active but to differ in their ability to mediate HIV-1 Env-mediated fusion (51).
Our results indicate that VVC-resistant viruses use the VVC-bound form of G-protein-coupled CCR5 more efficiently than the corresponding form of the uncoupled receptor. Thus, how VVC and similar antagonists inhibit these viruses depends largely on their ability to stabilize G-protein-coupled or -uncoupled forms of CCR5. In a recent study, MVC and TAK779 were shown to stabilize slightly different CCR5 conformations that vary in the ability to activate G proteins. More specifically, TAK779 preferentially stabilized G-protein-uncoupled, inactive conformations, while MVC bound equally well to both G-protein-coupled and -uncoupled receptors. As a result, TAK779 was more effective than MVC at stabilizing the G-protein-uncoupled, inactive state of CCR5 (25). Whether our resistant viruses would use TAK779-stabilized, G-protein-uncoupled receptors less efficiently than their MVC-bound active and inactive counterparts remains to be determined. The small-molecule inhibitor APL also stabilizes two distinct CCR5 conformations with properties that differ depending on their coupling state: APL enhances RANTES binding to uncoupled CCR5 but inhibits binding to the coupled form of the receptor (52). Overall, the various small-molecule inhibitors may stabilize different conformational subsets of CCR5 that also function differentially as coreceptors for resistant viruses.
An important unresolved issue is whether the abundances of the different forms of CCR5 (coupled or uncoupled) are cell type dependent or, alternatively, influenced by the activation state of a cell. The amounts of CCR5 expressed in some transfected cell lines may be greater than the levels of G proteins available for coupling, creating an excess of uncoupled forms of CCR5. That might not be the case in primary cells. The different context for CCR5 might explain why MPI values for resistant viruses are generally higher for transiently or stably transfected cell lines than for primary cells, as observed in our current and previous studies (24, 29). It is conceivable that the CCR5 conformation status could also have implications for the rates at which various stages of the fusion process take place in different cells, and hence for the potencies of inhibitors of different events in this process. Inhibitors affected in this way could include neutralizing antibodies (NAbs) that intervene against different stages of the virus-CD4-CCR5 binding axis, leading to cell-type-dependent skews in how different NAb specificities are quantified.
Resting and activated T cells may also have different proportions of CCR5 conformational/functional variants because of changes in the relative amounts of CCR5 and G proteins expressed under the two conditions. The formation of lipid rafts and the recruitment of signaling machinery to them are known to be more pronounced in activated cells, increasing the proportion of signaling-active CCR5 forms present compared to the case in resting cells (53, 54). Depending on the tissue compartment where infection occurs, the inhibitory actions of CCR5 inhibitors may be influenced by the abundances of resting and active T cells, and hence by the availability of G-protein-coupled and -uncoupled forms of CCR5. It has been argued that resting T cells may be involved in initial HIV-1 infections at mucosal sites, while infection of activated T cells plays a far greater role during systemic dissemination and propagation of the virus (55). In a recent study, a substantial proportion of chronic viruses (but not transmitted/founder viruses) were incompletely inhibited by a saturating concentration of MVC (56, 57). This viral phenotype was most clearly seen in a cell line expressing very high levels of both CD4 and CCR5, but it was also detectable using primary CD4+ T cells (56). Our hypothesis is that CCR5 inhibitors might be particularly effective against HIV-1 mucosal transmission. Thus, the frequency of incoming (i.e., transmitted/founder) viruses with reduced inhibitor susceptibility is lower than that seen with chronic viruses, and infections of resting T cells, which may express a large proportion of inhibitor-sensitive, G-protein-uncoupled CCR5 forms, predominate at such sites. Note that CCR5 inhibitors can indeed inhibit the vaginal transmission of test viruses to macaques when applied topically or orally (58–61).
ACKNOWLEDGMENTS
This work was supported by an NIH grant (R01 AI41420) to J.P.M. and by an amfAR Mathilde Krim fellowship to R.B. R.A., B.T., and W.A.G. received support from the NIH (R01 NS071112) and Sanofi-Aventis.
