Abstract
Replication-competent poxvirus vectors with an attenuation phenotype and with a high immunogenic capacity of the foreign expressed antigen are being pursued as novel vaccine vectors against different pathogens. In this investigation, we have examined the replication and immunogenic characteristics of two vaccinia virus (VACV) mutants, M65 and M101. These mutants were generated after 65 and 101 serial passages of persistently infected Friend erythroleukemia (FEL) cells. In cultured cells of different origins, the mutants are replication competent and have growth kinetics similar to or slightly reduced in comparison with those of the parental Western Reserve (WR) virus strain. In normal and immune-suppressed infected mice, the mutants showed different levels of attenuation and pathogenicity in comparison with WR and modified vaccinia Ankara (MVA) strains. Wide genome analysis after deep sequencing revealed selected genomic deletions and mutations in a number of viral open reading frames (ORFs). Mice immunized in a DNA prime/mutant boost regimen with viral vectors expressing the LACK (Leishmania homologue for receptors of activated C kinase) antigen of Leishmania infantum showed protection or a delay in the onset of cutaneous leishmaniasis. Protection was similar to that triggered by MVA-LACK. In immunized mice, both polyfunctional CD4+ and CD8+ T cells with an effector memory phenotype were activated by the two mutants, but the DNA-LACK/M65-LACK protocol preferentially induced CD4+ whereas DNA-LACK/M101-LACK preferentially induced CD8+ T cell responses. Altogether, our findings showed the adaptive changes of the WR genome during long-term virus-host cell interaction and how the replication competency of M65 and M101 mutants confers distinct biological properties and immunogenicity in mice compared to those of the MVA strain. These mutants could have applicability for understanding VACV biology and as potential vaccine vectors against pathogens and tumors.
INTRODUCTION
Poxvirus vectors have emerged as prominent vehicles for delivering antigens of pathogens from prevalent diseases. Different strains of vaccinia virus (VACV)-expressing antigens from different pathogen-causing diseases are used nowadays in preclinical and clinical trials against HIV, malaria, tuberculosis, and leishmaniasis and also against cancer (1). The most promising vectors used in vaccination trials are the attenuated canarypox, fowlpox, modified vaccinia Ankara (MVA), and NYVAC virus strains (2–5). While those viruses do not produce virus progeny in human cells, which ensures safety, some evidence points out that replication-competent viruses with a limited but amplified time of infection and expression of heterologous antigen could provide more immunogenic vaccines (6).
Traditional smallpox vaccines have relied on replication-competent and attenuated vaccinia virus (VACV) vectors, but the side effects, particularly in immunocompromised individuals, preclude their use as recombinant viral vectors for current vaccines. Recently, a replication-competent NYVAC vector from the Copenhagen smallpox strain, with reinserted host range genes and limited replication in tissues, was shown to be a candidate vaccine vector against HIV (7). It is unclear how many rounds of VACV vector replication are needed in vivo to activate effective immune responses leading to long-term protection after pathogen challenge. Hence, we decided to investigate VACV vectors that can replicate in vivo for several rounds for their capacity as recombinants to induce immune responses with protection against a pathogen. These vectors have been isolated during a persistent VACV infection (Western Reserve [WR] strain) in the Friend erythroleukemia cell (FEL) line (8). It has been previously reported that persistent infection with the IHD-W strain of VACV can be established in FEL cells and that the virus produced was indistinguishable from the parental virus (9). In the case of the WR strain, during persistent infection of FEL cells, mutants with large deletions of about 8 MDa at the left terminus of the viral genome (10), alterations in some of the structural proteins with roles in the morphogenetic pathway, a small-plaque-size phenotype compared with that of the WR parental virus, and alterations in replication capacity in some mammalian cell lines (11) were produced. Recombinants based on these mutants at early passages in FEL cells and expressing parasite antigens for malaria and leishmania have been shown in prime/boost regimens in mice to elicit protection after challenge with parasites (12, 13).
To continue the further exploration of mutants from the persistent FEL cell infection and to define their adaptive changes during long-term virus persistence and immunological properties as vaccine vectors, in this investigation we selected two mutants from infected FEL cells after 65 (referred to as mutant M65) and 101 (mutant M101) passages and have characterized their biological, genetic, and immune properties. Pathogenesis of both mutants was monitored in normal and immune-deficient mice by weight loss after systemic inoculation (14); the duration of gene expression and replication capacity were followed in mice infected with the mutants expressing the sensitive firefly luciferase gene as a marker and by virus yields, as described previously (6, 15). Genomic status (10) was defined by a wide genome analysis after deep DNA sequencing of M65 and M101. Moreover, we analyzed if the expression of a parasite antigen in a prime/boost regimen with M65 and M101 recombinant viruses could induce protection against experimental challenge. We focused on leishmaniasis as this is one of the most neglected tropical diseases, prevalent in 88 countries and presenting an estimated annual incidence of 2 million and about 12 million cases worldwide (16). Among all leishmania antigens used, we selected one of the most promising genes: Leishmania homologue for receptors of activated C kinase, or LACK (17). The model of infection chosen was the permissive BALB/c mouse model inoculated in the footpad, as mice of this strain are highly susceptible and develop cutaneous lesions after the inoculation of Leishmania major and L. amazonensis. Immunogenicity was evaluated by intracellular cytokine staining (ICS).
Overall, this study demonstrated how the VACV genomes get adapted after 65 and 101 passages during a persistent virus infection and how the different biological characteristics acquired for M65 and M101 impact the immune responses of the host. Moreover, comparative studies with the prototype vaccine vector MVA reveal differences in the replication capacity and immunological behavior of the mutants. These novel characteristics of M65 and M101 could be useful in the design of new poxvirus-based vaccine strains against prevalent diseases.
MATERIALS AND METHODS
Ethics statement.
The Ethical Committee of Animal Experimentation (CEEA-CNB) of Centro Nacional de Biotecnología (CNB-CSIC) approved the animal studies in accordance with national and international guidelines and with the Royal Decree (RD 1201/2005) (permit number 011045).
Cells, plasmids, and viruses.
Cells were maintained in a humidified air 5% CO2 atmosphere at 37°C. Primary chicken embryo fibroblasts (CEF) were obtained from specific-pathogen-free (SPF) 11-day-old eggs (Intervet, Salamanca, Spain) and were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS). African green monkey kidney cells (BSC-40) and human cells (HELA) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% newborn calf serum (NCS). 3T3 murine cells and baby hamster kidney cells (BHK-21) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% calf serum (CS). The origin of FEL cells (a subclone of line 745A) and conditions for the establishment of virus persistence with the WR strain of VACV have been previously described (18, 19). Briefly, on the second and third days of infection with 1 PFU/cell of purified WR strain, when about 90% of the cells have died, surviving cells were removed and resuspended in fresh medium. After recovery (2 to 3 weeks), the cells were subjected to passage every 4 to 5 days.
The mammalian expression plasmid vector pCI-neo-LACK was previously described (20). The empty plasmid pCI-neo (Promega) was used as a control (DNAϕ). Both constructs were purified using a Qiagen plasmid purification kit (Qiagen).
The viruses used in this study included the VACV WR parental strain, a WR strain recombinant for the luciferase gene of Photinus Pyralis (WR-Luc) (15), and an attenuated MVA strain recombinant for the luciferase gene (MVA-Luc) (21).
M65 and M101 were grown in BHK-21 cells and purified by sucrose gradient centrifugation for DNA extraction and in BSC-40 cells for the rest of the studies. M65-LACK, M101-LACK, M65-Luc, and M101-Luc were grown in BSC-40 cells. Purification and titration of viruses were performed as described before (22, 23). Plasmids and viruses were diluted for inoculation in endotoxin-free phosphate-buffered saline (PBS).
Construction of vaccinia virus recombinants expressing firefly luciferase gene.
Vaccinia virus insertion plasmid pSC11-luc derives from the pSC11 insertion plasmid for the thymidine kinase (TK) locus (J2R gene) and contains the gene encoding the firefly luciferase of Photinus pyralis under the control of the vaccinia virus early/late promoter p7.5. This plasmid also contains the Escherichia coli β-galactosidase (LacZ-encoding) gene under the control of the p11 late viral promoter (15). M65-Luc and M101-Luc were obtained by transfecting pSC11-luc plasmid BSC-40 cells infected with the M65 or M101 strain and were harvested 48 to 72 h postinfection (h.p.i.), and β-galactosidase-producing plaques were selected by the addition of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) to the agar. For the generation of MVA-LACK, the plasmid pHLZ-LACK was used and recombinants were selected after a plaque assay by the addition of 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (X-Gluc) to the agar. The β-glucuronic acid-producing plaques were picked, cloned by plaque isolation, and amplified following standard procedures (24). The purity of M65-Luc, M101-Luc, and MVA-LACK was assessed by PCR.
Construction of vaccinia virus recombinants expressing Leishmania infantum LACK antigen (M65-LACK and M101-LACK).
Vaccinia virus insertion plasmid pHLZ-LACK contains the gene encoding the L. infantum LACK protein cloned into the SmaI site of the pHLZ VACV insertion plasmid under the control of the synthetic early/late pE/L viral promoter and the hemagglutinin (HA) flanking sites (20). This plasmid contains the E. coli β-glucuronidase gene under the control of the p7.5 early/late viral promoter. M65-LACK and M101-LACK were obtained by transfection with pHLZ-LACK plasmid BSC-40 cells infected with M65 or M101 viruses and harvested 48 to 72 h postinfection (h.p.i.) and virus plaques selected after the addition of X-Gluc to the agar. The β-glucuronidase-producing plaques were picked, cloned by plaque isolation, and amplified following standard procedures (24). The purity of M65-LACK and M101-LACK was assessed by PCR.
Parasite strains and animals.
L. major (WHOM/IR/-173) was a kind gift from Nicholas Glaichenhaus (CNSR, Valbonne, France). Promastigotes were cultured at 27°C in Schneider's medium (Gibco BRL, United Kingdom) supplemented with 20% fetal calf serum (FCS; Gibco BRL, United Kingdom) and antibiotics. The virulence of the strain was preserved by periodic passage through BALB/c mice. Frozen stocks grown in culture until the stationary phase were used for the experiments. In order to purify only infective metacyclic promastigotes, stationary-phase cultures were resuspended in PBS at a final concentration of 1 × 108 to 5 × 108 promastigotes/ml and treated with 10 mg/ml of peanut agglutinin (PNA). After centrifugation at 1,000 rpm, metacyclic promastigotes were collected from the supernatant, washed twice in PBS, and resuspended in the same buffer for inoculation.
L. amazonensis (LTB0016 strain) was a kind gift from Diane McMahon-Pratt (Yale School of Medicine, New Haven, CT). Promastigotes were cultured at 27°C in Schneider's medium (Gibco BRL, United Kingdom) supplemented with 20% fetal calf serum (FCS; Gibco BRL, United Kingdom) and antibiotics. The virulence of the strain was preserved by periodic passage through BALB/c mice. Fresh aspirates were synchronized in culture until the stationary phase, passaged twice, and grown for 5 days before inoculation in animals.
All mice used in this study were female BALB/c mice ranging from 6 to 8 weeks of age purchased from Harlan Laboratories and housed in the Animal Facility of the Centro Nacional de Biotecnología-CSIC (Madrid, Spain) under specific-pathogen-free conditions.
DNA extraction from VACV-infected cells.
Cells (5 × 108 PFU) were resuspended in 1 ml of buffer containing 50 mM Tris-Hcl (pH 8), 50 mM NaCl, and 50 mM MgCl2. The suspension was incubated with 20 units of RNase-free DNase (Roche Applied Science) for 30 min at 37°C. After treatment with DNase, Sarkosyl was added at a final concentration of 0.5%. Samples were later treated for 1 h at 37°C with proteinase K (Qiagen) at a final concentration of 200 μg/ml. Equilibrated phenol (1 vol) was added, and the sample was mixed by inversion and centrifuged at 13,000 rpm for 5 min. The supernatant was collected, and 2 vol of chorophorm/isoamyl alcohol (1:1) was added. Samples were centrifuged at 12,000 rpm for 5 min. NaCl was added to 2 vol of 100% ethanol to reach a final concentration of 0.2 M, and samples were left overnight at −20°C. Samples were then centrifuged at 10,000 rpm for 5 min. Pellets were washed twice in 75% ethanol, air-dried, and resuspended in 200 μl of H2O. The DNA concentration was measured in a NanoDrop instrument (Thermo Scientific, Wilmington, DE).
Wide genome analysis and deep sequencing. (i) Generation of sequences.
Library preparation was performed according to the recommendations of Illumina. Briefly, 5 μg of genomic DNA was fragmented using a nebulizer to a peak size of 250 bases. Following end repair and A-tailing, internally indexed adapters were ligated to the DNA. Size selection after adapter ligation was performed on a 2% agarose gel, and a narrow-size fraction corresponding to an insertion size of 250 bases was recovered using GeneCatcher tips. The DNA was recovered from the gel using the QIAquick gel extraction method, and the DNA was amplified by 12 cycles of PCR using standard Illumina PE 1.1 and 2.0 primers. The quality of the libraries was confirmed with an Agilent 2100 Bioanalyzer. The quantified libraries were loaded as a pool onto one lane of an Illumina flow cell and sequenced on an Illumina Genome Analyzer IIx platform using a 36-cycle recipe.
(ii) Sequence alignment.
Short reads were aligned to viral genomes with Burrows-Wheeler alignment (BWA) (25) with default parameters for single-end data. Alignment files in the SAM format were transformed to the BAM format, sorted, and indexed with SAMtools (26).
(iii) Variant detection.
For detecting statistically sound single nucleotide polymorphisms (SNPs) and short indels, a Genome Analysis Toolkit (GATK) (27) was used (–stand_call_conf = 30.0 and –stand_emit_conf = 10.0). The IndelRealigner option was applied for refining alignments of reads based on misalignments due to the presence of indels.
(iv) Variant annotation.
PHP scripts developed in-house were used to classify the effect of SNPs as follows: Silent, substitutions that do not change any amino acid; Change, substitutions that change an amino acid; Upstr, substitutions that occur in the 200-bp upstream region of a gene; Downstr, substitutions that occur in the 200-bp downstream region of a gene. (Note that Copenhagen annotations are not available.)
