Abstract
The hepatitis C virus (HCV) genome contains numerous RNA elements that are required for its replication. Most of the identified RNA structures are located within the 5′ and 3′ untranslated regions (UTRs). One prominent RNA structure, termed the cis-acting replication element (CRE), is located within the NS5B coding region. Mutation of part of the CRE, the 5BSL3.2 stem-loop, impairs HCV RNA replication. This loop has been implicated in a kissing interaction with a complementary stem-loop structure in the 3′ UTR. Although it is clear that this interaction is required for viral replication, the function of the interaction, and its regulation are unknown. In order to gain insight into the CRE function, we isolated cellular proteins that preferentially bind the CRE and identified them using mass spectrometry. This approach identified EWSR1 as a CRE-binding protein. Silencing EWSR1 expression impairs HCV replication and infectious virus production but not translation. While EWRS1 is a shuttling protein that is extensively nuclear in hepatocytes, substantial amounts of EWSR1 localize to the cytosol in HCV-infected cells and colocalize with sites of HCV replication. A subset of EWRS1 translocates into detergent-resistant membrane fractions, which contain the viral replicase proteins, in cells with replicating HCV. EWSR1 directly binds the CRE, and this is dependent on the intact CRE structure. Finally, EWSR1 preferentially interacts with the CRE in the absence of the kissing interaction. This study implicates EWSR1 as a novel modulator of CRE function in HCV replication.
INTRODUCTION
Hepatitis C virus (HCV) is a major cause of chronic hepatitis, cirrhosis, and hepatocellular carcinoma. HCV is an enveloped, positive-strand RNA virus classified within the family Flaviviridae. The viral genome encodes an open reading frame of ∼3,011 codons that is translated as a single polyprotein, which is cleaved by viral and host proteases into at least 10 distinct products. The HCV RNA genome is flanked by the 5′ and 3′ untranslated regions (UTR). An internal ribosome entry site (IRES) within the 5′UTR is essential for translational initiation of the viral RNA (1, 2). Other RNA elements in the 5′UTR, stem-loop 1, and the 3′UTR, including a variable poly-U/UC tract and three stem-loops contained in the terminal 98 nucleotides that are referred to as the 3′X region, are indispensable for RNA replication (3–7). Multiple viral and host proteins have been suggested to interact with the HCV 3′UTR. Components of the viral replicase complex, NS3, NS5A, and NS5B, have each been shown biochemically to interact with the 3′UTR (8–10). Several cellular proteins have also been implicated in 3′UTR binding, including polypyrimidine tract binding protein (PTB), heterogeneous nuclear ribonucleoprotein C, glyceraldehyde dehydrogenase, HuR, and La (11–17). PTB, HuR, and La have also been shown to be required for efficient HCV RNA replication (18–21). These have been proposed to play multiple roles in HCV replication, including the regulation of translation versus the initiation of RNA replication and the circularization of the genome (22, 23).
A phylogenic analysis of nucleotide conservation within HCV isolates identified a highly conserved putative RNA structure within the NS5B coding region. Mutational analysis of the RNA structure, termed the cis-acting replication element (CRE), defined a stem-loop, 5BSL3.2, which is essential for HCV replication (24). 5BSL3.2 is approximately 50 bases in length and is part of a larger predicted cruciform structure (5BSL3). As confirmed by RNA structure probing, 5BSL3.2 consists of an 8-bp lower helix, a 6-bp upper helix, a 12-base terminal loop, and an 8-base internal loop (24, 25). The terminal loop of 5BSL3.2 forms a kissing interaction with a stem-loop structure in the 3′ UTR, SL2 (25, 26). Genetic analysis has shown that the stem structures are required for HCV replication but not the primary sequence of the stem. Truncation of the stem-loops severely impaired HCV replication, as well as mutation of the loop shown to pair with SL2. Importantly, compensatory mutations within SL2 restore replication of a 5BSL3.2 mutant (25, 26).
The internal bulge of 5BSL3.2 has also been shown to undergo a long-range interaction with a second RNA structure, SL9066, which is ∼200 nucleotides upstream of the CRE. This interaction is required for RNA replication and supports a model whereby the CRE structure could interact with either or both sequences upstream and downstream to form a complex extended pseudoknot (27).
Although it is clear that these RNA-RNA interactions are required for viral replication, the function of the interactions and their regulation by viral or cellular proteins is unknown. HCV NS5B has been reported to bind the CRE region, but to date no cellular proteins have been reported to modulate the interaction (28). In order to gain insight into the CRE function, we isolated cellular proteins that preferentially bind the CRE and identified them using mass spectrometry. We identified a novel interaction between the cellular RNA-binding protein EWSR1 and the HCV CRE. EWSR1 is a predominantly nuclear protein that shuttles between the nucleus and cytoplasm (29–31) and has been previously identified as a potential HCV cofactor in a genome-wide siRNA screen (32). It is a member of the TET family, which regulates transcription and RNA processing (33, 34). TET members contain an N-terminal transcription activation domain and a C-terminal RNA-binding domain. EWSR1 is involved in a wide variety of human solid tumors, including Ewing sarcoma, related primitive neuroectodermal tumors, malignant melanoma of soft parts, and desmoplastic small round cell tumors (35–37). In these tumors, the N terminus of EWSR1 is translocated and fused to genes encoding transcription factors.
A major function of native EWSR1 is the regulation of alternative splicing of pre-mRNAs involved in cell cycle progression and the DNA damage response (38, 39). It has been shown to associate with heterogeneous RNA-binding proteins (hnRNPs), such as RBM38 and RBM39, which are involved in splicing (40). In addition, recombinant, purified EWSR1 protein has shown to bind RNA in vitro (41). The RNA binding activity is localized to the carboxy terminus of the protein, which contains an RNA-recognition motif (RRM) and three RGG boxes (41). In addition, EWSR1 contains a tripartite nuclear localization signal (42).
Using a combination of small interfering RNA (siRNA) analysis, microscopy, biochemical fractionation, and an RNA-protein binding assay, we show that EWSR1 is required for HCV RNA replication, relocalizes to HCV replication complexes, and binds the CRE element. This implicates EWSR1 as a modulator of CRE function in HCV replication.
MATERIALS AND METHODS
Cells and virus.
The human hepatoma Huh-7.5 cell line (43), HEK 293T cells, and the Huh 7.5 cell line stably expressing a HCV 1b Con 1 replicon (43) were grown in Dulbecco modified Eagle medium-high glucose (+ glutamine, + sodium pyruvate) with 0.1 mM nonessential amino acids, 5% fetal bovine serum, and 1% penicillin-streptomycin (Invitrogen).
Infectious genotype 2a virus stock was synthesized by electroporating Huh-7.5 cells with viral RNA transcribed from the intragenotypic clone pJFHxJ6-CNS2C3 as previously described in detail (44–46). Infectious HCV was quantified via limiting dilution analysis and immunohistochemical staining of naive cells for NS5A as described previously (46). Subgenomic replicon RNA was transcribed from the previously constructed pSG-JFH1-Rluc (44) and electroporated into cells in the same manner as pJFHxJ6-CNS2C3. The translation reporter construct pFL-Rluc-J6/JFH1(GND) was a gift from Charles M. Rice (Rockefeller University) (47). It was transcribed and electroporated into cells in the same manner as pJFHxJ6-CNS2C3.
Plasmids.
