Abstract
A novel victorivirus, termed Rosellinia necatrix victorivirus 1 (RnVV1), was isolated from a plant pathogenic ascomycete, white root rot fungus Rosellinia necatrix, coinfected with a partitivirus. The virus was molecularly and biologically characterized using the natural and experimental hosts (chestnut blight fungus, Cryphonectria parasitica). RnVV1 was shown to have typical molecular victorivirus attributes, including a monopartite double-stranded RNA genome with two open reading frames (ORFs) encoding capsid protein (CP) and RNA-dependent RNA polymerase (RdRp), a UAAUG pentamer presumed to facilitate the coupled termination/reinitiation for translation of the two ORFs, a spherical particle structure ∼40 nm in diameter, and moderate levels of CP and RdRp sequence identity (34 to 58%) to those of members of the genus Victorivirus within the family Totiviridae. A reproducible transfection system with purified RnVV1 virions was developed for the two distinct fungal hosts. Transfection assay with purified RnVV1 virions combined with virus elimination by hyphal tipping showed that the effects of RnVV1 on the phenotype of the natural host were negligible. Interestingly, comparison of the RNA silencing-competent (standard strain EP155) and -defective (Δdcl-2) strains of C. parasitica infected with RnVV1 showed that RNA silencing acted against the virus to repress its replication, which was restored by coinfection with hypovirus or transgenic expression of an RNA silencing suppressor, hypovirus p29. Phenotypic changes were observed in the Δdcl-2 strain but not in EP155. This is the first reported study on the host range expansion of a Totiviridae member that is targeted by RNA silencing.
INTRODUCTION
The family Totiviridae comprises five genera—the Totivirus, Victorivirus, Giardiavirus, and Leishmaniavirus, and the recently established Trichomonasvirus—that are distinguishable from each other in terms of phylogeny and natural host organisms (1, 2). Commonly, the family members, which infect fungal or protozoan hosts, have a monopartite double-stranded RNA (dsRNA) genome encased in icosahedral virions of 30 to 40 nm in diameter with a T=1 icosahedral structure (1). These viruses largely show noncytopathic, persistent infections and appear to lack extracellular transmission (2, 3). Their genomes usually have two overlapping or contiguous open reading frames (ORFs) encoding a capsid protein (CP) of 74 to 99 kDa and an RNA-dependent RNA polymerase (RdRp) of 88 to 99 kDa or a CP-RdRp fusion protein of 156 to 210 kDa (4). The downstream RdRp ORF is expressed from the full-length viral transcript as a fusion product either by −1 or +1 ribosomal frameshift or as an independent nonfusion protein via termination/reinitiation mechanisms or presumably polyprotein processing, depending on genus.
Historically, studies of the prototype totivirus Saccharomyces cerevisiae virus L-A have greatly advanced our understanding of the structure, assembly, and replication of dsRNA viruses and yeast killer phenomena (5, 6). One major constraint on further progress with virological studies for the family in general has been the difficulty in developing infectivity systems such as transfection with infectious synthetic viral RNAs, complementary DNAs (cDNAs), or particles. In the 1980s, members of the family were reported to be infectious as particles, although with considerable inefficiency and difficulty, to spheroplasts of yeast and protoplasts of filamentous fungi (7–9). This situation is in contrast to that for giardiaviruses, which are exceptionally infectious as cell-free particles after virus egress and as full-length plus-sense RNA (10). However, reproducible transfection protocols with purified particles have been established for several other dsRNA viruses (see, for example, references 11, 12, and13) and also a single-stranded DNA (ssDNA) virus (14). The establishment of transfection protocols has led to better understanding of the etiology of mycovirus infection, exploration of mycovirus-host and virus-virus interactions, and the determination of experimental host ranges (15–17). These developments encouraged us to test a victorivirus from a filamentous fungus for infectivity as particles.
The chestnut blight fungus, Cryphonectria parasitica, has been established as a model filamentous ascomycete for studies of virus-host and virus-virus interactions on the basis of its several advantages over other fungi. These technical aspects include availability of reverse genetics systems for hypoviruses (family Hypoviridae) (18, 19), transfection protocols for mycoreoviruses (family Reoviridae) (12), and host genome manipulability, i.e., homologous-recombination-based gene disruption and multiple gene transformation techniques (20–23). Importantly, great advances have been made in our understanding of RNA silencing (RNAi) as an antiviral host defense mechanism in this fungus (24). Like many other filamentous ascomycetous fungi (25), C. parasitica carries two Dicer-like genes (dcl-1 and dcl-2), four Argonaute-like genes (agl-1 to agl-4), and four RdRp genes (rdr-1 to rdr-4) as possible key RNA silencing components. Among these, dcl-2 and agl-2 are required for antiviral defense (26, 27). The RNA silencing pathway is able to suppress replication of hypoviruses, mycoreoviruses (26, 27), and also a heterologous partitivirus (family Partitiviridae) (17). Furthermore, such RNA silencing is required for the generation and maintenance of defective interfering (DI)-RNAs of the prototype hypovirus, Cryphonectria hypovirus 1/EP713 (CHV1-EP713) (27, 28).
Another filamentous ascomycete Rosellinia necatrix is also an important soilborne pathogen that infects the roots of many perennial plants. Due to the lack of effective control measures, virocontrol (biological control) of the disease using mycoviruses that attenuate the fungal virulence is an attractive option (13, 29, 30). To this end, an extensive search of a large number of field isolates was conducted allowing the detection of diverse dsRNA viruses such as a bipartite dsRNA virus, Rosellinia necatrix megabirnavirus 1 (RnMBV1), a quadrivirus, Rosellinia necatrix quadrivirus 1 (RnQV1), the reovirus mycoreovirus 3 (MyRV3), and the Rosellinia necatrix partitiviruses 1 and 2 (RnPV1 and RnPV2) (13, 17, 31–35). Some of these viruses were shown to be able to confer hypovirulence under laboratory conditions. However, no victorivirus had been identified in this fungus until very recently (36), although victoriviruses are widely detectable in many filamentous fungi. In the present study, we carried out a molecular and biologic characterization of a novel victorivirus termed Rosellinia necatrix victorivirus 1-W1029 (RnVV1-W1029) that is infectious as particles to the natural host, R. necatrix, and a heterologous filamentous fungus, C. parasitica. Furthermore, we investigated RNA silencing against RnVV1 in the experimental host fungus.
MATERIALS AND METHODS
Fungal and viral strains.
Rosellinia necatrix strain W1029 was reisolated in 2009 from an experimental orchard of apple trees (Nagano Fruit Tree Experiment Station) at Suzaka, Nagano Prefecture, Japan, where virus-free W563 had been buried in 2007 (36). W1029 is believed to be one of the subisolates that has naturally acquired dsRNA segments of RnVV1 and a partitivirus termed Rosellinia necatrix partitivirus 3-W1029 (RnPV3-W1029) in the soil. The genomic segments of these viruses were referred to as S1 and S2 by Yaegashi et al. (36). The genotype and MCG (mycelial compatibility group) of W1029 were confirmed previously to be identical to the parental W563 (MCG139). Partitivirus-cured W1029 strains, i.e., W1029-T20 and W1029-T31, were obtained by hyphal tipping. These strains harbor only RnVV1. Because curing of RnVV1 was not successful, the parental W563 was used as a virus-free counterpart of W1029 or of W1029-T20 and W1029-T31. The standard virus-free R. necatrix strain W97 (MCG80) and W563 were used for transfection experiments (31). The C. parasitica standard strain EP155, a Dicer-like 2 gene-defective strain (Δdcl-2) (Segers et al. [26]), and the prototype hypovirus CHV1-EP713 (37) were generous gifts from Donald L. Nuss, and the two fungal strains were used as RnVV1 recipient host strains for transfection and hyphal fusion. A CHV1-EP713 mutant, Δp69b, was used in parallel (38). Transformants of EP155 expressing viral proteins were obtained as described bellow. All of the fungal and viral strains used in the present study are listed in Table 1. Fungal strains were grown on Difco potato dextrose agar (PDA; Becton, Dickinson and Company) plates in the dark (for R. necatrix) or on a benchtop (for C. parasitica) for phenotypic observation or in Difco potato dextrose broth (PDB; Becton, Dickinson and Company) liquid media for purification of viral particles, dsRNAs, or single-stranded RNAs (ssRNAs). For longer storage, fungi were preserved on solid regeneration media or a patch of glass wool at 4°C or −20°C, respectively.
Table 1.
Fungal and viral strains used in this study
| Strain(s) | Description | Source or reference |
|---|---|---|
| Fungal | ||
| W563 | Rosellinia necatrix field isolate (virus free) | 36 |
| W1029 | Rosellinia necatrix subisolate of W563 infected with RnVV1 and RnPV3 | 36 |
| W1029-T20, W1029-T31 | RnPV3-cured strains derived from W1029 via hyphal tipping | This study |
| W97 | Standard strain of Rosellinia necatrix (virus-free) | 31 |
| W97-Tf1-3 | W97 transfectants with RnVV1 | This study |
| EP155 | Standard strain of Cryphonectria parasitica (virus free) | ATCC 38755 |
| Δdcl-2 | dcl-2 knockout mutant of EP155 (RNA silencing defective, virus-free) | 26 |
| A12 | Δdcl-2 transfectant with RnVV1 | This study |
| A26 | Δdcl-2 transfectant with RnVV1 and RnPV3 | This study |
| A6 | Δdcl-2 transfectant with RnPV3 | This study |
| EP155/p19 | EP155 transformant with the TBSV p19 coding domain | This study |
| EP155/O-p19 | EP155 transformant with the codon-optimized TBSV p19 coding domain | This study |
| EP155/2b | EP155 transformant with the CMV 2b coding domain | This study |
| EP155/p29 | EP155 transformant with the CHV1 p29 coding domain | 16 |
| A-9 | Helminthosporium victoriae strain infected with HvV190S and HvV145S | ATCC 42018 |
| Viral | ||
| RnVV1-W1029 | Novel victorivirus (AB742454) | 36; this study |
| RnPV3-W1029 | Novel partitivirus (properties will be reported elsewhere) | 36; this study |
| CHV1-EP713 | Prototype of the family Hypoviridae | 18 |
| Δp69b (CHV1-Δp69) | ORF-A (p69; p29 + p40) deletion mutant of CHV1-EP713 | 38 |
| MyRV1 | Type species of the genus Mycoreovirus | 12, 51 |
Transformation of C. parasitica protoplasts.