Immunohistochemical studies.
Immunohistochemical staining were performed on 5-μm-thick 4% paraformaldehyde-fixed and paraffin-embedded lung sections using a Dako Autostainer (Dako Corp., Carpinteria, CA) with rabbit anti-WR primary antibody. After incubation with the secondary antibodies, positive cells were visualized using 3,3-diaminobenzidine tetrahydrochloride-plus as a chromogen. All sections were counterstained with hematoxylin. Images were taken using a Zeiss Axiophot microscope (Carl Zeiss Microimaging GmbH, Gottingen, Germany), converted into standard TIFF format, and analyzed with Nikon Digital Sight software.
Growth curves.
To determine virus yields produced intracellularly and released from cells in the different cell lines, confluent CEF, HeLa, or 3T3 cells grown in 24-well plates were infected in duplicate at a multiplicity of infection (MOI) of 0.01 or 5 PFU/cell. At 6, 24, and 48 h postinfection (h.p.i.), cells were recovered from the well, freeze-thawed three times, and sonicated and virus titration was performed in BSC-40 cells as previously described (23). Extracellular virus yields were determined by titrating fresh supernatants of cells infected for 24 and 48 h.p.i.
Measurement of luciferase activity in mouse tissue.
Gene expression of recombinant viruses was monitored by the luciferase assay to quantify heterologous gene expression in tissues from mice inoculated intraperitoneally (i.p.) with WR-Luc, M65-Luc, M101-Luc, or MVA-Luc. Tissues were collected at 6, 24, 48, or 72 h.p.i. At the indicated times postinoculation, animals were sacrificed, and spleens, livers, draining lymph nodes, and ovaries were dissected and stored at −80°C. Peritoneal cells were harvested from mouse peritoneal cavities, washed with 10 ml of sterile PBS, centrifuged for 5 min at 1,500 rpm, and stored at −80°C. Tissues from individual mice were homogenized in luciferase extraction buffer (Promega Corp., Madison, WI) (200 μl/spleen and 200 μl/ovary, lymph node, or peritoneal extracts) using an Eppendorf-fitted Ultra-Turrax T8 homogenizer (Janke & Kunkel, Staufen, Germany). Luciferase activity was measured in the presence of ATP and luciferin according to the manufacturer's instructions using a Lumat LB 9501 luminometer (Berthold, Nashua, NH) and was expressed as reference luciferase units (RLU) per milligram of protein. Protein in tissue extracts was measured with a bicinchoninic acid (BCA) protein assay kit (Pierce-Thermo Scientific, Rockford, IL). Tissues from individual mice were collected and homogenized in complete DMEM to test for the production of infectious virus by plaque assay in BSC-40 cells. The virus titer was expressed as PFU per milligram of protein.
Immunization and parasite challenge.
BALB/c mice, 6 to 8 weeks of age, were primed intradermally (i.d.) in the abdomen with 100 μg of DNA-LACK or with empty DNA-ϕ in a 100-μl volume per mouse. At day 14, mice were boosted i.p. with 2 × 107 PFU/mouse of MVA-LACK, M65-LACK, M101-LACK, or nonrecombinant vaccinia virus and PBS as a control. At 3 weeks or 8 weeks after boosting, six animals per group were challenged subcutaneously (s.c.) in the right hind footpad with 5 × 104 metacyclic PNA-purified L. major or 2 × 104 stationary-phase L. amazonensis promastigotes resuspended in 10 μl using BD Micro-Fine (BD Biosciences) 0.5-ml 30-gauge needles.
ICS assay.
The phenotypes of responding T cells were analyzed by intracellular cytokine staining (ICS) and fluorescence-activated cell sorting analysis as described elsewhere (28). After an overnight rest, 5 × 106 splenocytes (depleted of red blood cells) were stimulated with 2 μg/ml of LACK157–173 peptide or 5 × 105 A20 cells nucleofected with pCIneo-LACK plasmid (using a 4D Nucleofector; Lonza, Germany) during 6 h in RPMI 1640 medium supplemented with 10% FCS and containing 1 μl/well Golgiplug (BD Biosciences) to inhibit cytokine secretion. After stimulation, cells were washed, stained for the surface markers, fixed, permeabilized using a BD Cytofix/Cytoperm kit (Becton, Dickinson), and stained intracellularly using the appropriate fluorochromes. For memory analyses, the following antibodies were used: CD4-Alexa 700, CD8-V500, CD62L-fluorescein isothiocyanate (CD62L-FITC), CD44-SPRD, CD127-phycoerythrin Cy5.5 (CD127-PECy5.5), gamma interferon (IFN-γ)–PECy7, tumor necrosis factor alpha-PE (TNF-α-PE), and interleukin-2–allophycocyanin (IL-2-APC). All antibodies were from BD Biosciences. Cells were acquired using an LSRII flow cytometer (Becton, Dickinson) equipped with a high-throughput system. The number of events ranged between 105 and 106. Dead cells were excluded using a violet LIVE/DEAD stain kit (Invitrogen). Lymphocytes were gated on a forward-scatter-area versus side-scatter-area pseudocolor dot plot. CD4+ and CD8+ events were gated versus CD44 and CD62L or CD62L and CD127 to analyze the memory phenotype. IFN-γ, TNF-α, and IL-2 were gated in the different memory populations and then combined using the Boolean operator. Sample analysis was performed using FlowJo version 8.5.3 (Tree Star, Ashland, OR).
Measurement of vaccinia virus-specific antibodies by ELISA.
At 11 days and 8 weeks after immunization, serum was collected from each group of animals and the presence of specific anti-vaccinia virus antibodies was analyzed by enzyme-linked immunosorbent assay (ELISA). In brief, 96-well Maxisorp plates (Nunc, Denmark) were coated overnight at 4°C with extracts of BSC-40 cells infected with the Western Reserve (WR) VACV strain. Plates were washed with PBS–0.05% Tween 20 (PBS-T) and blocked with 5% milk–PBS (blocking buffer) overnight at 4°C. Serum samples were diluted 50-fold in PBS–0.1% Tween–1% milk, added in 50 μl per well, and incubated for 1 h at 37°C. Plates were washed five times. Peroxidase-conjugated goat anti-mouse total IgG (Sigma) was added, and the reaction mixture was incubated for 1 h at 37°C. Plates were then reacted with TMB (3,3′,5,5′-tetramethylbenzidine) peroxidase substrate (Sigma), and absorbance was read at 450 nm on a Labsystem Multiskan Plus Sunrise plate reader using Magellan software.
Statistical analysis.
The statistical significance (P < 0.05 [*], P < 0.005 [**], or P < 0.001 [***]) of differences between immunization groups of mice was determined by Student's t test (2 tail, type 3).
For the statistical analysis of ICS data, we used a novel approach that corrects measurements for the medium response (RPMI) and at the same time allows the calculation of confidence intervals and P values of hypothesis tests (29, 30).
The data analysis program, Simplified Presentation of Incredibly Complex Evaluations (SPICE, version 4.1.5; Mario Roederer, Vaccine Research Center, NIAID, NIH), was used to analyze and generate graphical representations of T cell responses detected by polychromatic flow cytometry. All values used for analyzing proportionate representation of responses were background subtracted.
RESULTS
M65 and M101 mutant viruses are replication competent in cell cultures of various origins.
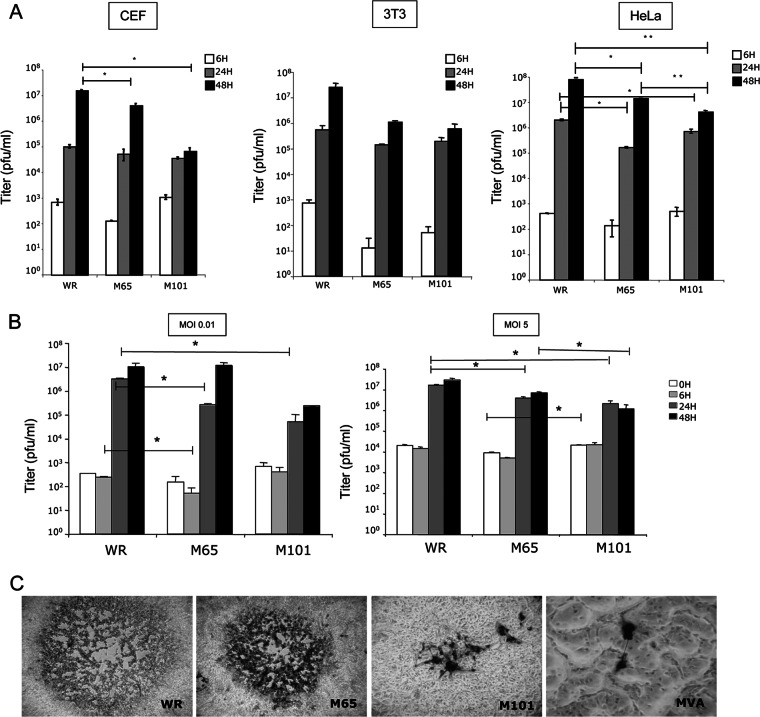
Established cultures of FEL cells infected with 1 PFU/cell of the WR strain of VACV were maintained for 65 and 101 passages in DMEM supplemented with 10% fetal calf serum, and individual virus plaques were isolated in monkey BSC-40 cells overlaid with agar. One representative virus plaque of each culture (referred to here as M65 and M101) was grown. Since mutants M65 and M101 were obtained after a long-term infection of FEL cells, it was of interest to know how the selective pressure during the virus-host cell interaction affected the biological characteristics of the mutant viruses. Thus, we first analyzed the replication capacity of these viruses (0.01 PFU/cell) in cells of different origins by determining virus-growth curves in chicken (CEF), mouse (3T3), and human (HeLa) cell lines. As depicted in Fig. 1 with whole-cell extracts, some differences were observed in CEF cells infected with parental WR strain and mutant viruses. M65 showed a small difference in growth in comparison with WR, and M101 presented a higher reduction than parental virus (P = 0.02) and also than M65 at 48 h.p.i. (P = 0.058). However, the difference in growth of M101 was minimized when CEF cells were infected with 5 PFU/cell (Fig. 1B). When supernatants of infected cells were titrated to estimate virus production, a similar tendency of virus production was observed (not shown). The classical virus plaque morphology of each virus in comparison with those of the WR and MVA strains is shown in Fig. 1C. Thus, mutant viruses M65 and M101 have plaque size morphologies different from those of WR and MVA and are able to replicate in cell lines of different origins, proving a wide host range capacity.
Fig 1.
Viral growth efficiency and plaque size phenotype in cultured cells. (A) CEF, 3T3, and HeLa cells were infected at an MOI of 0.01 PFU/cell, collected at 6, 24, and 48 h.p.i., frozen and thawed three times, and sonicated, and virus infectivity was titrated by a plaque assay in BSC-40 cells. (B) Comparison of virus yields in CEF cells infected at an MOI of 0.01 PFU/cell and an MOI of 5 PFU/cell. Data are represented as means ± standard deviations (SD). (C) Plaque size in BSC-40 cells at 48 h.p.i. after immunostaining, comparing the WR and MVA strains versus M65 and M101 mutants. Magnification, ×10.
Virus distribution and antigen expression in mice.
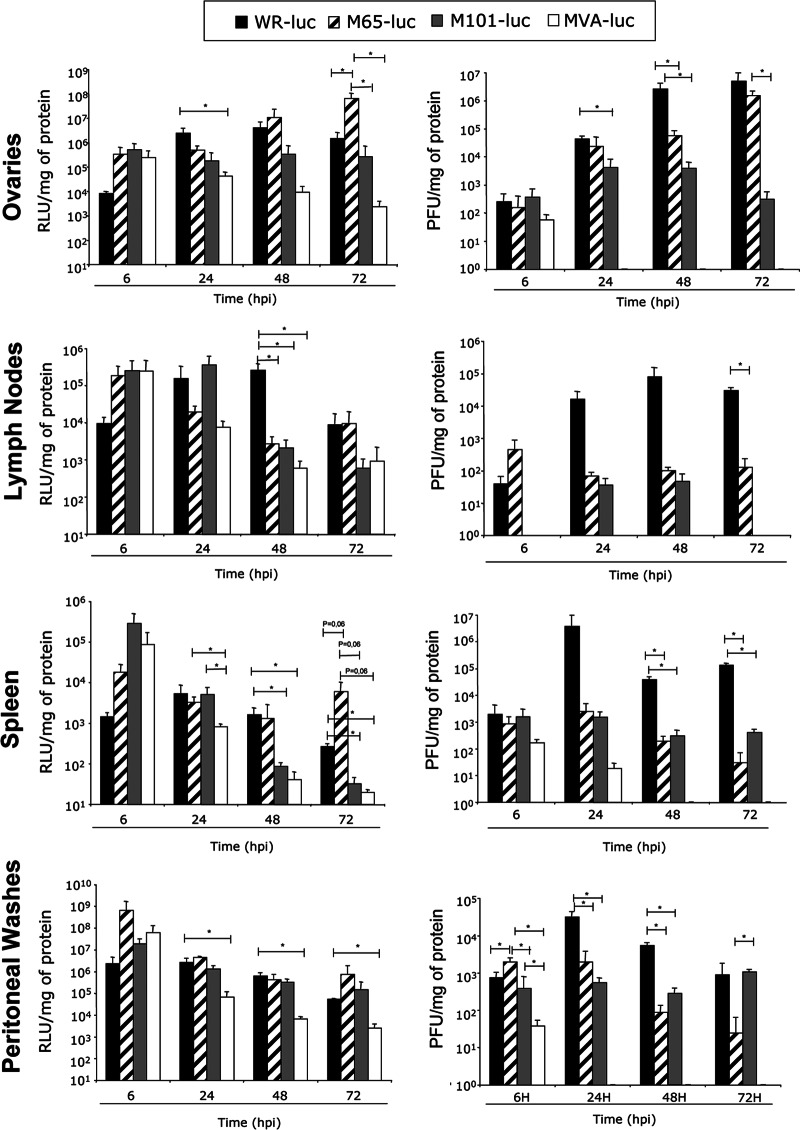
Next, we evaluated the replication capacity of the mutants in an in vivo model. We have previously reported the use of the luciferase marker to follow VACV replication and antigen expression in mice (6, 15, 21, 31). In order to estimate the replication capacity and the duration of heterologous antigen expression by M65 and M101, recombinant viruses expressing the luciferase firefly gene of Photinus pyralis were generated as described in Materials and Methods. As controls, we used the parental strain WR-Luc and the attenuated mutant virus MVA-Luc, respectively, as described previously (21). BALB/c mice were inoculated i.p. with 2 × 107 PFU of WR-Luc, M65-Luc, M101-Luc, or MVA-Luc. At 6, 24, 48, and 72 h postinoculation, three animals per group were sacrificed and the luciferase activity and virus production in different tissues were assayed in cell homogenates from peritoneal washes, spleen, lymph nodes, and ovaries. As reported previously (21), luciferase activity was clearly observed in all tissues from WR-Luc-infected mice. Similar behavior was observed in M65-Luc-infected mice that showed similar or increased levels of luciferase in all the organs at 72 h.p.i. compared to WR-Luc mice, particularly in ovaries and spleen (Fig. 2). M101-Luc-infected animals showed a decrease in luciferase activity with time in the organs analyzed compared to WR-Luc-infected animals, except in ovaries or peritoneal washes, where expression was maintained. In general, in terms of viral gene expression over the 72 h, the mutants gave more expression than the highly attenuated MVA-Luc vector.
