Abstract
Severe fever with thrombocytopenia syndrome is an emerging infectious disease caused by a novel bunyavirus (SFTSV). Lack of vaccines and inadequate therapeutic treatments have made the spread of the virus a global concern. Viral nucleocapsid protein (N) is essential for its transcription and replication. Here, we present the crystal structures of N from SFTSV and its homologs from Buenaventura (BUE) and Granada (GRA) viruses. The structures reveal that phleboviral N folds into a compact core domain and an extended N-terminal arm that mediates oligomerization, such as tetramer, pentamer, and hexamer of N assemblies. Structural superimposition indicates that phleboviral N adopts a conserved architecture and uses a similar RNA encapsidation strategy as that of RVFV-N. The RNA binding cavity runs along the inner edge of the ring-like assembly. A triple mutant of SFTSV-N, R64D/K67D/K74D, almost lost its ability to bind RNA in vitro, is deficient in its ability to transcribe and replicate. Structural studies of the mutant reveal that both alterations in quaternary assembly and the charge distribution contribute to the loss of RNA binding. In the screening of inhibitors Suramin was identified to bind phleboviral N specifically. The complex crystal structure of SFTSV-N with Suramin was refined to a 2.30-Å resolution. Suramin was found sitting in the putative RNA binding cavity of SFTSV-N. The inhibitory effect of Suramin on SFTSV replication was confirmed in Vero cells. Therefore, a common Suramin-based therapeutic approach targeting SFTSV-N and its homologs could be developed for containing phleboviral outbreaks.
INTRODUCTION
Severe fever with thrombocytopenia syndrome (SFTS) is an emerging infectious disease caused by SFTS virus (SFTSV), a novel virus belonging to the genus Phlebovirus of Bunyaviridae family. SFTS is characterized by high fever, thrombocytopenia, leukocytopenia, gastrointestinal symptoms, and multiorgan dysfunction, with an average case fatality rate of 10 to 12% (1, 2). Recently, a new phlebovirus, Heartland virus, was isolated from two patients suffering with severe febrile illness in Missouri (3). The genomic sequences of Heartland virus share high similarity with SFTSV. Therefore, SFTSV infections appear to be a global public threat. Currently, specific medical countermeasures and vaccines against SFTSV are lacking. Hence, there is an urgent need to find effective means to combat the newly emerging SFTSV.
Like other members of the family Bunyaviridae, SFTSV is a negative-strand RNA virus containing three single-stranded RNA genome segments designated as large (L), medium (M), and small (S). The L segment encodes a multifunctional protein that comprises an N-terminal endonuclease domain (4), which is also reported in Arenavirus (5), and a large RNA-dependent RNA polymerase domain. The M segment encodes glycoproteins Gn and Gc that are inserted in the viral lipidic envelope and are responsible for cellular attachment and entry. The S segment encodes a nucleocapsid protein (N) and a nonstructural protein (NSs) via an ambisense coding strategy (2, 6). N is the most abundant viral protein in virions and infected cells. It encapsidates the viral genomic RNA (vRNA), resulting in the formation of ribonucleoprotein complexes (RNPs) (7, 8). In addition to its critical role in protecting the vRNA, N also plays an active role in RNA transcription and replication, as well as in virion assembly (7–10). N is the most conserved protein in the Phlebovirus genus.
The structures of the Rift Valley fever virus N (RVFV-N), solved by two groups independently, were the first representative structures for phleboviral N (11, 12). RVFV-N is composed of a C-terminal core domain for binding viral RNA and an N-terminal arm that mediates oligomerization. More recently, the crystal structures of RVFV-N in complex with RNA depicting the mode of RNA sequestration by N have been reported (13). Another structure of N from Crimean-Congo hemorrhagic fever virus belonging to Bunyaviridae family but from a different genus, Nairovirus genus, revealed a fold (14–16) that was surprisingly similar to N of Lassa virus belonging to the relatively distant Arenaviridae family (17, 18). These results highlight the limitations and pitfalls of classification based on primary sequence homology.
To develop specific antiviral drugs targeting SFTSV and to extend our knowledge of the biological role of SFTSV-N, comprehensive three-dimensional structures, mutagenesis studies, in vitro and in vivo functional analysis are urgently needed. Here, we report the structures of SFTSV-N and its homologs from Buenaventura and Granada viruses (BUEV and GRAV). Further, screening for small molecule inhibitors of SFTSV-N identified Suramin as a potent inhibitor, which could influence N′s RNA binding ability and block the life cycle of the SFTSV. Crystal structure of the binary complex of SFTSV-N with Suramin coupled with structures of BUEV-N and GRAV-N shed light on the molecular details of a conserved mechanism of tethering RNA by phleboviral N and provide first glimpse into the nature of potential inhibitors that could be designed for containing phleboviral outbreaks.
MATERIALS AND METHODS
Constructs.
The full-length N genes of three phleboviruses—severe fever with thrombocytopenia syndrome virus (SFTSV), Buenaventura virus (BUEV; strain Co Ar 3319), and Granada virus (GRAV; strain GR25)—were codon optimized using JCat (19) and synthesized by Sangon Biotech (Shanghai) Co., Ltd. All the N genes were subcloned into pMCSG7 (20). The resultant plasmid pMCSG7-N was used as a template for generating mutants using QuikChange site-directed mutagenesis kit according to the manufacturer's instructions (Stratagene). All the plasmids were sequenced in order to verify the sequence.
Protein expression and purification.
The plasmids harboring the target gene, encoding 6×His-TEV-Ns, were transformed into E. coli BL21(DE3) (Tiangen Biotechnology). The cells were grown in Luria-Bertani (LB) medium containing ampicillin (100 μg/ml) at 37°C until the optical density at 600 nm reached 0.8. The culture was then induced with 0.2 mM isopropyl-β-d-thiogalactoside (IPTG) for 20 h at 16°C. Cells were harvested by centrifugation and resuspended in phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 50 mM Na2HPO4, 10 mM KH2PO4 [pH 7.4]). After lysis by sonication and clarification by centrifugation, the supernatant was applied to a nickel-nitrilotriacetic acid (Ni-NTA) resin gravity column (Qiagen) that had been previously equilibrated with PBS. The column was first washed with 50 ml of PBS, followed by washing with 50 ml of PBS containing 20 mM imidazole, and finally eluted with PBS containing 300 mM imidazole. After buffer exchange, the His tag was removed by tobacco etch virus (TEV) treatment. Uncut protein was separated by a second Ni-affinity chromatography step. Fractions containing the protein were pooled, concentrated, and loaded on Superdex G200 size exclusion chromatography column (Amersham) equilibrated with 20 mM Tris-HCl (pH 8.0), 200 mM NaCl, and 2 mM dithiothreitol (DTT). Fractions containing pure N were pooled and stored at −80°C. RNA-free wild-type (WT) Ns were also purified by adding 1 M NaCl and 3 M urea to the lysis buffer. Binary complex of SFTSV-N with Suramin (Sigma) was prepared by mixing purified SFTSV-N and Suramin in a 1:2 ratio. Excess Suramin was removed by SEC using a Superdex G200 column. The resulting SFTSV-N–Suramin complex was stored at −80°C until further use.
Crystallization.
Initial crystallization conditions were screened by hanging-drop vapor diffusion method at 25°C. Crystals were obtained by mixing equal volumes of the protein solution with the reservoir solution (1:1) in 2-μl drops and equilibrating the drops against 300 μl of reservoir solution. Crystals for SFTSV-N, and its binary complex with Suramin were grown in a solution containing 5 to 8% PEG8000, 0.1 M sodium acetate (pH 4.3 to 4.7), and 0.2 M zinc acetate. Hexagonal crystals (P6 space group) of SFTSV-N appeared in higher concentrations of PEG8000, while cube-shaped crystals (C2 space group) for both SFTSV-N and its complex with Suramin were obtained at lower concentrations of PEG8000. Diffraction-quality crystals of the triple mutant of SFTSV-N were grown in a reservoir solution containing 2 to 3.5 M NaCl, 0.1 M sodium acetate (pH 4.1 to 5.1), and 0.2 M Li2SO4. Crystals for WT BUEV-N, WT GRAV-N, and the triple mutant of GRAV-N were obtained in three different conditions: (i) 47% (wt/vol) PEG200 and 0.1 M cacodylate (pH 6.5), (ii) 0.1 M MgCl2, 21% PEG1000, and 0.1 M imidazole (pH 8.0), and (iii) 0.1 M calcium acetate, 12% (wt/vol) PEG3350, 0.04 M citric acid, and 0.06 M Bis-tris (pH 6.0), respectively.