The EWSR1 bacterial expression construct was generated as follows: KV290-EWSR1-WT was made by PCR amplification of EWSR1 from a cDNA clone (Open Biosystems), which was cloned into a N-terminal MBP fusion vector, construct, generously provided by David Kovar (University of Chicago) (48). High-fidelity Phusion DNA polymerase (Finnzymes) was used. EWSR1 was cloned into KV290 using In-Fusion Cloning kit (Clontech) making use of a BamHI restriction site. The EWSR1 mammalian expression construct (designated pLPCX-EWSR1) was made by PCR amplification of EWSR1 from a cDNA clone using high-fidelity Phusion DNA polymerase. EWSR1 was cloned into pLPCX mammalian expression vector (Clontech) using In-Fusion Cloning Kit making use of XhoI and HindIII restriction sites. This was used to make Huh-7.5-EWSR1 cells.
RNA pulldown constructs were made by PCR amplification from HCV genotype 2a. Primers were designed with a BamHI site at the 5′ end of the fragment, and an XbaI site at the 3′end of the fragment. Taq polymerase (New England BioLabs) was used to generate “A” overhangs amenable to TOPO vector cloning. PCR fragments were ligated and inserted into pCR2.1-TOPO vector (Invitrogen). The fragments were excised from pCR2.1-TOPO with BamHI and XbaI and ligated into the BamHI and XbaI sites of pBluescript SKII (−) to generate plasmids for RNA transcription. The cloned CRE consists of HCV 2a nucleotides 9277 to 9422, the cloned 3′UTR consists of HCV 2a nucleotides 9432 to 9678, and the cloned CRE + 3′UTR consists of HCV 2a nucleotides 9277 to 9678. The mutant CRE and CRE plus 3′UTR del SL2 constructs were generated using a QuikChange II XL kit (Stratagene) and sequenced to confirm mutation.
RNA-protein interaction analysis.
For the RNA pulldown assay, we adapted a protocol used previously to identify cellular proteins interacting with the HCV 3′UTR (12). Briefly, a biotin-labeled DNA oligonucleotide that is complementary to the multiple-cloning site sequence that is common between the three RNAs was annealed to the HCV RNA runoff transcript. We used 20 μl of 293T cytoplasmic extract, isolated as previously described (49), 75 μl of the streptavidin-coated paramagnetic bead suspension (Dynal, Inc.), and 100 pmol of RNA-oligonucleotide (RNA-oligo) duplex. The streptavidin bead preservative solution was removed by magnetic separation, and the beads were washed two times with binding buffer containing 1× protease inhibitor mixture (mini-EDTA-free; Roche Applied Science), 1 mM dithiothreitol, 100 mm NaCl, 20 mm HEPES (pH 7.5), 20 U of SUPERaseIN (Ambion)/ml and then mixed with the RNA-oligo duplex. This mixture was incubated on ice in binding buffer for 30 min to immobilize the RNA-oligo duplex on the beads. The buffer solution was removed, and 293T cytoplasmic extracts were then mixed with the beads to capture proteins binding the HCV RNA. Beads were incubated with the cell extracts on ice for a total of 1 h with occasional gentle vortexing. After the incubation, the beads were washed three times with the binding buffer. Elution was done by adding 30 μl of 4× sodium dodecyl sulfate (SDS) gel loading dye to washed beads, followed by heating at 95°C for 10 min. Samples were resolved by 8 to 16% gradient SDS-PAGE, and the gel was silver stained. Mass spectrometry was performed by the Taplin Mass Spectrometry Facility (Harvard University).
For the RNase protection assay, 32P-labeled RNA probes (50 pmol) were incubated for 30 min at 30°C with 1.0–10 μg of purified protein in 20 μl of RNA EMSA buffer (50 mM Tris-HCl [pH 8.0], 0.1 M NaCl, 14.4 mM 2-mercapoethanol). After 30 min, they were exposed to 120 mJ of UV using a Stratalinker (Stratagene). RNase T1 was then added to a final concentration of 50 U/μl and incubated at 37°C for an additional 15 min. Unlabeled competitor RNAs were added at 25- and 50-fold molar excess. SDS-PAGE loading buffer was added, and samples were subjected to gradient SDS-PAGE (4 to 20% gels). Gels were dried down on 3MM blotting paper (Whatman) and visualized using a phosphorimager (FLA2000; Fuji).
Cellular fractionation and membrane floatation analysis.
Fractionation of HCV-Con1 replicon cells and Huh-7.5 cells was performed as described previously (50). Briefly, cells were first lysed in 1 ml of hypotonic buffer (10 mM Tris-HCV [pH 7.5], 10 mM KCl, 5 mM MgCl2). Nuclei and unbroken cells were removed by centrifugation at 1,000 × g for 5 min in microcentrifuge at 4°C. Cell lysates were then mixed with 3 ml of 72% sucrose in low-salt buffer (LSB; comprising 50 mM Tris-HCl [pH 7.5], 25 mM KCl, and 5 mM MgCl2) and overlaid with 4 ml of 55% sucrose in LSB, followed by 1.5 ml of 10% sucrose in LSB. The sucrose gradient was centrifuged at 38,000 rpm in a Beckman SW41 Ti rotor for 14 h for 4°C. After centrifugation, 1-ml fractions were taken from the top of the gradient.
To characterize detergent resistant membranes, supernatants of cell lysates were treated with 1% TX-100 (Sigma-Aldrich, St. Louis, MO) at 4°C for 1 h and followed by centrifugation as described above. Pooled gradient fractions consisted of 400 μl each of individual 1-ml fractions. Pooled fractions were concentrated to 150 μl using a YM-10 Centricon (Millipore).
Purification of recombinant protein.
N-terminally MBP-tagged EWSR1 was expressed in Escherichia coli Rosetta strain [BL21(DE3)/(pLysS); Invitrogen] from the KV290 (48) construct by induction of a 2-liter culture at an optical density at 600 nm (OD600) of 0.6 with 0.1 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) at 25°C for 16 h. Pelleted bacteria were resuspended in 25 ml of BPER buffer (Thermo Science) and incubated at room temperature with mild rotation for 30 min. Cell debris was pelleted by centrifugation at 10,000 rpm using a Sorvall SLA600TC rotor at 4°C for 15 min. Supernatants were brought to 20 ml in BPER buffer and sonicated on ice five times for 15 s each time. The mixture was incubated with 1 ml of Amylose-agarose (Qiagen; 50% slurry) for 16 h at 4°C. The beads were washed two times with 10 ml of BPER buffer and two times with 10 ml of amylose buffer (20 mM Tris-HCl [pH 7.4], 0.2 M NaCl, 1 mM EDTA). MBP-EWSR1 was eluted with 25 ml of amylose buffer plus 10 mM maltose. Protein was concentrated by using Amicon Ultra-4 filters (Millipore) and buffer exchanged with 25 mM Tris-HCl (pH 7.5)–25 mM NaCl.
Western blot analysis.
Adherent cells were washed twice in 1× phosphate-buffered saline (PBS), lysed in 1× Laemmli buffer (250 mM Tris-HCl [pH 6.8], 2% SDS, 10% glycerol, 2.5% β-mercaptoethanol, 0.0025% bromophenol blue), and sonicated. Proteins were separated on 4 to 20% SDS-PAGE gels (Lonza, Inc.) and transferred to nitrocellulose. After being blocked in 10% dry, nonfat milk (1× PBS, 0.1% Tween 20), primary antibodies were added overnight at 4°C. Horseradish peroxidase-conjugated secondary antibodies were added for 30 min in 5% dry milk and included goat anti-rabbit (catalog no. 31462; Thermo Scientific) and rabbit anti-mouse (catalog no. 31452; Thermo Scientific), which were detected using SuperSignal-Femto chemiluminescent substrate (Pierce-Thermo Scientific) and exposure to film. The primary antibodies used included mouse anti-NS5A (9E10; a gift from Charles Rice, Rockefeller University), rabbit anti-EWSR1 (ab93837; Abcam), rabbit anti-calnexin (Stressgen), rabbit anti-caveolin-2 (BD Biosciences), rabbit anti-actin (Sigma), and rabbit anti-NS5B (Abcam).