Protoplasts were prepared from mycelia of C. parasitica virus-free strain EP155 (see below). Transformation of the resulting protoplasts with a vector, pCPXHY2 (39), carrying a hygromycin B phosphotransferase gene (hph) as a selectable marker, was performed as described previously (40). Four transformant lines with each of the viral suppressor of RNA silencing (VSR) genes, i.e., p19 of tomato bushy stunt virus (TBSV; a plant tombusvirus, EP155/p19), codon-optimized TBSV p19 (EP155/O-p19), 2b of cucumber mosaic virus (CMV; a plant cucumovirus) (EP155/2b), or p29 of CHV1 (EP155/p29) (16), were generated. The cDNA clones for the coding domains of CMV 2b and TBSV p19 were a generous gift from David Baulcombe. A codon-optimized version of the TBSV p19 cDNA was synthesized by Genewiz, Inc. (South Plainfield, NJ), and the sequence is available upon request.
Virus purification and electron microscopy.
Viral particles were purified as described by Lin et al. (33). Basically, homogenized mycelia of W1029 or W1029-T20 in liquid nitrogen were mixed with 4 volumes of extraction buffer (0.1 M sodium phosphate [pH 7.0] containing 0.1% [wt/vol] β-mercaptoethanol) and clarified with 20% carbon tetrachloride. After centrifugation (∼2,000 × g), 0.3 M sodium chloride and 6% (wt/vol) polyethylene glycol (PEG) 6000 solution were added to the aqueous phase and stirred for at least 1 h on ice. The resulting low-speed centrifugation (∼3,600 × g) pellets were resuspended and subjected to differential centrifugation and subsequent sucrose density gradient (20 to 50%) centrifugation at high speed (∼66,000 × g) for 2 h. Viral fractions were subjected to further high-speed centrifugation, and then the pellet was resuspended in distilled water. This preparation was used for molecular analysis, transfection, and electron microscopy. For electron microscopy observations, purified virus particles were negatively strained with 2% (wt/vol) uranyl acetate. Prepared specimens were observed in a Hitachi model H-7650 transmission electron microscope (Hitachi, Tokyo, Japan) and photographed as digital images.
Sequence and bioinformatics analyses.
Purification of the total dsRNA fraction was carried out using CC41 Sepharose (Whatman). After agarose gel electrophoresis, the RnVV1 genomic dsRNA was extracted from gel slices using an RNaid kit (MP Biomedicals, LLC, Illkrich, France). The full-length sequence of the RnVV1 genome was determined by sequencing cDNA clones obtained by non-PCR and PCR-based methods, and 3′-RACE (rapid amplification of cDNA ends) clones were used for the terminal regions as described by Chiba et al. (13). Some reverse transcription-PCR (RT-PCR) fragments were used for confirmation of unsecured portions of a contig unless otherwise mentioned. A partial sequence of the RnPV3 dsRNA1 genomic segment was obtained from cDNA clones, showing high similarity to partitivirus as documented by Yaegashi et al. (36) (GenBank accession no. AB698491). The sequences of all oligonucleotides used in the present study are available upon request. Prediction of RNA secondary structures was performed using the online programs HPKNOTTER (41) and DotKnot (42) for detecting an H-type pseudoknot near the UAAUG pentanucleotide (2,662 to 2,666 nucleotides [nt]) at the CP-RdRp junction of the RnVV1 coding strand (see Fig. 1D), and Mfold (43) was used for detection of the stem-loop hairpin downstream of the pentamer. Phylogenetic analysis was performed as described by Chiba et al. (34). An amino acid sequence spanning the conserved RdRp motifs (I to VIII) of RnVV1 (amino acid positions 244 to 582, adopted from Bruenn [44]), and related sequences from selected members of the Totiviridae were extracted and aligned using the online version of MAFFT version 6 (45). Gaps in the alignment (32.7%) were eliminated in MEGA version 4.02 (46), and an optimal substitution model for cured alignment, LG+I+G, was determined by using the Akaike information criterion as implemented in the PlotTest server (47). A maximum-likelihood tree was generated by PhyML 3.0 at the ATGC website (http://www.atgc-montpellier.fr/phyml/). Settings for other parameters were as follows: BIONJ as a starting tree, subtree pruning and regrafting for the type of tree improvement (48), and the approximate likelihood ratio test (aLRT) using a Shimodaira-Hasegawa-like procedure for branch support calculation (49). The tree obtained was visualized using FigTree software (http://tree.bio.ed.ac.uk/software/) in a midpoint-rooted form and depicted by the drawing software maintaining the relative node lengths.
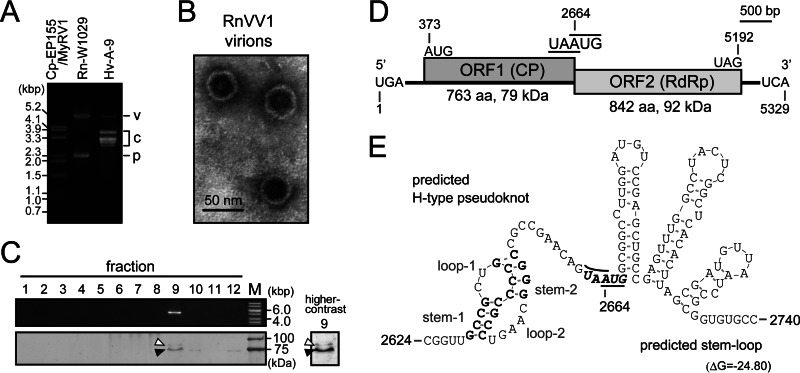
Fig 1.
Molecular properties of Rosellinia necatrix victorivirus 1 (RnVV1). (A) Electrophoretic profile of dsRNAs isolated from R. necatrix W1029. dsRNAs purified form R. necatrix W1029 (genome of a victorivirus and a partitivirus), C. parasitica EP155 infected with MyRV1-9B21 (51), and H. victoriae A-9 (genome of a victorivirus and a chrysovirus) (62) were analyzed comparatively by 1.0% agarose gel electrophoresis (78). The mobilities of dsRNAs are specified to the left of the panel, respectively. Positions of dsRNA genomes: v, victoriviruses; c, chrysovirus; p, partitivirus. (B) Micrograph of RnVV1 virions. For virus purification, partitivirus-free mycelia from strain W1029-T20 were used as starting material. The sucrose gradient fraction containing 5.2-kbp (S1) dsRNA (fraction 9 in panel C), but not 2.3-kbp (S2) dsRNA, was negatively stained with uranyl acetate and examined by transmission electron microscopy. (C) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and agarose gel electrophoresis of sucrose gradient fractions (lanes 1 to 12, top to bottom) during virus purification. Fraction 9, probably containing viral genomic dsRNA (upper panel) and structural proteins (lower panel), was visualized by ethidium bromide (EtBr) and silver staining. The 1-kb ladder and precision size standard (lane M) are shown on the right. Black and white arrowheads indicate the major and faint polypeptide bands, respectively. A high-contrast image of lane 9 is independently presented. (D) Schematic representation of the RnVV1 genome structure. The coding strand of 5,329-bp W1029-dsRNA, typical of a victorivirus genome, is shown. Large ORFs for the putative CP (ORF1) and RdRp (ORF2) overlap by a single nucleotide “A” positioned at nucleotide 2664, being included in the stop and start codons for ORF1 and 2, respectively. (E) Unique secondary RNA structures surrounding the ORF-junction region. The RNA structures were computer predicted and depicted.
Protoplast isolation and viral particle-mediated transfection.
Protoplasts isolated from the R. necatrix strains W97 and W563 and the C. parasitica EP155 and Δdcl-2 strains were prepared and transfected with purified virus particles as described previously (11, 12). The purified virus fractions were passed through Ultrafree-MC sterile centrifugal filter units (Millipore, Tokyo, Japan) and introduced into protoplasts in a PEG-calcium chloride-mediated manner. Treated cells were regenerated in or on liquid (R. necatrix) or solid (C. parasitica) regeneration medium and allowed to form a colony in a dish of 9 cm in diameter in the dark for R. necatrix and on a benchtop for C. parasitica and transferred to fresh PDA plates from multiple sites. These individuals were analyzed for RnVV1 infection by dsRNA purification.
Biological characterization.
The effect of virus infection on fungal phenotype and mycelial growth rate was observed on PDA solid medium. A plug of parental mycelia was transferred to PDA and cultured at 25°C in the dark (R. necatrix) or at 24 to 27°C on a benchtop for 1 week (C. parasitica). The resulting colonies were compared among virus-free and virus-carrying strains and photographed. The mycelial growth rate was evaluated from five replicates per strain.
Transmission of RnVV1 in a C. parasitica system via hyphal anastomosis (horizontal transmission) or asexual conidial spores (vertical transmission) was evaluated as described by Suzuki and Nuss (38). Subcultures grown on PDA plates from conidial isolates or hyphal fusants were subsequently grown on PDA-cellophane plates or in PDB, and dsRNA and ssRNA fractions were extracted and analyzed. The rate of dsRNA-positive progeny was scored as the vertical transmission rate from a total of 48 individuals. Horizontal transmission was examined by Northern analysis using ssRNA fractions.
RNA sampling and assays.
RNA samples were obtained from mycelia using the method of Chiba et al. (17), and Northern blot analysis was conducted according to the method of Suzuki et al. (50). After phenol-chloroform extraction of fungal homogenate, total RNA was obtained by ethanol precipitation, whereas ssRNA was obtained by lithium chloride precipitation. dsRNA fraction was further purified from total RNA by using a CC41 Sepharose column. For the detection of viruses and endogenous mRNAs or transgene transcripts, ca. 2- to 5-μg and 10- to 15-μg portions of the ssRNA samples were used, respectively. Northern and RNA-dot blots were prepared by using dsRNA or total RNA fractions during transfection and vertical transmission. RNAs in the agarose gels were denatured with formaldehyde (ssRNA) or sodium hydroxide (dsRNA) and transferred to nylon membranes (Hybond-N+; Amersham Bioscience). For RNA dot blotting, heat-denatured (68°C) dsRNA or total RNA samples containing formaldehyde and formamide were directly spotted onto nylon membranes. The cDNA probes for RnVV1, RnPV3, CHV1, dcl-2, and VSRs were prepared using a PCR DIG probe synthesis kit (Roche) with the specific primer sets, whose sequences are available upon request, and hybridized. The alkaline phosphatase-DIG antibody conjugate was applied to probe-hybridized membranes, and signals were detected using CDP star (Roche).
GenBank/EMBL/DDBJ accession number.
The sequence of RnVV1 reported here is deposited in the GenBank/EMBL/DDBJ data bank under accession number AB742454.
RESULTS
W1029 is infected with a novel partitivirus and a novel victorivirus.