Data collection, processing, and structure determination.
Data for the WT SFTSV-N (pentamer and hexamer) and its triple mutant, WT BUEV-N, and the triple mutant of GRAV-N were collected at 100 K using an ADSC Q315 charge-coupled device (CCD) detector on beamline BL17U1 at the Shanghai Synchrotron Radiation Facility (SSRF). Diffraction data for the binary complex of SFTSV-N with Suramin was collected at 100 K using an ADSC Q315 CCD detector on beamline BL17A at the Photon Factory (KEK, Japan). The data for WT GRAV-N was collected at beamline BL5.0.1 (Advanced Light Source, Lawrence Berkeley National Lab, USA). In order to obtain more accurate and higher signal-to-noise ratio diffraction data for the binary complex of SFTSV-N with Suramin, data were collected using MDS data collection strategy (21). The data sets were indexed, integrated, and scaled using the HKL2000 software (22). The WT SFTSV-N structure (P6 space group) was initially solved by molecular replacement method using coordinates of RVFV-N (PDB code 3OV9) (11) as a template, and the structures for WT SFTSV-N (C2 space group), WT SFTSV-N–Suramin complex, WT BUEV-N, and WT GRAV-N were solved by using the new WT SFTSV-N coordinates as the search model. The triple-mutant structures were solved by using corresponding WT coordinates as the search model. Molecular replacement was performed using Phaser (23). Model building and iterative refinement were carried out using Coot (24), Phenix_refine (25), and Refmac (26). Molecular graphics were generated using Pymol (27). Statistics for data collection and refinement are listed in Table 1.
Table 1.
Data collection and refinement statistics of SFTSV-N, GRAV-N, and BUE-Na
| Parameter | WT SFTSV-N pentamer | WT SFTSV-N hexamer | Triple mutant SFTSV-N | SFTSV-N:Suramin | WT GRAV-N | Triple mutant GRAV-N | WT BUEV-N |
|---|---|---|---|---|---|---|---|
| Diffraction data statistics | |||||||
| X-ray source | SSRF | KEK | SSRF | KEK | ALS | SSRF | SSRF |
| Distance (mm) | 380 | 290 | 350 | 340 | 350 | 300 | 300 |
| No. of images | 360 | 360 | 720 | 720 | 720 | 360 | 180 |
| Oscillation width (°) | 0.5 | 0.5 | 0.25 | 0.5 | 0.5 | 0.5 | 1 |
| Wavelength (Å) | 0.98 | 0.97 | 0.98 | 0.97 | 1 | 0.98 | 0.98 |
| Space group | C2 | P6 | C2221 | C2 | P1 | P1 | I422 |
| a, b, c (Å) | 111.19, 155.57, 97.65 | 104.00, 104.00, 48.87 | 95.49, 108.48, 221.41 | 103.15, 154.89, 93.29 | 51.29, 96.34, 100.80 | 54.16, 76.80, 77.15 | 97.11, 97.11, 176.64 |
| α, β, γ (°) | 90.00, 107.52, 90.00 | 90.00, 90.00, 120.00 | 90.00, 90.00, 90.00 | 90.00, 102.12, 90.00 | 64.57, 81.72, 85.10 | 82.79, 69.51, 69.36 | 90.00, 90.00, 90.00 |
| Mosaicity (°) | 0.57 | 0.57 | 0.53 | 0.73 | 0.79 | 1.44 | 0.54 |
| No. of molecules/ASU | 5 | 1 | 4 | 5 | 6 | 4 | 1 |
| Resolution range (Å) | 50.00–2.80 (2.90–2.80) | 50.00–1.90 (1.97–1.90) | 50.00–2.45 (2.54–2.45) | 50.00–2.30 (2.38–2.30) | 50.00–2.50 (2.54–2.50) | 50.00–2.50 (2.59–2.50) | 50.00–2.66 (2.76–2.66) |
| Total no. of reflections | 93,382 (8,918) | 255,730 (18,320) | 291,435 (25,445) | 364,560 (12,014) | 178,030 (6,438) | 72,164 (6,475) | 176,574 (17,618) |
| No. of unique reflections | 35,916 (3,567) | 23,900 (2,290) | 42,237 (4,104) | 60,760 (5,006) | 57,429 (2,575) | 37,981 (3,597) | 12,523 (1,215) |
| Rsym (%) | 10.7 (39.7) | 7.5 (37.3) | 8.0 (35.0) | 10.3 (36.3) | 7.9 (37.7) | 3.4 (34.1) | 7.4 (42.5) |
| Mean I/σ (I) | 10.17 (2.00) | 30.42 (4.16) | 34.78 (5.35) | 24.78 (2.00) | 20.92 (1.87) | 23.88 (2.40) | 51.00 (8.42) |
| Completeness (%) | 92.7 (92.4) | 99.7 (96.9) | 99.8 (98.5) | 96.3 (79.1) | 97.4 (87.2) | 97.2 (91.7) | 99.9 (100.0) |
| Redundancy | 2.6 (2.5) | 10.7 (8.0) | 6.9 (6.2) | 6.0 (2.4) | 3.1 (2.5) | 1.9 (1.8) | 14.1 (14.5) |
| Refinement statistics | |||||||
| Resolution range (Å) | 43.81–2.80 | 45.03–1.90 | 43.81–2.45 | 42.72–2.30 | 49.64–2.51 | 28.83–2.45 | 32.67–2.66 |
| Rwork/Rfree (%) | 22.57/25.99 | 17.64/21.33 | 21.74/25.97 | 22.33/26.06 | 19.71/27.61 | 20.16/27.66 | 21.14/25.40 |
| No. of atoms | |||||||
| Protein | 9,480 | 1,914 | 7,429 | 9,554 | 11,558 | 7,734 | 1,871 |
| Water | 69 | 257 | 78 | 250 | 195 | 115 | 30 |
| B factor (Å2) | |||||||
| Protein | 43.81 | 22.10 | 72.34 | 51.19 | 59.10 | 59.23 | 55.32 |
| Suramin | 77.78 | ||||||
| Water | 30.45 | 29.24 | 55.57 | 47.25 | 51.10 | 48.28 | 49.02 |
| RMSD | |||||||
| Bond length (Å) | 0.006 | 0.007 | 0.010 | 0.007 | 0.013 | 0.017 | 0.009 |
| Bond angle (°) | 0.982 | 1.093 | 1.191 | 1.052 | 1.599 | 2.001 | 1.047 |
| Ramachandran plot (%) | |||||||
| Favored regions | 97.78 | 98.37 | 98.21 | 98.75 | 97.77 | 95.76 | 95.44 |
| Allowed regions | 2.22 | 1.63 | 1.79 | 1.25 | 1.89 | 3.23 | 4.56 |
| Outliers | 0 | 0 | 0 | 0 | 0.34 | 1.01 | 0 |
Numbers in parentheses represent values for the highest-resolution shell.
SAXS.
Small angle X-ray scattering (SAXS) data for SFTSV-N and its triple mutant were collected at beam line 12-ID-B, APS, Argonne National Lab. Each sample was measured with three exposures—0.5, 1.0, and 6.0 s—at 10°C at three concentrations: 2.5, 5.0, and 10.0 mg/ml in a buffer composed of 20 mM Tris-HCl (pH 8.0), 200 mM NaCl, and 2 mM DTT. The scattering intensity I(q) was measured for q values (q = 4πsinθ/λ, where 2θ is the scattering angle) ranging from 0.01 to 0.3 Å−1. The resulting scattering curves for each sample were radially averaged and buffer subtracted. The data were integrated, scaled, and buffer subtracted to get the standard scattering curves. Multiple curves with different concentrations and different exposure times were scaled and averaged in PRIMUS (28). The initial Rg values were analyzed by PRIMUS (28), from the Guinier plot analysis. The P(r) distribution function was calculated with the program GNOM (29). The maximum particle dimension (Dmax) was estimated from the P(r). The molecular weight was estimated directly using the P(r) distribution function from the web server (http://www.if.sc.usp.br/∼saxs/saxsmow.html) (30). CRYSOL (31) was used to calculate the fit between coordinates and the scattering curve. The low-resolution shape reconstruction were modeled by GASBOR (32) from the calculated P(r) distribution curve. Several individual ab initio models agree well and were averaged by DEMAVER (33). The high-resolution crystal structure model was superimposed into the low-resolution shape model by SUPCOMB (34).
FTS assay.