Fig 2.
Kinetics of luciferase expression and viral production in mice. BALB/c mice (3 per group) were inoculated i.p. with 2 × 107 PFU/mouse of WR-Luc, M65-Luc, M101-Luc, or MVA-Luc. Tissues were collected at 6, 24, 48, and 72 h.p.i., and luciferase expression and viral titers were determined as described in Materials and Methods. Results represent mean values of luciferase from samples of three animals per time expressed as RLU per mg of protein (left panels) or viral production expressed as PFU per mg of protein (right panels). Data are representative of the results of three different experiments.
Together, the results of three independent experiments revealed that M65 and M101 are replication-competent vectors in mice. While M65 was able to replicate and to produce high levels of luciferase comparable to the levels seen with WR-Luc in all the organs studied, M101 showed a more restricted phenotype, but compared to MVA-Luc, it maintained replication in organs such as ovaries and in cells of the peritoneal cavity.
M65 and M101 viruses showed different degrees of attenuation after inoculation by the i.n. route of infection.
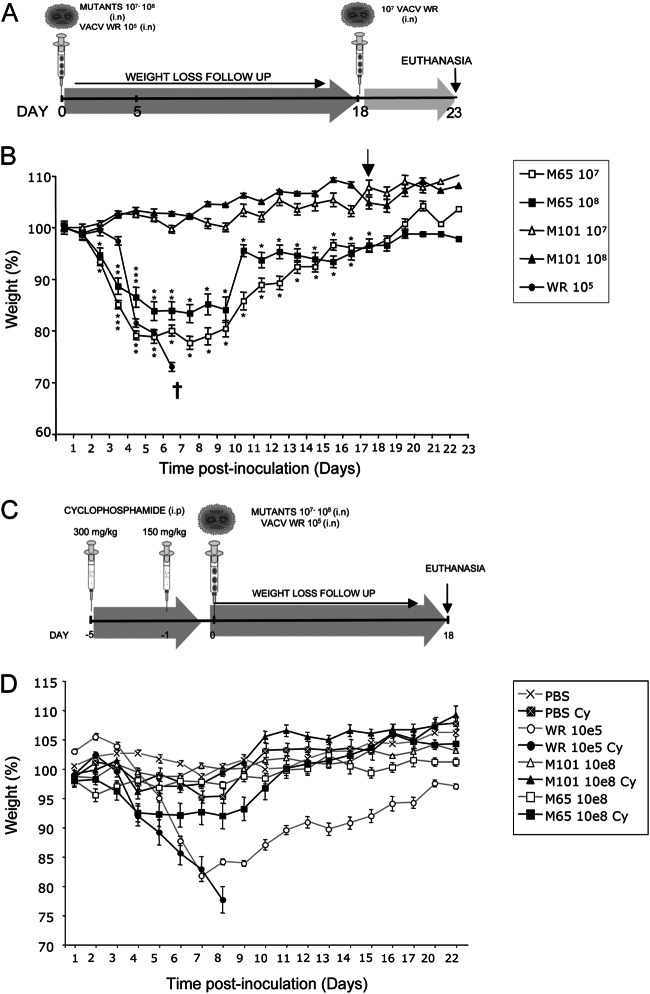
To assess the attenuation profile of M65 and M101 viruses, groups of three BALB/c mice per dose were inoculated intranasally (i.n.) with 107 and 108 PFU, respectively. Control animals were infected with 105 PFU of wild-type (wt) virus. Loss of weight and other signs of illness (absence of grooming, rough coat hair, loss of mobility, inflammation of the eye membrane, and hunched posture) were monitored daily after infection with the different viruses. A scheme of the immunization schedule is represented in Fig. 3A. As shown in Fig. 3B, all the animals infected with 105 PFU of parental WR exhibited severe loss of weight and developed signs of illness such as rough coat hair, loss of mobility, and hunched posture (not shown) and had to be sacrificed at day 6 postinfection. M65, at the different doses tested, caused mildly rough coat hair, loss of mobility, or hunched posture compared with the wt virus (not shown). A marked loss of weight was also observed until day 6, with a peak at 5 days postinfection, when animals started to gain weight and signs of illness disappeared gradually. M101 at different doses tested did not produce any sign of illness or weight loss with time.
Fig 3.
Weight loss of mice inoculated with different viruses. (A) Immunization schedule. BALB/c mice (6 to 8 weeks old; 3 per group) received 107 or 108 PFU of M65-wt or M101-wt virus or 105 PFU of WR virus by the intranasal (i.n.) route. At 18 days after the first inoculation, 107 PFU, equivalent to 100 lethal doses (LD) of WR virus, was delivered i.n. to all the groups. (B) Monitoring of BALB/c mouse weight loss with time. Asterisks show P values of differences between M65 and M101 at the same dose. (C) Immune system-suppressed mouse schedule. C57BL6 mice (12 weeks old; 4 per group) received 300 mg/kg of cyclophosphamide by the i.p. route 5 days before the immunization and a second dose of 150 mg/kg 24 h before the inoculation of the different viruses. Animals treated with cyclophosphamide or mock treated with PBS received 108 PFU of M65 or M101 virus or 105 of WR virus by the i.n. route. (D) Monitoring of C57BL6 mouse weight loss with time. Symbols are given for each virus.
To further prove an attenuation phenotype of M65 and M101, we used an immunocompromised mouse model. This is based on C57BL6 female mice first treated with 300 mg of cyclophosphamide/kg of body weight administered i.p. and 5 days later treated with a second dose of the drug but at 150 mg/kg (32). Twenty-four hours later, untreated and drug-treated mice were inoculated i.p. with different doses of each virus and weight loss and survival were followed daily. A scheme is shown in Fig. 3C, and pathogenicity results are shown in Fig. 3D. As expected, drug-treated mice were more susceptible to WR infection than untreated animals and died by 8 days postinfection with a dose of 105 PFU. In contrast, all drug-treated animals infected with M65 or M101 survived to a dose 1,000-fold higher than the lethal dose of wt WR virus.
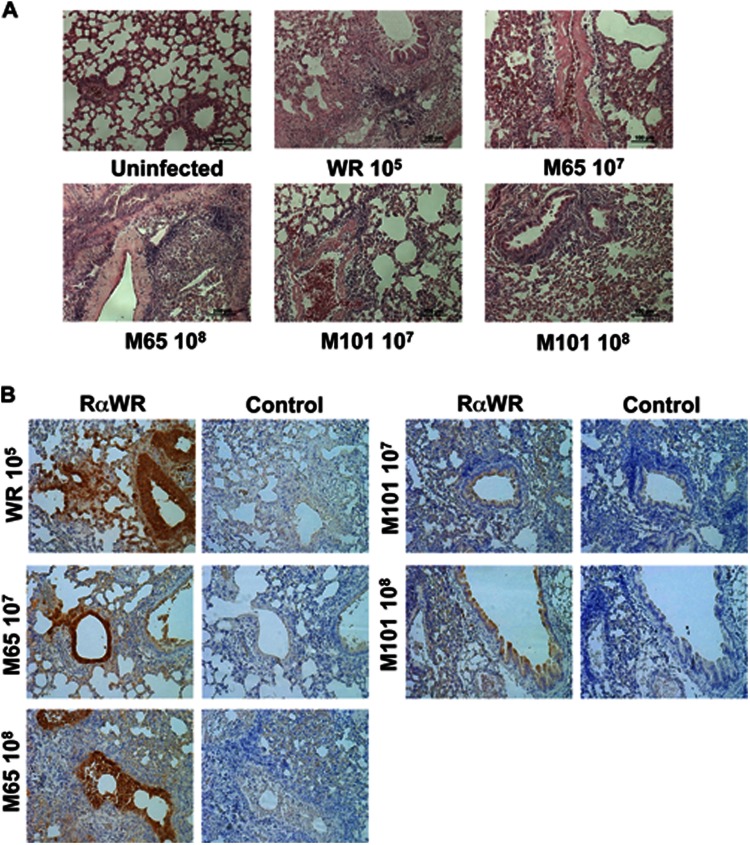
To define the degree of pathological changes induced by the mutant viruses, at day 5 postinfection we analyzed the immunopathology of lung tissue. We performed hematoxylin & eosin staining of different sections of the lungs in order to identify signs of tissue infection, such as leukocyte extravasations and accumulation, bronchiole epithelium modification, and the presence of interstitial hemorrhage. We also studied the presence of VACV antigens by immunohistochemistry. As depicted in Fig. 4A, lungs of animals infected i.n. with WR showed high levels of leukocyte extravasations and accumulation, a severe inflammation of the bronchiole epithelium, and interstitial hemorrhages. The presence of VACV proteins as revealed by immunohistochemistry in areas of the lungs that presented a pathological phenotype pointed out the role of infection in the disease outcome. Similar results were observed in animals inoculated i.n. with 108 PFU of M65, but the inflammation of bronchiole epithelium was markedly reduced in comparison with the wt virus results. At the lower dose of M65, the modification of the epithelium was less aggressive, as leukocyte accumulation and extravasations were also less pronounced than with a higher dose. Both doses of M101 showed leukocyte accumulation and extravasations, but the modification of the epithelium was mild and bronchioles were not as collapsed as in the case of infections with M65 or WR viruses. The specific staining with VACV antibodies is an indicator of the presence of viral proteins in the lung, as shown in Fig. 4B, after reactivity with anti-WR serum. Clearly, the viral antigen reactivity is greater in WR and M65 infections, while for M101 a lower reactivity is observed around the bronchiole, as expected from limited replication in the tissue.
Fig 4.
Histopathological studies in lungs of animals infected with mutant viruses M65 and M101. At day 5 postinfection of mice inoculated as described for Fig. 3A, one animal of each group was sacrificed and lungs were extracted for histological studies (magnification, ×10). (A) Hematoxilin-eosin staining was performed in order to identify signs of tissue infection such as leukocyte extravasations and accumulation, bronchiole epithelium modification, and presence of interstitial hemorrhage. Uninfected and doses of virus infection are indicated. (B) The presence of VACV proteins in the lung was analyzed by immunohistochemistry using rabbit polyclonal antibodies against VACV (panels indicated as RαWR). The brown color denotes virus-specific antigens. Sections incubated with secondary antibody were used as control staining (panels indicated as “Control”).
Since the animals recovered after M65 and M101 infection, we next tested whether they maintained an immune protective status against a lethal dose of the virulent WR virus. Thus, groups of 3 animals each were inoculated with doses of 107or 108 PFU and 18 days later were challenged with 107 PFU of WR (equivalent to 100 lethal doses [LD]) administered i.n., and we followed protection over time (as shown in Fig. 3). None of the animals developed signs of disease or weight loss at any time, and all were fully protected.
The results shown in Fig. 3 and 4 and described above demonstrate the attenuation phenotype of M65 and M101 in both normal and immune-suppressed animals, with M101 showing a higher degree of attenuation; also, both viruses triggered a protective immune response to the virulent WR strain.
Genomic features of M65 and M101.
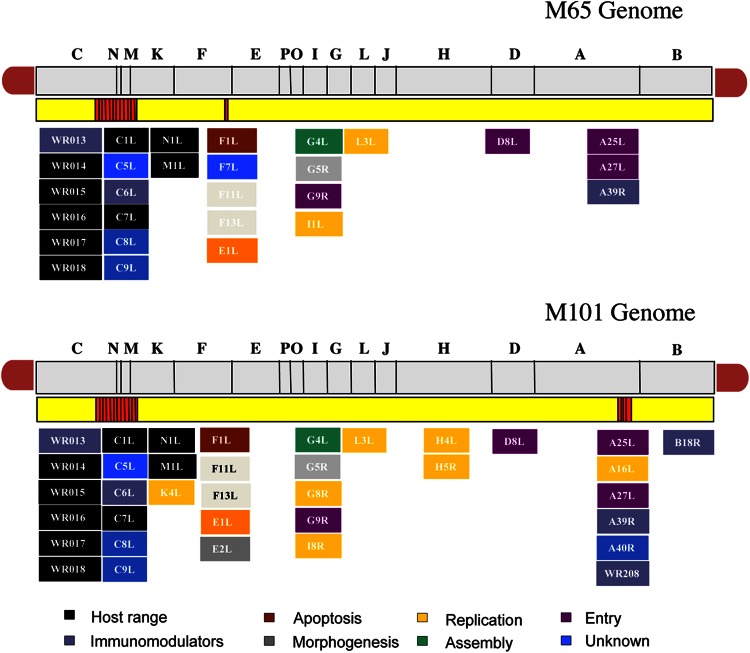
Next, we defined the nucleotide sequence of the entire genomes of M65 and M101 by deep genome sequencing. The sequence of the linear, double-stranded DNA molecule of the M65 and M101 mutants showed percent GC identical to that of the parental WR virus (33.3% GC and 66.7% AT). As shown in Fig. 5, M65 and M101 share a deletion in the left end of the genome that includes genes C5L, C6L, C7L, C8L, C9L, VACWR013, VACWR014, VACWR015, VACWR016, VACWR017, and VACWR018. The genes C5L, C8L, and C9L encode proteins of unknown function or hypothetical proteins. C6L encodes an immunomodulatory protein that blocks the induction of IFN-β (33, 34). C7L encodes a host range protein (6), and VACWR013—also called C12L—encodes vaccinia virus IL-18 binding protein (35). VACWR014, VACWR015, VACWR016, VACWR017, and VACWR018 encode ankyrin host range proteins.