A fungal strain W1029 was initially identified as being dsRNA positive, with the presence of two segments (S1 dsRNA and S2 dsRNA) resolved in agarose gel (36). As shown in Fig. 1A (lane Rn-W1029), W1029 carried dsRNAs that migrated at the same position (∼5.2 kbp in size) as Helminthosporium victoriae virus 190S (HvV190S) and at the same position (∼2.3 kbp in size) as mycoreovirus 1 segment 4 (MyRV1 S4) (51). The slowly migrating dsRNA (S1 dsRNA) is the main subject of the present study and was found to be the genome of a novel victorivirus termed RnVV1-W1029 as stated below. The 2.3-kbp dsRNA(s) represents the genome of a novel strain of the partitivirus (RnPV3-W1029), and details of its characterization will be reported elsewhere. We successfully obtained some cured strains of RnPV3 by hyphal tipping (see blow), and one of them (W1029-T20) was used for virion purification. A purified virus fraction (fraction 9 of the sucrose gradient) contained spherical particles (∼40 nm in diameter) (Fig. 1B) enclosing the 5.2-kbp dsRNA and comprised a major and a minor protein of ∼75 and ∼90 kDa (Fig. 1C, lane 9), probably encoded by ORF1 and ORF2 (see below), respectively. These characteristics conform to those of the genus Victorivirus in the family Totiviridae (52).
Molecular and phylogenetic properties of a novel victorivirus (RnVV1) isolated from R. necatrix strain W1029.
A cDNA library for the RnVV1 genome of the 5-kbp segment was constructed, and approximately 25 clones of >1.5 kbp were sequenced, resulting in a single long contig of ∼4.7 kbp. Its sequence displayed high sequence similarity to the members of the genus Victorivirus. Subsequent RACE analysis gave the full-length sequence of the dsRNA element (RnVV1) of 5,329 bp with two potential ORFs encoding the putative CP (2,292 nt; 763 amino acids; 79 kDa) and the putative RdRp (2,529 nt; 842 amino acids; 92 kDa), respectively (Fig. 1D). The H-type pseudoknot structure was found upstream of the UAAUG penta-nucleotide (nt 2662 to 2666) functioning as the stop codon (underlined) for the 5′-proximal ORF and as the start codon (italicized) for the 3′-proximal ORF (Fig. 1E). This pentamer is considered to be a key motif for translation of the downstream ORF by coupled translation termination and reinitiation (53, 54). Furthermore, this motif is followed by three of hairpin structures (Fig. 1E), which may contribute to the coupled translation. For the terminal sequences, a potentially formed inverted repeat between the 5′ and 3′ ends was detected (data not shown). The extreme three terminal nucleotides in particular seemed to have strict pairing.
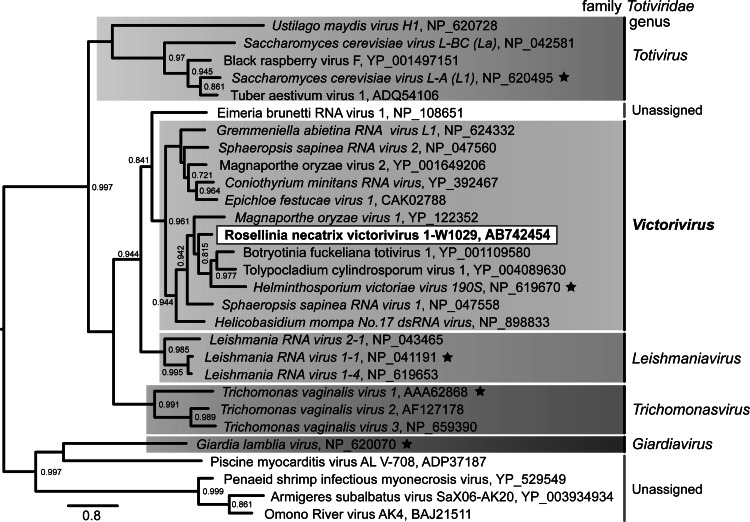
A BLAST database search (http://www.ncbi.nlm.nih.gov/BLAST/) using the RnVV1 proteins reveled moderate levels of CP and RdRp sequence identity (34 to 58%) to those of members of the genus Victorivirus within the family Totiviridae. Phylogenetic analysis based on the amino acid sequences of RdRps spanning core motifs I to VIII clearly showed that RnVV1 clustered with some selected members of the genus Victorivirus (Fig. 2). RnVV1 is closely related to the type species of the genus, HvV190S. As reported earlier, the genus Victorivirus is more closely related phylogenetically to its immediate ancestor, the genus Leishmaniavirus infecting protozoa, than the genus Totivirus, including members infecting Saccharomyces cerevisiae.
Fig 2.
Maximum-likelihood (ML) phylogenetic analysis of RnVV1. RdRp sequences spanning the core motif (I to VIII) from members of the families Totiviridae and related viruses were analyzed (see Materials and Methods). An ML tree is shown in midpoint-rooted form with the branch-supporting aLRT scores (0 to 1) on the nodes, where significant (>0.7). The clusters are differentiated by genera. Full virus names and their accession numbers are incorporated in the tree. Definitive virus members are italicized, and those marked by stars indicate the type species of the genera.
Biological properties of RnVV1 in the original and heterologous R. necatrix host strains.
To investigate the effects of the virus on its host fungus, two sets of experiments using two host backgrounds were conducted. The first set involved partial virus curing for the W1029 fungal strain. Asexual sporulation, widely used for this purpose because of its simplicity, is not applicable for cure of mycovirus infections in R. necatrix since it rarely sporulates under laboratory conditions (55). Instead, we used a hyphal tipping method and successfully obtained some RnPV3-cured strains of W1029, as demonstrated by Northern (Fig. 3A, W1029-T20 and W1029-T31) and RT-PCR analyses (data not shown). However, no virus-free strain of W1029 could be obtained. As described by Yaegashi et al. (36), W563 (virus-free), believed to be the isogenic parental strain of W1029 (see Materials and Methods), was included as a reference strain for comparative assay. We compared the colony morphologies of four strains with the same host background: W563, W1029, W1029-T20, and W1029-T31. No effects on growth rate or appearance were observed regardless of whether the host fungus was uninfected or infected with RnVV1 alone or in combination with RnPV3 (Fig. 3C and D).
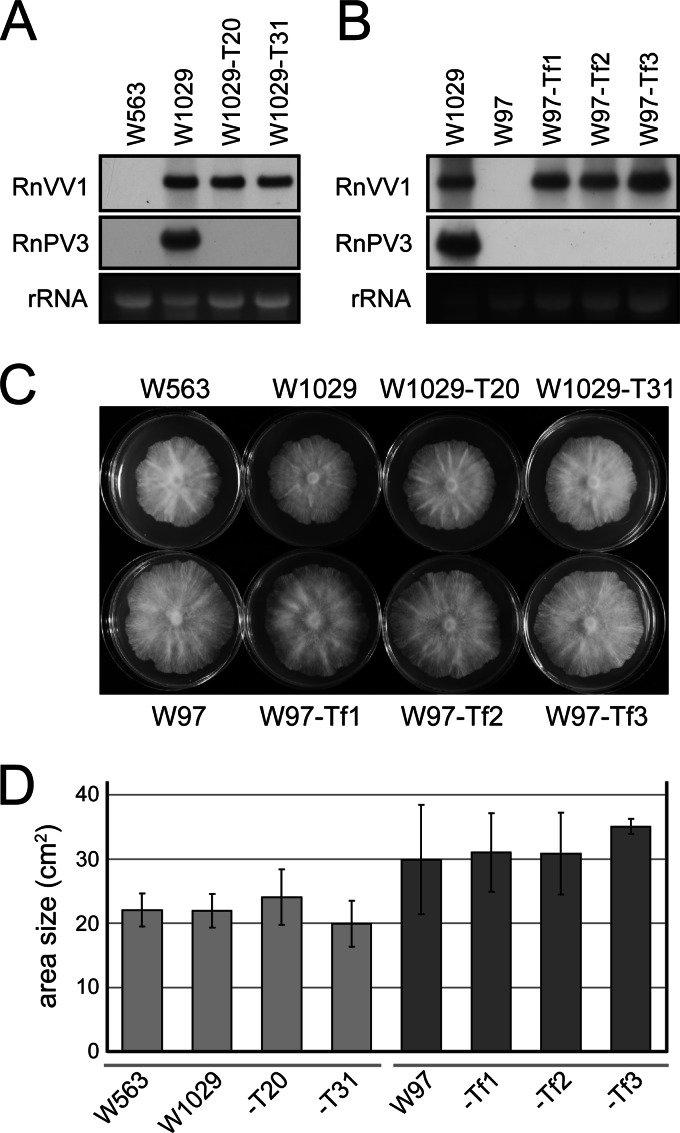
Fig 3.
Biological properties of RnVV1 in R. necatrix strains. (A) Preparation of partitivirus-cured strains. Hyphal tipping of W1029 revealed two strains that were cured for partitivirus (RnPV3) infection (W1029-T20 and W1029-T31), but no victorivirus (RnVV1)-cured strains. These strains, W1029, and W563, a parental virus-free strain of W1029, were analyzed for virus infection by Northern blotting. The cDNA probes for RnVV1 and RnPV3-dsRNA1 (36) were used. rRNA stained with EtBr is shown as a loading control (A and B). (B) Transfection of R. necatrix standard strain W97 with purified RnVV1 virions. Transfectant strains W97-Tf1, W97-Tf2, and W97-Tf3 were subjected to Northern analysis as in panel A. W1029 and virus-free W97 were included as positive and negative controls. (C) Colony morphology of RnVV1-infected R. necatrix strains. Colonies of fungal strains obtained in panels A and B were grown on PDA for 1 week in the dark and photographed. (D) Evaluation of fungal growth rate. Fungal colonies of five replicates for each strain were measured for area size (cm2), and their growth rates are presented in the graph. No obvious effect of RnVV1 infection on host phenotype or growth rate was observed (C and D).
For further confirmation (as a second set of experiments), we attempted transfection with RnVV1 particles of the well-defined standard strain W97 (virus-free) of R. necatrix that belongs to a different MCG (MCG80) than W1029 (MCG139) or W563 (MCG139) (Table 1), thus allowing no horizontal virus transfer between these fungal strains. A total of 13 subcultures were obtained from a single transfection plate (regeneration media), all of which were shown to be transfected by dsRNA gel analysis (data not shown). Three representative transfectants (W97-Tf1, W97-Tf2, and W97-Tf3) were analyzed and compared to the parental strain W97 (Fig. 3B). As shown in Fig. 3C and D, all strains displayed similar appearance and growth rates, regardless of RnVV1 infection. Likewise, strain W563 was transfected with RnVV1, resulting in no phenotypic alterations (data not shown). Therefore, based on these results, we concluded that RnVV1 infects R. necatrix asymptomatically under laboratory conditions.
Transfection of the heterologous fungus C. parasitica with RnVV1 particles.