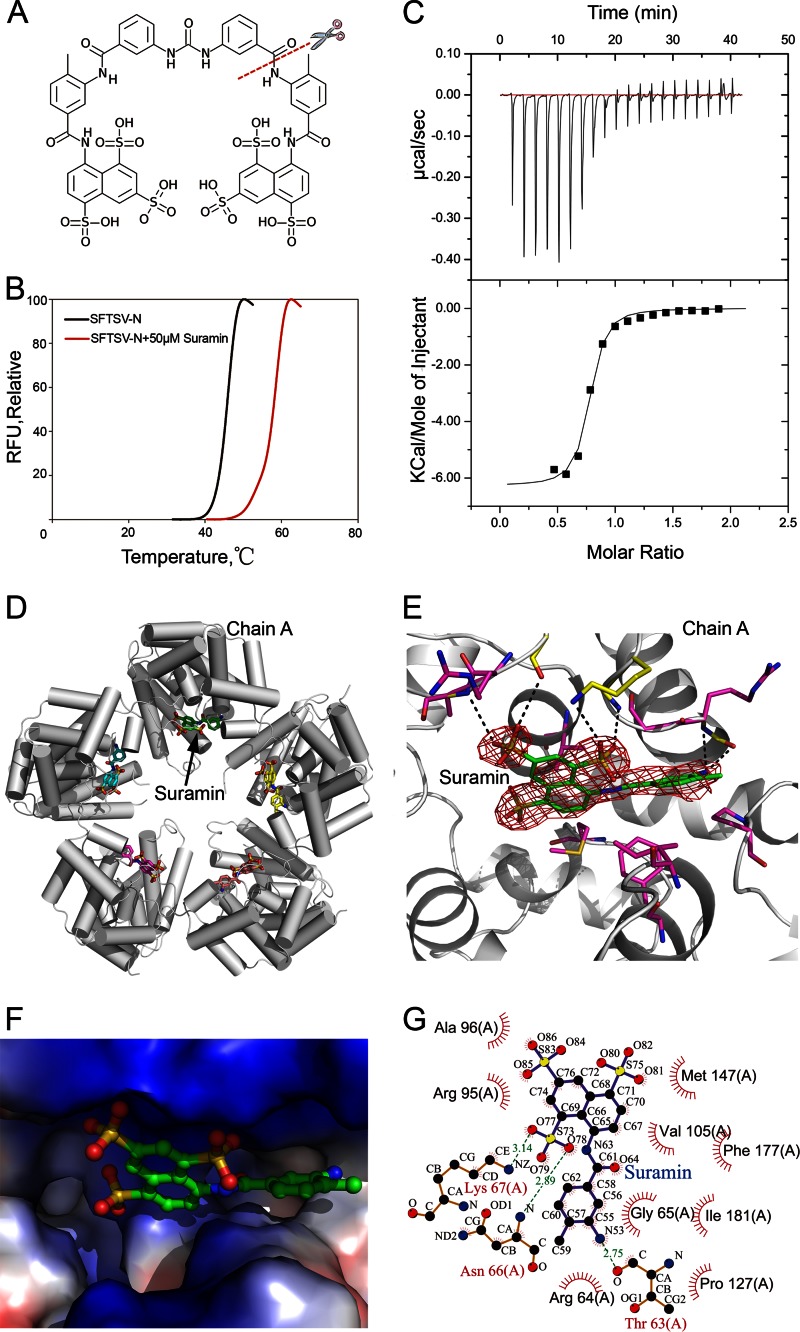
A robotic pipeline in the High Throughput Analysis Laboratory of Northwestern University (NU-HTAL) was used for carrying out small molecule screening by fluorescence-based thermal shift (FTS) assay. The pipeline uses an Echo550 acoustic transfer robot (Labcyte) for compound addition and a Mosquito robot (TTP LabTech) for protein dispensing, followed by thermal scanning coupled with fluorescence detection which is performed on a real-time PCR machine CFX384 (Bio-Rad Laboratories). The assay was run in 384-well PCR plates, using 1 μg of protein and 10 nl of 5000× Sypro Orange (Invitrogen) per well in a 10-μl assay with HEPES buffer (20 mM HEPES [pH 7.5], 150 mM NaCl). The assay concentration for protein was 3.6 μM, and that for Sypro Orange was 5×. After compound addition, the plate was sealed with an optical seal, shaken, and centrifuged. The thermal scan ranged from 10 to 95°C with a temperature ramp rate of 1.5°C/min. The fluorescence was recorded every 10 s. The data analysis and report generation were performed by using the in-house software excel-FTS of the NU-HTAL. A total 1,600 of the 2,000 molecules in the Spectrum collection (MicroSource Discovery Systems, Inc.) were screened for binding SFTSV-N in a multicompound pooled format. Six hit compounds with positive melting-temperature (Tm) shifts were identified in the screen. The most prominent hit was Suramin. In a dose-response test, the Tm shift reached 12°C for a concentration of 5 μM, after which it began to plateau.
Isothermal titration calorimetry (ITC).
Proteins and Suramin were suspended in Tris buffer (20 mM Tris-HCl [pH 8.0], 200 mM NaCl, and 2 mM DTT). Protein concentration was measured using absorbance at 280 nm. Prior to titration, both protein and Suramin were centrifuged at 18,000 × g at 4°C for more than 10 min to remove any debris and air bubbles. The calorimetric titrations were performed on a MicroCal ITC200 instrument (Microcal, Inc., Northampton, MA) at 25°C. The reactant (50 μM N) was placed in the 300-μl sample chamber and Suramin (500 μM) was added using the syringe with 20 successive additions of 2 μl (with an initial injection of 0.5 μl), spaced 120 s apart. The data was analyzed using the ORIGIN software. The association constant (Ka), enthalpy change (ΔH), and the stoichiometry (N) were calculated by fitting the thermo grams to one set of binding sites.
Effect of Suramin on SFTSV replication.
Vero cells were seeded into 96-well plates. When the cells were grown ca. 85% confluent, 100 50% tissue culture infective doses (TCID50) of SFTSV (HB29 strain) in 100 μl were added into each well. After incubation for 1 h, infected cells were washed to remove unbound virus. Then Suramin or heparin was added into the SFTSV preabsorbed cells in four replicate wells at final concentrations of 0.8, 0.4, 0.2, 0.1, 0.05, and 0 μg/μl. The medium was replaced every 3 days, when the production of SFTSV in culture supernatant was detected by enzyme-linked immunosorbent assay (ELISA).
ELISA.
A plate was coated with purified rabbit anti-SFTSV polyclonal antibody for 12 h at 4°C. After washing the plate with PBST, 80 μl of culture supernatant was added into each well, followed by addition of 20 μl of lysis buffer. After incubation for 1 h, the plate was washed and 100 μl of horseradish peroxidase-conjugated rabbit anti-SFTSV-N antibody (diluted in 5% skim milk) was added into each well. The plate was incubated for 1 h at 37°C and then washed. Subsequently, 100 μl of 3,3′,5,5′-tetramethylbenzidine (TMB) substrate was added to develop color. The plate was read at an optical density at 450 nm by a microplate reader.
Luciferase reporter assay.
Three plasmids, including L protein expression plasmid (VR1012-L), M reporter plasmid constructed by substitution of the ORF of SFTSV glycoprotein with firefly luciferase (Phh21-luc/M), and the Renilla luciferase expression plasmid (pGL4.74) to normalize transfection efficiency, were cotransfected with WT SFTSV-N encoding plasmid (VR1012-N) or site-directed N mutants (VR1012-Nmutants) into 293T cell in 24-well plates. Empty vector (VR1012) was used as negative control for N. Each group of plasmids was transfected in four replicate wells. After culture for 24 h, cells were lysed. The firefly luciferase activity was measured by adding 20 μl of cell lysate into a luminometer tube containing LAR II and read by the luminometer. Further, Renilla luciferase activity was measured by adding 100 μl of Stop&Glo reagent into the tube and then read by the luminometer. The ratio of firefly luciferase activity to Renilla luciferase activity was presented in the results.
Statistical analysis.
The significance of differences between groups showing similar variance was evaluated by the Student t test.
Protein structure accession numbers.
The atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB) under accession codes 4J4U and 4J4R for WT SFTSV-N, 4J4S for triple-mutant SFTSV-N, 4J4V for WT SFTSV-N complex with Suramin, 4J4X for WT GRAV-N, 4J4Y for triple-mutant GRAV-N, and 4J4W for WT BUEV-N.
RESULTS
Production of WT RNA-free SFTSV-N and its homologs.