Fig 5.
Diagrams of the genomes of M65 and M101 viruses. Genes deleted or mutated in both genomes are shown in boxes. In the scheme, the deletion in the left end of the genome in both viruses and the small deletion in the right end of M101 virus (both deletions shown in red) can be observed. Different colors are used to denote gene functions.
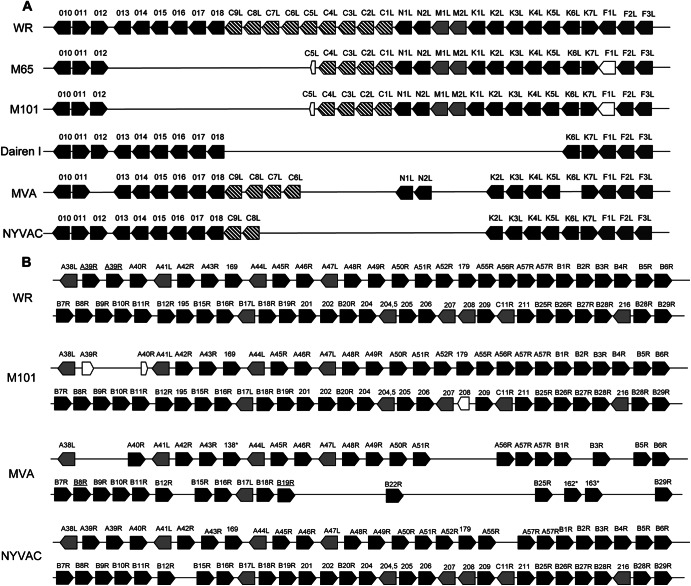
In addition to the left flank deletions, M101 showed a small deletion in the right flank of the genome that included A39R (the remaining sequence in the mutant genomes represents only 70.72% A39R in the WR genome) encoding a proinflammatory semaphorine (36) and A40R (the remaining sequence represents only 1.46% of the gene in the WR genome); the encoded protein has an unknown function, and it is the first example of a vaccinia virus protein that acquires SUMO-1 (37). It has been previously shown that disruption of the A40R gene did not affect virus plaque size, in vitro growth rate and virus titer, virus envelope (EV) formation, or virulence in a murine intranasal model (38). A schematic representation of the genes present in the M65 and M101 mutants in comparison with the WR and MVA strains is shown in Fig. 6A (for the left end of the genome) and Fig. 6B (for the right end of the genome).
Fig 6.
Schematic representation of the deletions present in the left end (A) or right end (B) of the genome of different strains of vaccinia virus. The gene nomenclature corresponds to the Copenhagen genome wherever possible. Numbers represent the names of the genes in WR nomenclature. Numbers with asterisks are in MVA nomenclature. Designations of truncated genes are underlined. Truncated genes are represented by reduced-size empty arrows.
Genes mutated in the M65 and M101 genomes.
A total of 59 genes in M65 and 61 genes in M101 presented at least 1 missense mutation compared with the WR consensus sequence. Homologies of sequences and numbers of single nucleotide polymorphisms (SNPs), silent mutations, and missense mutations for M65 and M101 genes are shown in Table 1 and Table 2, respectively. Based on these findings, we next analyzed the different mutations in the viral open reading frames (ORFs) by the biological functions known.
Table 1.
Mutations present in M65 genome in comparison with WR sequencea
| Gene ID | %Sequence | No. of SNPs | No. of silent mutations | No. of changes | Function | Annotation |
|---|---|---|---|---|---|---|
| D8L | 100 | 8 | 1 | 6 | Entry, carbonic anhydrase | GeneID:3707569 | PBR:42551 |
| F1L | 99.85 | 5 | 1 | 4 | Apoptosis, caspase 9 inhibitor | GeneID:3707497 | PBR:42477 |
| C1L | 100 | 7 | 2 | 3 | Host range | GeneID:3707642 | PBR:42464 |
| E1L | 100 | 7 | 2 | 3 | Replication, polymerase large subunit | GeneID:3707514 | PBR:42495 |
| F11L | 100 | 6 | 2 | 3 | Morphogenesis, cell motility | GeneID:3707507 | PBR:42487 |
| G9R | 100 | 5 | 2 | 3 | Entry fusion complex | GeneID:3707543 | PBR:42525 |
| M1L | 100 | 4 | 0 | 3 | Ankyrin | GeneID:3707645 | PBR:42467 |
| VACWR148 | 100 | 6 | 3 | 3 | Inclusion protein | GeneID:3707678 | PBR:42586 |
| A27L | 100 | 2 | 0 | 2 | Entry fusion protein | GeneID:3707680 | PBR:42588 |
| A39R | 100 | 3 | 1 | 2 | Immunomodulator, semaphorin | GeneID:3707693 | PBR:42602 |
| F7L | 100 | 5 | 0 | 2 | Unknown | GeneID:3707503 | PBR:42483 |
| F13L | 100 | 5 | 1 | 2 | Morphogenesis, EEV phospholipase | GeneID:3707509 | PBR:42489 |
| G4L | 100 | 2 | 0 | 2 | Assembly, glutarredoxin 2 | GeneID:3707537 | PBR:42519 |
| G5R | 100 | 8 | 4 | 2 | Morphogenesis | GeneID:3707538 | PBR:42520 |
| I1L | 100 | 3 | 1 | 2 | DNA binding protein | GeneID:3707603 | PBR:42508 |
| L3L | 100 | 4 | 2 | 2 | Transcription, internal virion protein | GeneID:3707546 | PBR:42528 |
| N1L | 100 | 6 | 0 | 2 | Host range virokine/NfκB inhibitor | GeneID:3707643 | PBR:42465 |
| A4L | 100 | 1 | 0 | 1 | Core protein | GeneID:3707521 | PBR:42561 |
| A11R | 100 | 4 | 3 | 1 | Membrane formation | GeneID:3707528 | PBR:42568 |
| A14.5L* | 100 | 1 | 0 | 1 | Virulence factor | GeneID:3707532 | PBR:42572 |
| A16L | 100 | 1 | 0 | 1 | Entry complex | GeneID:3707666 | PBR:42574 |
| A20R | 100 | 1 | 0 | 1 | Replication, DNA processivity factor | GeneID:3707671 | PBR:42579 |
| A34R | 100 | 1 | 0 | 1 | Morphogenesis, virus spread | GeneID:3707687 | PBR:42596 |
| B12R* | 100 | 1 | 0 | 1 | Serine/threonine kinase | GeneID:3707665 | PBR:42633 |
| VACWR-B15R* | 100 | 1 | 0 | 1 | Immunomodulator, IL-1 binding protein | GeneID:3707574 | PBR:42636 |
| B18R | 100 | 1 | 0 | 1 | Ankyrin | GeneID:3707576 | PBR:42638 |
| B19R | 100 | 3 | 2 | 1 | Immunomodulator, type I IFN receptor | GeneID:3707577 | PBR:42639 |
| C4L | 100 | 1 | 0 | 1 | Immunomodulator, NfκB inhibitor | GeneID:3707639 | PBR:42461 |
| C5L | 34.47 | 2 | 1 | 1 | Unknown | GeneID:3707638 | PBR:42460 |
| D1R | 100 | 8 | 5 | 1 | Large capping enzyme | GeneID:3707562 | PBR:42544 |
| D2L | 100 | 2 | 1 | 1 | Virion core structural protein | GeneID:3707563 | PBR:42545 |
| D11L* | 100 | 3 | 2 | 1 | Replication, helicase | GeneID:3707708 | PBR:42554 |
| D13L* | 100 | 3 | 1 | 1 | Rifampin resistance | GeneID:3707516 | PBR:42556 |
| E2L | 100 | 5 | 1 | 1 | Morphogenesis, virus spread | GeneID:3707591 | PBR:42496 |
| E8R | 100 | 1 | 0 | 1 | Transcription, ER membrane protein | GeneID:3707597 | PBR:42502 |
| E9L | 100 | 3 | 2 | 1 | Replication, DNA polymerase | GeneID:3707598 | PBR:42503 |
| F4L | 100 | 5 | 3 | 1 | Replication, ribonucleotide reductase small subunit | GeneID:3707500 | PBR:42480 |
| F8L | 100 | 6 | 1 | 1 | Unknown, cytoplasmic protein | GeneID:3707504 | PBR:42484 |
| F14.5L* | 100 | 1 | 0 | 1 | Entry, cell adhesion | GeneID:4557754 | PBR:42491 |
| F17R | 100 | 1 | 0 | 1 | DNA binding phosphoprotein | GeneID:3707513 | PBR:42494 |
| G3L* | 100 | 1 | 0 | 1 | Entry fusion complex | GeneID:3707535 | PBR:42517 |
| G6R* | 100 | 1 | 0 | 1 | Virus-host interaction | GeneID:3707540 | PBR:42522 |
| G8R | 100 | 5 | 2 | 1 | Replication, late transcription factor | GeneID:3707542 | PBR:42524 |
| H3L* | 100 | 2 | 1 | 1 | Heparin binding surface protein | GeneID:3707557 | PBR:42539 |
| H5R | 100 | 2 | 1 | 1 | Late transcription factor | GeneID:3707559 | PBR:42541 |
| I3L | 100 | 2 | 0 | 1 | Replication, DNA binding phosphoprotein | GeneID:3707605 | PBR:42510 |
| I7L | 100 | 10 | 9 | 1 | Virion core protease | GeneID:3707609 | PBR:42514 |
| I8R | 100 | 1 | 0 | 1 | Replication, RNA helicase | GeneID:3707610 | PBR:42515 |
| J5L | 100 | 1 | 0 | 1 | Unknown, membrane protein | GeneID:3707553 | PBR:42535 |
| K1L | 100 | 1 | 0 | 1 | Host range, ankyrin, NfκB inhibitor | GeneID:3707647 | PBR:42469 |
| K2L | 100 | 2 | 1 | 1 | Serpin, cell-cell fusion inhibitor | GeneID:3707648 | PBR:42470 |
| K4L | 100 | 1 | 0 | 1 | Replication, nicking joining enzyme | GeneID:3707650 | PBR:42472 |
| L1R | 100 | 4 | 2 | 1 | IMV membrane protein | GeneID:3707544 | PBR:42526 |
| L2R | 100 | 3 | 0 | 1 | Morphogenesis, crescent formation | GeneID:3707545 | PBR:42527 |
| O1L* | 100 | 2 | 0 | 1 | Virulence factor | GeneID:3707601 | PBR:42506 |
| VACWR192* | 100 | 1 | 0 | 1 | Kelch-like protein | GeneID:3707663 |
| VACWR201 | 100 | 6 | 0 | 1 | Unknown | GeneID:3707578 |
| VACWR202 | 100 | 6 | 4 | 1 | Ankyrin | GeneID:3707579 |
| VACWR203 | 100 | 2 | 0 | 1 | Ankyrin | GeneID:3707580 |
Gene ID, gene names (asterisks highlight genes that are not mutated in the M101 genome); %Sequence, percentages of sequencing coverage in comparison with WR genome; SNPs, single nucleotide polymorphisms; No. of changes, number of missense mutations; Function, described functions of the different genes; EEV, equine encephalitis virus; ER, endoplasmic reticulum.
Table 2.