RNA interference.
EWSR1 siRNAs are Silencer Select predesigned siRNAs s4886 and s4887 (Ambion). Sense sequences for each siRNA are as follows (s4886, AGAUUUUCAAGGGAGCAAtt; s4887, GAGUAGCUAUGGUCAACAAtt). The EWSR1 3′UTR siRNA is AUUGUUUCUUCACAAAUGGtt. Irrelevant (IRR) and HCV-specific siRNAs sequences are described elsewhere (51) and were used as controls. RNA interference (RNAi) assays were performed as described previously (21, 51). Briefly, 106 Huh-7.5 cells in 0.05 ml of cold 1× PBS (pH 7.4) were electroporated with 1 nmol of siRNA using an ECM 830 electroporator (BTX Genetronics) with a 96-well attachment. Cells (∼8,000) were plated in 96 wells.
Real-time RT-PCR.
RNA was extracted from 96-well tissue culture cells by using an RNeasy 96 kit (Qiagen) and eluted in ∼130 μl of RNase-free water. Extracts (2 μl) were reverse transcribed and PCR amplified by using the SuperScript III Platinum One-Step qRT-PCR system with Platinum Taq (Invitrogen). HCV genotype 2a RNAs were amplified using 260 nM forward primer (5′-ACT TCA TTA GCG GCA TCC AAT AC-3′), 260 nM reverse primer (5′-CGG CAC TGA ATG CCA TCA T-3′), and 180 nM probe (5′-6FAM-CAG GAT TGT CAA CAC TGC CAG GGA ACC-IowaBlack-3′) (Integrated DNA Technologies), which recognizes NS4B of HCV JFH-1. Reactions were multiplexed with a 0.5× amount of 18S rRNA TaqMan gene expression assay (4319413E; Applied Biosystems) as an internal control. Reverse transcription-PCR (RT-PCR) was programmed for 50°C for 30 min, 95°C for 6 min, and then 50 cycles of 95°C for 15 s and 60°C for 1 min using an ABI 7300 system (Applied Biosystems). HCV genotype 1b replicon RNAs were amplified using 260 nM forward primer (5′-CCG GGA GAG CCA TAG TGG TCT-3′), 260 nM reverse primer (CCA AAT CTC CAG GCA TTG AGC-3′), and 180 nM probe (5′-6FAM-CAC CGG AAT TGC CAG GAC GAC CGG-MGBNFQ-3′). RT-PCR was programmed for 60°C for 30 min, 95°C for 7 min, and then 40 cycles of 95°C for 15 s and 60°C for 1:15 min using an ABI 7300 system (Applied Biosystems). Reactions were normalized using 18S rRNA TaqMan gene expression assay (4319413E; Applied Biosystems). The data were analyzed with SDS v1.4 software (Applied Biosystems) and normalized to internal controls. Relative quantitation was calculated by comparing the cycle threshold (CT) values using 2ΔΔCT.
Immunofluorescence microscopy.
Glass coverslips in 24-well dishes were first coated with 100 μg of poly-l-lysine/ml for 10 min, washed with sterile water, and air dried before seeding 50,000 cells. All washes and reagents were prepared in 1× PBS (pH 7.5) and used at room temperature. The cells were fixed with 4% paraformaldehyde (15 min) and permeabilized with 0.1% Triton X-100. Coverslips were then blocked for 30 min in 10% normal goat serum (Millipore). Primary antibodies in 5% goat serum and 0.1% Triton X-100 were incubated overnight at 4°C. EWSR1 and double-stranded RNA (dsRNA) antibodies (J2 monoclonal antibody; English and Scientific Consulting Bt.) were used at a dilution of 1:1000. Alexa Fluor 488 or Alexa Fluor 594 secondary antibodies (Invitrogen) were used at 1:1,200 in 5% goat serum for 30 min. Coverslips were mounted in ProLong Gold AntiFade with DAPI (4′,6′-diamidino-2-phenylindole) nuclear stain (Invitrogen). The samples were imaged using an Olympus DSU spinning disc confocal microscope equipped with a Photometrics Evolve EMCCD camera. Digital images were taken using Slidebook v5.0 software and processed using ImageJ (National Institutes of Health). Quantification of fluorescence intensity was determined from multiple images taken from duplicate coverslips using ImageJ.
Statistical analysis.
Data are presented as means ± the standard deviations. To assess statistical significance, two-tailed, paired Student t tests were performed.
RESULTS
EWSR1 binds HCV RNA elements.
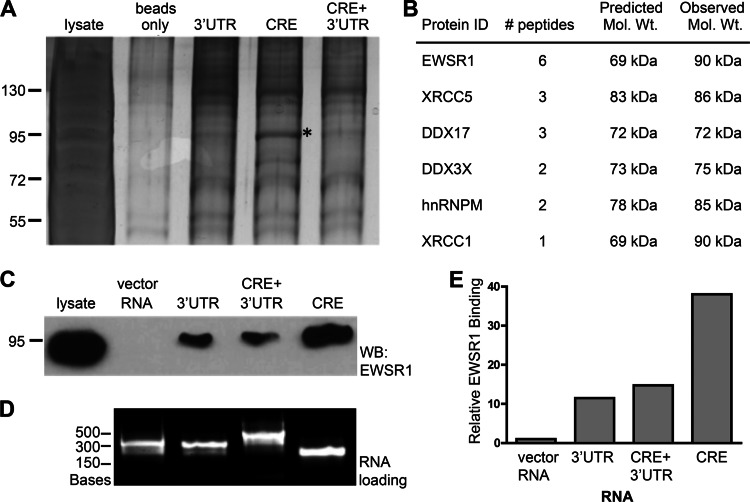
In order to better understand the function of HCV RNA elements in HCV replication, we initiated efforts to identify cellular proteins binding to the 3′UTR and/or CRE regions of the HCV JFH1 genome. The 263-nucleotide region of the 3′UTR, the 117-nucleotide region of the CRE and a 398-nucleotide region containing the CRE and 3′UTR were cloned into in vitro transcription vectors. We first performed RNA-protein binding experiments using the respective in vitro-transcribed RNAs, a biotin-labeled DNA oligonucleotide that is complementary to the multiple-cloning site sequence that is common between the three RNAs, and 293T cellular cytoplasmic lysates. 293T cell lysates were used since they support HCV JFH1 replication, albeit at lower levels than Huh7 cells derivatives (52). RNA-protein complexes were captured by streptavidin beads, washed, separated by SDS-PAGE, and silver stained. A prominent band, migrating at ∼92 kDa was selectively enriched in the CRE-RNA-protein complexes (Fig. 1A, marked by an asterisk).
Fig 1.