We thank Marina Alexander for helpful discussions and Julie Strizki for supplying CCR5 inhibitors.
Footnotes
Published ahead of print 6 March 2013
REFERENCES
- 1. Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Trkola A, Dragic T, Arthos J, Binley JM, Olson WC, Allaway GP, Cheng-Mayer C, Robinson J, Maddon PJ, Moore JP. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184–187 [DOI] [PubMed] [Google Scholar]
- 3. Hill CM, Deng H, Unutmaz D, Kewalramani VN, Bastiani L, Gorny MK, Zolla-Pazner S, Littman DR. 1997. Envelope glycoproteins from human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus can use human CCR5 as a coreceptor for viral entry and make direct CD4-dependent interactions with this chemokine receptor. J. Virol. 71:6296–6304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang CC, Lam SN, Acharya P, Tang M, Xiang SH, Hussan SS, Stanfield RL, Robinson J, Sodroski J, Wilson IA, Wyatt R, Bewley CA, Kwong PD. 2007. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science 317:1930–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tsamis F, Gavrilov S, Kajumo F, Seibert C, Kuhmann S, Ketas T, Trkola A, Palani A, Clader JW, Tagat JR, McCombie S, Baroudy B, Moore JP, Sakmar TP, Dragic T. 2003. Analysis of the mechanism by which the small-molecule CCR5 antagonists SCH-351125 and SCH-350581 inhibit human immunodeficiency virus type 1 entry. J. Virol. 77:5201–5208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dragic T, Trkola A, Thompson DA, Cormier EG, Kajumo FA, Maxwell E, Lin SW, Ying W, Smith SO, Sakmar TP, Moore JP. 2000. A binding pocket for a small molecule inhibitor of HIV-1 entry within the transmembrane helices of CCR5. Proc. Natl. Acad. Sci. U. S. A. 97:5639–5644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kuhmann SE, Pugach P, Kunstman KJ, Taylor J, Stanfield RL, Snyder A, Strizki JM, Riley J, Baroudy BM, Wilson IA, Korber BT, Wolinsky SM, Moore JP. 2004. Genetic and phenotypic analyses of human immunodeficiency virus type 1 escape from a small-molecule CCR5 inhibitor. J. Virol. 78:2790–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Westby M, Smith-Burchnell C, Mori J, Lewis M, Mosley M, Stockdale M, Dorr P, Ciaramella G, Perros M. 2007. Reduced maximal inhibition in phenotypic susceptibility assays indicates that viral strains resistant to the CCR5 antagonist maraviroc utilize inhibitor-bound receptor for entry. J. Virol. 81:2359–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Berro R, Klasse PJ, Jakobsen MR, Gorry PR, Moore JP, Sanders RW. 2012. V3 determinants of HIV-1 escape from the CCR5 inhibitors maraviroc and vicriviroc. Virology 427:158–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yao XJ, Velez Ruiz G, Whorton MR, Rasmussen SG, DeVree BT, Deupi X, Sunahara RK, Kobilka B. 2009. The effect of ligand efficacy on the formation and stability of a GPCR-G protein complex. Proc. Natl. Acad. Sci. U. S. A. 106:9501–9506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ye S, Zaitseva E, Caltabiano G, Schertler GF, Sakmar TP, Deupi X, Vogel R. 2010. Tracking G-protein-coupled receptor activation using genetically encoded infrared probes. Nature 464:1386–1389 [DOI] [PubMed] [Google Scholar]
- 13. Brady AE, Limbird LE. 2002. G protein-coupled receptor interacting proteins: emerging roles in localization and signal transduction. Cell Signal. 14:297–309 [DOI] [PubMed] [Google Scholar]