Mutations present in M101 genome in comparison with WR sequencea
| Gene ID | %Sequence | No. of SNPs | No. of silent mutations | No. of changes | Function | Annotation |
|---|---|---|---|---|---|---|
| D8L | 100 | 9 | 1 | 7 | Entry, carbonic anhydrase | GeneID:3707569 | PBR:42551 |
| A39R | 70.72 | 5 | 1 | 4 | Immunomodulator, semaphorin | GeneID:3707693 | PBR:42602 |
| C1L | 100 | 8 | 2 | 4 | Host range | GeneID:3707642 | PBR:42464 |
| F1L | 99.85 | 5 | 1 | 4 | Apoptosis, caspase 9 inhibitor | GeneID:3707497 | PBR:42477 |
| F11L | 100 | 7 | 2 | 4 | Morphogenesis, cell motility | GeneID:3707507 | PBR:42487 |
| E1L | 100 | 8 | 2 | 3 | Replication. polymerase large subunit | GeneID:3707514 | PBR:42495 |
| F13L | 100 | 6 | 1 | 3 | Morphogenesis, EEV phospholipase | GeneID:3707509 | PBR:42489 |
| M1L | 100 | 4 | 0 | 3 | Ankyrin | GeneID:3707645 | PBR:42467 |
| VACWR148 | 100 | 7 | 3 | 3 | Inclusion protein | GeneID:3707678 | PBR:42586 |
| A16L | 100 | 2 | 0 | 2 | Entry complex | GeneID:3707666 | PBR:42574 |
| A27L | 100 | 3 | 0 | 2 | Entry fusion protein | GeneID:3707680 | PBR:42588 |
| B18R | 100 | 2 | 0 | 2 | Ankyrin | GeneID:3707576 | PBR:42638 |
| E2L | 100 | 7 | 1 | 2 | Morphogenesis, virus spread | GeneID:3707591 | PBR:42496 |
| F8L | 100 | 5 | 1 | 2 | Unknown, cytoplasmic protein | GeneID:3707504 | PBR:42484 |
| G4L | 100 | 2 | 0 | 2 | Assembly, glutarredoxin 2 | GeneID:3707537 | PBR:42519 |
| G5R | 100 | 8 | 4 | 2 | Morphogenesis | GeneID:3707538 | PBR:42520 |
| G8R | 100 | 4 | 1 | 2 | Replication, late transcription factor | GeneID:3707542 | PBR:42524 |
| G9R | 100 | 5 | 2 | 2 | Entry fusion complex | GeneID:3707543 | PBR:42525 |
| H4L* | 99.96 | 5 | 3 | 2 | Replication, RNA polymerase-associated protein | GeneID:3707558 | PBR:42540 |
| H5R | 100 | 3 | 1 | 2 | Late transcription factor | GeneID:3707559 | PBR:42541 |
| I1L | 100 | 3 | 1 | 2 | DNA binding protein | GeneID:3707603 | PBR:42508 |
| I8R | 100 | 2 | 0 | 2 | Replication, RNA helicase | GeneID:3707610 | PBR:42515 |
| J5L | 100 | 2 | 0 | 2 | Unknown, membrane protein | GeneID:3707553 | PBR:42535 |
| K4L | 100 | 2 | 0 | 2 | Replication, nicking joining enzyme | GeneID:3707650 | PBR:42472 |
| L2R | 100 | 3 | 0 | 2 | Morphogenesis , crescent formation | GeneID:3707545 | PBR:42527 |
| L3L | 100 | 4 | 2 | 2 | Transcription, internal virion protein | GeneID:3707546 | PBR:42528 |
| N1L | 100 | 6 | 0 | 2 | Host range virokine/NfκB inhibitor | GeneID:3707666 | PBR:42574 |
| A4L | 100 | 1 | 0 | 1 | Core protein | GeneID:3707521 | PBR:42561 |
| A11R | 100 | 4 | 3 | 1 | Membrane formation | GeneID:3707528 | PBR:42568 |
| A14L* | 100 | 1 | 0 | 1 | Morphogenesis, IMV membrane protein | GeneID:3707531 | PBR:42571 |
| A17L* | 100 | 2 | 0 | 1 | Morphogenesis, IMV membrane protein | GeneID:3707667 | PBR:42575 |
| A20R | 100 | 1 | 0 | 1 | Replication, DNA processivity factor | GeneID:3707671 | PBR:42579 |
| A24R* | 100 | 1 | 0 | 1 | RNA polymerase subunit 132 | GeneID:3707674 | PBR:42582 |
| A28L* | 100 | 3 | 0 | 1 | Entry, IMV virus entry | GeneID:3707681 | PBR:42589 |
| A34R | 100 | 1 | 0 | 1 | Morphogenesis, virus spread | GeneID:3707687 | PBR:42596 |
| A40R* | 1.46 | 4 | 0 | 1 | Unknown, lectin homolog | GeneID:3707695 | PBR:42604 |
| A53R* | 100 | 1 | 0 | 1 | Immunomodulator, TNF receptor | GeneID:3707710 | PBR:42618 |
| B2R* | 100 | 1 | 0 | 1 | Schlafen protein | GeneID:3707655 | PBR:42623 |
| B19R | 100 | 3 | 2 | 1 | Immunomodulator, type I interferon receptor | GeneID:3707577 | PBR:42639 |
| C4L | 100 | 1 | 0 | 1 | Immunomodulator, NfκB inhibitor | GeneID:3707639 | PBR:42461 |
| C5L | 34.31 | 2 | 1 | 1 | Unknown | GeneID:3707638 | PBR:42460 |
| D1R | 100 | 8 | 5 | 1 | Large capping enzyme | GeneID:3707562 | PBR:42544 |
| D2L | 100 | 2 | 1 | 1 | Virion core structural protein | GeneID:3707563 | PBR:42545 |
| E8R | 100 | 1 | 0 | 1 | Transcription, ER membrane protein | GeneID:3707597 | PBR:42502 |
| E9L | 100 | 3 | 2 | 1 | Replication, DNA polymerase | GeneID:3707598 | PBR:42503 |
| F4L | 100 | 5 | 3 | 1 | Replication, RNase reductase small subunit | GeneID:3707500 | PBR:42480 |
| F7L | 100 | 4 | 0 | 1 | Unknown | GeneID:3707503 | PBR:42483 |
| F16L* | 100 | 4 | 1 | 1 | Unknown | GeneID:3707512 | PBR:42493 |
| F17R | 100 | 1 | 0 | 1 | DNA binding phosphoprotein | GeneID:3707513 | PBR:42494 |
| H1L* | 100 | 2 | 0 | 1 | Viral transcription | GeneID:3707555 | PBR:42537 |
| I3L | 100 | 2 | 0 | 1 | Replication, DNA binding phosphoprotein | GeneID:3707605 | PBR:42510 |
| I4L* | 100 | 2 | 1 | 1 | Ribonucleotide reductase large subunit | GeneID:3707606 | PBR:42511 |
| I7L | 100 | 10 | 9 | 1 | Virion core protease | GeneID:3707609 | PBR:42514 |
| J2R | 100 | 1 | 0 | 1 | Thymidine kinase | GeneID:3707550 | PBR:42532 |
| K2L | 100 | 2 | 1 | 1 | Serpin, cell-cell fusion inhibitor | GeneID:3707648 | PBR:42470 |
| L1R | 100 | 4 | 2 | 1 | IMV membrane protein | GeneID:3707544 | PBR:42526 |
| VACWR011* | 100 | 1 | 0 | 1 | Immunomodulator, ubiquitin ligase | GeneID:3707626 | PBR:42448 |
| VACWR169* | 100 | 1 | 0 | 1 | Unknown | GeneID:3707699 | PBR:42608 |
| VACWR201 | 100 | 6 | 0 | 1 | Unknown | GeneID:3707578 |
| VACWR202 | 100 | 6 | 4 | 1 | Ankyrin | GeneID:3707579 |
| VACWR203 | 100 | 2 | 0 | 1 | Ankyrin | GeneID:3707580 |
Gene ID, gene names (asterisks highlight genes that are not mutated in M65 genome); %Sequence, percentages of sequencing coverage in comparison with WR genome; SNPs, single nucleotide polymorphism; No. of changes, number of missense mutations; Function, described functions of the different genes; EEV, equine encephalitis virus; ER, endoplasmic reticulum.
Mutations in genes involved in virus entry.
D8L accumulates the maximum number of missense mutations in each virus, 6 (L21W, H67R, W96L, E105A, K108T, and H213Y) in M65 and 7 (Q3R, L21W, H67R, Y104H, N175D, H213N, and P222L) in M101, and 2 (L21W and H67R) are shared in the two viruses. D8L has been described as a viral envelope protein responsible for binding to chondroitin sulfate on the surface of the cells. Inactivation of D8L in the WR strain results in a smaller viral plaques in CEF and decreases the viral ability to propagate in mouse brain (39). A27L, a gene that encodes a protein that binds to heparan sulfate and forms triple-coiled-coil trimers (40), presents two different missense mutations in each virus (A24N and K26E in M65 and F6S and A25D in M101).
Other than A16L—in the case of M101—G9R is the only gene that contains missense mutations among the genes in the vaccinia virus entry complex (formed by A28L, A21L, L5R, A16L, H2R, G3L, J5R, and G9R) (41). This gene presents 3 missense mutations (V5A, D21N, and H192Y) in M65 and 2 (D21N and H192Y) in M101. The missense mutation present in A16L is A36T.
Mutations in genes involved in late morphogenesis and plaque size.
Five proteins of VACV A33, A34, B5, F12, and F13 are present in the envelope of the virus, and the deletion of any of them causes a reduction in plaque size on cell monolayers (42). Among all those proteins, only A34 and F13L presented missense mutations in the genome of M65 and M101 and both proteins were mutated in both viruses.
M65 and M101 showed one missense mutation in the A34R gene (required for efficient intracellular trafficking from the endoplasmic reticulum to the site of wrapping) (42), in two different positions, S45I and Stop169R, respectively. The mutation in the stop codon adds 6 amino acids (R, Q, Q, K, M, and N) before reaching the next stop codon.
F13L is required for extracellular virus envelope (EV) formation and also plays a critical role in cell-to-cell transmission (43). F13 is needed for wrapping of intracellular mature virions by cisternae derived from trans-Golgi membranes or membranes from endosomes. Different mutations in F13L diminished EV production to various degrees and produced small plaque sizes (44, 45). Neither of the 2 mutations present in the M65 genome (A183V and R291K) and none of the 3 present in M101 (D235E, I236L, or R291K) coincide with the ones described previously (amino acids from 153 to 156 or K314R or D319E).
Another protein described as crucial in EV formation and virus spread is E2. The E2L gene presents one mutation in M65 (N690D) and 2 different mutations in M101 (V237A and N690D). Although no punctual missense mutations are correlated with a small-plaque phenotype, null E2L mutants produced small plaques and showed diminished production of EVs (46).
Alterations in genes involved in actin tail formation could cause an impact on plaque size as well. VACV gene F11L encodes a protein that inhibits RhoA-mDia signaling to facilitate the actin-dependent exocytosis of the virus (47). M65 presents one missense mutation (K327N), while M101 accumulates 3 different missense mutations (L89R, E328D, and I329V). The two viruses share a missense mutation generating a stop codon in position 332 that results in a truncation of 16 amino acids.
Mutations in genes involved in host defense or apoptosis modulation.
Genes C1L, N1L, M1L, and F1L accumulate a broad number of missense mutations in both M65 and M101. C1L encodes a hypothetical protein which shares a BCL-2 domain present in proteins involved in host defense (48). A frameshift mutation have been identified in the C1L sequence of both mutants in the 23rd nucleotide of the gene that encodes the 8th amino acid. Additional mutations have been identified, but none of them were present in the conserved BCL-2 domains described. Nonetheless, N1L, another member of the BCL-2 family (49) that acts as an NFκ-B inhibitor (50), showed the same two mutations in both viruses: D23N, present in the conserved BCL-2 α2 domain, and G84C, present at the end of the α5 domain.
The M1L deletion is enough to attenuate VACV (51) and shares 3 missense mutations (H130Q, N131H, and F377S) in the two viruses. The B18R gene, which encodes type I interferon binding protein (52), presents an I293M mutation in both viruses and an extra mutation in M101 (L468N). In addition to those genes, M65 virus presents two missense mutations (A15D and K21N) in F7L, a gene that encodes 3-beta-hydroxysteroid dehydrogenase and whose deletion causes attenuation in vivo (53).
Mutations in genes involved in DNA replication and transcription.
Some missense mutations were found in genes involved in DNA synthesis, replication, or transcription; most of them are present in the central region of the genome. K4L, which encodes a vaccinia virus nicking-joining enzyme (18), presents one mutation in M65 (A265S) and two in M101 (L35V and A265S). The E1L gene, encoding poly(A) polymerase (PAP) VP-55 (19), contains 3 identical mutations (L10F, N68D, and R199L) in the two viruses. Nuclease G5R (54) also presents identical missense mutations in the two viruses (E124K and N254T). G8R, which encodes structural orthologs of proliferating cell nuclear antigen (55), presents one missense mutation in M65 (A187V) and two in M101 (A87V and N224D). I8R, the gene encoding DNA and RNA helicases (56), contains one mutation (N217D) in the M65 genome and two (N217D and D296Y) in the M101 genome. The L3L gene encodes a transcription factor for early genes (57) and presents two mutations (Q34K and I275T) in both viruses. H4L RNA polymerase (58) presents two different mutations (Y623C and I624V) in the M101 genome. The H5R late gene transcription factor (59), which is phosphorylated in the threonine residue by the B1R complex (60), showed one missense mutation (R53G) in both viruses, and the M101 mutant presented another missense mutation (T15A) in one threonine.
Recombinants based on M65 and M101 mutant viruses elicit protection in a Leishmania-mouse model of infection.
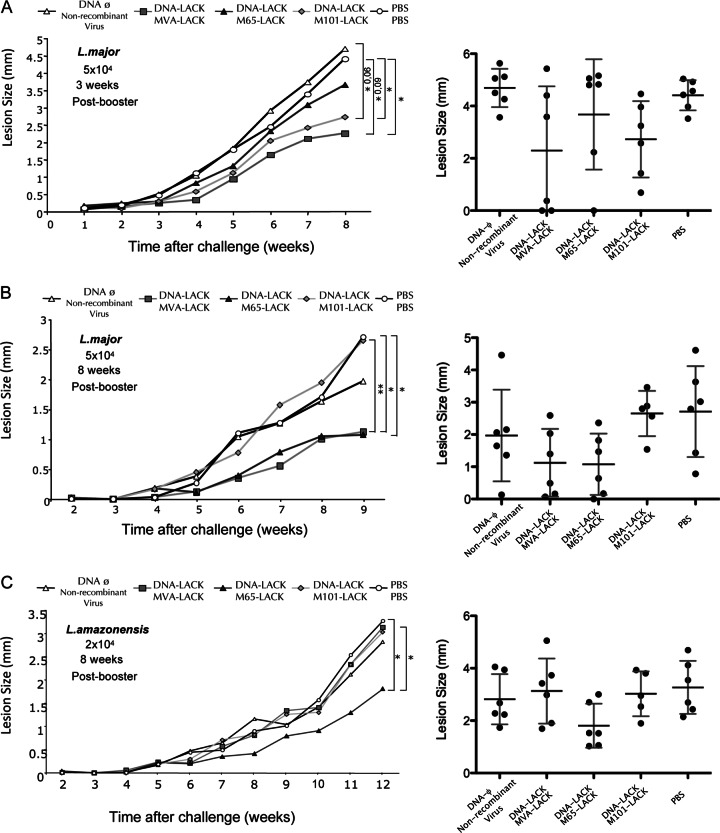
Since mutants M65 and M101 had acquired during persistence in FEL cells a number of properties (reduced plaque size phenotype, attenuation profile, limited replication capacity in an organism, and the presence of multiple mutations in viral genes with important biological functions), it was of interest to test whether these mutants could be of significant value as recombinant vaccine candidates. This was tested in a mouse model of cutaneous leishmaniosis after priming with DNA and boosting with M65 and M101 recombinants in comparison with the nonreplicating MVA strain, with all vectors expressing the Leishmania infantum LACK antigen, as this protocol is widely used to enhance antigen-specific T cell immune responses and to evaluate efficacy (5). With this aim, recombinant viruses that expressed the LACK antigen were generated as described in Materials and Methods. BALB/c mice (6 per group), 6 to 8 weeks of age, were primed i.d. in the abdomen with 100 μg of DNA-LACK or DNA-ϕ in a 100-μl volume per mouse. At day 14, mice were boosted i.p. with 2 × 107 PFU/mouse of M65-LACK, M101-LACK, or MVA-LACK or with nonrecombinant virus used as a control; PBS/PBS was also used as another control. Different conditions, short- and long-term, were designed for the challenge with the parasites: 5 × 104 metacyclic promastigotes of L. major were inoculated 3 weeks or 8 weeks after the booster (Fig. 7, panels A and B), and lesion size was followed weekly as described previously (20).