EWSR1 precipitates with HCV RNA replication elements. HCV RNAs corresponding to either the CRE, the 3′UTR or the CRE plus+ 3′UTR were incubated with uninfected cellular protein extracts and precipitated via a complementary biotinylated oligonucleotide and streptavidin agarose beads. (A) Silver-stained SDS-PAGE of precipitate. An asterisk (*) denotes the CRE-specific band that was excised for mass spectrometry analysis. (B) Peptide identities, quantities, predicted, and observed molecular mass from the band in panel A that were identified by mass spectrometry. (C) Immunoblot of HCV RNA pull-downs probed for EWSR1. (D) Loading of RNA as bound to beads per pulldown. (E) Quantitation of band densitometry in panel C. This RNA pulldown experiment (C to E) was representative of three independent experiments.
This band was excised and analyzed by matrix-assisted laser desorption ionization–mass spectrometry to identify the enriched protein(s) (Fig. 1B). The analysis identified six proteins, four of which are known RNA-binding proteins (EWSR1, DDX17, DDX3X, and hnRNPM). We examined whether EWSR1, for which the greatest number of peptides were identified, was enriched in CRE RNA-protein complexes by immunoblot. The RNA pull-downs were repeated as before, this time including 302 nucleotides of nonspecific vector RNA as a negative control (Fig. 1C). RNA-protein complexes were analyzed by EWSR1 immunoblot. EWSR1 bound specifically to HCV RNA, and not the vector RNA, and was enriched in the CRE RNA-protein complex (Fig. 1C and E). The RNA loading for each sample was equivalent (Fig. 1D).
EWSR1 is required for efficient HCV replication.
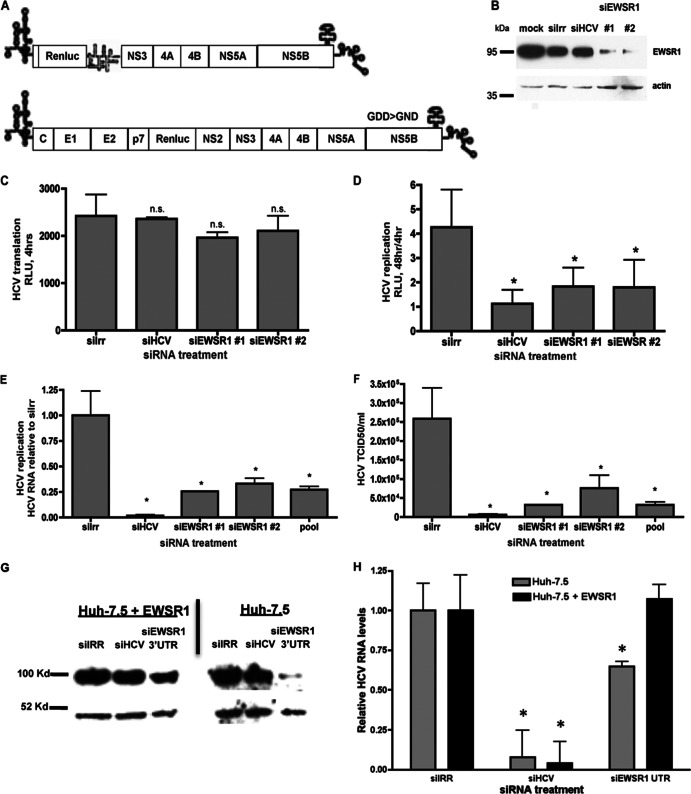
To determine the biological role of EWSR1 in the HCV viral life cycle, we investigated the effects of EWSR1 silencing on HCV RNA translation, replication, and infectious virus production. We electroporated siRNAs targeting an irrelevant sequence (Irr), HCV, or two distinct sequences of EWSR1 into Huh-7.5 cells and then transfected these cells with either genomic HCV-luciferase polymerase-defective (GDD→GND) RNAs to monitor translation. Replication was measured by transfection of subgenomic HCV-luciferase RNAs (Fig. 2A). Compared to an irrelevant siRNA (siIRR), the siRNAs targeting ESWR1 resulted in a reduction of endogenous EWSR1 protein, indicating effective silencing (Fig. 2B). EWSR1 siRNAs had little effect on the translation of genomic pol- (GDD→GND)-HCV RNAs (Fig. 2C); however, they significantly reduced HCV subgenomic replication (Fig. 2D).
Fig 2.
EWSR1 is required for HCV efficient RNA replication and infectious virus production. (A) HCV reporter constructs used to measure replication (sg-neo-luciferase) or input translation (the polymerase defective HCV-luciferase GDD→GND). Huh-7.5 cells were electroporated with the indicated siRNAs, and maintained for 48 h. (B to D) Protein was harvested, separated by SDS-PAGE, and immunoblotted for EWSR1 or actin (B) or transfected with polymerase-defective HCV RNAs (C) for 4 h or subgenomic HCV-luciferase RNAs (D) and assayed for luciferase activity at 4 h for input RNA translation and 48 h for replication-associated reporter activity. (E to G) Alternatively, the siRNA-treated cells were infected with HCV for 48 h, and intracellular HCV RNA (E) or infectious HCV (F) production was quantified. (G and H) Huh-7.5 or Huh-7.5-EWSR1 cells were treated with the indicated siRNAs for 72 h, and then protein lysates were harvested, separated by SDS-PAGE, and immunoblotted for EWSR1 or actin (G) or infected with HCV for 48 h (H), and the HCV RNA was quantified. *, P ≤ 0.05.
We next examined the significance of EWSR1 in the viral life cycle using infectious HCV. Huh-7.5 cells that were treated for 2 days with the indicated siRNAs were infected for 48 h, and then supernatants and cellular RNA were harvested. Viral RNA was quantified by quantitative real-time RT-PCR, virus titers were determined via limiting dilution analysis and compared to cells silenced with an irrelevant siRNA (siIrr). We find that individual siRNAs targeting EWSR1 significantly inhibit HCV RNA accumulation (Fig. 2E) and infectious virus production (Fig. 2F). These results indicate that EWSR1 is required for efficient HCV RNA replication and infectious virus production, but not protein translation.
To rule out off target effects of siRNAs, we next determined whether HCV replication could be restored by expression of a siRNA-resistant EWSR1. Huh-7.5 cells or Huh-7.5 cells ectopically expressing EWSR1 were treated with the indicated siRNAs. siEWSR1-3′UTR targets the 3′UTR, thus the endogenous EWSR1 is silenced by this siRNA, but not the ectopically expressed EWSR1 (Fig. 2G). Overexpression of siRNA-resistant EWSR1 restored HCV replication in cells treated with the EWSR1-3′UTR (Fig. 2H). Thus, the antiviral activity of siEWSR1 is attributable to EWSR1 (and not an off-target effect).
We also examined the significance of EWSR1 in the replication of the HCV 1b genotype. Huh 7.5 cells stably expressing an HCV 1b Con 1 replicon (43) were treated with siRNAs directed against EWSR1 for 72 h, and RNA and protein were harvested. Replicon RNA was quantified by quantitative real-time RT-PCR, and compared to cells silenced with an irrelevant siRNA (siIrr). We find that individual siRNAs targeting EWSR1 significantly inhibit HCV 1b genotype replicon RNA accumulation compared to irrelevantly treated cells (Fig. 3A). We also see a significant decrease in viral NS5B protein expression in EWSR1 silenced cells (Fig. 3B). These results indicate that EWSR1 is required for efficient HCV RNA replication of the 1b viral genotype and that the requirement for EWSR1 in HCV replication is conserved between genotypes 1 and 2.
Fig 3.