- 14. Hepler JR, Gilman AG. 1992. G proteins. Trends Biochem. Sci. 17:383–387 [DOI] [PubMed] [Google Scholar]
- 15. Kaslow HR, Burns DL. 1992. Pertussis toxin and target eukaryotic cells: binding, entry, and activation. FASEB J. 6:2684–2690 [DOI] [PubMed] [Google Scholar]
- 16. Wu Y, Yoder A. 2009. Chemokine coreceptor signaling in HIV-1 infection and pathogenesis. PLoS Pathog. 5:e1000520 doi: 10.1371/journal.ppat.1000520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cocchi F, DeVico AL, Garzino-Demo A, Cara A, Gallo RC, Lusso P. 1996. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat. Med. 2:1244–1247 [DOI] [PubMed] [Google Scholar]
- 18. Farzan M, Choe H, Martin KA, Sun Y, Sidelko M, Mackay CR, Gerard NP, Sodroski J, Gerard C. 1997. HIV-1 entry and macrophage inflammatory protein-1beta-mediated signaling are independent functions of the chemokine receptor CCR5. J. Biol. Chem. 272:6854–6857 [DOI] [PubMed] [Google Scholar]
- 19. Amara A, Vidy A, Boulla G, Mollier K, Garcia-Perez J, Alcami J, Blanpain C, Parmentier M, Virelizier JL, Charneau P, Arenzana-Seisdedos F. 2003. G protein-dependent CCR5 signaling is not required for efficient infection of primary T lymphocytes and macrophages by R5 human immunodeficiency virus type 1 isolates. J. Virol. 77:2550–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yoder A, Yu D, Dong L, Iyer SR, Xu X, Kelly J, Liu J, Wang W, Vorster PJ, Agulto L, Stephany DA, Cooper JN, Marsh JW, Wu Y. 2008. HIV envelope-CXCR4 signaling activates cofilin to overcome cortical actin restriction in resting CD4 T cells. Cell 134:782–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harmon B, Ratner L. 2008. Induction of the Galpha(q) signaling cascade by the human immunodeficiency virus envelope is required for virus entry. J. Virol. 82:9191–9205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Harmon B, Campbell N, Ratner L. 2010. Role of Abl kinase and the Wave2 signaling complex in HIV-1 entry at a post-hemifusion step. PLoS Pathog. 6:e1000956 doi: 10.1371/journal.ppat.1000956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang W, Guo J, Yu D, Vorster PJ, Chen W, Wu Y. 2012. A dichotomy in cortical actin and chemotactic actin activity between human memory and naive T cells contributes to their differential susceptibility to HIV-1 infection. J. Biol. Chem. 287:35455–35469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Berro R, Klasse PJ, Lascano D, Flegler A, Nagashima KA, Sanders RW, Sakmar TP, Hope TJ, Moore JP. 2011. Multiple CCR5 conformations on the cell surface are used differentially by human immunodeficiency viruses resistant or sensitive to CCR5 inhibitors. J. Virol. 85:8227–8240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Garcia-Perez J, Rueda P, Staropoli I, Kellenberger E, Alcami J, Arenzana-Seisdedos F, Lagane B. 2011. New insights into the mechanisms whereby low molecular weight CCR5 ligands inhibit HIV-1 infection. J. Biol. Chem. 286:4978–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Alvarez Arias D, Navenot JM, Zhang WB, Broach J, Peiper SC. 2003. Constitutive activation of CCR5 and CCR2 induced by conformational changes in the conserved TXP motif in transmembrane helix 2. J. Biol. Chem. 278:36513–36521 [DOI] [PubMed] [Google Scholar]
- 27. Seibert C, Ying W, Gavrilov S, Tsamis F, Kuhmann SE, Palani A, Tagat JR, Clader JW, McCombie SW, Baroudy BM, Smith SO, Dragic T, Moore JP, Sakmar TP. 2006. Interaction of small molecule inhibitors of HIV-1 entry with CCR5. Virology 349:41–54 [DOI] [PubMed] [Google Scholar]