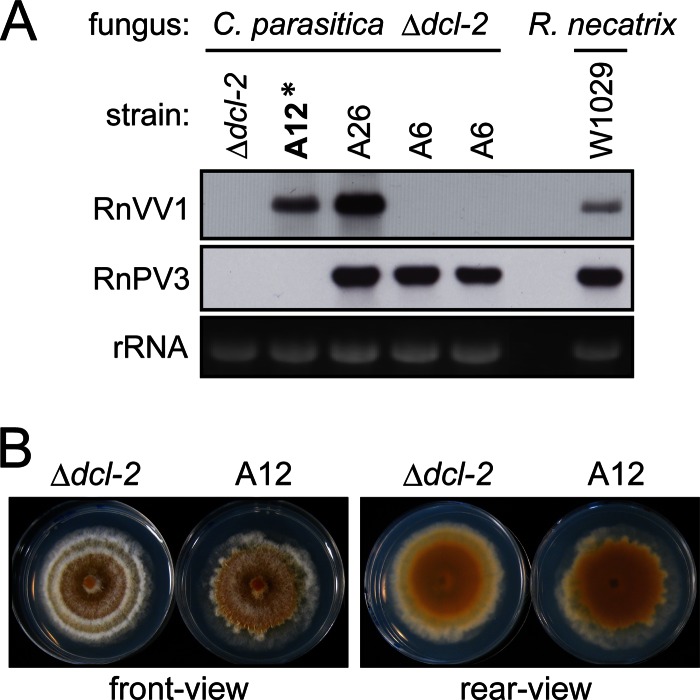
Little had been known about the host ranges of victoriviruses because of the lack of a reproducible inoculation method for them (52). The transfection protocol developed above provided an opportunity to address this long-standing question. We tested the chestnut blight fungus, C. parasitica, which is the best-studied filamentous fungal host, for its ability to support the replication of RnVV1. We first used the standard fungal strain EP155 (virus-free) to examine its susceptibility, but repeated transfection tests failed (data not shown). It was anticipated that a low transfection efficiency in EP155 might be responsible for this failure. Therefore, we switched the fungal strain to the RNA silencing-defective Δdcl-2 strain (a dicer-like gene deletion mutant of the EP155 strain) which had previously shown higher susceptibility to a heterologous partitivirus RnPV2 than the EP155 strain (17). This mutant strain was successfully transfected with the heterologous victorivirus RnVV1. Because we used virus fractions from W1029 (RnVV1 + RnPV3) as the inoculum source, three types of transfectants were obtained: Δdcl-2 strains doubly infected by RnVV1 and RnPV3 (strain A26) and singly infected by either RnVV1 (strain A12) or RnPV3 (strain A6) (Fig. 4). The transfections were reproducibly successful for the Δdcl-2 strain in repeated transfection experiments. The representative transfectant (A12) showed an altered phenotype characterized by a slightly reduced growth rate and a slightly irregular margin. An independent transfectant with RnVV1 manifested an indistinguishable phenotype (data not shown). However, the transfectants were unstable phenotypically, and their phenotype was inclined to become less pronounced after successive subculture.
Fig 4.
Transfection of an RNA silencing-deficient strain (Δdcl-2) of the heterologous experimental host C. parasitica. (A) Virus infection profile of transfectants. The ssRNA samples extracted from transfectant strains (denoted at the top) were subjected to Northern analysis using an RnVV1-specific or an RnPV3 dsRNA1-specific cDNA probe. R. necatrix W1029 was included as a control experiment. The loading control is represented by rRNA stained with EtBr. (B) Colony morphology of the C. parasitica Δdcl-2 strain infected with RnVV1. The virus-free Δdcl-2 strain and its RnVV1-infected derivative (A12 strain, asterisked in panel A) were grown on PDA for 1 week on a benchtop and photographed.
RNA silencing-competent strain EP155 of C. parasitica supports lower levels of RnVV1 replication.
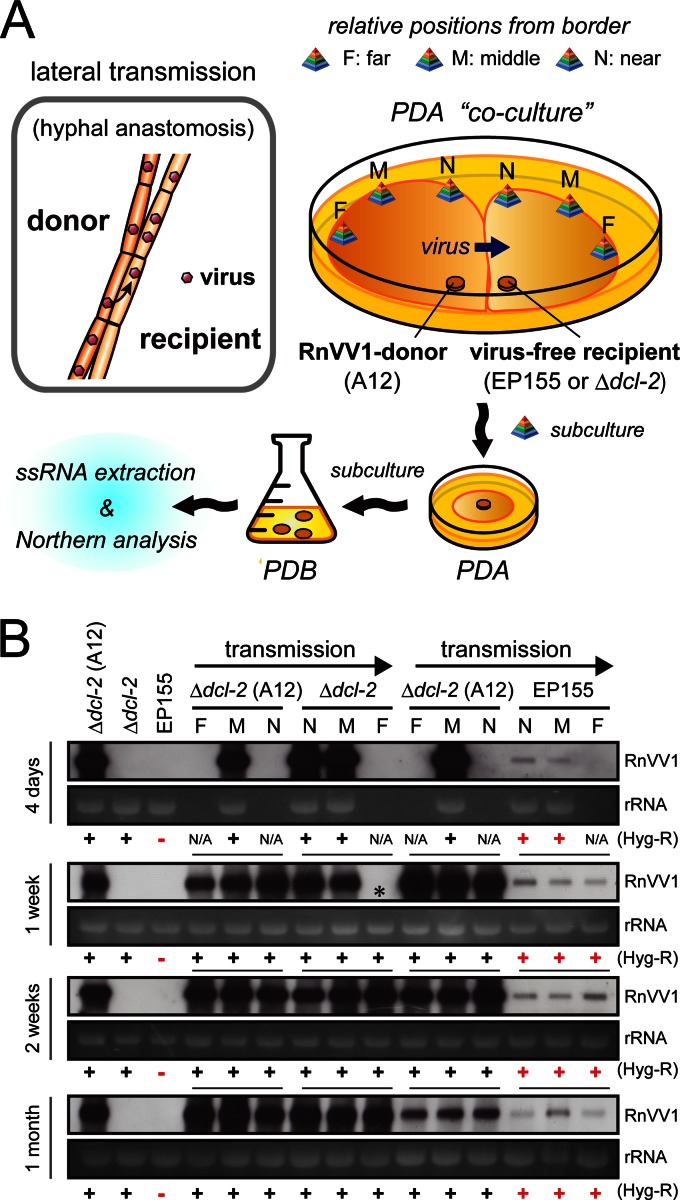
It was of interest to determine whether the failure of the EP155 wild-type fungal strain to be infected was attributable to its intrinsic antiviral defense or to an experimental artifact. To this end, horizontal transmission of RnVV1 from the donor strain A12 (Δdcl-2 background) to the virus-free EP155 strain was conducted via anastomosis. The experimental procedures used are shown schematically in Fig. 5A. Hyphal anastomosis of two independent fungal mycelia (Fig. 5A, top left box) facilitates lateral transmission of a mycovirus, although it may occasionally involve heterokaryon formation. In the present study, mycelial plugs from the virus donor strain A12 and virus-free EP155 or Δdcl-2 recipient strains were inoculated side by side on PDA plates in order to allow hyphal fusion and virus transmission (Fig. 5A, top right). After certain periods of coculture, mycelial plugs were taken from three different positions (F, far from border; M, middle; N, near border) on each of the donor and recipient sides and subcultured onto new PDA plates, followed by further cultures in liquid PDB media for RNA sample preparation in order to monitor the level of RnVV1 accumulation by Northern analysis (Fig. 5A, bottom).
Fig 5.
Lateral RnVV1 transmission into the RNA silencing-competent EP155 wild-type strain of C. parasitica. (A) Concept of horizontal virus transfer and experimental flow. Virus can be transmitted laterally from donor mycelium to the recipient during hyphal anastomosis (gray box). This event is undertaken during “coculture” on PDA where the donor and recipient mycelia are inoculated side by side. To trace the viral movement, samples from three positions on each side (F, M, and N; pyramid) were subcultured onto new PDA and subsequently inoculated in PDB media for RNA sample preparation. (B) Northern analysis of RnVV1 in the daughter strains of cocultures. Cocultures were independently conducted for 4 days, 5 days, 1 week, 2 weeks, 3 weeks, and 1 month, and the daughter strains were obtained. The presence of RnVV1 was detected using a virus-specific cDNA probe, and representative blots are presented. EtBr-stained rRNA is shown as a loading control. Arrows above the panels represent directions of virus movement. Susceptibility (−) (Hyg-S) and resistance (+) (Hyg-R) to hygromycin B of each fungal strain are indicated below the panels (Hyg-R). Red “–” and “+” symbols indicate the Hyg-S phenotype of EP155 and conversion of the phenotype of EP155-derived recipient colonies from Hyg-S to Hyg-R due to heterokaryon formation, respectively. Note that only the Δdcl-2 strain has the hygromycin B resistance gene, hph. An asterisk indicates failure of transmission. N/A denotes that samples were not obtained from the corresponding positions.
Consequently, the overexposed Northern blots showed that RnVV1 was transmitted to and accumulated in the recipient EP155 at very low levels, being faintly detectable irrespective of where the agar plugs had been taken from (N, M, and F) (Fig. 5B). Hygromycin B resistance is conferred to the Δdcl-2 strain during disruption of the dcl-2 gene (26). In order to eliminate possible effects of heterokaryon formation in recipient strains that may have occurred after anastomosis, it was necessary to determine the recipient karyotypes. EP155 virus-recipients grew on PDA containing hygromycin B (PDA-Hyg), although their growth was slower and less in extent than that of the A12 donor strain, indicating spread of the Δdcl-2 karyons to the recipient side (Fig. 5B, Hyg-R red “+”). It was noteworthy that a period of coculture as short as 4 days resulted in successful virus transmission along with heterokaryon formation, suggesting that viruses and nuclei in the donor A12 were readily transferred to the recipient side (Fig. 5B, 4 days). To confirm this phenomenon, EP155 RnVV1 recipients taken from the M position of a 4-day-old coculture (EP/RnVV1) were subjected to further transmission assay as a donor strain, because this strain showed slightly weaker resistance to the drug than the others (data not shown). After 10 days of coculture, subcultures from the recipient EP155 were obtained (EP/RnVV1/EP) and were found to be susceptible to hygromycin B, suggesting that the effect of Δdcl-2 karyons had become minimal (Fig. 6A). Northern analysis demonstrated similar levels of RnVV1 accumulation between EP/RnVV1 and EP/RnVV1/EP (Fig. 6B), strongly suggesting infectivity of RnVV1 to RNA silencing-competent C. parasitica. It should be noted that dcl-2 expression levels were also similar between these strains. Interestingly, subcultures from the donor A12 sides (N, M, and F) of an old (>1 month) coculture with the recipient EP155 showed considerably reduced RnVV1 accumulation in comparison with subcultures from equivalent coculture with the recipient Δdcl-2 strain (Fig. 5B, 1 month). This phenomenon may have been due to transfer of the EP155 karyon to the donor side as in the case of the reverse situation described above (Fig. 5B). Heterokaryon formation during horizontal virus transmission needs to be taken into consideration, particularly when two karyotypes show different susceptibility to a given virus. Overall, the data indicated that RnVV1 is able to infect EP155 by lateral transmission and, accordingly, it is unlikely that RNA silencing defines its experimental host range.