Three Ns from SFTSV and its homologs (GRAV-N and BUEV-N) (Fig. 1) were produced using E. coli. During purification, the results of size-exclusion chromatography (SEC) suggested that Ns formed oligomers of different sizes (see Fig. S1A in the supplemental material). The A260/A280 absorbance ratio (>1.2) indicated that the purified proteins contained nucleic acids (see Fig. S1B in the supplemental material). N probably bound bacterial nucleic acids during expression. An RNase treatment could lower the A260/A280 ratio, suggesting that the protein bound RNA predominantly. We adopted two different approaches to producing N devoid of nucleic acids. In one approach, several mutants of SFTSV-N were constructed and expressed in E. coli. Interestingly, mutation of 3 amino acids absolutely conserved across the Phlebovirus genus, R64D/K67D/K74D (11), resulted in a triple mutant that almost lost its ability to bind nucleic acids when expressed using E. coli (A260/A280 ≈ 0.5) (Fig. S1B) and could be purified as a homogenous nucleic acid-free N oligomer (see Fig. S1A in the supplemental material). Similar triple mutations of equivalent residues in GRAV-N (R69D/K72E/K79E) and BUEV-N (R63D/K66E/K73E) also resulted in Ns devoid of nucleic acids. The second approach involved partial unfolding of proteins in the presence of urea. Specifically, 3 M urea and 1 M NaCl were added to the lysis buffer during recovery and purification of the protein (12). Such wild-type (WT) N preps contained almost no nucleic acids (A260/A280 ≈ 0.6) (see Fig. S1B in the supplemental material).
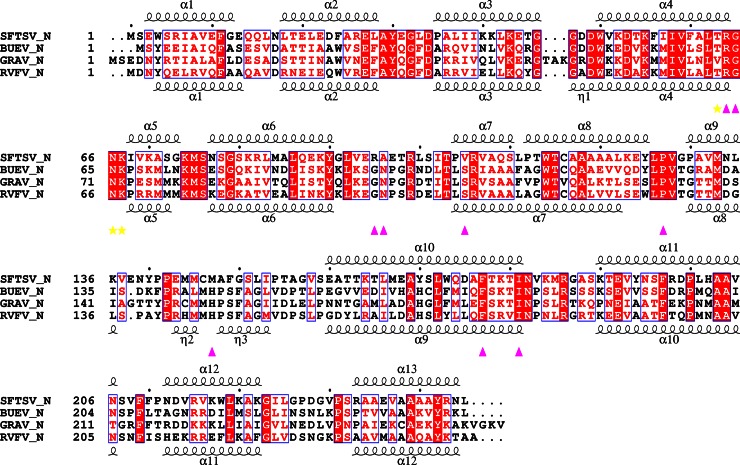
Fig 1.
Sequence comparison of the SFTSV-N, GRAV-N, and BUEV-N with secondary structural elements indicated. Strictly conserved residues are boxed in white on a red background, and highly conserved residues are boxed in red on a white background. Every tenth residue is indicated with a dot (.). The α-helix is depicted by a coil. The Suramin binding residues of SFTSV-N are indicated by yellow stasr (hydrogen bond interaction) and filled magenta triangles (hydrophobic interaction), respectively. The secondary structure of RVFV-N is according to Ferron et al. (11). Alignment was performed by CLUSTAL W (43). The figure was generated by ESPript (44).
Crystal structures of N from SFTSV and its homologs.
Crystallization screening was performed using commercially available sparse matrix screens for all three soluble WT Ns, as well as their triple mutants. After optimization, diffraction quality crystals for all three WT Ns and triple mutants of SFTSV-N and GRAV-N were obtained. Preliminary crystallographic analysis results are summarized in Table 1.
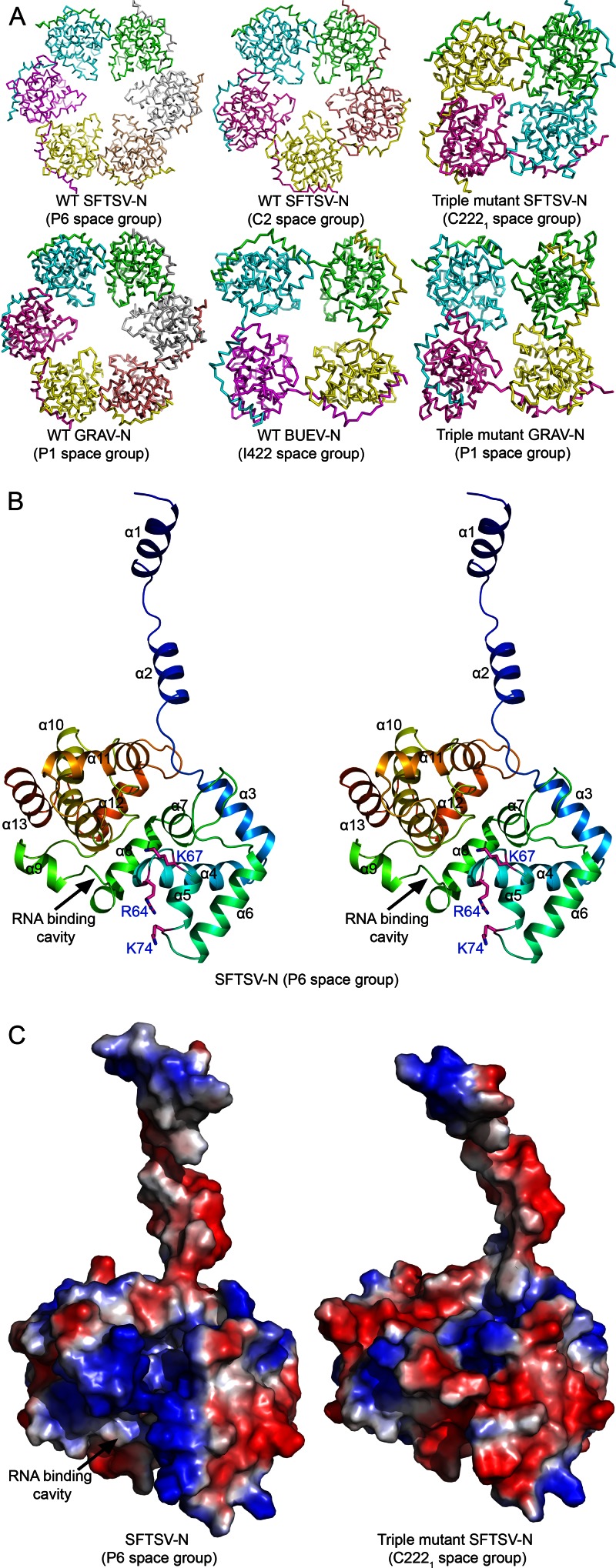
WT SFTSV-N crystallized in two different space groups: P6 and C2. In P6 space group, the asymmetric unit (ASU) contains a single molecule of SFTSV-N. However, inspection of the symmetry mates revealed that the protein had crystallized as a hexamer. Six molecules of SFTSV-N are arranged in a hexameric ring-like structure with the crystallographic 6-fold axis passing through the center (Fig. 2A). The hexameric structure of SFTSV-N could be refined to a 1.90-Å resolution with acceptable statistics (Table 1). In C2 space group, the ASU contains five molecules of SFTSV-N arranged in a pentameric ring-like structure (Fig. 2A). Statistics for the final model refined to a 2.80-Å resolution are listed in Table 1. Structures of both hexameric and pentameric SFTSV-N, crystallized in different space groups probably due to the crystallization conditions preference, provide a consensus on the molecular details of the architecture of the SFTSV-N.
Fig 2.
Overall structure of phleboviral N. (A) Cα trace of the structures of the three WT phleboviral N and two triple mutants. All of the WT structures form a ring-like oligomeric assembly. Four molecules of the triple mutant SFTSV-N are arranged in two dimers. Each dimer participates in the formation of a tetramer through the crystallographic 2-fold axis. One of the tetramers (formed by chains A and B) is shown. In the tetrameric structures of the triple mutants of SFTSV-N and GRAV-N the central cavity is blocked. (B) Wall-Eye Stereo view of ribbon representation of the crystal structure of the SFTSV-N showing the N terminus forming an arm that extends from the core domain (residues 35 to 245). The α-helices and key residues (magenta) are labeled. (C) The putative RNA binding cavity of WT and the triple mutant of SFTSV-N are shown by corresponding electrostatic potential surfaces (blue represents positive potential, red negative potential).