- 28. Berro R, Sanders RW, Lu M, Klasse PJ, Moore JP. 2009. Two HIV-1 variants resistant to small molecule CCR5 inhibitors differ in how they use CCR5 for entry. PLoS Pathog. 5:e1000548 doi: 10.1371/journal.ppat.1000548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Anastassopoulou CG, Ketas TJ, Klasse PJ, Moore JP. 2009. Resistance to CCR5 inhibitors caused by sequence changes in the fusion peptide of HIV-1 gp41. Proc. Natl. Acad. Sci. U. S. A. 106:5318–5323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Marozsan AJ, Kuhmann SE, Morgan T, Herrera C, Rivera-Troche E, Xu S, Baroudy BM, Strizki J, Moore JP. 2005. Generation and properties of a human immunodeficiency virus type 1 isolate resistant to the small molecule CCR5 inhibitor, SCH-417690 (SCH-D). Virology 338:182–199 [DOI] [PubMed] [Google Scholar]
- 31. Japour AJ, Mayers DL, Johnson VA, Kuritzkes DR, Beckett LA, Arduino JM, Lane J, Black RJ, Reichelderfer PS, D'Aquila RT. 1993. Standardized peripheral blood mononuclear cell culture assay for determination of drug susceptibilities of clinical human immunodeficiency virus type 1 isolates. Antimicrob. Agents Chemother. 37:1095–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Trkola A, Pomales AB, Yuan H, Korber B, Maddon PJ, Allaway GP, Katinger H, Barbas CF, 3rd, Burton DR, Ho DD. 1995. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J. Virol. 69:6609–6617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Decaillot FM, Kazmi MA, Lin Y, Ray-Saha S, Sakmar TP, Sachdev P. 2011. CXCR7/CXCR4 heterodimer constitutively recruits beta-arrestin to enhance cell migration. J. Biol. Chem. 286:32188–32197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pugach P, Marozsan AJ, Ketas TJ, Landes EL, Moore JP, Kuhmann SE. 2007. HIV-1 clones resistant to a small molecule CCR5 inhibitor use the inhibitor-bound form of CCR5 for entry. Virology 361:212–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moore JP, Kuritzkes DR. 2009. A piece de resistance: how HIV-1 escapes small molecule CCR5 inhibitors. Curr. Opin. HIV AIDS 4:118–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ogert RA, Wojcik L, Buontempo C, Ba L, Buontempo P, Ralston R, Strizki J, Howe JA. 2008. Mapping resistance to the CCR5 co-receptor antagonist vicriviroc using heterologous chimeric HIV-1 envelope genes reveals key determinants in the C2-V5 domain of gp120. Virology 373:387–399 [DOI] [PubMed] [Google Scholar]
- 37. Mangmool S, Kurose H. 2011. G(i/o) protein-dependent and -independent actions of pertussis toxin (PTX). Toxins (Basel) 3:884–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Alfano M, Schmidtmayerova H, Amella CA, Pushkarsky T, Bukrinsky M. 1999. The B-oligomer of pertussis toxin deactivates CC chemokine receptor 5 and blocks entry of M-tropic HIV-1 strains. J. Exp. Med. 190:597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chijiwa T, Mishima A, Hagiwara M, Sano M, Hayashi K, Inoue T, Naito K, Toshioka T, Hidaka H. 1990. Inhibition of forskolin-induced neurite outgrowth and protein phosphorylation by a newly synthesized selective inhibitor of cyclic AMP-dependent protein kinase, N-[2-(p-bromocinnamylamino)ethyl]-5-isoquinolinesulfonamide (H-89), of PC12D pheochromocytoma cells. J. Biol. Chem. 265:5267–5272 [PubMed] [Google Scholar]