Fig 7.
Evaluation of protection conferred by MVA-LACK, M65-LACK, and M101-LACK recombinant viruses in heterologous prime/boost vaccination regimen. BALB/c mice (6 per group), 6 to 8 weeks of age, were primed intradermally (i.d.) in the abdomen with 100 μg of DNA-LACK or DNA-φ in a 100-μl volume per mouse. At day 14, mice were boosted intraperitoneally (i.p.) with 2 × 107 PFU/mouse of M65-LACK, M101-LACK, MVA-LACK, nonrecombinant parental virus, or PBS/PBS. Graphs represent development of lesions after different challenges with leishmania parasites: 5 × 104 L. major metacyclic promastigotes 3 weeks after the booster (A), 5 × 104 L. major metacyclic promastigotes 8 weeks after the booster (B), and 2 × 104 L. amazonensis stationary-phase synchronized promastigotes 8 weeks after the booster (C). Asterisks show P values for the different groups. Right panels represent each mouse lesion size measurement (closed symbol) at the experiment endpoint. The SD and mean values are indicated by vertical and horizontal bars, respectively.
In the short-term challenge (Fig. 7A), with parasite inoculation 3 weeks after vector booster, there was a 42% reduction in lesion size in the group that received DNA-LACK/M101-LACK in comparison with the DNA-ϕ/nonrecombinant virus group (P = 0.01) or a 38% reduction in comparison with the PBS/PBS group (P = 0.018). No significant reduction was observed in the group vaccinated with DNA-LACK/M65-LACK. The group boosted with MVA-LACK showed a reduction in lesion size similar to that seen with M101-LACK. In the long-term challenge (Fig. 6B), with parasite inoculation 8 weeks after booster, we observed a 59% reduction in lesion size in the group that received DNA-LACK/M65-LACK in comparison with the group that received PBS/PBS (P = 0.043) and a 60% reduction in comparison with the group that received DNA-LACK/M101-LACK (P = 0.011), which failed to protect. It is also remarkable that in the group that received DNA-LACK/M65-LACK, 33% of animals healed the initial inflammation postchallenge and did not develop lesions. The group boosted with MVA-LACK showed a reduction in lesion size similar to that seen with M65-LACK.
To confirm if vaccine protection against other strains of Leishmania that caused cutaneous leishmaniasis, as with Leishmania amazonensis, could be observed, groups of 6 animals each were immunized by prime/boost following the same protocol as described above and animals challenged 8 weeks after booster with 2 × 104 stationary-phase synchronized promastigotes of L. amazonensis inoculated in the footpad. As shown in Fig. 7C, the combination of DNA-LACK and M65-LACK delayed the development of lesions and animals presented a 44% reduction in lesion size in comparison with animals that received PBS/PBS (P = 0.02), a 35% reduction in comparison with animals that received nonrecombinant virus (P = 0.08), and a 40% reduction in comparison with animals that received DNA-LACK/M101-LACK vaccination (P = 0.04). The group boosted with MVA-LACK did not show a reduction in lesion size and behaved in a manner similar to that seen with the group administered M101-LACK.
Overall, mouse vaccination and pathogenicity studies revealed that recombinants based on M65 or M101 can induce protection against different strains of leishmania. Moreover, these mutants exhibit distinct behavior in triggering immediate and delayed protective immune responses. The highly attenuated nonreplicative MVA-LACK strain protected during short and long times after the booster against L. major challenge but not after L. amazonensis challenge. The M101-LACK mutant conferred protection at short times after the booster but not at long times after the booster; however, M65-LACK triggered protection when animals were challenged at long times after a booster with both L. major and L. amazonensis strains.
Differential activation of memory immune responses in animals immunized with recombinant MVA-LACK, M65-LACK, or M101-LACK virus.
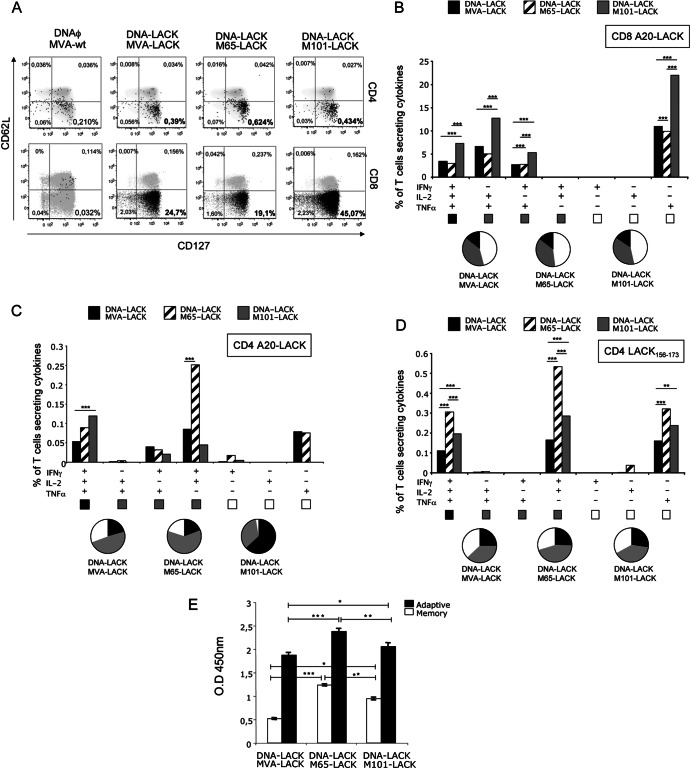
Since memory T cell responses are critical for induction of long-term protection against parasitic infection, we assessed at 8 weeks postboost the memory profile elicited by the heterologous prime/boost immunization using the different vectors. We defined long-term responses from the phenotype of the LACK-specific memory T cells. Splenocytes from mice immunized as described for Fig. 8 were stained for CD4, CD8, CD127, and CD62L surface markers in order to differentiate between different memory populations. With those markers, we can distinguish between central memory T cells (CD127+ CD62L+), effector memory T cells (CD127+ CD62L−), and activated effector T cells (CD127− CD62L−). We also evaluated IFN-γ, TNF-α, and IL-2 secretion by ICS after in vitro stimulation with LACK157–173 peptide (a crucial CD4+ T cell-restricted peptide involved in the development of a Th2 response and susceptibility [61]) or A20 cells nucleofected with the mammalian expression plasmid pCI-Neo-LACK.
Fig 8.
Memory immune response in mice immunized with the different mutant viruses. BALB/c mice (4 per group) were immunized as described for Fig. 7. (A) Analysis of the phenotype of memory antigen-specific CD4+ and CD8+ T cells in splenocytes restimulated with A20 cells nucleofected with pCINeo-LACK. Memory T cells were classified as central memory (CD62L+ CD127+), effector memory (CD62L− CD127+), or effector (CD62L− CD127−). Percentages represent the frequencies of T cells secreting IFN-γ and/or TNF-α and/or IL-2. (B to D) Within the lymphocyte population, T cells were gated and analyzed for IFN-γ, TNF-α, and/or IL-2 production. Cytokine production by LACK-specific CD8+ T cells (B), LACK-specific CD4+ T cells (C), and LACK157-173 peptide-specific CD4+ T cells (D) was analyzed. The different combinations of cytokines are indicated on the x axis; percentages of T cells producing any cytokine are indicated on the y axis. The different pies show the quality of the response measured as the relative quantities of single-, double-, or triple-cytokine-producing cells. (E) Humoral response to the viral vector, measured by ELISA in serum (1:50 dilution) of infected mice at 11 days (adaptive) or 8 weeks (memory) post-booster immunization. O.D, optical density.
The sum of the frequencies of antigen-specific T cells secreting IFN-γ and/or IL-2 and/or TNF-α was determined. At 8 weeks postboost, there was a specific induction of CD4+ and CD8+ T cells in the groups of mice that received DNA-LACK/MVA-LACK, DNA-LACK/M65-LACK, or DNA-LACK/M101-LACK in comparison with animals immunized with DNAϕ/MVA-wt (Fig. 8A); both the CD4+ and the CD8+ antigen-specific T cell responses had mainly an effector memory phenotype (CD127+ CD62L−). The higher magnitude of the CD4+ T cell response was observed in splenocytes from animals immunized with DNA-LACK/M65-LACK (0.624% versus 0.39% in splenocytes from animals immunized with DNA-LACK/MVA-LACK or 0.434% in splenocytes from animals immunized with DNA-LACK/M101-LACK). Similar differences were observed when splenocytes were restimulated with LACK157–173 peptide. The highest CD8+ T cell response was observed in splenocytes from animals immunized with DNA-LACK/M101-LACK (45.07% versus 24.7% obtained with DNA-LACK/MVA-LACK or 19.1% in splenocytes from animals immunized with DNA-LACK/M65-LACK).
The simultaneous measurements of three cytokines allowed the assessment of the quality of the vaccine-induced CD4+ and CD8+ T cell responses. On the basis of the analysis of IFN-γ, TNF-α, and IL-2 secretion, seven distinct LACK-specific CD4+ and CD8+ T cell populations were identified. As shown in Fig. 8B, the heterologous vaccination protocol consisting of DNA-LACK/MVA-LACK, DNA-LACK/M65-LACK, or DNA-LACK/M101-LACK induces polyfunctional CD8+ T cell responses, with around 50% of cells secreting two or more cytokines. Although all the different immunizations triggered the same polyfunctional profile, the magnitude of the response was markedly higher in splenocytes from animals immunized with the combination of DNA-LACK and M101-LACK.
CD4+ T cells also showed a high polyfunctional profile against A20 cells nucleofected with pCINeo-LACK plasmid (Fig. 8C). Splenocytes from animals receiving DNA-LACK/M101-LACK showed the highest percentage of triple-positive T cells (61.3% versus 48% in splenocytes from animals immunized with DNA-LACK/M65-LACK or 18.9% in splenocytes from animals immunized with DNA-LACK/M65-LACK). Splenocytes from animals that received DNA-LACK/M65-LACK showed the highest magnitude of double-positive T cells (61.3%, represented by cells that secreted IFN-γ and IL-2). Nevertheless, when splenocytes were stimulated with LACK157–173 peptide, all groups exhibited the same polyfunctional profile but the highest magnitude of the different antigen-specific CD4+ T cells was detected in animals immunized with DNA-LACK/M65-LACK (Fig. 8D).
The overall antibody levels in sera specific for VACV proteins were slightly reduced in the recombinant vector MVA versus the mutant viruses M65 and M101 (Fig. 8E).
The findings described above showed that, at the memory phase, both polyfunctional CD4+ and CD8+ T cells with an effector memory phenotype were activated by the different virus vectors, but DNA-LACK/M65-LACK preferentially induced CD4+ T cell responses, while DNA-LACK/M101-LACK preferentially induced CD8+ T cell responses.
DISCUSSION
Among viral vectors, poxviruses have become among the most promising candidates for development of recombinant vaccines against prevalent diseases. The first generation of vaccines against cancer, HIV/AIDS, and other infectious diseases was based on replication-competent strains of VACV such as WR, Wyeth, Lister, Copenhagen, and Tian Tan (62, 63); due to safety concerns, however, most of the candidates currently assayed in vaccine studies are nonreplicative VACV vectors such as canarypox, fowlpox, MVA, or NYVAC (3, 5). Although highly attenuated vectors are capable of inducing protective immunity against a variety of pathogens, their limitation in replication capacity reduces their potential to induce immune responses similarly to replication-competent counterparts (64–67). In order to find an equilibrium between safety and replication, different poxviruses with an intermediate phenotype are being sought.
In this investigation, we have taken advantage of two VACV mutants, M65 and M101, which are attenuated variants generated after 65 and 101 serial passages of persistently infected Friend erythroleukemia (FEL) cells (11). This cell line was also used by Pogo and Friend to establish persistent infections with the VACV IHD-W strain and to isolate deletion mutants, but viruses were not characterized in detail (9). Although the mutants present different attenuation levels, as demonstrated in this study, the pathogenicity of both viruses is strongly reduced in mice in comparison with the WR parental strain. In vitro replication studies in cells of various origins showed that viral titers of the mutants were slightly reduced in comparison with those of WR, depending on the cell line. In mice, the mutant virus M65-Luc showed a replication capacity similar to that of WR-Luc virus. In the case of M101-Luc, it showed a more restricted phenotype, comparable to that of MVA-Luc in lymph nodes and spleen, although M101-Luc expression persisted longer than MVA-Luc expression in organs such as ovaries or in cells of the peritoneal cavity.
Wide genome analysis after deep sequencing revealed that the two viruses shared identical deletions in the left end of the genome, comprehending some immunomodulatory but mainly host range genes (from the gene encoding IL-18 binding protein to C5L; Fig. 5). This is a common phenomenon that occurs in the VACV genome after serial passages in cultured cells or eggs, as was described previously (68). Other strains of vaccinia virus generated after serial passages such as Dairen I (obtained after 13 serial passages of the parental Dairen vaccine strain in 1-day-old eggs (69) and MVA (generated after more than 500 passages in CEF cells) present large deletions in the left end of the genome (70). In the case of the Dairen I strain, the 15.4-Kb deletion involves genes from C9L to K5L. The left end of the MVA genome lacks genes from C16L to C23L, from C8L to K1L, and K6L.
Among other vaccine strains, NYVAC (genetically engineered by the removal of 18 genes from the Copenhagen strain) lacks genes from C7L to K1L. The principal difference with these 3 strains of viruses with the left-end deletions is that M65 and M101 virus maintain intact N1L, N2L, M1L, M2L, and K1L genes that are deleted in Dairen I, MVA, and NYVAC. This region was also removed from the Tian Tan strain for increased attenuation, leading to reduced replication capacity in rabbit and human cell lines (51). A comparison of the different genes deleted in the left end of the genome is summarized in Fig. 6A.