The triple mutant of SFTSV-N crystallized as a tetramer in C2221 space group and diffracted X-rays to a 2.45-Å resolution. The ASU contains four molecules that are arranged as two dimers. Instead of the pentameric or hexameric assembly observed in WT SFTSV-N crystals, each dimer of the triple mutant participates in the formation of a tetramer through the crystallographic 2-fold symmetry axis. The secondary structural elements of the triple mutant of SFTSV-N are similar to that of the WT protein. The core domain of monomers of the triple mutant within a tetramer could be superimposed on each other (with maximal Cα RMSD value of 2.6 Å), as well as on monomers of WT SFTSV-N (with maximal Cα RMSD value of 2.2 Å).
Structures of WT GRAV-N and BUEV-N were refined to a 2.50-Å (P1 space group) and a 2.66-Å (I422 space group) resolution, respectively. Although six molecules of GRAV-N are observed forming a ring-like hexamer in one ASU, there is only one BUEV-N molecule in ASU forming a tetramer through crystallographic symmetry operation. The triple mutant of GRAV-N crystallized as a tetramer in P1 space group. The structure was solved and refined to a 2.50-Å resolution.
The monomers of GRAV-N and BUEV-N could be superimposed on that of SFTSV-N. The RMSD value for matching Cα atoms of SFTSV-N with GRAV-N was 1.3 Å, while that for Cα atoms overlaid over BUEV-N was 1.9 Å. In addition, structures of GRAV-N and BUEV-N look similar with the main chain Cα atoms overlapping with an RMSD value of 1.7 Å. Thus, the overall structure of the SFTSV-N homologs seems to be evolutionarily conserved.
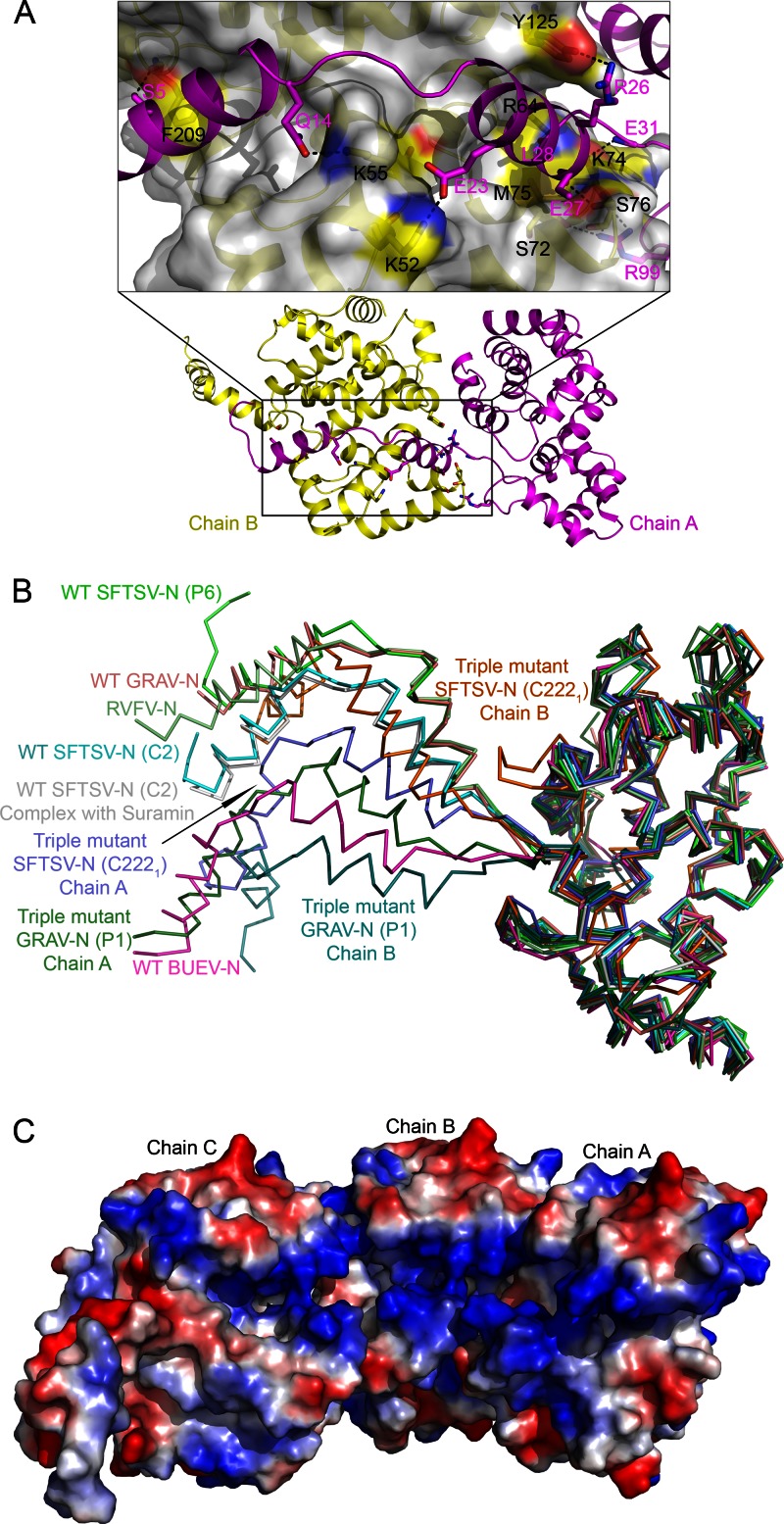
All of the Ns and their mutants reported in the present study adopt the same tertiary architecture consisting of a compact C-terminal core with an extended N-terminal arm (Fig. 2B and C; see Movie S1 in the supplemental material). In hexameric, pentameric, and tetrameric WT N structures, the N-terminal extension arm from one monomer fits into a groove formed on the surface of the core domain of an adjacent monomer (see Fig. S2A in the supplemental material), linking the monomers together by hydrogen bonds and hydrophobic interactions (Fig. 3A; see Table S1 in the supplemental material), as observed previously for RVFV-N (11). The major difference between the monomers from different oligomeric assemblies of N molecules and RVFV-N lies in the position of the N-terminal extension arm (Fig. 3B). The arm is extended farther outward to accommodate additional units of N while constructing larger oligomers.
Fig 3.
Structural characterization of SFTSV-N and its homologs. (A) SFTSV-N protomer binds adjacent N primarily through hydrogen bonds and hydrophobic interactions. Hydrogen bonds within 3.5 Å are indicated as dashed black lines. The key residues are shown as sticks with labels (black for chain A, magenta for chain B). Details of the interactions are listed in Table S1 in the supplemental material. (B) Structural comparison of nine protomers solved in the present study with RVFV-N (3OV9-A). Cα trace of each protomer is labeled individually as indicated in parenthesis. (C) The electrostatic potential surface representation of three adjacent WT SFTSV-N molecules. The putative RNA binding cavity runs along the inner edge of the ring-like assembly of SFTSV-N.
Solution structures determined by SAXS.
To characterize structures of N in solution, we performed small angle X-ray scattering (SAXS) experiments (see Fig. S3 in the supplemental material). The radius of gyration (Rg) derived from the SAXS data for WT SFTSV-N is slightly larger than that of its triple mutant (38.46 ± 0.16 Å versus 34.60 ± 0.10 Å, respectively) (see Fig. S3G in the supplemental material). The shapes of molecular envelope were constructed by SAXS ab initio modeling. Without symmetry restraint, the model for WT SFTSV-N assumes a ring-like shape with a hole in the center. This arrangement of the monomers was in agreement with those of the pentameric or hexameric assembly observed in P6 or C2 crystal forms. CRYSOL (31) was then used to confirm the solution assembly. As a result, pentameric SFTSV-N could fit with the experimental curve with a chi value of 2.1, which was much better than the fit (chi = 7.5) between hexameric SFTSV-N and the experimental curve (see Fig. S2D in the supplemental material). Thus, WT SFTSV-N expressed and purified from E. coli is likely to adopt a pentameric state primarily. Ten individual GASBOR (32) calculations with P5 symmetry as a restraint were performed to construct shapes for WT SFTSV-N. The resultant pentameric model of WT SFTSV-N in solution had an acceptable chi value of 0.44 ± 0.02 (see Fig. S3E in the supplemental material). For the triple mutant of SFTSV-N, a P4 symmetry constraint was used, and the final chi value was 0.93 ± 0.04. The envelope of triple mutant SFTSV-N shows a compact cube-shaped tetramer, where the central cavity is blocked (see Fig. S3F in the supplemental material).
Structural basis for loss of RNA binding function of triple mutants.