Fig 6.
Effect of karyon-movement on RnVV1 infection in C. parasitica EP155. (A and B) Effect of the presence of the Δdcl-2 karyon in the EP155 recipient on RnVV1 infection. Virus-infected EP155 recipient strains obtained from independent cocultures, namely, EP/RnVV1 (donor: A12, recipient: EP155, 4 days coculture, Fig. 5B) and EP/RnVV1/EP (donor: EP/RnVV1, recipient: EP155, 10 days coculture), were grown on hygromycin B-free (PDA panel) or hygromycin B-containing (PDA-Hyg panel) media for 4 days (A). RnVV1 infection in the fungal strains shown in panel A was analyzed by Northern blotting of ssRNA with specific cDNA probes for RnVV1 and host dcl-2 (26) (B). EtBr-stained rRNAs are shown as loading controls. (C) Lateral transmission experiment using a Δdcl-2 karyon-free EP155 as a donor strain. The EP/RnVV1/EP strain was subjected to coculture with virus-free EP155 (10 days), and the virus transmission was analyzed by Northern blotting as in panel B. The growth of each subculture on PDA-Hyg plates are presented below the panel (Hyg-R). The conversion of colony phenotype from Hyg-R to Hyg-S (A and B) and from Hyg-S to Hyg-R (C) after coculturing are highlighted by red “–” and “+” symbols, respectively.
RnVV1 is stably maintained in RNA silencing-competent strain EP155.
Although RnVV1 infects C. parasitica EP155 by lateral transmission, its accumulation levels were significantly restricted compared to those in a Δdcl-2 background. Hence, there was a possibility that RnVV1 infection in EP155 might be unstable under the pressure of RNA silencing. Therefore, we monitored the stability of RnVV1 infection during successive subcultures of EP/RnVV1/EP with an EP155 genetic background and infected with RnVV1. Northern analysis detected RnVV1 in all subcultured isolates taken from randomly selected positions of a large colony, at least until the fourth round of subculture (data not shown). Furthermore, lateral transmission of the virus from EP/RnVV1/EP to virus-free EP155 was also achieved successfully, as shown in Fig. 6C. Although these results were beyond our study focus, they clearly indicated the stability of RnVV1 in the standard strain of C. parasitica. These indicate that RNA silencing severely restricts the replication of RnVV1 but fails to eliminate the virus. However, the vertical transmission rate appeared to be extremely low, because none of the 48 conidial isolates, derived from RnVV1-infected Δdcl-2 strain, carried the virus (data not shown), which should lower the ecological fitness in the nature.
Coinfection with the prototype hypovirus restores the replication level of RnVV1.
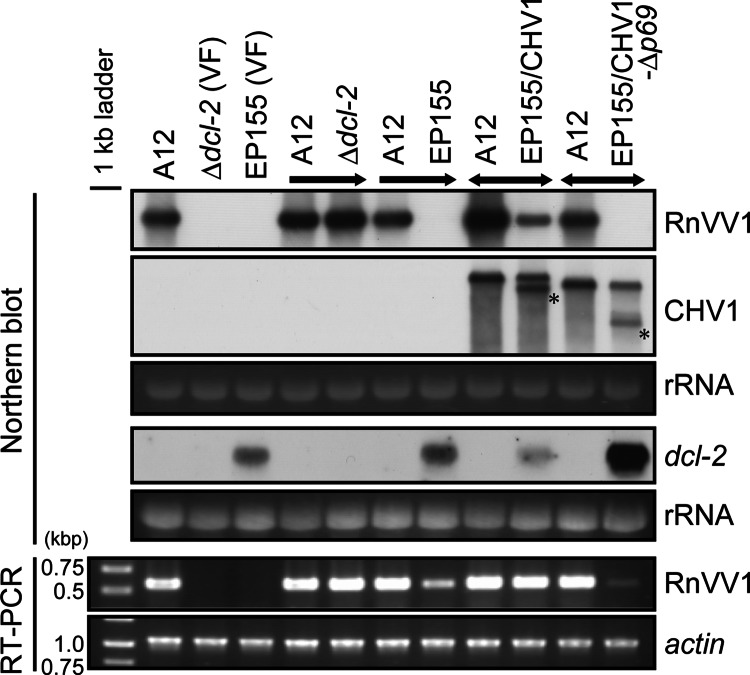
A one-way synergistic interaction between the prototypes of hypovirus and mycoreovirus (CHV1 and MyRV1) has been noted (Sun et al. [16]). The mechanism of this synergism involves elevation of the MyRV1 accumulation level by CHV1, conferred by a multifunctional hypoviral RNA silencing suppressor, p29, which was the first of its kind identified (56). Thus, it was anticipated that the very low level of RnVV1 in EP155 might be similarly elevated by coinfection with CHV1. To test this possibility, the A12 fungal strain (the Δdcl-2 strain infected by RnVV1) and EP155 infected with either of two CHV1 strains, the wild type (CHV1-EP713) or a mutant virus Δp69b (CHV1-Δp69) lacking most of the ORF A, encoding the 69-kDa polyprotein processed into p29 and p40 (57), were subjected to anastomosis. Viruses were mutually transferred between the two fungal sides, as shown schematically in Fig. 5A. Because the heterokaryon formation between EP155 and the Δdcl-2 strain had minimal impact on viral and dcl-2 mRNA accumulations as long as coculture period was <2 weeks (Fig. 5B), subcultures from 10-day-old cocultures were analyzed. Comparison of doubly and singly infected fungal colonies showed that CHV1, but not CHV1-Δp69, enhanced the in trans replication of RnVV1 in an EP155 background (Fig. 7, RnVV1). The RnVV1 replication level in virus-free or CHV1-Δp69b-infected EP155 was below the limit of detection on the Northern blot, but subsequent RT-PCR did detect virus infection (Fig. 7, RT-PCR). In contrast, RnVV1 moved into CHV1-infected EP155 and accumulated at a higher level than in the former examples, albeit at a lower level than in the Δdcl-2 strain, suggesting that CHV1 suppressed RnVV1-targeted RNA silencing. This situation is similar to that reported for coinfection of CHV1 and MyRV1 (16) and provides the second example of synergistic CHV1 infection in another mycovirus (victorivirus).
Fig 7.
Elevation of RnVV1 replication by the prototype hypovirus in EP155. A synergistic effect of CHV1 coinfection on RnVV1 replication in EP155 was observed. Virus-free Δdcl-2 or EP155 strain and CHV1- or CHV1-Δp69-infected EP155 were subjected to coculture with A12 (Δdcl-2+RnVV1), and daughter strains from each “M” position (see Fig. 5A) were obtained. The ssRNA fractions from these were recovered and analyzed by Northern blotting with cDNA probes specific for RnVV1, CHV1, and host dcl-2. Loading controls are indicated by staining of rRNA with EtBr. Failure to detect RnVV1 in the first panel (Northern blotting) was further verified by RT-PCR using a specific primer set for RnVV1 (lower panels). Control PCR was conducted for the host actin transcript. A 1-kb ladder was loaded as a size standard on the left. Arrows above the panels represent the directions of virus movement. Asterisks denote defective interfering RNAs of CHV1, which hardly moved to the Δdcl-2 host.
Nuss and coworkers demonstrated that the RNA silencing components DCL-2 and AGL-2 are required for the generation of CHV1 DI-RNA species (27, 28), and our previous study showed that the existing DI-RNA in EP155 rarely moved to the Δdcl-2 recipient side along with helper CHV1 in the lateral transmission assay (17). In the present study, we confirmed this with a virus mutant, CHV1-Δp69, during RnVV1 coinfection assay in which DI-RNAs appeared only in the EP155 host backgrounds (see the asterisked DI-RNAs in Fig. 7).
p29, the viral suppressor of RNA silencing (VSR) of CHV1, increases RnVV1 replication.
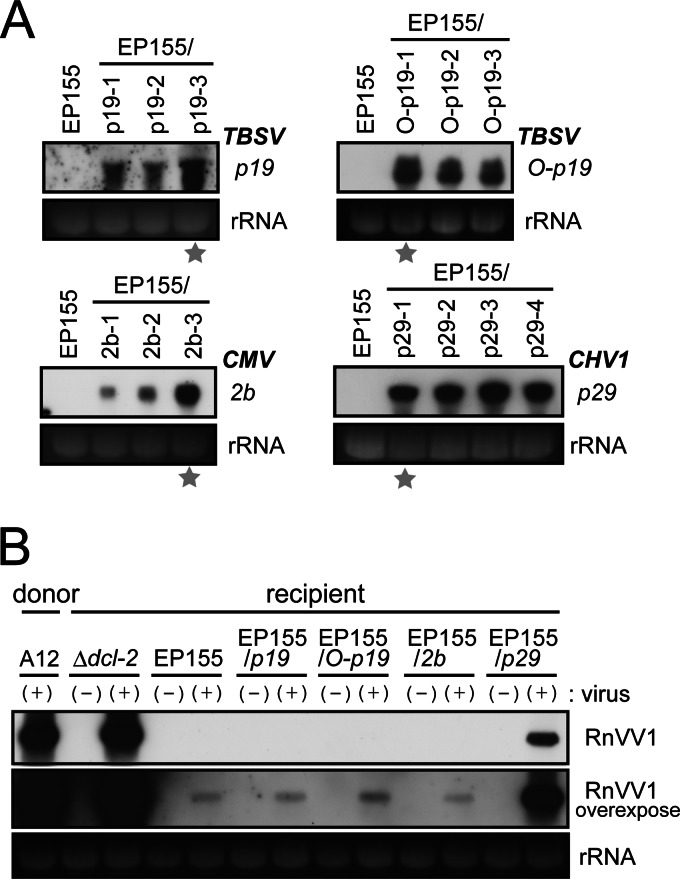
To test whether this synergism is attributable to the VSR activities of CHV1 and is reproduced by VSRs from plant viruses, we analyzed the levels of RnVV1 accumulation in EP155 strains transgenically expressing known VSRs. For this purpose, four transformants of EP155 with each of the coding domains for TBSV p19 (EP155/p19), codon-optimized p19 (EP155/O-p19), CMV 2b (EP155/2b), and CHV1 p29 (EP155/p29) were prepared. Homokaryotic isolates for each transformant strain were obtained by single conidial isolation (Fig. 8A), and representatives were subjected to coculture with strain A12 for 1 week. Reduced pigmentation in EP155/p29 was described previously (16, 39), whereas no phenotypic changes were observed in other transformants (data not shown). Subcultures from both donor and recipient sides were obtained, and the levels of RnVV1 accumulation were analyzed by Northern blotting. None of the plant viral VSRs enhanced RnVV1 replication (EP155/p19, EP155/O-p19, and E155P/2b) (data not shown), whereas CHV1 p29 increased the level of RnVV1 accumulation (Fig. 8B). These assays clearly revealed that although RnVV1 replication is impaired by host RNA silencing, it can be restored by disruption or suppression of this antiviral defense mechanism.