EWSR1 is required for HCV efficient RNA replication and viral protein production of a genotype 1b replicon. (A and B) A cell line stably expressing a HCV 1b Con1 replicon was transfected with the indicated siRNAs and maintained for 72 h. After 72 h, cellular RNA (A) or protein (B) was harvested. Replicon RNA was quantified using quantitative RT-PCR. Cellular protein was separated by SDS-PAGE and immunoblotted for NS5B or actin.
EWSR1 relocalizes to HCV replication complexes.
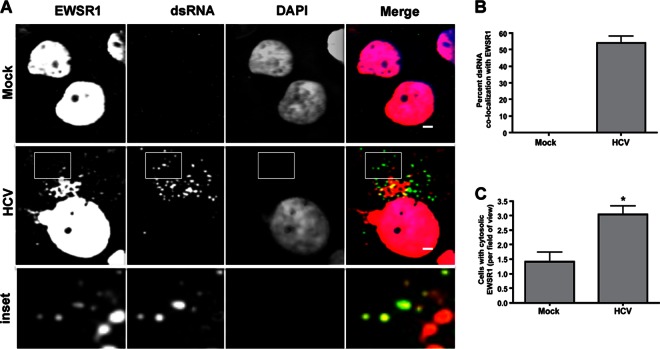
EWSR1 is an RNA-binding protein that shuttles between the nucleus and cytoplasm, but is predominantly nuclear (29–31). Given that HCV replicates in the cytoplasm, we next examined the localization of EWSR1 in HCV-infected cells. Huh-7.5 cells were infected with HCV for 48 h, fixed, and probed with antibodies to EWSR1 and dsRNA, the latter being the HCV replication intermediate and thus a marker of viral replication complexes. The subcellular localization of EWSR1 was altered significantly by HCV infection. EWSR1 is predominantly nuclear in mock-infected cells, whereas a significant proportion of HCV-infected cells have enhanced cytoplasmic EWSR1 localization (Fig. 4A and C). EWSR1 colocalized with dsRNA, and thus the sites of HCV replication (Fig. 4A). Quantification of EWSR1 colocalization with dsRNA showed that ca. 60% of all cytoplasmic EWSR1 in infected cells is localized with dsRNA (Fig. 4B). From these analyses, we conclude that the EWSR1 is relocalized to sites of active HCV replication.
Fig 4.
Localization of HCV replication complexes with EWSR1. (A) Huh-7.5 cells were infected with HCV for 48 h. Cells were fixed and probed with antibodies to EWSR1 (red) and dsRNA (green), the latter detecting HCV replication complexes. DAPI (blue) was used as a nuclear marker. The indicated color is relevant to the merged image. (B) ImageJ quantification of percent cytoplasmic EWSR1 staining localizing with dsRNA replication marker. (C) Quantification of the number of cells positive for cytoplasmic staining in infected and uninfected cell fields of view. Scale, 1 μm. *, P ≤ 0.05.
We next examined the localization of EWSR1 and HCV replication complexes using biochemical fractionation. HCV replication protein complexes associate with detergent-resistant membranes (DRMs), which are thought to be similar to lipid rafts (50). We isolated DRMs from Huh-7.5 cells with or without HCV replicons and tested for enrichment of caveolin-2, which is a marker of DRMs; in addition to the selective loss of calnexin, which does not specifically associate with DRMs (Fig. 5). Having satisfied these criteria, we probed for EWSR1 and found that a population of EWSR1 becomes associated with DRMs in HCV replicon cells. EWSR1 is exclusively contained in detergent-sensitive fractions in Huh-7.5 cells, whereas a substantial proportion of EWSR1 is found in DRMs in HCV replicon cells. The EWSR1 in DRMs cofractionates with the HCV replicase marker NS5A (Fig. 5). Thus, we observe EWSR1 localizing to sites of HCV replication, as defined by colocalization with dsRNA and relocalization to HCV replicase-containing DRMs.
Fig 5.
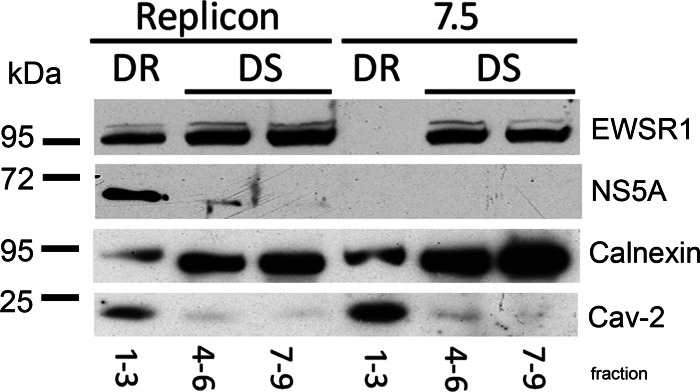
Cofractionation of ESWR1 with HCV replication complexes. Detergent-resistant membranes were prepared from Huh-7.5 cells with or without HCV replicons by Triton X-100 treatment at 4°C, followed by ultracentrifugation. Detergent-resistant (DR) membranes remain at the top of the sucrose gradient (fractions 1 to 3), whereas detergent-sensitive (DS) membranes sediment in the lower fractions (fractions 4 to 9). Protein lysates from pooled fractions were run on SDS-PAGE and probed for EWSR1, HCV NS5A, calnexin, and caveolin-2 (Cav-2).
EWSR1 binding to the CRE requires an intact 5BSL3.2 structure.
The previous experiments indicate an association of EWSR1 with the CRE but do not demonstrate direct RNA binding. In order to test whether EWSR1 directly binds the CRE, recombinant maltose-binding protein (MBP) fused to the N terminus of EWSR1, was purified and tested for CRE binding in vitro using an RNase protection assay. MBP or MBP-EWSR1 was incubated with 32P-labeled CRE RNA, UV-cross-linked, and then digested with RNase T1. Samples were denatured and run on an SDS-PAGE gel, dried, and visualized by autoradiography. MBP-EWSR1, but not MBP binds the CRE in vitro. The binding reaction was also performed in the presence of excess cold competitor wild-type (wt) or mut CRE RNA, consisting of the wild-type 5BSL3.2, but with the stem-loop replaced by an A bulge so as to maintain the stem structure. We observed effective competition for EWSR1-CRE binding by the wt CRE, but not the mut CRE (Fig. 6). This indicates that EWSR1 directly binds the CRE and that this requires the 5BSL3.2 loop, which is also required for the kissing loop interaction and HCV replication.
Fig 6.

Purified recombinant EWSR1 directly and selectively binds the HCV CRE RNA in vitro. (A) MBP or MBP-EWSR1 was incubated with 32P-labeled CRE RNA for 30 min either alone, or in the presence on increasing amounts of cold competitor RNA (either wt CRE or mut CRE), then subjected to UV cross-linking and RNase T1 treatment. The product was separated on an SDS-PAGE gel and visualized by autoradiography. (B) Quantitation of EWSR1/CRE binding in panel A. The ratio of EWSR1/CRE to digested CRE band quantitation is shown.