The main difference between the M65 and M101 genomes is a small deletion in the right end of the genome of M101 virus that is not present in M65 genome and covers A39R and A40R genes. The absence of the A39R (which is already truncated in WR strain) or A40R gene cannot explain the phenotypical differences observed between M65 and M101, such as plaque size, attenuation, or replication capacity. MVA has also deleted the A39R gene and accumulated other deletions in the right flank of the genome. For other strains, such as Dairen I, there is no information about the right flank of the genome. A comparison between virus strains of the different genes deleted in the right end of the genome is summarized in Fig. 6B.
The key biological differences between M65 and M101 could be explained by the increased number of missense mutations present in the M101 genome, particularly in genes involved in virus entry or in late morphogenesis and plaque size. Although none of the particular mutations identified in those genes have been described as crucial for virus replication, the particular roles of different mutations in virus-host cell interactions, morphogenesis, and plaque size remain to be determined. In the latter case, A34R showed a missense mutation in its natural stop codon that can provide, in part, an explanation for the smaller plaque size mentioned in Table 3 and for the in vivo attenuation findings.
Table 3.
Comparative biological characteristics of M65 and M101 mutant virusesa
| Property | Characteristic | M65 | M101 |
|---|---|---|---|
| Replication | Plaque size | Intermediate | Small |
| In vitro replication | Similar to that of WR virus | Slightly reduced in comparison with WR | |
| In vivo replication | Similar to that of WR but maintaining higher levels of the marker luciferase 72 h postinfection in ovaries and spleen cells | Similar to that of MVA but maintaining higher levels of the marker luciferase in ovaries and cells from the peritoneal cavity | |
| Pathogenesis | Weight loss | Transient in BALB/c mice inoculated with 107 PFU/mouse or 108 PFU/mouse; transient in immunocompromised mice inoculated with 108 PFU/mouse | Not observed in either BALB/c mice or C57BL6 immunocompromised mice |
| Signs of disease | Transient rough hair coat, loss of mobility, and hunched posture | Not observed | |
| Histopathology | Leukocyte extravasation and accumulation; inflammation of lung tissue; significant presence of viral proteins in affected areas | Reduced leukocyte extravasation and accumulation; reduced inflammation of lung tissue; reduced presence of viral proteins in affected areas | |
| Genome | Deletions | 7-kb left-end deletion comprising C5L, C6L, C7L, C8L, C9L,VACWR013, VACWR014, VACWR015,VACWR016, VACWR017, and VACWR018 | 7-kb left-end deletion comprising C5L, C6L, C7L, C8L, C9L, VACWR013, VACWR014, VACWR015, VACWR016, VACWR017, and VACWR018 plus small deletion of A39R-A40R in right end |
| Mutations | 59 genes containing at least one missense mutation | 61 genes containing at least one missense mutation; higher no. of missense mutations per gene than in M65 genome | |
| Protection against leishmania | Challenge 3 wks postbooster | Protection not observed against L. major | Reduction in lesion size in animals infected with L. major |
| Challenge 8 wks postbooster | 33% complete protection and 59% reduction of footpad lesion size against L .major; reduction in footpad lesion development in animals challenged with L. amazonensis | Protection not observed against both L. major or L. amazonensis challenge | |
| Immune response | Memory immune responses | Preferentially induced high quality CD4+ LACK-specífic T cell responses | Preferentially induced high-quality CD8+ LACK-specífic T cell responses |
Specific properties corresponding to in vitro or in vivo virus replication, pathogenesis, genetic sequences, protection against leishmaniasis, and immune responses triggered are indicated.
It is significant that the viral gene K3L, involved in the inhibition of the antiviral protein kinase PKR (71), remained intact after over 100 passages, indicating the likely requirement of that gene to counteract the host antiviral response for the viruses to remain viable. In this regard, the phenomenon of the presence of “gene-accordions” for VACV to rapidly adapt in culture to defeat host defenses in spite of low mutation rates has been described recently (72). Clearly, this phenomenon has not been observed in the persistent infection of FEL cells, as the antiviral factors E3L and K3L were not modified by mutations.
To further evaluate the potential of M65 and M101 as vaccine vectors against diseases, recombinant viruses that expressed the protective Leishmania LACK antigen were generated and tested in heterologous DNA prime/poxvirus boost regimens for protective efficacy against leishmaniasis. Different degrees of protection or delay in the onset of infection were observed with the two viruses depending on the timing between booster and challenge. When animals were challenged with L. major at 3 weeks after the virus boost, higher levels of protection were observed in the group that received DNA-LACK/M101-LACK compared with groups boosted with M65-LACK. Notwithstanding, when parasites were administered 8 weeks after the virus boost, protection was mainly observed in mice that received DNA-LACK/M65-LACK. The same results seen with M65-LACK were obtained in animals challenged 8 weeks after the booster with the L. amazonensis strain. These results suggest that replication competence could play an important role in disease prevention. The mutant with a limited replication phenotype, M101, is more effective in protection than a more active replication-competent virus when parasite infection occurs within few weeks after vaccination; in contrast, more effective parasite protection is obtained with a more active replication-competent virus, M65, when infection occurs at least 2 months after vaccination. In the case of the MVA virus strain, protection after vaccination was observed at early and late parasite challenges. This could be explained by a higher number of immunomodulatory genes missing in the MVA genome versus the genomes of mutants M65 and M101. For this reason, we also analyzed in this work the memory immune response elicited by the heterologous vaccination approach in order to determine if replication capacity could have an impact on the immune response to the heterologous antigen. We evaluated the antigen-specific CD4+ and CD8+ T cell responses, as they have a major role in the immunology of cutaneous and visceral leishmaniasis (73).
High-quality CD4+ and CD8+ antigen-specific T cells with an effector memory phenotype were activated by the different viruses in a heterologous prime/boost regimen. The importance of these polyfunctional T cells might be explained by the fact that each single cell is capable of a broader repertoire of functions. The DNA-LACK/M65-LACK regimen preferentially induced CD4+ T cell responses, while the DNA-LACK/M101-LACK regimen preferentially induced CD8+ T cell responses. DNA-LACK/MVA-LACK induced an intermediate phenotype. It was recently described that CD8+ effector memory T cells are the most relevant subset for long-term protective immunity against parasites (74, 75), but high-quality effector memory CD4+ T cells might also play a relevant role in protection. Darrah et al. demonstrated for L. major that expansion of multifunctional CD4+ T cells secreting IFN-γ, IL-2, and TNF-α strongly correlates with protection against subsequent challenge (76). Furthermore, the presence of high-quality, multifunctional CD4+ T cells in peripheral blood mononuclear cells of patients that healed American cutaneous leishmaniasis, thus supporting the significance of our findings, has also been reported (77). The increase in the frequency of CD4+ T cells observed in the animals inoculated with M65-LACK virus that can be explained by a higher persistence of heterologous antigen due to an increased replication capacity is remarkable (78). This also points out that different vector replication capacities can trigger the activation and proliferation of different immune populations.
Altogether, our findings showed the adaptive changes introduced in the VACV genome during a long-term persistent virus infection and the biological consequences, as derived from studies of two viral mutants isolated after 65 and 101 passages in FEL cells. We found that M65 and M101 mutants of VACV have biological and immunological characteristics in both in vitro and in vivo systems that are different from those of the parental WR and MVA strains, based on safety, replication competency, pathogenicity, immune profiling, and protective efficacy (Table 3). M101 could be considered a vaccine candidate when more antigen-specific CD8+ T cell activation is needed, while M65 could be used when more CD4+ T cells are required. Nonetheless, although both viruses are based on the VACV WR strain and clinical application to use this strain has been done only for the treatment of advanced tumors as a compassion measure (79), considering future clinical approval, the vectors developed in this work will need to be further modified by selective removal of immunomodulatory genes, as we have proposed and performed (5, 31, 33). M65 could also be used as an oncolytic vector when more active replication competency is required, while M101 could be used as a priming and/or booster component when more CD8+ T cell responses to selected antigens are needed.
Overall, our results provide clear differences between these mutants (M65 and M101) and the widely studied MVA strain. Specifically, differences included wider host range replication competency, longer persistence of gene expression in tissues, and differential impacts on LACK-specific T (CD4+ and CD8+) cell populations and on protective efficacy against Leishmania species. These findings identify novel biological and immunological attributes of VACV mutants during the selective pressure in cultured FEL cells that are different from those arising during the generation of MVA in CEF cells, which are relevant observations for VACV evolution and vaccine development.
ACKNOWLEDGMENTS
This investigation was supported by research grant SAF2008-02036. L.S.-S. and E.M.P. were supported by Ph.D. Fellowships from the Ministry of Economy.
We are grateful to Victoria Jiménez for tissue culture and virus growth. We thank Diane McMahon-Pratt for providing the L. amazonensis LTB0016 strain and Heinz Himmelbauer of the Ultrasequencing Unit at CRG (Spain) for DNA sequencing.
Footnotes
Published ahead of print 17 April 2013
REFERENCES
- 1. Gómez CE, Najera JL, Krupa M, Perdiguero B, Esteban M. 2011. MVA and NYVAC as vaccines against emergent infectious diseases and cancer. Curr. Gene Ther. 11:189–217 [DOI] [PubMed] [Google Scholar]
- 2. Gómez CE, Najera JL, Krupa M, Esteban M. 2008. The poxvirus vectors MVA and NYVAC as gene delivery systems for vaccination against infectious diseases and cancer. Curr. Gene Ther. 8:97–120 [DOI] [PubMed] [Google Scholar]
- 3. Esteban M. 2009. Attenuated poxvirus vectors MVA and NYVAC as promising vaccine candidates against HIV/AIDS. Hum. Vaccin. 5:867–871 [DOI] [PubMed] [Google Scholar]
- 4. Pantaleo G, Esteban M, Jacobs B, Tartaglia J. 2010. Poxvirus vector-based HIV vaccines. Curr. Opin. HIV AIDS 5:391–396 [DOI] [PubMed] [Google Scholar]
- 5. Gómez CE, Perdiguero B, Garcia-Arriaza J, Esteban M. 2012. Poxvirus vectors as HIV/AIDS vaccines in humans. Hum. Vaccin. Immunother. 8:1192–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nájera JL, Gómez CE, García-Arriaza J, Sorzano CO, Esteban M. 2010. Insertion of vaccinia virus C7L host range gene into NYVAC-B genome potentiates immune responses against HIV-1 antigens. PLoS One 5:e11406 doi: 10.1371/journal.pone.0011406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kibler KV, Gomez CE, Perdiguero B, Wong S, Huynh T, Holechek S, Arndt W, Jimenez V, Gonzalez-Sanz R, Denzler K, Haddad EK, Wagner R, Sekaly RP, Tartaglia J, Pantaleo G, Jacobs BL, Esteban M. 2011. Improved NYVAC-based vaccine vectors. PLoS One 6:e25674 doi: 10.1371/journal.pone.0025674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lai AC, Pogo BG. 1989. Characterization of vaccinia virus deletion mutants isolated from persistently infected Friend erythroleukemia cells. Virus Res. 12:239–250 [DOI] [PubMed] [Google Scholar]
- 9. Pogo BG, Friend C. 1982. Persistent infection of Friend erythroleukemia cells with vaccinia virus. Proc. Natl. Acad. Sci. U. S. A. 79:4805–4809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Paez E, Dallo S, Esteban M. 1987. Virus attenuation and identification of structural proteins of vaccinia virus that are selectively modified during virus persistence. J. Virol. 61:2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Paez E, Dallo S, Esteban M. 1985. Generation of a dominant 8-MDa deletion at the left terminus of vaccinia virus DNA. Proc. Natl. Acad. Sci. U. S. A. 82:3365–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McMahon-Pratt D, Rodriguez D, Rodriguez JR, Zhang Y, Manson K, Bergman C, Rivas L, Rodriguez JF, Lohman KL, Ruddle NH. 1993. Recombinant vaccinia viruses expressing GP46/M-2 protect against Leishmania infection. Infect. Immun. 61:3351–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li S, Rodrigues M, Rodriguez D, Rodriguez JR, Esteban M, Palese P, Nussenzweig RS, Zavala F. 1993. Priming with recombinant influenza virus followed by administration of recombinant vaccinia virus induces CD8+ T-cell-mediated protective immunity against malaria. Proc. Natl. Acad. Sci. U. S. A. 90:5214–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bloom DC, Edwards KM, Hager C, Moyer RW. 1991. Identification and characterization of two nonessential regions of the rabbitpox virus genome involved in virulence. J. Virol. 65:1530–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rodriguez JF, Rodriguez D, Rodriguez JR, McGowan EB, Esteban M. 1988. Expression of the firefly luciferase gene in vaccinia virus: a highly sensitive gene marker to follow virus dissemination in tissues of infected animals. Proc. Natl. Acad. Sci. U. S. A. 85:1667–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. World Health Organization 2004. Report of the Scientific Working Group meeting on leishmaniasis. TDR Research Publications, Geneva, Switzerland [Google Scholar]