Fig 8.
Effects of VSR expression on the level of RnVV1 replication. (A) Preparation of EP155 transformants expressing VSRs. Protoplasts of EP155 were transformed with each of the coding domains of VSRs, CHV1 p29 (EP155/p29), CMV 2b (EP155/2b), TBSV p19 (EP155/p19), and its codon-optimized version (EP155/O-p19). For each transformation, several homokaryotic transformants were obtained by single conidial isolation, and three or four selected transformants for each VSR were analyzed for transgene expression by Northern blotting with specific cDNA probes for VSR sequences. Strains marked by stars, considered to express transgenes abundantly, were used for subsequent analysis, as shown in panel B. (B) Evaluation of RnVV1 replication levels in VSR-expressing fungal strains. RnVV1 was introduced into the transformants by coculture with A12. The ssRNA fractions, obtained from virus-free (−) and virus-carrying (+) transformant colonies, were analyzed by Northern blotting using an RnVV1-specific cDNA probe. EP155, Δdcl-2, and A12 strains (see Table 1) were analyzed in parallel. EtBr-stained rRNAs were used as loading controls.
RnVV1 infection dose not induce host dcl-2 expression.
It is known that CHV1 interferes with the host RNA silencing associated with the suppression of dcl-2 and agl-2 expression, possibly through the action of p29 of ORF A (26, 27). We compared the levels of dcl-2 expression among EP155 colonies infected with different virus strains. RnVV1 failed to induce dcl-2 expression in EP155 and showed levels similar to that in virus-free EP155 (Fig. 6B and 7, dcl-2). In the fungal strain doubly infected with wild-type CHV1 and RnVV1, slightly reduced expression was observed relative to RnVV1-infected EP155. In contrast, coinfection of EP155 with CHV1-Δp69 and RnVV1 resulted in a significantly elevated level of dcl-2 mRNA (Fig. 7, dcl-2), as reported for EP155 infected with CHV1-Δp29 (27). Interestingly, RT-PCR showed that the level of RnVV1 accumulation in EP155 coinfected with CHV1-Δp69 was lower than that in singly infected EP155, suggesting a trend opposite to that of dcl-2 mRNA (Fig. 7). This causal link warrants further careful exploration. These combined results suggest that RnVV1 infection alone has no significant effect on dcl-2 levels.
DISCUSSION
Although a variety of viruses have been detected in R. necatrix (58, 59), RnVV1 is the first Victorivirus member to have been identified in this fungus, emerging as a good host system for virus-host and virus-virus studies (29). RnVV1 is phylogenetically placed in a group comprising other victoriviruses (Fig. 2) and has features typical of a victorivirus: a monopartite genome of 5,329 bp with two contiguous ORFs (Fig. 1D) and an icosahedral particle structure ∼40 nm in diameter (Fig. 1B). Like other members of the genus Victorivirus, RnVV1 likely expresses RdRp as an independent protein product probably via the termination-reinitiation mechanism. As shown by Li et al. (54), many victoriviruses have a tetranucleotide (AUGA) in which the first triplet is considered to serve as the start codon of the downstream RdRp ORF and the last triplet serves as the stop codon of the upstream CP ORF. The same authors demonstrated that the 32-nt sequence immediately upstream of the tetramer that potentially forms a pseudoknot structure is important for the reinitiation translation of HvV190S RdRp (54). Although the positive strand of the RnVV1 genome possesses the pentamer (UAAUG) separating the two ORFs, in place of the tetramer, it has the stem I and stem II (Fig. 1E) that are well conserved in victoriviruses (54). Furthermore, three stem-loop structures can be formed immediately downstream of the pentamer, which may also be important for the coupled translation. Of note is that the pentamer (UAAUG) and adjacent sequences of CHV1 have been reported to serve as a facilitator of coupled termination/reinitiation (53) in fungal cells as in the case for influenza B virus segment 7 (60) and the ZART1 retrotransposon (61).
Hillman et al. (12) developed a reproducible transfection protocol for a C. parasitica-infecting mycoreovirus, MyRV1, within the family Reoviridae. Subsequently, similar techniques were applied for several other mycoviruses belonging to the same and different families such as Reoviridae (15), Partitiviridae (11), and Megabirnaviridae (13) and even for a gemini-like ssDNA virus (14). The present study investigated the transfection of two different fungal hosts, the natural one and an experimental one, with the novel victorivirus RnVV1 belonging to the genus Victorivirus of the family Totiviridae, for which no reproducible, efficient infectivity system has been available hitherto. The protocol developed for RnVV1 should contribute to studies of the effects of other victoriviruses in filamentous fungi that are often coinfected with some other viruses (52). Examples of mixed infections include R. necatrix strains W1028 and W1030 (36). In addition, H. victoriae strain A-9 is infected with the prototype victorivirus HvV190S and the chrysovirus Helminthosporium victoriae virus 145S (HvV145S), which has been highlighted as an agent associated with the debilitation phenotype of a plant-pathogenic fungus (3, 4). The role of the individual viruses in disease development is currently being investigated (S. A. Ghabrial, personal communication). It is generally difficult to determine viral etiology in mixed infections. Here, we were able to establish that RnVV1, coinfecting W1029 with RnPV3, infects the natural host asymptomatically using a combined approach involving selective virus elimination (RnPV3) and virus reintroduction (RnVV1) (Fig. 3).
Little is known about mycoviral host ranges. However, experiments using infectious cDNA clones or synthetic transcripts have shown that the prototype hypovirus replicates in fungal species of the same family (63) or a different one within the same order as that of its natural host, the chestnut blight fungus C. parasitica (64). The virion transfection protocol has also provided chance to expand the host range of mycoviruses, such as RnPV1 and MyRV3 (65). In the present study, the protocol has been applied to RnVV1, the first victorivirus whose host range has been expanded experimentally. As in the case for the partitiviruses (RnPV1 and RnPV2) from R. necatrix (17, 65), C. parasitica, belonging to an order Diaporthales different from the Xylariales accommodating R. necatrix, was shown by transfection assay to support the replication of RnVV1. We are now attempting to determine whether RnVV1 can infect organisms differing at taxon levels higher than class.
C. parasitica can naturally host a variety of viruses belonging to five families, including a chrysovirus recently found in a closely related fungus, C. nitschkei (66). However, no victoriviruses have been reported to naturally infect C. parasitica (67). RnVV1 has been added to an array of viruses that are able to replicate in this versatile fungus. This has helped to establish the standard EP155 strain of C. parasitica as a model filamentous fungal host. Viruses of C. parasitica were originally detected from different field fungal isolates and are most likely able to infect the standard C. parasitica strain EP155 by a combination of protofusion of different C. parasitica strains and transfection by infectious synthetic RNA, cloned viral cDNAs, and purified virions. This situation is in stark contrast to Neurospora crassa, a model filamentous fungus for genetics and molecular biology, for which no replicating virus is reported (68). It is interesting that some distinct viruses induce similar phenotypic alterations in EP155 or its RNA silencing-defective mutants, as exemplified by RnVV1, RnPV2, and RnMBV1 (data not shown). In addition, a recent report showed different symptom induction profiles caused by different CHV1 strains in EP155 and ∆dcl-2 (69). These may be good candidates for omics analyses aimed at identification of host components involved in symptom induction. The publicly available draft sequence of the EP155 genome (http://genome.jgi-psf.org/Crypa2/Crypa2.home.html) and an efficient homologous recombination-based gene disruption system (22) will definitely accelerate such studies.
Previously, we showed that a partitivirus, RnPV2, from R. necatrix carried a DI-RNA that was eliminated using our transfection protocol and obtained two variants of C. parasitica Δdcl-2 transfected with either DI-RNA-carrying or DI-RNA-lacking virus, RnPV2-DI(+) or RnPV2-DI(−) (17). We observed that the viral replication and symptom severity of both viruses were considerably enhanced in the Δdcl-2 strain relative to those in EP155 and that the DI-RNA strongly reduced viral replication and alleviated the symptoms. The phenotypic alteration and replication patterns exhibited in C. parasitica infected with RnVV1 are similar to those of RnPV2-DI(+)-infected C. parasitica (Fig. 4 and 5). As shown in Fig. 4, RnVV1 induced mild symptoms in the Δdcl-2 strain, altered the colony morphology of the Δdcl-2 strain, similarly to the changes induced in the Δdcl-2 strain by RnPV2-DI(+), a less replicative and less pathogenic variant. However, as in the natural host fungus (Fig. 3), RnVV1 exerted no overt phenotypic effects in EP155 (data not shown), similarly to RnPV2-DI(+) but unlike RnPV2-DI(−).
Drinnenberg et al. (70) reported that totiviruses with killer satellite RNAs can be retained only in RNA silencing-defective yeasts that have lost their key component genes during the course of evolution and that no totiviruses are detectable in RNA silencing-competent yeasts across major fungal groups such as Ascomycota, Basidiomycota, and Zygomycota. This fact, and the low levels of RnVV1 replication in EP155, suggested that RnVV1 would be eliminated during repeated subculture. This possibility is supported by previous reports indicating that even in natural hosts, some mycoviruses are distributed unevenly and are easily lost during subculture (15, 71). However, RnVV1 was readily transmitted via anastomosis (Fig. 5 to 8), was stably retained after repeated subculture, and was even detectable in samples taken from any location within an old EP155 colony (data not shown). Furthermore, victoriviruses are detectable in a wide variety of fungi that are known, or considered, to be RNA silencing competent (25). Given these considerations, victoriviruses may have evolved a way of coping with RNA silencing pressure to ensure their long-term survival.
RNA silencing in filamentous fungi as an antiviral host defense and viral counterattack mechanism is an increasingly important research area, as is the case in plant/virus and invertebrate animal/virus systems. However, only a few examples of fungal RNA silencing as an antiviral defense mechanism are known. Although R. necatrix is more amenable to virus research than many other filamentous fungi, C. parasitica is much more suitable as a virus host in this sense because of the availability of mutant host strains with defects in RNA silencing. In C. parasitica, it is known that one each of the two or four homologues of Dicer (dcl-2) and Argonaute (agl-2) are required for host defense and that a multifunctional hypoviral protein, p29, suppresses it (26, 27). Thus far, RNA silencing has been shown to target hypoviruses, a mycoreovirus, and a partitivirus (17, 26, 69). It is evident that RnVV1 is targeted by RNA silencing in the C. parasitica standard strain EP155. This has been substantiated by a few observations: replication levels of RnVV1 are enhanced in an RNA-silencing mutant, Δdcl-2, relative to the silencing-competent strain EP155 (Fig. 5 and 6), and RnVV1 levels in EP155 are elevated by coinfection by the wild-type CHV1 but not by its mutant CHV1-Δp69 lacking the RNA-silencing suppressor (VSR), p29, and by transgenic expression of p29 (Fig. 7). These findings suggest that RnVV1 may have very weak suppressor activity against RNA silencing in the heterologous host, or as hypothesized by Himeno et al. (72) for a victorivirus (Magnaporthe oryzae virus 2) from Magnaporthe oryzae, it may be able to escape, to some extent, from host RNA silencing. The latter possibility is supported by minimal effects of RnVV1 infection on dcl-2 mRNA accumulation (Fig. 7) which is induced by the p29-defective CHV1 strain, MyRV1 (26), and dsRNA (73). Generally, the replication levels of viruses with potent suppressors are not augmented drastically by coinfecting viruses or by suppressors from different viruses. However, viruses with deficient or defective VSRs can replicate in RNA silencing-defective host organisms to levels that are comparable to those of viruses possessing fully competent VSRs. RnVV1 apparently behaves like the latter.