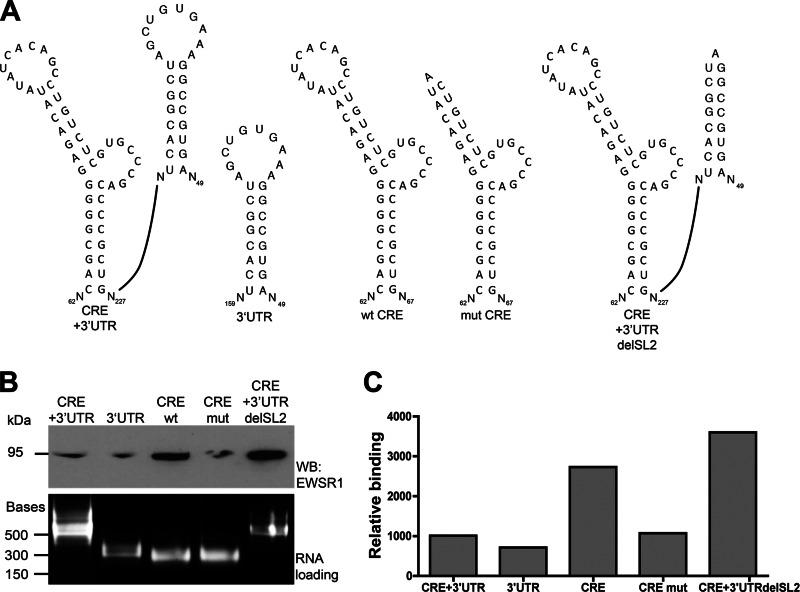
We next examined the requirement of the 5BSL3.2 RNA structure, which mediates the kissing loop interaction, for EWSR1 binding of the CRE. A mutant was generated consisting of the wild-type 5BSL3.2, but with the stem-loop replaced by an A bulge so as to maintain the stem structure, but abolishing the exposed loop (mut CRE, Fig. 7A). We tested this construct, compared to the wild-type CRE (wt CRE), in the same RNA pulldown assay described in Fig. 1. We next examined the requirement of the 5BSL3.2 RNA structure, which mediates the kissing loop interaction, for EWSR1 binding of the CRE. (Fig. 7A). We tested this construct, compared to the wild-type CRE (wt CRE), in the same RNA pulldown assay. We once again observe a preference for EWSR1 association with the CRE, compared to the 3′UTR or the CRE plus 3′ UTR. Deletion of the 5BSL3.2 loop decreased EWSR1 association with the CRE (Fig. 4.7B). Thus, EWSR1 preferentially binds the CRE with an intact 5BSL3.2 loop.
Fig 7.
EWSR1 preferentially interacts with the CRE in the absence of a potential kissing interaction. (A) A deletion of the loop in stem-loop 5BSL3.2 in the CRE structure was created, as well as a deletion of the loop in stem-loop II of the 3′UTR, and then used in the RNA-protein interaction assay as described in Fig. 1 (top). (B) Ethidium bromide-stained agarose gel of probe RNAs that were used in the RNA-protein interaction pulldown assay (bottom). Nucleotides marked with an asterisk (*) are required for replication, and underlined nucleotides participate in 3′UTR kissing loop interaction, as determined previously (24, 25). The lower panel shows loading of RNA as bound to beads per pulldown. (C) Quantitation of band densitometry in panel B (top panel).
EWSR1 preferentially interacts with the CRE in the absence of a potential kissing interaction.
The decreased interaction of EWSR1 with the CRE plus 3′UTR compared to the CRE alone could indicate that a preformed kissing interaction between the CRE and SL2 in the 3′UTR precludes EWSR1 binding. To test this possibility, we constructed a mutant version of the CRE plus 3′ UTR RNA probe containing a deletion in the SL2 loop such that the kissing interaction between the CRE and SL2 was abrogated (Fig. 7A). EWSR1 binds the CRE plus 3′UTR with a mutated SL2 significantly more that the wild-type CRE + 3′UTR. Indeed, the binding of the CRE + 3′UTR with mutated SL2 is similar to that of the CRE alone (Fig. 7B and C). This indicates that EWSR1 preferentially interacts with the CRE in the absence of the kissing interaction.
DISCUSSION
By taking an unbiased approach to identify host proteins that bind HCV RNA replication elements, we defined EWSR1 as an interaction partner of the HCV CRE. RNAi analysis showed a requirement for EWSR1 in HCV RNA replication and virus production, but not translation. Confocal microscopy and biochemical fractionation analysis show that EWSR1 redistributes to the cytoplasm of infected cells and associates with viral replication complex markers. CRE mutational analysis showed that the CRE 5BSL3.2 loop, which is required for the HCV 3′UTR kissing loop interaction and viral replication, is also required for the interaction of EWSR1 with the CRE. Finally, using purified components, EWSR1 was shown to directly bind the CRE in vitro.
DDX3X was also identified as a potential CRE binding partner via our mass spectrometry analysis (Fig. 1). DDX3X has been previously identified as required for HCV replication (21, 53) and interacts with the HCV core protein (54), however, that interaction is not required for HCV infection (55). DDX3X has also been reported to play a role in viral translation (56). The significance (if any) of DDX3X binding the HCV CRE will be the subject of another study.
While ablation of the CRE leads to a drastic reduction in the ability of the virus to replicate, a compensatory mutation of 3′ SL2 was shown to support partial viral replication in the absence of the complete CRE structure (24). This virus was incapable of wild-type levels of replication, suggesting an important role for the structure of 5BSL3.2 in optimizing the kissing loop interaction. The inability of EWSR1 to bind a disrupted CRE structure (Fig. 4 and 5), as well as the conservation of 5BSL3.2 in multiple genotypes of HCV, also suggests an important role for the correct structure of 5BSL3.2 in viral replication. It should be noted that while EWSR1 binding to the CRE requires to 5BSL3.2 structure, we do not have evidence that EWSR1 binds the loop of 5BSL3.2. Indeed, given that mutations in the 5BSL3.2 loop can be rescued by complementary mutations in 3′UTR SL2 loop, we doubt that EWSR1 specifically recognizes nucleotides within the loop. It is more likely that EWSR1 recognizes structural components of the CRE.
The role of the kissing loop interaction in HCV replication remains unclear. It has been postulated to function as a molecular switch from viral translation to replication (27). Translation and RNA replication cannot occur at the same time on the same positive-stranded RNA viral genome (57), and the switch from one process to another must be regulated. We propose that EWSR1 binding to 5BSL3.2 may regulate this process, possibly by the following model. The genomic HCV RNA forms a pseudoknot at its 3′ end, with 5BSL3.2 interacting with SL2 of the 3′UTR and SL9066 upstream. In this state, translation of the virus would proceed, in part because the HCV RNA replication elements in the 3′ of the genome are not accessible to viral replicase binding. After initial translation, the kissing interaction is disrupted, and EWSR1 binds to 5BSL3.2. This sterically hinders the kissing loop interaction of the CRE with SL2 of the 3′UTR and thus prevents pseudoknot formation. In the absence of pseudoknot formation, the viral replicase can bind RNA elements in the 3′UTR and initiate minus-strand RNA synthesis. In support of this model, EWSR1 depletion does not impact translation, in which the pseudoknot would form, but is required for HCV replication, which would rely on disruption of the pseudoknot. Alternatively, EWSR1 may impact HCV replication by affecting replicase formation, minus-strand initiation, or genomic RNA trafficking.
Our model draws some parallels with a proposed model for poliovirus translation and replication regulation, whereby viral protein 3CD and cellular protein PCBP2 interact with the 5′ cloverleaf RNA structure. PCBP2 has an essential function in IRES-dependent translation of poliovirus (58–60) and also forms a RNP complex with the viral protein 3CD, which is a critical step in the initiation of negative-strand RNA synthesis (59, 61, 62). Cleavage of PCBP2 by viral proteases inhibits the translational function of PCBP2, while the ternary complex still forms and RNA replication proceeds (23). Thus, the regulated interactions of a host cell protein with an RNA structure can modulate viral translation versus RNA replication.