We mapped the location of the mutations (R64D, K67D, and K74D) on the structures of SFTSV-N. Examination of the structures of the WT SFTSV-N reveals that R64 and K74 are involved in intermolecular hydrogen bonding interactions with an adjacent subunit of SFTSV-N molecule (Fig. 3A; see Table S1 in the supplemental material), while the side chain of K67 is protruding inside the cavity formed at the center of the quaternary assembly (Fig. 2B). The structure of the triple mutant reveals that the hydrogen bonds of the side chain of R64 with the backbone carbonyl oxygen of L29 are broken upon mutation to aspartate. Further, the K74D mutation breaks the hydrogen bond between the side chain of K74 and E31 (see Table S1 in the supplemental material). Instead, some new hydrogen bonds were observed in the triple mutant (see Table S1 in the supplemental material). Thus, although the triple mutations alter a few intermolecular interactions, they do not seem to affect the overall secondary structure of N.
However, there are differences in the quaternary assembly of the WT N versus that of the triple mutant. First, both WT SFTSV-N and WT GRAV-N formed a hexamer, while the triple mutants crystallized as a tetramer (Fig. 2A). Second, while WT Ns were arranged in a circular pattern with a large central hole (Fig. 2A; see Movie S2 in the supplemental material), even in the tetrameric WT BUEV-N, the monomers of the two triple mutants pack as a compact tetramer with the central hole being blocked. Lastly, the positive electrostatic potential in the putative RNA binding cavity is dramatically reduced due to the introduction of negatively charged residues (SFTSV-N: K67D; GRAV-N: K72E) (Fig. 2C). Thus, the triple mutations perturb the quaternary arrangement of the Ns.
Putative RNA binding cavity analysis.
In RVFV-N, RNAs are accommodated in a cavity by extensive hydrophobic interactions between bases and residues of the RNA-binding cavity and by base stacking, which involves side chains of Y30, F33, A109, A110, P147, I180, P199, A202, L128, F178, P127, and G65 (13). In addition, a network of polar interactions between RNA 5′ phosphates and residues such as N66, K67, R70, R99, and R106 were described (13). There are 17 residues that are involved in RNA binding in RVFV-N. The structures of SFTSV-N, GRAV-N, and BUEV-N are homologous to RVFV-N (Fig. 1; Fig. 3B). Superimposition of those structures on RVFV-N: RNA complex revealed the presence of a similar RNA binding cavity for the three N homologs. In the cavities of all three N homologs, 12 of 17 corresponding residues are strictly conserved, while 4 of them are conserved (see Table S2 in the supplemental material). In addition, most of the side chains of those 17 residues in the three Ns are in positions similar to that of the corresponding residues in RVFV-N (see Fig. S2B in the supplemental material). Thus, the RNA encapsidation mechanism by different species of phleboviruses is likely to be similar.
RNA binding residues are important for SFTSV transcription and replication.
To further investigate the functional role of residues involved in vRNA binding, we examined the ability of a panel of SFTSV-N mutants to promote expression of M reporter gene. Although the activities of N mutants R64Q and K67Q decreased marginally by 20%, the activity of the mutant K74Q did not change significantly (see Fig. S4 in the supplemental material). However, the activity was reduced by 80 or 60% when all three residues—R64, K67, and K74—of SFTSV-N were simultaneously mutated to Q or D (see Fig. S4 in the supplemental material). Therefore, while individual mutations showed a modest decrease in vRNA binding, a synergic effect was observed when all three vRNA binding residues were mutated. Since the N homologs from BUE and GRA viruses use a similar conserved mechanism for tethering RNA, equivalent residues from these proteins—R63, K66, and K73 for BUEV-N and R69, K72, and K79 for GRAV-N—are likely to play an important role in the transcription and replication of the respective viruses.
Identification of Suramin as an inhibitor of SFTSV-N function.
To screen small molecules which could inhibit SFTSV-N from encapsidating vRNA, we performed fluorescence-based thermal shift assays. Several hits were scored based on the increase in melting temperature (Tm), with Suramin (Fig. 4A) being the most prominent hit, showing a 12°C shift in Tm of the protein (Fig. 4B). The binding of Suramin to SFTSV-N was confirmed by ITC, where Suramin showed an affinity of Kd = 0.44 μM for N (Fig. 4C). In comparison, binding of Suramin to either GRAV-N or BUEV-N resulted in a 6°C shift in Tm (see Fig. S5A in the supplemental material). Although BUEV-N displayed a Kd of 2.02 μM for Suramin, GRAV-N bound Suramin with a Kd of 2.48 μM, as suggested by ITC assays (see Fig. S5E and F in the supplemental material). Thus, Suramin could bind all three WT phleboviral N with high affinity. Further, we tested the specificity of Suramin for N from other genus of the Bunyaviridae family. Suramin bound RVFV-N and refolded SFTSV-N (see Fig. S5C in the supplemental material), although it did not bind the Ns from Orthobunyavirus, Nairovirus and Hantavirus (see Fig. S5D in the supplemental material). Suramin did not bind the Ns of influenza A virus, rabies virus, Lassa virus, and human immunodeficiency virus (HIV) capsid protein either (see Fig. S5D in the supplemental material). Therefore, Suramin seems to specifically bind SFTSV-N and its homologs within the Phlebovirus genus.
Fig 4.
Structure of the binary complex of SFTSV-N with Suramin. (A) Chemical structure of Suramin with only the naphthalene-1,3,5-trisulfonic acid head modeled in the structure in the present study. (B) Interaction of SFTSV-N with Suramin detected by thermal shift assay. (C) ITC titration curves of SFTSV-N with Suramin. The top panels shows raw data of heat changes upon addition of Suramin (500 μM) into the cell containing 50 μM RNA-free SFTSV-N. The bottom panel shows processed data corresponding to the heat of each injection plotted against the molar ratio of total Suramin to total SFTSV-N after subtraction of the heat of control. The affinity constant (Kd = 0.44 ± 0.095 μM) was derived at 1:1 fixed stoichiometry. (D) Pentameric arrangement of the SFTSV-N–Suramin complex. SFTSV-N molecules are represented by tubes, and Suramin is indicated as sticks. (E) Closeup view of the interface between SFTSV-N and Suramin. Electron density of a 2Fo-Fc simulated annealing (SA) omit map for Suramin in the cavity of the SFTSV-N binding site contoured at 1.0 σ. Residues involved in hydrogen bonding (within 3.5 Å) (yellow) and hydrophobic interactions (magenta) with Suramin are shown as sticks. (F) Closeup view of the interface between SFTSV-N and Suramin with the protein surface colored according to electrostatic potential surface. (G) LIGPLOT representation of the SFTSV-N–Suramin complex depicting bonding atoms and bond lengths. Interacting atoms are connected by green dashed lines with bond lengths indicated (in Å). Residues involved in direct hydrophobic interactions with Suramin are shown as red semicircles with radiating spokes.
Structural analysis of the SFTSV-N in complex with Suramin.
The crystal structure of the binary complex of SFTSV-N with Suramin was solved in C2 space group and refined to a 2.30-Å resolution. Similar to the WT SFTSV-N, the binary complex of SFTSV-N with Suramin crystallized as a pentamer (Fig. 4D; see Movie S3 in the supplemental material). Clear electron density for the naphthalene-1,3,5-trisulfonic acid head of Suramin permitted unambiguous modeling of the moiety into the structure of SFTSV-N (Fig. 4E). However, density for the remainder of the ligand was weak, fragmented and could not be improved by B-factor sharpening (35, 36). One naphthalene-1,3,5-trisulfonic acid head of Suramin bound one molecule of SFTSV-N, which was consistent with the 1:1 stoichiometry suggested by ITC (Fig. 4C). All five SFTSV-N molecules present in the ASU bound the Suramin moiety using similar interactions. The naphthalene-1,3,5-trisulfonic acid moiety sits in a deep pocket located at the center of the protein (Fig. 4F; see Fig. S2C in the supplemental material). In addition, Suramin is anchored by three hydrogen bonds (K67-NZ with O77-Suramin, T63-O with N53-Suramin, and N66-N with O78-Suramin) and electrostatic interactions (Fig. 4F and G).
We compared the structure of Suramin bound SFTSV-N with the structures of BUEV-N and GRAV-N by superimposing the three structures. In particular, we inspected the composition of amino acids within 5.0 Å of the ligand (see Fig. S5B and Table S3 in the supplemental material). Interestingly, within the three N homologs, the majority of the residues which are involved in binding Suramin are conserved. Of 16 conserved residues that interact with Suramin, 8 are strictly conserved. Only one residue is not conserved (see Table S3 in the supplemental material). Thus, except for a few minor differences, the Suramin binding site is made up of highly conserved residues, suggesting a very similar mode of Suramin-binding for the phleboviral N. Since Suramin binds to the putative RNA binding cavity in SFTSV-N (see Fig. S2B and C in the supplemental material), Suramin was predicted to compromise viral RNA replication.