CHV1 p29 is the only example of a VSR derived from mycoviruses and working in fungi. When supplied transgenically, CHV1 p29 enhances in trans replication of homologous virus mutants lacking p29 (50) and heterologous viruses (16) in their natural host fungus, C. parasitica. Although it remains unclear how CHV1 p29 acts as a VSR, it has been suggested previously that its activity suppresses the transcription levels of host dcl-2 and agl-2 genes (26, 27). This suggestion is based on the observation that infection by Δp29 mutant virus results in highly elevated accumulation of dcl-2 mRNA relative to the wild-type CHV1. This notion was confirmed using different CHV1 strains that induce distinct symptoms (68). Comparison among EP155 colonies infected by RnVV1 alone or together with CHV1 or CHV1-Δp69 showed a reverse correlation between the level of RnVV1 and dcl-2 expression (see Fig. 7, Northern blot for dcl-2 and RT-PCR for RnVV1). Some VSRs from plant and animal viruses, including TBSV p19, have been known to be functional across their host kingdoms (74–76). This fact allowed us to assume that at least p19, which targets small interfering RNA to inhibit RNA silencing (77), could be functional in C. parasitica. However, none of plant viral VSRs tested in the present study enhanced RnVV1 replication in EP155 (Fig. 8). Further investigation is needed to explain their failure in elevation of the heterologous virus replication in this fungus, C. parasitica.
In summary, we have established a transfection protocol using purified particles for a novel victorivirus, RnVV1, that originally coinfected a field strain, W1029, of the phytopathogenic fungus, R. necatrix with a partitivirus, RnPV3. This allowed us to carry out a thorough characterization of the virus, expansion of its experimental host range, and targeting of the RNA silencing that operates against it.
ACKNOWLEDGMENTS
This study was supported in part by Yomogi, Inc. (to N.S.), the Naito Foundation, a Grant-in-Aid for Scientific Research from the Japanese Ministry of Education, Culture, Sports, Science, and Technology (KAKENHI to N.S.), and the Program for Promotion of Basic and Applied Researches for Innovations in Bio-Oriented Industries (to H.K. and S.K.).
We are grateful to Donald L. Nuss and Said A. Ghabrial for the generous gift of the fungal EP155, Δdcl-2, and A-9 strains. We thank David Baulcombe for the generous gifts of plasmid clones pBin-p19 and pBin-2b. We also thank Said A. Ghabrial and Ana Eusebio-Cope for fruitful discussions and critical readings of the manuscript. Technical support provided by Natsuko Nakayama and Kazuyuki Maruyama during this study is appreciated.
Footnotes
Published ahead of print 3 April 2013
REFERENCES
- 1. Wickner RB, Ghabrial SA, Nibert ML, Patterson JL, Wang CC. 2011. Family Totiviridae, p 639–650 In King AMQ, Adams MJ, Carstens EB, Lefkowits EJ. (ed), Virus taxonomy. Ninth report of the International Committee for the Taxonomy of Viruses Elsevier/Academic Press, Inc., New York, NY [Google Scholar]
- 2. Goodman RP, Ghabrial SA, Fichorova RN, Nibert ML. 2011. Trichomonasvirus: a new genus of protozoan viruses in the family Totiviridae. Arch. Virol. 156:171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ghabrial S, Suzuki N. 2009. Viruses of plant pathogenic fungi. Annu. Rev. Phytopathol. 47:353–384 [DOI] [PubMed] [Google Scholar]
- 4. Ghabrial S. 2008. Totiviruses, p 163–174 In Mahy BWJ, Van Regenmortel MHV. (ed), Encyclopedia of virology, 3rd ed, vol 5 Elsevier, Oxford, England [Google Scholar]
- 5. Wickner RB. 1996. Double-stranded RNA viruses of Saccharomyces cerevisiae. Microbiol. Rev. 60:250–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wickner RB, Ribas JC, Searfos A. 2002. The double-stranded RNA viruses of Saccharomyces cerevisiae, p 67–108 In Tavantzis S. (ed), Molecular biology of double-stranded RNA: concepts and applications in agriculture, forestry, and medicine. CRC Press, Boca Raton, FL [Google Scholar]
- 7. Ghabrial SA. 1986. Transmissible disease of Helminthosporium victoriae: evidence for a viral etiology, p 163–176 In Buck KW. (ed), Fungal virology. CRC Press, Boca Raton, FL [Google Scholar]
- 8. el-Sherbeini M, Bostian KA. 1987. Viruses in fungi: infection of yeast with the K1 and K2 killer viruses. Proc. Natl. Acad. Sci. U. S. A. 84:4293–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stanway CA, Buck KW. 1984. Infection of protoplasts of the wheat take-all fungus, Gaeumannomyces graminis var. tritici, with double-stranded-RNA viruses J. Gen. Virol. 65:2061–2065 [Google Scholar]
- 10. Furfine ES, Wang CC. 1990. Transfection of the Giardia lamblia double-stranded RNA virus into Giardia lamblia by electroporation of a single-stranded RNA copy of the viral genome. Mol. Cell. Biol. 10:3659–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sasaki A, Kanematsu S, Onoue M, Oyama Y, Yoshida K. 2006. Infection of Rosellinia necatrix with purified viral particles of a member of Partitiviridae (RnPV1-W8). Arch. Virol. 151:697–707 [DOI] [PubMed] [Google Scholar]
- 12. Hillman BI, Supyani S, Kondo H, Suzuki N. 2004. A reovirus of the fungus Cryphonectria parasitica that is infectious as particles and related to the Coltivirus genus of animal pathogens. J. Virol. 78:892–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chiba S, Salaipeth L, Lin YH, Sasaki A, Kanematsu S, Suzuki N. 2009. A novel bipartite double-stranded RNA mycovirus from the white root rot fungus Rosellinia necatrix: molecular and biological characterization, taxonomic considerations, and potential for biological control. J. Virol. 83:12801–12812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yu X, Li B, Fu Y, Jiang D, Ghabrial SA, Li G, Peng Y, Xie J, Cheng J, Huang J, Yi X. 2010. A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc. Natl. Acad. Sci. U. S. A. 107:8387–8392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sasaki A, Kanematsu S, Onoue M, Oikawa Y, Nakamura H, Yoshida K. 2007. Artificial infection of Rosellinia necatrix with purified viral particles of a member of the genus Mycoreovirus reveals its uneven distribution in single colonies. Phytopathology 97:278–286 [DOI] [PubMed] [Google Scholar]
- 16. Sun L, Nuss DL, Suzuki N. 2006. Synergism between a mycoreovirus and a hypovirus mediated by the papain-like protease p29 of the prototypic hypovirus CHV1-EP713. J. Gen. Virol. 87:3703–3714 [DOI] [PubMed] [Google Scholar]
- 17. Chiba S, Lin YH, Kondo H, Kanematsu S, Suzuki N. 2013. Effects of defective-interfering RNA on symptom induction by, and replication of a novel partitivirus from a phytopathogenic fungus Rosellinia necatrix. J. Virol. 87:2330–2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Choi GH, Nuss DL. 1992. Hypovirulence of chestnut blight fungus conferred by an infectious viral cDNA. Science 257:800–803 [DOI] [PubMed] [Google Scholar]
- 19. Chen BS, Choi GH, Nuss DL. 1994. Attenuation of fungal virulence by synthetic infectious hypovirus transcripts. Science 264:1762–1764 [DOI] [PubMed] [Google Scholar]
- 20. Gao S, Nuss DL. 1996. Distinct roles for two G protein alpha subunits in fungal virulence, morphology, and reproduction revealed by targeted gene disruption. Proc. Natl. Acad. Sci. U. S. A. 93:14122–14127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kasahara S, Nuss DL. 1997. Targeted disruption of a fungal G-protein beta subunit gene results in increased vegetative growth but reduced virulence. Mol. Plant-Microbe Interact. 10:984–993 [DOI] [PubMed] [Google Scholar]
- 22. Lan X, Yao Z, Zhou Y, Shang J, Lin H, Nuss DL, Chen B. 2008. Deletion of the cpku80 gene in the chestnut blight fungus, Cryphonectria parasitica, enhances gene disruption efficiency. Curr. Genet. 53:59–66 [DOI] [PubMed] [Google Scholar]
- 23. Choi GH, Dawe AL, Churbanov A, Smith ML, Milgroom MG, Nuss DL. 2012. Molecular characterization of vegetative incompatibility genes that restrict hypovirus transmission in the chestnut blight fungus Cryphonectria parasitica. Genetics 190:113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nuss D. L. Mycoviruses, RNA silencing, and viral RNA recombination. Adv. Virus Res. 80:25–48, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nakayashiki H, Nguyen QB. 2008. RNA interference: roles in fungal biology. Curr. Opin. Microbiol. 11:494–502 [DOI] [PubMed] [Google Scholar]
- 26. Segers GC, Zhang X, Deng F, Sun Q, Nuss DL. 2007. Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. Proc. Natl. Acad. Sci. U. S. A. 104:12902–12906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sun Q, Choi GH, Nuss DL. 2009. A single Argonaute gene is required for induction of RNA silencing antiviral defense and promotes viral RNA recombination. Proc. Natl. Acad. Sci. U. S. A. 106:17927–17932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang X, Nuss DL. 2008. A host dicer is required for defective viral RNA production and recombinant virus vector RNA instability for a positive sense RNA virus. Proc. Natl. Acad. Sci. U. S. A. 105:16749–16754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kondo H, Kanematsu S, Suzuki N. Viruses of the white root rot fungus, Rosellinia necatrix. Adv. Virus Res. 86:177–214 [DOI] [PubMed] [Google Scholar]
- 30. Matsumoto N. 1998. Biological control of root diseases with dsRNA based on population structure of pathogens. JARQ 32:31–35 [Google Scholar]
- 31. Kanematsu S, Arakawa M, Oikawa Y, Onoue M, Osaki H, Nakamura H, Ikeda K, Kuga-Uetake Y, Nitta H, Sasaki A, Suzaki K, Yoshida K, Matsumoto N. 2004. A reovirus causes hypovirulence of Rosellinia necatrix. Phytopathology 94:561–568 [DOI] [PubMed] [Google Scholar]
- 32. Sasaki A, Miyanishi M, Ozaki K, Onoue M, Yoshida K. 2005. Molecular characterization of a partitivirus from the plant pathogenic ascomycete Rosellinia necatrix. Arch. Virol. 150:1069–1083 [DOI] [PubMed] [Google Scholar]