In contrast to the poliovirus model, we observed that EWSR1 is required for RNA replication of HCV, but not initial translation of the virus. It is not yet known whether EWSR1 interacts with other viral or cellular proteins, while at the CRE, but since it does not appear to have functions in both translation and RNA replication, this hints that a higher order molecular structure, including EWSR1 may form at the HCV CRE and help regulate the switch from viral translation to RNA replication. Future studies will test the proposed model of kissing loop modulation by EWSR1. This includes examining whether EWSR1 prevents the kissing loop interaction and if this increases the accessibility of the 3′UTR to the HCV replicase to initiate minus-strand synthesis.
ACKNOWLEDGMENTS
We thank Ana Shulla and Kelly Coller Metzinger for critically reading the manuscript. We thank Charles Rice and Takaji Wakita for reagents. We thank the University of Chicago Light Microscopy Facility and director Vytas Bindokas.
This study was funded by NIAID (1R01AI080703) and Susan and David Sherman. T.E.O. was funded by NIH training grant T32 GM007183.
Footnotes
Published ahead of print 3 April 2013
REFERENCES
- 1. Fukushi S, Katayama K, Kurihara C, Ishiyama N, Hoshino FB, Ando T, Oya A. 1994. Complete 5′ noncoding region is necessary for the efficient internal initiation of hepatitis C virus RNA. Biochem. Biophys. Res. Commun. 199:425–432 [DOI] [PubMed] [Google Scholar]
- 2. Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66:1476–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Friebe P, Bartenschlager R. 2002. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J. Virol. 76:5326–5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kolykhalov AA, Agapov EV, Blight KJ, Mihalik K, Feinstone SM, Rice CM. 1997. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277:570–574 [DOI] [PubMed] [Google Scholar]
- 5. Kolykhalov AA, Feinstone SM, Rice CM. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 70:3363–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kolykhalov AA, Mihalik K, Feinstone SM, Rice CM. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lyons AJ, Lytle JR, Gomez J, Robertson HD. 2001. Hepatitis C virus internal ribosome entry site RNA contains a tertiary structural element in a functional domain of stem-loop II. Nucleic Acids Res. 29:2535–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang L, Hwang J, Sharma SD, Hargittai MR, Chen Y, Arnold JJ, Raney KD, Cameron CE. 2005. Hepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding protein. J. Biol. Chem. 280:36417–36428 [DOI] [PubMed] [Google Scholar]
- 9. Kanai A, Tanabe K, Kohara M. 1995. Poly(U) binding activity of hepatitis C virus NS3 protein, a putative RNA helicase. FEBS Lett. 376:221–224 [DOI] [PubMed] [Google Scholar]
- 10. Luo G, Hamatake RK, Mathis DM, Racela J, Rigat KL, Lemm J, Colonno RJ. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74:851–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gontarek RR, Gutshall LL, Herold KM, Tsai J, Sathe GM, Mao J, Prescott C, Del Vecchio AM. 1999. hnRNP C and polypyrimidine tract-binding protein specifically interact with the pyrimidine-rich region within the 3′NTR of the HCV RNA genome. Nucleic Acids Res. 27:1457–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harris D, Zhang Z, Chaubey B, Pandey VN. 2006. Identification of cellular factors associated with the 3′-nontranslated region of the hepatitis C virus genome. Mol. Cell. Proteomics 5:1006–1018 [DOI] [PubMed] [Google Scholar]
- 13. Luo G. 1999. Cellular proteins bind to the poly(U) tract of the 3′ untranslated region of hepatitis C virus RNA genome. Virology 256:105–118 [DOI] [PubMed] [Google Scholar]
- 14. Petrik J, Parker H, Alexander GJ. 1999. Human hepatic glyceraldehyde-3-phosphate dehydrogenase binds to the poly(U) tract of the 3′ non-coding region of hepatitis C virus genomic RNA. J. Gen. Virol. 80(Pt 12):3109–3113 [DOI] [PubMed] [Google Scholar]
- 15. Spangberg K, Goobar-Larsson L, Wahren-Herlenius M, Schwartz S. 1999. The La protein from human liver cells interacts specifically with the U-rich region in the hepatitis C virus 3′ untranslated region. J. Hum. Virol. 2:296–307 [PubMed] [Google Scholar]
- 16. Spangberg K, Wiklund L, Schwartz S. 2001. Binding of the La autoantigen to the hepatitis C virus 3′ untranslated region protects the RNA from rapid degradation in vitro. J. Gen. Virol. 82:113–120 [DOI] [PubMed] [Google Scholar]
- 17. Spangberg K, Wiklund L, Schwartz S. 2000. HuR, a protein implicated in oncogene and growth factor mRNA decay, binds to the 3′ ends of hepatitis C virus RNA of both polarities. Virology 274:378–390 [DOI] [PubMed] [Google Scholar]
- 18. Chang KS, Luo G. 2006. The polypyrimidine tract-binding protein (PTB) is required for efficient replication of hepatitis C virus (HCV) RNA. Virus Res. 115:1–8 [DOI] [PubMed] [Google Scholar]
- 19. Domitrovich AM, Diebel KW, Ali N, Sarker S, Siddiqui A. 2005. Role of La autoantigen and polypyrimidine tract-binding protein in HCV replication. Virology 335:72–86 [DOI] [PubMed] [Google Scholar]
- 20. Korf M, Jarczak D, Beger C, Manns MP, Kruger M. 2005. Inhibition of hepatitis C virus translation and subgenomic replication by siRNAs directed against highly conserved HCV sequence and cellular HCV cofactors. J. Hepatol. 43:225–234 [DOI] [PubMed] [Google Scholar]
- 21. Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, Landthaler M, Landgraf P, Kan S, Lindenbach BD, Chien M, Weir DB, Russo JJ, Ju J, Brownstein MJ, Sheridan R, Sander C, Zavolan M, Tuschl T, Rice CM. 2007. Cellular cofactors affecting hepatitis C virus infection and replication. Proc. Natl. Acad. Sci. U. S. A. 104:12884–12889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Daijogo S, Semler BL. 2011. Mechanistic intersections between picornavirus translation and RNA replication. Adv. Virus Res. 80:1–24 [DOI] [PubMed] [Google Scholar]
- 23. Perera R, Daijogo S, Walter BL, Nguyen JH, Semler BL. 2007. Cellular protein modification by poliovirus: the two faces of poly(rC)-binding protein. J. Virol. 81:8919–8932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. You S, Stump DD, Branch AD, Rice CM. 2004. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J. Virol. 78:1352–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Friebe P, Boudet J, Simorre JP, Bartenschlager R. 2005. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 79:380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. You S, Rice CM. 2008. 3′ RNA elements in hepatitis C virus replication: kissing partners and long poly(U). J. Virol. 82:184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Diviney S, Tuplin A, Struthers M, Armstrong V, Elliott RM, Simmonds P, Evans DJ. 2008. A hepatitis C virus cis-acting replication element forms a long-range RNA-RNA interaction with upstream RNA sequences in NS5B. J. Virol. 82:9008–9022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee H, Shin H, Wimmer E, Paul AV. 2004. cis-Acting RNA signals in the NS5B C-terminal coding sequence of the hepatitis C virus genome. J. Virol. 78:10865–10877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leemann-Zakaryan RP, Pahlich S, Grossenbacher D, Gehring H. 2011. Tyrosine phosphorylation in the C-terminal nuclear localization and retention signal (C-NLS) of the EWS protein. Sarcoma 2011:218483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Leemann-Zakaryan RP, Pahlich S, Sedda MJ, Quero L, Grossenbacher D, Gehring H. 2009. Dynamic subcellular localization of the Ewing sarcoma proto-oncoprotein and its association with and stabilization of microtubules. J. Mol. Biol. 386:1–13 [DOI] [PubMed] [Google Scholar]