Suramin compromises SFTSV viral replication in Vero cells.
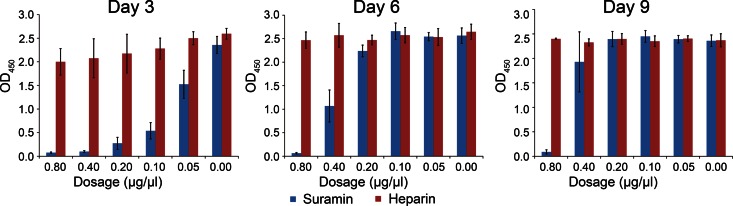
The inhibitory effect of Suramin on viral replication was examined in Vero cells by adding the small molecule at 1 h after viral infection. Another negatively charged molecule, heparin, was used as a control. On day 3, Suramin exerted an obvious repression on viral replication in a dose-dependent manner (Fig. 5). On day 6, higher dosages of Suramin ranging from 0.4 to 0.8 μg/μl showed a repressive effect on viral replication, but the effect of lower dosages ranging from 0.05 to 0.2 μg/μl was comparable to heparin. On day 9, only the dosage of 0.8 μg/μl had a repressive effect on viral replication (Fig. 5). Therefore, Suramin at the dosage of 0.8 μg/μl could exert a persistent inhibitive effect on viral replication. The virus growth assay demonstrated that the small molecule drug Suramin resulted in an impaired production of SFTSV, presumably by obstructing vRNA binding as indicated by structural studies. Since the structure of SFTSV-N is conserved, this effect of Suramin could be extrapolated to other N homologs belonging to the Phlebovirus genus.
Fig 5.
Suramin inhibits the replication of SFTSV in vivo. Vero cells were infected by 100 TCID50 SFTSV for 1 h, and then a gradient of Suramin or heparin was added to the SFTSV preabsorbed cells in four replicate wells at final concentrations as indicated in the figure. The medium was replaced every 3 days. The release of SFTSV in culture supernatant was detected by ELISA.
DISCUSSION
Two conformations of the RVFV-N, closed dimeric and open hexameric, and electron microscopic images of heterogeneous RNP-like particles comprised of 4 to 7 U of RVFV-N have been reported (11, 12). More recently, complex structures of RVFV-N with RNA have also been described (13). These structures of the N from the Phlebovirus genus revealed a new fold for binding vRNA (13). Although SFTSV belongs to the same genus, the pathogenesis of SFTSV infections is different. Therefore, structural studies of proteins vital to the life cycle of the virus are essential for developing structure-guided therapeutics to contain SFTSV outbreaks. N shields genomic vRNA from the host defense machinery. An ensemble of crystal structures of N from SFTSV and related viruses BUEV and GRAV solved by us have revealed different oligomeric states of the N (tetramer, pentamer, and hexamer), which enabled us to not only compare the structural conservation but also understand the mode of oligomerization, a process vital for encapsidation and protection of vRNA of the progeny. Structures of the tetramer, pentamer, and hexamer of SFTSV-N and its homologs reported here reveal that the N-terminal arm plays an important role in the assembly of the oligomers as discussed previously (13).
To map the location of the putative RNA binding site, we superimposed structures of SFTSV-N, GRAV-N, and BUEV-N onto that of RVFV-N–RNA complex (PDB code 4H5O). A conserved RNA binding cavity is found in phleboviral N running along the inner edge of the ring-like quaternary assembly. Most of the residues forming this cavity are conserved (see Table S2 in the supplemental material). Thus, SFTSV-N, GRAV-N, BUEV-N and possibly other N in Phlebovirus genus are very likely to use a similar mechanism to encapsidate RNA as the RVFV-N.
In the structure of SFTSV-N triple mutant, a similar RNA binding cavity could also be found, except a positively charged amino acid K67 was substituted with a negatively charged residue (aspartic acid, D), which changed the charge distribution of the cavity. The triple mutations resulted in the alterations of the quaternary assembly of SFTSV-N triple mutant as observed by SAXS analysis of the WT and triple mutant N (see Fig. S3F in the supplemental material). Thus, alterations in the quaternary assembly and the opposite charge introduced via substituted amino acid (e.g., Arg-to-Asp mutation) contributed to the reduction of nucleic acid binding ability of the triple mutants (see Fig. S1B in the supplemental material).
The identified small molecule inhibitors of SFTSV-N RNA binding function suggest that small molecules with negative charge could serve as potential ligands. Among the resulting ligands, Suramin showed highest affinity for SFTSV-N. Suramin is a drug used for the treatment of trypanosomiasis and helminthiasis (37, 38). Recently, its potential as an antineoplastic agent in the treatment of cancers and as an antiviral agent is being evaluated (39–41). Interestingly, while Suramin could bind SFTSV-N homologs such as RVFV-N (42), GRAV-N, and BUEV-N, it did not bind the Ns from other genera of the Bunyaviridae family (Orthobunyavirus, Nairovirus, and Hantavirus) or other viruses such as influenza A virus, Rabies virus, Lassa virus, and HIV capsid protein. Thus, Suramin shows specificity for phleboviral N. The implication of the ability of Suramin to compete with RNA binding by SFTSV-N in vitro was further illustrated by the inhibition of viral replication in Vero cells. Clearly, in this context, the crystal structure of the binary complex of SFTSV-N with Suramin is valuable and can aid further development of this compound as a drug for the containment of SFTSV outbreaks.
In the Suramin-bound structure of SFTSV-N, Suramin is found in the cavity encircling the central hole of the ring-like assembly. As expected, the Suramin binding pocket in SFTSV-N overlaps with its putative RNA binding cavity as that of RVFV-N. Six residues of SFTSV-N involved in binding Suramin—G65, N66, K67, I181, P127, and F177—are identical with the corresponding conserved residues found in RVFV-N for binding RNA (see Fig. S2C, Table S2, and Table S3 in the supplemental material). Further, the naphthalene-1,3,5-trisulfonic acid head of Suramin is buried deeper in the Suramin binding pocket of SFTSV-N compared to the RNA bound to RVFV-N. This was further supported by Suramin's inhibitory effects on SFTSV replication in Vero cells.
In summary, our structural and functional studies on SFTSV-N and its homologs shed light on the overall structural conservation and suggest a shared mechanism for encapsidating vRNA. We have identified Suramin as an inhibitor of phleboviral N′s RNA binding function that affects their vRNA binding and viral replication. Since Suramin could bind N in Phlebovirus genus specifically, it is possible to design an inhibitor with a broad spectrum of activity against phleboviruses.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the staff at synchrotron beamlines (17U1 of SSRF, 17A of KEK, 5.0.1 of ALS, and 12-ID-B of APS) for help in the X-ray diffraction and solution X-ray scattering data collection. We thank Y. Y. Chen and Y. Wang at the Protein Science Core Facility of IBP for technical help with the ITC experiments and automatic crystallizations, respectively.
This study is supported by the Ministry of Health of China (grant 2013ZX10004-602), the Ministry of Science and Technology of China (grants 2013CB911103, 2009CB918803, and 2011CB911103), and the National Natural Science Foundation of China (grants 31270795, 31200559, 31070660, and 31021062). We also received support from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contracts HHSN272200700058C and HHSN272201200026C.
Footnotes
Published ahead of print 10 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00672-13.