- 33. Lin YH, Chiba S, Tani A, Kondo H, Sasaki A, Kanematsu S, Suzuki N. 2012. A novel quadripartite dsRNA virus isolated from a phytopathogenic filamentous fungus, Rosellinia necatrix. Virology 426:42–50 [DOI] [PubMed] [Google Scholar]
- 34. Chiba S, Kondo H, Tani A, Saisho D, Sakamoto W, Kanematsu S, Suzuki N. 2011. Widespread endogenization of genome sequences of non-retroviral RNA viruses into plant genomes. PLoS Pathog. 7:e1002146 doi: 10.1371/journal.ppat.1002146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lin YH, Hisano S, Yaegashi H, Kanematsu S, Suzuki N. A second quadrivirus strain from the phytopathogenic filamentous fungus Rosellinia necatrix. Arch. Virol. [Epub ahead of print.] doi: 10.1007/s00705-012-1580-8 [DOI] [PubMed] [Google Scholar]
- 36. Yaegashi H, Nakamura H, Sawahata T, Sasaki A, Iwanami Y, Ito T, Kanematsu S. 2013. Appearance of mycovirus-like double-stranded RNAs in the white root rot fungus, Rosellinia necatrix, in an apple orchard. FEMS Microbiol. Ecol. 83:49–62 [DOI] [PubMed] [Google Scholar]
- 37. Shapira R, Choi GH, Nuss DL. 1991. Virus-like genetic organization and expression strategy for a double-stranded RNA genetic element associated with biological control of chestnut blight. EMBO J. 10:731–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Suzuki N, Nuss DL. 2002. Contribution of protein p40 to hypovirus-mediated modulation of fungal host phenotype and viral RNA accumulation. J. Virol. 76:7747–7759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Craven MG, Pawlyk DM, Choi GH, Nuss DL. 1993. Papain-like protease p29 as a symptom determinant encoded by a hypovirulence-associated virus of the chestnut blight fungus. J. Virol. 67:6513–6521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Faruk MI, Eusebio-Cope A, Suzuki N. 2008. A host factor involved in hypovirus symptom expression in the chestnut blight fungus, Cryphonectria parasitica. J. Virol. 82:740–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huang CH, Lu CL, Chiu HT. 2005. A heuristic approach for detecting RNA H-type pseudoknots. Bioinformatics 21:3501–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sperschneider J, Datta A. 2010. DotKnot: pseudoknot prediction using the probability dot plot under a refined energy model. Nucleic Acids Res. 38:e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bruenn JA. 1993. A closely related group of RNA-dependent RNA polymerases from double-stranded RNA viruses. Nucleic Acids Res. 21:5667–5669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Katoh K, Toh H. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 9:286–298 [DOI] [PubMed] [Google Scholar]
- 46. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 47. Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105 [DOI] [PubMed] [Google Scholar]
- 48. Hordijk W, Gascuel O. 2005. Improving the efficiency of SPR moves in phylogenetic tree search methods based on maximum likelihood. Bioinformatics 21:4338–4347 [DOI] [PubMed] [Google Scholar]
- 49. Anisimova M, Gascuel O. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55:539–552 [DOI] [PubMed] [Google Scholar]
- 50. Suzuki N, Maruyama K, Moriyama M, Nuss DL. 2003. Hypovirus papain-like protease p29 functions in trans to enhance viral double-stranded RNA accumulation and vertical transmission. J. Virol. 77:11697–11707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Suzuki N, Supyani S, Maruyama K, Hillman BI. 2004. Complete genome sequence of mycoreovirus-1/Cp9B21, a member of a novel genus within the family Reoviridae, isolated from the chestnut blight fungus Cryphonectria parasitica. J. Gen. Virol. 85:3437–3448 [DOI] [PubMed] [Google Scholar]
- 52. Ghabrial SA, Nibert ML. 2009. Victorivirus, a new genus of fungal viruses in the family Totiviridae. Arch. Virol. 154:373–379 [DOI] [PubMed] [Google Scholar]
- 53. Guo LH, Sun L, Chiba S, Araki H, Suzuki N. 2009. Coupled termination/reinitiation for translation of the downstream open reading frame B of the prototypic hypovirus CHV1-EP713. Nucleic Acids Res. 37:3645–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li H, Havens WM, Nibert ML, Ghabrial SA. 2011. RNA sequence determinants of a coupled termination-reinitiation strategy for downstream open reading frame translation in Helminthosporium victoriae virus 190S and other victoriviruses (Family Totiviridae). J. Virol. 85:7343–7352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nakamura H, Ikeda K, Arakawa M, Matsumoto N. 2002. Conidioma production of the white root rot fungus in axenic culture under near-ultraviolet light radiation. Mycoscience 43:251–254 [Google Scholar]
- 56. Segers GC, van Wezel R, Zhang X, Hong Y, Nuss DL. 2006. Hypovirus papain-like protease p29 suppresses RNA silencing in the natural fungal host and in a heterologous plant system. Eukaryot. Cell 5:896–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Choi GH, Shapira R, Nuss DL. 1991. Cotranslational autoproteolysis involved in gene expression from a double-stranded RNA genetic element associated with hypovirulence of the chestnut blight fungus. Proc. Natl. Acad. Sci. U. S. A. 88:1167–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Arakawa M, Nakamura H, Uetake Y, Matsumoto N. 2002. Presence and distribution of double-stranded RNA elements in the white root rot fungus Rosellinia necatrix. Mycoscience 43:21–26 [Google Scholar]
- 59. Ikeda K, Nakamura H, Arakawa M, Matsumoto N. 2004. Diversity and vertical transmission of double-stranded RNA elements in root rot pathogens of trees, Helicobasidium mompa and Rosellinia necatrix. Mycol. Res. 108:626–634 [DOI] [PubMed] [Google Scholar]
- 60. Powell ML, Napthine S, Jackson RJ, Brierley I, Brown TD. 2008. Characterization of the termination-reinitiation strategy employed in the expression of influenza B virus BM2 protein. RNA 14:2394–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kojima KK, Matsumoto T, Fujiwara H. 2005. Eukaryotic translational coupling in UAAUG stop-start codons for the bicistronic RNA translation of the non-long terminal repeat retrotransposon SART1. Mol. Cell. Biol. 25:7675–7686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sanderlin RS, Ghabrial SA. 1978. Physicochemical properties of two distinct types of virus-like particles from Helminthosporium victoriae. Virology 87:142–151 [DOI] [PubMed] [Google Scholar]
- 63. Chen B, Chen CH, Bowman BH, Nuss DL. 1996. Phenotypic changes associated with wild type and mutant hypovirus RNA transfection of plant pathogenic fungi phylogenetically related to Cryphonectria parasitica. Phytopathology 86:301–310 [Google Scholar]
- 64. Sasaki A, Onoue M, Kanematsu S, Suzaki K, Miyanishi M, Suzuki N, Nuss DL, Yoshida K. 2002. Extending chestnut blight hypovirus host range within diaporthales by biolistic delivery of viral cDNA. Mol. Plant-Microbe Interact. 15:780–789 [DOI] [PubMed] [Google Scholar]
- 65. Kanematsu S, Sasaki A, Onoue M, Oikawa Y, Ito T. 2010. Extending the fungal host range of a partitivirus and a mycoreovirus from Rosellinia necatrix by inoculation of protoplasts with virus particles. Phytopathology 100:922–930 [DOI] [PubMed] [Google Scholar]
- 66. Liu YC, Dynek JN, Hillman BI, Milgroom MG. 2007. Diversity of viruses in Cryphonectria parasitica and C. nitschkei in Japan and China, and partial characterization of a new chrysovirus species. Mycol. Res. 111:433–442 [DOI] [PubMed] [Google Scholar]
- 67. Hillman BI, Suzuki N. 2004. Viruses of the chestnut blight fungus, Cryphonectria parasitica. Adv. Virus Res. 63:423–472 [DOI] [PubMed] [Google Scholar]
- 68. Dang Y, Yang Q, Xue Z, Liu Y. 2011. RNA interference in fungi: pathways, functions, and applications. Eukaryot. Cell 10:1148–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang X, Shi D, Nuss DL. 2012. Variations in hypovirus interactions with the fungal-host RNA-silencing antiviral-defense response. J. Virol. 86:12933–12939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Drinnenberg IA, Fink GR, Bartel DP. 2011. Compatibility with killer explains the rise of RNAi-deficient fungi. Science 333:1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yaegashi H, Sawahata T, Ito T, Kanematsu S. 2011. A novel colony-print immunoassay reveals differential patterns of distribution and horizontal transmission of four unrelated mycoviruses in Rosellinia necatrix. Virology 409:280–289 [DOI] [PubMed] [Google Scholar]
- 72. Himeno M, Maejima K, Komatsu K, Ozeki J, Hashimoto M, Kagiwada S, Yamaji Y, Namba S. 2010. Significantly low level of small RNA accumulation derived from an encapsidated mycovirus with dsRNA genome. Virology 396:69–75 [DOI] [PubMed] [Google Scholar]
- 73. Choudhary S, Lee HC, Maiti M, He Q, Cheng P, Liu Q, Liu Y. 2007. A double-stranded-RNA response program important for RNA interference efficiency. Mol. Cell. Biol. 27:3995–4005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Qian S, Zhong X, Yu L, Ding B, de Haan P, Boris-Lawrie K. 2009. HIV-1 Tat RNA silencing suppressor activity is conserved across kingdoms and counteracts translational repression of HIV-1. Proc. Natl. Acad. Sci. U. S. A. 106:605–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Maliogka VI, Calvo M, Carbonell A, Garcia JA, Valli A. 2012. Heterologous RNA-silencing suppressors from both plant- and animal-infecting viruses support plum pox virus infection. J. Gen. Virol. 93:1601–1611 [DOI] [PubMed] [Google Scholar]
- 76. Wu Q, Wang X, Ding SW. 2010. Viral suppressors of RNA-based viral immunity: host targets. Cell Host Microbe 8:12–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Burgyan J, Havelda Z. 2011. Viral suppressors of RNA silencing. Trends Plant Sci. 16:265–272 [DOI] [PubMed] [Google Scholar]
- 78. Sun L, Suzuki N. 2008. Intragenic rearrangements of a mycoreovirus induced by the multifunctional protein p29 encoded by the prototypic hypovirus CHV1-EP713. RNA 14:2557–2571 [DOI] [PMC free article] [PubMed] [Google Scholar]