- 40. Lagane B, Ballet S, Planchenault T, Balabanian K, Le Poul E, Blanpain C, Percherancier Y, Staropoli I, Vassart G, Oppermann M, Parmentier M, Bachelerie F. 2005. Mutation of the DRY motif reveals different structural requirements for the CC chemokine receptor 5-mediated signaling and receptor endocytosis. Mol. Pharmacol. 67:1966–1976 [DOI] [PubMed] [Google Scholar]
- 41. Mettling C, Desmetz C, Fiser AL, Reant B, Corbeau P, Lin YL. 2008. Galphai protein-dependent extracellular signal-regulated kinase-1/2 activation is required for HIV-1 reverse transcription. AIDS 22:1569–1576 [DOI] [PubMed] [Google Scholar]
- 42. Marozsan AJ, Torre VS, Johnson M, Ball SC, Cross JV, Templeton DJ, Quinones-Mateu ME, Offord RE, Arts EJ. 2001. Mechanisms involved in stimulation of human immunodeficiency virus type 1 replication by aminooxypentane RANTES. J. Virol. 75:8624–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tilton JC, Amrine-Madsen H, Miamidian JL, Kitrinos KM, Pfaff J, Demarest JF, Ray N, Jeffrey JL, Labranche CC, Doms RW. 2010. HIV type 1 from a patient with baseline resistance to CCR5 antagonists uses drug-bound receptor for entry. AIDS Res. Hum. Retroviruses 26:13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Abrol R, Griffith AR, Bray JK, Goddard WA., 3rd 2012. Structure prediction of G protein-coupled receptors and their ensemble of functionally important conformations. Methods Mol. Biol. 914:237–254 [DOI] [PubMed] [Google Scholar]
- 45. Labrecque J, Metz M, Lau G, Darkes MC, Wong RS, Bogucki D, Carpenter B, Chen G, Li T, Nan S, Schols D, Bridger GJ, Fricker SP, Skerlj RT. 2011. HIV-1 entry inhibition by small-molecule CCR5 antagonists: a combined molecular modeling and mutant study using a high-throughput assay. Virology 413:231–243 [DOI] [PubMed] [Google Scholar]
- 46. Kondru R, Zhang J, Ji C, Mirzadegan T, Rotstein D, Sankuratri S, Dioszegi M. 2008. Molecular interactions of CCR5 with major classes of small-molecule anti-HIV CCR5 antagonists. Mol. Pharmacol. 73:789–800 [DOI] [PubMed] [Google Scholar]
- 47. Grunbeck A, Huber T, Abrol R, Trzaskowski B, Goddard WA, 3rd, Sakmar TP. 2012. Genetically encoded photo-cross-linkers map the binding site of an allosteric drug on a G protein-coupled receptor. ACS Chem. Biol. 7:967–972 [DOI] [PubMed] [Google Scholar]
- 48. Billick E, Seibert C, Pugach P, Ketas T, Trkola A, Endres MJ, Murgolo NJ, Coates E, Reyes GR, Baroudy BM, Sakmar TP, Moore JP, Kuhmann SE. 2004. The differential sensitivity of human and rhesus macaque CCR5 to small-molecule inhibitors of human immunodeficiency virus type 1 entry is explained by a single amino acid difference and suggests a mechanism of action for these inhibitors. J. Virol. 78:4134–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Govaerts C, Blanpain C, Deupi X, Ballet S, Ballesteros JA, Wodak SJ, Vassart G, Pardo L, Parmentier M. 2001. The TXP motif in the second transmembrane helix of CCR5. A structural determinant of chemokine-induced activation. J. Biol. Chem. 276:13217–13225 [DOI] [PubMed] [Google Scholar]
- 50. Govaerts C, Bondue A, Springael JY, Olivella M, Deupi X, Le Poul E, Wodak SJ, Parmentier M, Pardo L, Blanpain C. 2003. Activation of CCR5 by chemokines involves an aromatic cluster between transmembrane helices 2 and 3. J. Biol. Chem. 278:1892–1903 [DOI] [PubMed] [Google Scholar]