- 17. Ahmed SBH, Bahloul C, Robbana C, Askri S, Dellagi K. 2004. A comparative evaluation of different DNA vaccine candidates against experimental murine leishmaniasis due to L. major. Vaccine 22:1631–1639 [DOI] [PubMed] [Google Scholar]
- 18. Eckert D, Williams O, Meseda CA, Merchlinsky M. 2005. Vaccinia virus nicking-joining enzyme is encoded by K4L (VACWR035). J. Virol. 79:15084–15090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gershon PD. 2004. Studying vaccinia virus RNA processing in vitro. Methods Mol. Biol. 269:151–168 [DOI] [PubMed] [Google Scholar]
- 20. Pérez-Jiménez E, Kochan G, Gherardi MM, Esteban M. 2006. MVA-LACK as a safe and efficient vector for vaccination against leishmaniasis. Microbes Infect. 8:810–822 [DOI] [PubMed] [Google Scholar]
- 21. Gómez CE, Najera JL, Domingo-Gil E, Ochoa-Callejero L, Gonzalez-Aseguinolaza G, Esteban M. 2007. Virus distribution of the attenuated MVA and NYVAC poxvirus strains in mice. J. Gen. Virol. 88:2473–2478 [DOI] [PubMed] [Google Scholar]
- 22. Didierlaurent A, Ramirez JC, Gherardi M, Zimmerli SC, Graf M, Orbea HA, Pantaleo G, Wagner R, Esteban M, Kraehenbuhl JP, Sirard JC. 2004. Attenuated poxviruses expressing a synthetic HIV protein stimulate HLA-A2-restricted cytotoxic T-cell responses. Vaccine 22:3395–3403 [DOI] [PubMed] [Google Scholar]
- 23. Ramsay AJ, Leong KH, Ramshaw IA. 1997. DNA vaccination against virus infection and enhancement of antiviral immunity following consecutive immunization with DNA and viral vectors. Immunol. Cell Biol. 75:382. [DOI] [PubMed] [Google Scholar]
- 24. Chakrabarti S, Brechling K, Moss B. 1985. Vaccinia virus expression vector: coexpression of beta-galactosidase provides visual screening of recombinant virus plaques. Mol. Cell. Biol. 5:3403–3409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mooij P, Balla-Jhagjhoorsingh SS, Koopman G, Beenhakker N, van Haaften P, Baak I, Nieuwenhuis IG, Kondova I, Wagner R, Wolf H, Gomez CE, Najera JL, Jimenez V, Esteban M, Heeney JL. 2008. Differential CD4+ versus CD8+ T-cell responses elicited by different poxvirus-based human immunodeficiency virus type 1 vaccine candidates provide comparable efficacies in primates. J. Virol. 82:2975–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. García-Arriaza J, Najera JL, Gomez CE, Sorzano CO, Esteban M. 2010. Immunogenic profiling in mice of a HIV/AIDS vaccine candidate (MVA-B) expressing four HIV-1 antigens and potentiation by specific gene deletions. PLoS One 5:e12395 doi: 10.1371/journal.pone.0012395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nájera JL, Gomez CE, Garcia-Arriaza J, Sorzano CO, Esteban M. 2010. Insertion of vaccinia virus C7L host range gene into NYVAC-B genome potentiates immune responses against HIV-1 antigens. PLoS One 5:e11406 doi: 10.1371/journal.pone.0011406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ramírez JC, Gherardi MM, Esteban M. 2000. Biology of attenuated modified vaccinia virus Ankara recombinant vector in mice: virus fate and activation of B- and T-cell immune responses in comparison with the Western Reserve strain and advantages as a vaccine. J. Virol. 74:923–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rodriguez D, Rodriguez JR, Rodriguez JF, Trauber D, Esteban M. 1989. Highly attenuated vaccinia virus mutants for the generation of safe recombinant viruses. Proc. Natl. Acad. Sci. U. S. A. 86:1287–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. García-Arriaza J, Najera JL, Gomez CE, Tewabe N, Sorzano CO, Calandra T, Roger T, Esteban M. 2011. A candidate HIV/AIDS vaccine (MVA-B) lacking vaccinia virus gene C6L enhances memory HIV-1-specific T-cell responses. PLoS One 6:e24244 doi: 10.1371/journal.pone.0024244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Unterholzner L, Sumner RP, Baran M, Ren H, Mansur DS, Bourke NM, Randow F, Smith GL, Bowie AG. 2011. Vaccinia virus protein C6 is a virulence factor that binds TBK-1 adaptor proteins and inhibits activation of IRF3 and IRF7. PLoS Pathog. 7:e1002247 doi: 10.1371/journal.ppat.1002247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Symons JA, Adams E, Tscharke DC, Reading PC, Waldmann H, Smith GL. 2002. The vaccinia virus C12L protein inhibits mouse IL-18 and promotes virus virulence in the murine intranasal model. J. Gen. Virol. 83:2833–2844 [DOI] [PubMed] [Google Scholar]
- 36. Gardner JD, Tscharke DC, Reading PC, Smith GL. 2001. Vaccinia virus semaphorin A39R is a 50–55 kDa secreted glycoprotein that affects the outcome of infection in a murine intradermal model. J. Gen. Virol. 82:2083–2093 [DOI] [PubMed] [Google Scholar]
- 37. Palacios S, Perez LH, Welsch S, Schleich S, Chmielarska K, Melchior F, Locker JK. 2005. Quantitative SUMO-1 modification of a vaccinia virus protein is required for its specific localization and prevents its self-association. Mol. Biol. Cell 16:2822–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wilcock D, Duncan SA, Traktman P, Zhang WH, Smith GL. 1999. The vaccinia virus A4OR gene product is a nonstructural, type II membrane glycoprotein that is expressed at the cell surface. J. Gen. Virol. 80(Pt 8):2137–2148 [DOI] [PubMed] [Google Scholar]
- 39. Chernos VI, Vovk TS, Ivanova ON, Antonova TP, Loparev VN. 1993. Insertion mutants of the vaccinia virus. The effect of inactivating E7R and D8L genes on the biological properties of the virus. Mol. Gen. Mikrobiol. Virusol. 1993:30–34 (In Russian.) [PubMed] [Google Scholar]
- 40. Hsiao JC, Chung CS, Chang W. 1999. Vaccinia virus envelope D8L protein binds to cell surface chondroitin sulfate and mediates the adsorption of intracellular mature virions to cells. J. Virol. 73:8750–8761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Senkevich TG, Ojeda S, Townsley A, Nelson GE, Moss B. 2005. Poxvirus multiprotein entry-fusion complex. Proc. Natl. Acad. Sci. U. S. A. 102:18572–18577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Earley AK, Chan WM, Ward BM. 2008. The vaccinia virus B5 protein requires A34 for efficient intracellular trafficking from the endoplasmic reticulum to the site of wrapping and incorporation into progeny virions. J. Virol. 82:2161–2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Blasco R, Moss B. 1991. Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000-Dalton outer envelope protein. J. Virol. 65:5910–5920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Honeychurch KM, Yang G, Jordan R, Hruby DE. 2007. The vaccinia virus F13L YPPL motif is required for efficient release of extracellular enveloped virus. J. Virol. 81:7310–7315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Roper RL, Moss B. 1999. Envelope formation is blocked by mutation of a sequence related to the HKD phospholipid metabolism motif in the vaccinia virus F13L protein. J. Virol. 73:1108–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Domi A, Weisberg AS, Moss B. 2008. Vaccinia virus E2L null mutants exhibit a major reduction in extracellular virion formation and virus spread. J. Virol. 82:4215–4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Arakawa Y, Cordeiro JV, Schleich S, Newsome TP, Way M. 2007. The release of vaccinia virus from infected cells requires RhoA-mDia modulation of cortical actin. Cell Host Microbe 1:227–240 [DOI] [PubMed] [Google Scholar]
- 48. González JM, Esteban M. 2010. A poxvirus Bcl-2-like gene family involved in regulation of host immune response: sequence similarity and evolutionary history. Virol. J. 7:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cooray S, Bahar MW, Abrescia NG, McVey CE, Bartlett NW, Chen RA, Stuart DI, Grimes JM, Smith GL. 2007. Functional and structural studies of the vaccinia virus virulence factor N1 reveal a Bcl-2-like anti-apoptotic protein. J. Gen. Virol. 88(Pt 6):1656–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. DiPerna G, Stack J, Bowie AG, Boyd A, Kotwal G, Zhang Z, Arvikar S, Latz E, Fitzgerald KA, Marshall WL. 2004. Poxvirus protein N1L targets the I-kappaB kinase complex, inhibits signaling to NF-kappaB by the tumor necrosis factor superfamily of receptors, and inhibits NF-kappaB and IRF3 signaling by toll-like receptors. J. Biol. Chem. 279:36570–36578 [DOI] [PubMed] [Google Scholar]
- 51. Zhu W, Fang Q, Zhuang K, Wang H, Yu W, Zhou J, Liu L, Tien P, Zhang L, Chen Z. 2007. The attenuation of vaccinia Tian Tan strain by the removal of the viral M1L-K2L genes. J. Virol. Methods 144:17–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Colamonici OR, Domanski P, Sweitzer SM, Larner A, Buller RM. 1995. Vaccinia virus B18R gene encodes a type I interferon-binding protein that blocks interferon alpha transmembrane signaling. J. Biol. Chem. 270:15974–15978 [DOI] [PubMed] [Google Scholar]
- 53. Moore JB, Smith GL. 1992. Steroid hormone synthesis by a vaccinia enzyme: a new type of virus virulence factor. EMBO J. 11:1973–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Da Silva M, Shen L, Tcherepanov V, Watson C, Upton C. 2006. Predicted function of the vaccinia virus G5R protein. Bioinformatics 22:2846–2850 [DOI] [PubMed] [Google Scholar]
- 55. Da Silva M, Upton C. 2009. Vaccinia virus G8R protein: a structural ortholog of proliferating cell nuclear antigen (PCNA). PLoS One 4:e5479 doi: 10.1371/journal.pone.0005479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bayliss CD, Smith GL. 1996. Vaccinia virion protein I8R has both DNA and RNA helicase activities: implications for vaccinia virus transcription. J. Virol. 70:794–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Resch W, Moss B. 2005. The conserved poxvirus L3 virion protein is required for transcription of vaccinia virus early genes. J. Virol. 79:14719–14729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mohamed MR, Niles EG. 2001. The viral RNA polymerase H4L subunit is required for Vaccinia virus early gene transcription termination. J. Biol. Chem. 276:20758–20765 [DOI] [PubMed] [Google Scholar]
- 59. Kovacs GR, Moss B. 1996. The vaccinia virus H5R gene encodes late gene transcription factor 4: purification, cloning, and overexpression. J. Virol. 70:6796–6802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Beaud G, Beaud R, Leader DP. 1995. Vaccinia virus gene H5R encodes a protein that is phosphorylated by the multisubstrate vaccinia virus B1R protein kinase. J. Virol. 69:1819–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Launois P, Pingel S, Himmelrich H, Locksley R, Louis J. 2007. Different epitopes of the LACK protein are recognized by V beta 4 V alpha 8 CD4+ T cells in H-2b and H-2d mice susceptible to Leishmania major. Microbes Infect. 9:1260–1266 [DOI] [PubMed] [Google Scholar]
- 62. Perkus ME, Tartaglia J, Paoletti E. 1995. Poxvirus-based vaccine candidates for cancer, AIDS, and other infectious diseases. J. Leukoc. Biol. 58:1–13 [DOI] [PubMed] [Google Scholar]
- 63. Jacobs BL, Langland JO, Kibler KV, Denzler KL, White SD, Holechek SA, Wong S, Huynh T, Baskin CR. 2009. Vaccinia virus vaccines: past, present and future. Antiviral Res. 84:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Abaitua F, Rodriguez JR, Garzon A, Rodriguez D, Esteban M. 2006. Improving recombinant MVA immune responses: potentiation of the immune responses to HIV-1 with MVA and DNA vectors expressing Env and the cytokines IL-12 and IFN-gamma. Virus Res. 116:11–20 [DOI] [PubMed] [Google Scholar]
- 65. Dai K, Liu Y, Liu M, Xu J, Huang W, Huang X, Liu L, Wan Y, Hao Y, Shao Y. 2008. Pathogenicity and immunogenicity of recombinant Tiantan Vaccinia Virus with deleted C12L and A53R genes. Vaccine 26:5062–5071 [DOI] [PubMed] [Google Scholar]
- 66. Karkhanis LU, Ross TM. 2007. Mucosal vaccine vectors: replication-competent versus replication-deficient poxviruses. Curr. Pharm. Des. 13:2015–2023 [DOI] [PubMed] [Google Scholar]
- 67. Weyer J, Rupprecht CE, Mans J, Viljoen GJ, Nel LH. 2007. Generation and evaluation of a recombinant modified vaccinia virus Ankara vaccine for rabies. Vaccine 25:4213–4222 [DOI] [PubMed] [Google Scholar]
- 68. Kotwal GJ, Moss B. 1988. Analysis of a large cluster of nonessential genes deleted from a vaccinia virus terminal transposition mutant. Virology 167:524–537 [PubMed] [Google Scholar]
- 69. Tagaya I, Kitamura T, Sano Y. 1961. A new mutant of dermovaccinia virus. Nature 192:381–382 [DOI] [PubMed] [Google Scholar]
- 70. Meyer H, Sutter G, Mayr A. 1991. Mapping of deletions in the genome of the highly attenuated vaccinia virus MVA and their influence on virulence. J. Gen. Virol. 72(Pt 5):1031–1038 [DOI] [PubMed] [Google Scholar]
- 71. García MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C, Esteban M. 2006. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. 70:1032–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Elde NC, Child SJ, Eickbush MT, Kitzman JO, Rogers KS, Shendure J, Geballe AP, Malik HS. 2012. Poxviruses deploy genomic accordions to adapt rapidly against host antiviral defenses. Cell 150:831–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Seder RA, Darrah PA, Roederer M. 2008. T-cell quality in memory and protection: implications for vaccine design. Nat. Rev. Immunol. 8:247–258 [DOI] [PubMed] [Google Scholar]
- 74. Reyes-Sandoval A, Wyllie DH, Bauza K, Milicic A, Forbes EK, Rollier CS, Hill AV. 2011. CD8+ T effector memory cells protect against liver-stage malaria. J. Immunol. 187:1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Rigato PO, de Alencar BC, de Vasconcelos JR, Dominguez MR, Araujo AF, Machado AV, Gazzinelli RT, Bruna-Romero O, Rodrigues MM. 2011. Heterologous plasmid DNA prime-recombinant human adenovirus 5 boost vaccination generates a stable pool of protective long-lived CD8(+) T effector memory cells specific for a human parasite, Trypanosoma cruzi. Infect. Immun. 79:2120–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Darrah PA, Patel DT, De Luca PM, Lindsay RW, Davey DF, Flynn BJ, Hoff ST, Andersen P, Reed SG, Morris SL, Roederer M, Seder RA. 2007. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat. Med. 13:843–850 [DOI] [PubMed] [Google Scholar]
- 77. Macedo AB, Sanchez-Arcila JC, Schubach AO, Mendonca SC, Marins-Dos-Santos A, de Fatima Madeira M, Gagini T, Pimentel MI, De Luca PM. 2012. Multifunctional CD4 T cells in patients with American cutaneous leishmaniasis. Clin. Exp. Immunol. 167:505–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Obst R, van Santen HM, Mathis D, Benoist C. 2005. Antigen persistence is required throughout the expansion phase of a CD4(+) T cell response. J. Exp. Med. 201:1555–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Thorne SH. 2008. Oncolytic vaccinia virus: from bedside to benchtop and back. Curr. Opin. Mol. Ther. 10:387–392 [PubMed] [Google Scholar]