- 31. Pahlich S, Zakaryan RP, Gehring H. 2008. Identification of proteins interacting with protein arginine methyltransferase 8: the Ewing sarcoma (EWS) protein binds independent of its methylation state. Proteins 72:1125–1137 [DOI] [PubMed] [Google Scholar]
- 32. Li Q, Brass AL, Ng A, Hu Z, Xavier RJ, Liang TJ, Elledge SJ. 2009. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc. Natl. Acad. Sci. U. S. A. 106:16410–16415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Law WJ, Cann KL, Hicks GG. 2006. TLS, EWS, and TAF15: a model for transcriptional integration of gene expression. Brief. Funct. Genomics Proteomics 5:8–14 [DOI] [PubMed] [Google Scholar]
- 34. Tan AY, Manley JL. 2009. The TET family of proteins: functions and roles in disease. J. Mol. Cell Biol. 1:82–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Crozat A, Aman P, Mandahl N, Ron D. 1993. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature 363:640–644 [DOI] [PubMed] [Google Scholar]
- 36. Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, Kovar H, Joubert I, de Jong P, Rouleau G. 1992. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature 359:162–165 [DOI] [PubMed] [Google Scholar]
- 37. Ohno T, Rao VN, Reddy ES. 1993. EWS/Fli-1 chimeric protein is a transcriptional activator. Cancer Res. 53:5859–5863 [PubMed] [Google Scholar]
- 38. Paronetto MP, Minana B, Valcarcel J. 2011. The Ewing sarcoma protein regulates DNA damage-induced alternative splicing. Mol. Cell 43:353–368 [DOI] [PubMed] [Google Scholar]
- 39. Sanchez G, Bittencourt D, Laud K, Barbier J, Delattre O, Auboeuf D, Dutertre M. 2008. Alteration of cyclin D1 transcript elongation by a mutated transcription factor up-regulates the oncogenic D1b splice isoform in cancer. Proc. Natl. Acad. Sci. U. S. A. 105:6004–6009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zinszner H, Albalat R, Ron D. 1994. A novel effector domain from the RNA-binding protein TLS or EWS is required for oncogenic transformation by CHOP. Genes Dev. 8:2513–2526 [DOI] [PubMed] [Google Scholar]
- 41. Ohno T, Ouchida M, Lee L, Gatalica Z, Rao VN, Reddy ES. 1994. The EWS gene, involved in Ewing family of tumors, malignant melanoma of soft parts and desmoplastic small round cell tumors, codes for an RNA binding protein with novel regulatory domains. Oncogene 9:3087–3097 [PubMed] [Google Scholar]
- 42. Shaw DJ, Morse R, Todd AG, Eggleton P, Lorson CL, Young PJ. 2009. Identification of a tripartite import signal in the Ewing sarcoma protein (EWS). Biochem. Biophys. Res. Commun. 390:1197–1201 [DOI] [PubMed] [Google Scholar]
- 43. Blight KJ, McKeating JA, Rice CM. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001–13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Berger KL, Cooper JD, Heaton NS, Yoon R, Oakland TE, Jordan TX, Mateu G, Grakoui A, Randall G. 2009. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc. Natl. Acad. Sci. U. S. A. 106:7577–7582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mateu G, Donis RO, Wakita T, Bukh J, Grakoui A. 2008. Intragenotypic JFH1 based recombinant hepatitis C virus produces high levels of infectious particles but causes increased cell death. Virology 376:397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Randall G, Chen L, Panis M, Fischer AK, Lindenbach BD, Sun J, Heathcote J, Rice CM, Edwards AM, McGilvray ID. 2006. Silencing of USP18 potentiates the antiviral activity of interferon against hepatitis C virus infection. Gastroenterology 131:1584–1591 [DOI] [PubMed] [Google Scholar]
- 47. Jones CT, Murray CL, Eastman DK, Tassello J, Rice CM. 2007. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 81:8374–8383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Neidt EM, Skau CT, Kovar DR. 2008. The cytokinesis formins from the nematode worm and fission yeast differentially mediate actin filament assembly. J. Biol. Chem. 283:23872–23883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Abmayr SM, Yao T, Parmely T, Workman JL. 2006. Preparation of nuclear and cytoplasmic extracts from mammalian cells. Curr. Protoc. Mol. Biol. Chapter 12:Unit 12 11 [DOI] [PubMed] [Google Scholar]
- 50. Aizaki H, Lee KJ, Sung VM, Ishiko H, Lai MM. 2004. Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology 324:450–461 [DOI] [PubMed] [Google Scholar]
- 51. Randall G, Grakoui A, Rice CM. 2003. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc. Natl. Acad. Sci. U. S. A. 100:235–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kato T, Date T, Miyamoto M, Zhao Z, Mizokami M, Wakita T. 2005. Nonhepatic cell lines HeLa and 293 support efficient replication of the hepatitis C virus genotype 2a subgenomic replicon. J. Virol. 79:592–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ariumi Y, Kuroki M, Abe K, Dansako H, Ikeda M, Wakita T, Kato N. 2007. DDX3 DEAD-box RNA helicase is required for hepatitis C virus RNA replication. J. Virol. 81:13922–13926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Owsianka AM, Patel AH. 1999. Hepatitis C virus core protein interacts with a human DEAD box protein DDX3. Virology 257:330–340 [DOI] [PubMed] [Google Scholar]
- 55. Angus AG, Dalrymple D, Boulant S, McGivern DR, Clayton RF, Scott MJ, Adair R, Graham S, Owsianka AM, Targett-Adams P, Li K, Wakita T, McLauchlan J, Lemon SM, Patel AH. 2010. Requirement of cellular DDX3 for hepatitis C virus replication is unrelated to its interaction with the viral core protein. J. Gen. Virol. 91:122–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee CS, Dias AP, Jedrychowski M, Patel AH, Hsu JL, Reed R. 2008. Human DDX3 functions in translation and interacts with the translation initiation factor eIF3. Nucleic Acids Res. 36:4708–4718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gamarnik AV, Andino R. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Blyn LB, Swiderek KM, Richards O, Stahl DC, Semler BL, Ehrenfeld E. 1996. Poly(rC) binding protein 2 binds to stem-loop IV of the poliovirus RNA 5′ noncoding region: identification by automated liquid chromatography-tandem mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 93:11115–11120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gamarnik AV, Andino R. 1997. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA 3:882–892 [PMC free article] [PubMed] [Google Scholar]
- 60. Sean P, Semler BL. 2008. Coxsackievirus B RNA replication: lessons from poliovirus. Curr. Top. Microbiol. Immunol. 323:89–121 [DOI] [PubMed] [Google Scholar]
- 61. Andino R, Rieckhof GE, Baltimore D. 1990. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell 63:369–380 [DOI] [PubMed] [Google Scholar]
- 62. Parsley TB, Towner JS, Blyn LB, Ehrenfeld E, Semler BL. 1997. Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 3:1124–1134 [PMC free article] [PubMed] [Google Scholar]