- 51. de Voux A, Chan MC, Folefoc AT, Madziva MT, Flanagan CA. 2013. Constitutively active CCR5 chemokine receptors differ in mediating HIV envelope-dependent fusion. PLoS One 8:e54532 doi: 10.1371/journal.pone.0054532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gonsiorek W, Strizki JM, Hesk D, Lundell D, Hipkin RW. 2006. Analysis of CCR5 binding kinetics and coupling states reveals distinctive binding properties of small molecule CCR5 antagonists, as reported by Levin J. 46th Intersci. Conf. Antimicrob. Agents Chemother, 27 to 30 September 2006 http://www.natap.org/2006/ICAAC/ICAAC_57.htm [Google Scholar]
- 53. Tuosto L, Parolini I, Schroder S, Sargiacomo M, Lanzavecchia A, Viola A. 2001. Organization of plasma membrane functional rafts upon T cell activation. Eur. J. Immunol. 31:345–349 [DOI] [PubMed] [Google Scholar]
- 54. Cardaba CM, Kerr JS, Mueller A. 2008. CCR5 internalisation and signalling have different dependence on membrane lipid raft integrity. Cell Signal. 20:1687–1694 [DOI] [PubMed] [Google Scholar]
- 55. Zhang ZQ, Wietgrefe SW, Li Q, Shore MD, Duan L, Reilly C, Lifson JD, Haase AT. 2004. Roles of substrate availability and infection of resting and activated CD4+ T cells in transmission and acute simian immunodeficiency virus infection. Proc. Natl. Acad. Sci. U. S. A. 101:5640–5645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Parker ZF, Iyer SS, Wilen CB, Parrish NF, Chikere KC, Lee FH, Didigu CA, Berro R, Klasse PJ, Lee B, Moore JP, Shaw GM, Hahn BH, Doms RW. 2013. Transmitted/founder and chronic HIV-1 envelope proteins are distinguished by differential utilization of CCR5. J. Virol. 87:2401–2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ping L-H, Joseph SB, Anderson JA, Abrahams M-R, Salazar-Gonzalez JF, Kincer LP, Treurnicht FK, Arney L, Ojeda S, Zhang M, Keys J, Potter EL, Chu H, Moore P, Salazar-Gonzalez M, Iyer S, Jabara C, Kirchherr J, Mapanje C, Ngandu N, Seoighe C, Hoffman I, Gao F, Tang Y, Labranche C, Lee B, Saville A, Vermeulen M, Fiscus S, Morris L, Karim SA, Haynes BF, Shaw GM, Korber BT, Hahn BH, Cohen MS, Montefiori D, Williamson C, Swanstrom R. 24 April 2013. Comparison of viral Env proteins from acute and chronic infections with subtype C human immunodeficiency virus type 1 identifies differences in glycosylation and CCR5 utilization and suggests a new strategy for immunogen design. J. Virol. doi: 10.1128/JVI.03577-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Veazey RS, Springer MS, Marx PA, Dufour J, Klasse PJ, Moore JP. 2005. Protection of macaques from vaginal SHIV challenge by an orally delivered CCR5 inhibitor. Nat. Med. 11:1293–1294 [DOI] [PubMed] [Google Scholar]
- 59. Veazey RS, Ketas TJ, Dufour J, Moroney-Rasmussen T, Green LC, Klasse PJ, Moore JP. 2010. Protection of rhesus macaques from vaginal infection by vaginally delivered maraviroc, an inhibitor of HIV-1 entry via the CCR5 co-receptor. J. Infect. Dis. 202:739–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Malcolm RK, Veazey RS, Geer L, Lowry D, Fetherston SM, Murphy DJ, Boyd P, Major I, Shattock RJ, Klasse PJ, Doyle LA, Rasmussen KK, Goldman L, Ketas TJ, Moore JP. 2012. Sustained release of the CCR5 inhibitors CMPD167 and maraviroc from vaginal rings in rhesus macaques. Antimicrob. Agents Chemother. 56:2251–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Malcolm RK, Forbes CJ, Geer L, Veazey RS, Goldman L, Klasse PJ, Moore JP. 2013. Pharmacokinetics and efficacy of a vaginally administered maraviroc gel in rhesus macaques. J. Antimicrob. Chemother. 68:678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]