REFERENCES
- 1. Liu Y, Li Q, Hu W, Wu J, Wang Y, Mei L, Walker DH, Ren J, Yu XJ. 2012. Person-to-person transmission of severe fever with thrombocytopenia syndrome virus. Vector Borne Zoonotic Dis. 12:156–160 [DOI] [PubMed] [Google Scholar]
- 2. Yu XJ, Liang MF, Zhang SY, Liu Y, Li JD, Sun YL, Zhang L, Zhang QF, Popov VL, Li C, Qu J, Li Q, Zhang YP, Hai R, Wu W, Wang Q, Zhan FX, Wang XJ, Kan B, Wang SW, Wan KL, Jing HQ, Lu JX, Yin WW, Zhou H, Guan XH, Liu JF, Bi ZQ, Liu GH, Ren J, Wang H, Zhao Z, Song JD, He JR, Wan T, Zhang JS, Fu XP, Sun LN, Dong XP, Feng ZJ, Yang WZ, Hong T, Zhang Y, Walker DH, Wang Y, Li DX. 2011. Fever with thrombocytopenia associated with a novel bunyavirus in China. N. Engl. J. Med. 364:1523–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McMullan LK, Folk SM, Kelly AJ, MacNeil A, Goldsmith CS, Metcalfe MG, Batten BC, Albarino CG, Zaki SR, Rollin PE, Nicholson WL, Nichol ST. 2012. A new phlebovirus associated with severe febrile illness in Missouri. N. Engl. J. Med. 367:834–841 [DOI] [PubMed] [Google Scholar]
- 4. Reguera J, Weber F, Cusack S. 2010. Bunyaviridae RNA polymerases (L-protein) have an N-terminal, influenza-like endonuclease domain, essential for viral cap-dependent transcription. PLoS Pathog. 6:e1001101 doi: 10.1371/journal.ppat.1001101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Morin B, Coutard B, Lelke M, Ferron F, Kerber R, Jamal S, Frangeul A, Baronti C, Charrel R, de Lamballerie X, Vonrhein C, Lescar J, Bricogne G, Gunther S, Canard B. 2010. The N-terminal domain of the arenavirus L protein is an RNA endonuclease essential in mRNA transcription. PLoS Pathog. 6:e1001038 doi: 10.1371/journal.ppat.1001038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Elliott RM. 1990. Molecular biology of the Bunyaviridae. J. Gen. Virol. 71(Pt 3):501–522 [DOI] [PubMed] [Google Scholar]
- 7. Pekosz A, He B, Lamb RA. 1999. Reverse genetics of negative-strand RNA viruses: closing the circle. Proc. Natl. Acad. Sci. U. S. A. 96:8804–8806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Walter CT, Bento DF, Alonso AG, Barr JN. 2011. Amino acid changes within the Bunyamwera virus nucleocapsid protein differentially affect the mRNA transcription and RNA replication activities of assembled ribonucleoprotein templates. J. Gen. Virol. 92:80–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eifan SA, Elliott RM. 2009. Mutational analysis of the Bunyamwera orthobunyavirus nucleocapsid protein gene. J. Virol. 83:11307–11317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pinschewer DD, Perez M, de la Torre JC. 2003. Role of the virus nucleoprotein in the regulation of lymphocytic choriomeningitis virus transcription and RNA replication. J. Virol. 77:3882–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ferron F, Li Z, Danek EI, Luo D, Wong Y, Coutard B, Lantez V, Charrel R, Canard B, Walz T, Lescar J. 2011. The hexamer structure of Rift Valley fever virus nucleoprotein suggests a mechanism for its assembly into ribonucleoprotein complexes. PLoS Pathog. 7:e1002030 doi: 10.1371/journal.ppat.1002030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Raymond DD, Piper ME, Gerrard SR, Smith JL. 2010. Structure of the Rift Valley fever virus nucleocapsid protein reveals another architecture for RNA encapsidation. Proc. Natl. Acad. Sci. U. S. A. 107:11769–11774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Raymond DD, Piper ME, Gerrard SR, Skiniotis G, Smith JL. 2012. Phleboviruses encapsidate their genomes by sequestering RNA bases. Proc. Natl. Acad. Sci. U. S. A. 109:19208–19213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carter SD, Surtees R, Walter CT, Ariza A, Bergeron E, Nichol ST, Hiscox JA, Edwards TA, Barr JN. 2012. Structure, function, and evolution of the Crimean-Congo hemorrhagic fever virus nucleocapsid protein. J. Virol. 86:10914–10923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guo Y, Wang W, Ji W, Deng M, Sun Y, Zhou H, Yang C, Deng F, Wang H, Hu Z, Lou Z, Rao Z. 2012. Crimean-Congo hemorrhagic fever virus nucleoprotein reveals endonuclease activity in bunyaviruses. Proc. Natl. Acad. Sci. U. S. A. 109:5046–5051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang Y, Dutta S, Karlberg H, Devignot S, Weber F, Hao Q, Tan YJ, Mirazimi A, Kotaka M. 2012. Structure of Crimean-Congo hemorrhagic fever virus nucleoprotein: superhelical homo-oligomers and the role of caspase-3 cleavage. J. Virol. 86:12294–12303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hastie KM, Kimberlin CR, Zandonatti MA, MacRae IJ, Saphire EO. 2011. Structure of the Lassa virus nucleoprotein reveals a dsRNA-specific 3′ to 5′ exonuclease activity essential for immune suppression. Proc. Natl. Acad. Sci. U. S. A. 108:2396–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Qi X, Lan S, Wang W, Schelde LM, Dong H, Wallat GD, Ly H, Liang Y, Dong C. 2010. Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature 468:779–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grote A, Hiller K, Scheer M, Munch R, Nortemann B, Hempel DC, Jahn D. 2005. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 33:W526–W531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stols L, Gu M, Dieckman L, Raffen R, Collart FR, Donnelly MI. 2002. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr. Purif. 25:8–15 [DOI] [PubMed] [Google Scholar]
- 21. Liu ZJ, Chen L, Wu D, Ding W, Zhang H, Zhou W, Fu ZQ, Wang BC. 2011. A multi-dataset data-collection strategy produces better diffraction data. Acta Crystallogr. A 67:544–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 23. McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 25. Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255 [DOI] [PubMed] [Google Scholar]
- 27. Delano WL. 2002. PyMOL. Schrodinger, Inc., New York, NY: http://www.pymol.org [Google Scholar]
- 28. Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. 2003. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36:1277–1282 [Google Scholar]
- 29. Svergun D. 1992. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25:495–503 [Google Scholar]
- 30. Fischer H, de Oliveira Neto M, Napolitano HB, Polikarpov I, Craievich AF. 2010. Determination of the molecular weight of proteins in solution from a single small-angle X-ray scattering measurement on a relative scale. J. Appl. Crystallogr. 43:101–109 [Google Scholar]
- 31. Svergun D, Barberato C, Koch MHJ. 1995. CRYSOL: a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28:768–773 [Google Scholar]
- 32. Svergun DI, Petoukhov MV, Koch MH. 2001. Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 80:2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Volkov VV, Svergun DI. 2003. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36:860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kozin MB, Svergun DI. 2001. Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 34:33–41 [Google Scholar]
- 35. DeLaBarre B, Brunger AT. 2003. Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nat. Struct. Biol. 10:856–863 [DOI] [PubMed] [Google Scholar]
- 36. Su J, Li Y, Shaw N, Zhou W, Zhang M, Xu H, Wang BC, Liu ZJ. 2010. Crystal structure of a novel non-Pfam protein PF2046 solved using low resolution B-factor sharpening and multi-crystal averaging methods. Protein Cell 1:453–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Anderson J, Fuglsang H. 1978. Further studies on the treatment of ocular onchocerciasis with diethylcarbamazine and suramin. Br. J. Ophthalmol. 62:450–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Darsaud A, Chevrier C, Bourdon L, Dumas M, Buguet A, Bouteille B. 2004. Megazol combined with Suramin improves a new diagnosis index of the early meningo-encephalitic phase of experimental African trypanosomiasis. Trop. Med. Int. Health 9:83–91 [DOI] [PubMed] [Google Scholar]
- 39. Ahles TA, Herndon JE, 2nd, Small EJ, Vogelzang NJ, Kornblith AB, Ratain MJ, Stadler W, Palchak D, Marshall ME, Wilding G, Petrylak D, Holland JC. 2004. Quality of life impact of three different doses of Suramin in patients with metastatic hormone-refractory prostate carcinoma: results of Intergroup O159/Cancer and Leukemia Group B 9480. Cancer 101:2202–2208 [DOI] [PubMed] [Google Scholar]
- 40. Mastrangelo E, Pezzullo M, Tarantino D, Petazzi R, Germani F, Kramer D, Robel I, Rohayem J, Bolognesi M, Milani M. 2012. Structure-based inhibition of norovirus RNA-dependent RNA polymerases. J. Mol. Biol. 419:198–210 [DOI] [PubMed] [Google Scholar]
- 41. Mukherjee S, Hanson AM, Shadrick WR, Ndjomou J, Sweeney NL, Hernandez JJ, Bartczak D, Li K, Frankowski KJ, Heck JA, Arnold LA, Schoenen FJ, Frick DN. 2012. Identification and analysis of hepatitis C virus NS3 helicase inhibitors using nucleic acid binding assays. Nucleic Acids Res. 40:8607–8621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ellenbecker M, Lanchy JM, Lodmell JS. 2012. Identification of Rift Valley fever virus nucleocapsid protein-RNA binding inhibitors using a high-throughput screening assay. J. Biomol. Screen. 17:1062–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. CLUSTAL W and CLUSTAL X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 44. Gouet P, Robert X, Courcelle E. 2003. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31:3320–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.