Abstract
Since April 2012, there have been 17 laboratory-confirmed human cases of respiratory disease associated with newly recognized human betacoronavirus lineage C virus EMC (HCoV-EMC), and 7 of them were fatal. The transmissibility and pathogenesis of HCoV-EMC remain poorly understood, and elucidating its cellular tropism in human respiratory tissues will provide mechanistic insights into the key cellular targets for virus propagation and spread. We utilized ex vivo cultures of human bronchial and lung tissue specimens to investigate the tissue tropism and virus replication kinetics following experimental infection with HCoV-EMC compared with those following infection with human coronavirus 229E (HCoV-229E) and severe acute respiratory syndrome coronavirus (SARS-CoV). The innate immune responses elicited by HCoV-EMC were also investigated. HCoV-EMC productively replicated in human bronchial and lung ex vivo organ cultures. While SARS-CoV productively replicated in lung tissue, replication in human bronchial tissue was limited. Immunohistochemistry revealed that HCoV-EMC infected nonciliated bronchial epithelium, bronchiolar epithelial cells, alveolar epithelial cells, and endothelial cells. Transmission electron microscopy showed virions within the cytoplasm of bronchial epithelial cells and budding virions from alveolar epithelial cells (type II). In contrast, there was minimal HCoV-229E infection in these tissues. HCoV-EMC failed to elicit strong type I or III interferon (IFN) or proinflammatory innate immune responses in ex vivo respiratory tissue cultures. Treatment of human lung tissue ex vivo organ cultures with type I IFNs (alpha and beta IFNs) at 1 h postinfection reduced the replication of HCoV-EMC, suggesting a potential therapeutic use of IFNs for treatment of human infection.
INTRODUCTION
Coronavirus (CoV) infections in humans are generally mild and self-limited. Until the outbreak of severe acute respiratory syndrome (SARS) caused by severe acute respiratory syndrome coronavirus (SARS-CoV) in 2003, there was limited research on the tissue tropism and host response following human infection with coronaviruses. In comparison to human betacoronavirus 229E (HCoV-229E), SARS-CoV was found to be deficient at eliciting beta interferon (IFN-β) innate immune responses in primary human macrophages and dendritic cells (1, 2) since SARS-CoV encodes several antagonists of innate immune-sensing and signaling pathways (3, 4). The tropism of SARS-CoV in the respiratory tract was primarily restricted to differentiated human airway epithelium (5) and alveolar type II pneumocytes (6–8), with limited tropism for alveolar type I pneumocytes (9).
In 2012, a novel coronavirus was detected in two patients from Saudi Arabia and Qatar (10, 11). Thereafter, more cases were identified both prospectively and retrospectively in Saudi Arabia, Qatar, Jordan, and the United Kingdom, and as of February 2013, a total of 13 laboratory-confirmed cases and seven deaths have been reported (12). The apparent severity of this novel coronavirus contrasts with those seen for other human coronaviruses (HCoVs), with the exception of SARS-CoV (10, 13). The first reported case, which was fatal, occurred in June 2012 in a 60-year-old man in Saudi Arabia (11); the second reported case occurred in a 49-year-old Qatari man who was treated and discharged in the United Kingdom. From early case descriptions, it appeared that pneumonia leading to acute respiratory distress syndrome is the primary manifestation of the disease, but renal dysfunction was also observed in some cases. The WHO has provided a working case definition of the disease (12). The disease appears to have an incubation period of up to 10 days and is not easily transmitted between humans. The retrospective investigation of an outbreak of severe respiratory illness in Zarqa, Jordan, in April 2012 (14) confirmed that at least some of these cases were also caused by human betacoronavirus lineage C virus EMC (HCoV-EMC). Of the 13 cases confirmed to date, some have given a history of contact with animals (e.g., camels and sheep) (13), and a zoonotic origin of the infection is considered likely. More recently, an index case who acquired infection in the Middle East transmitted infection to two family contacts in the United Kingdom (15), providing evidence for limited human-to-human transmission, thereby raising the level of public health concern.
The virus isolate from the Saudi patient has been fully sequenced, and the sequencing identifies the virus as a novel human virus within betacoronavirus lineage C. Phylogenetically, it clusters in the same group as viruses isolated from Pipistrellus (Pipistrellus pipi/VM314/2008/NLD, bat CoV [BtCoV]/355A/2005, BtCoV/A434/2005, HKU4) and Tylonycteris (BtCoV/133/2005, HKU-5) bats in Hong Kong/China and the Netherlands (16, 17). These bat coronaviruses have not been cultured in vitro. The partial sequence of the open reading frame 1 (ORF1) region of the isolate from the patient from Qatar is more than 99% identical to that of the virus reported from Saudi Arabia (HCoV-EMC) (10, 18). Previously known HCoVs are alphacoronaviruses HCoV-229E and HCoV-NL63, betacoronavirus lineage A (HCoV-OC43 and HKU1), and betacoronavirus lineage B (SARS-CoV) (19, 20).
We have previously used ex vivo cultures of human bronchial and lung tissues to investigate the tropism of influenza viruses (21) and showed that productive infection of the upper airways may correlate with pandemic potential in swine influenza viruses (22). We now utilize ex vivo cultures of human bronchial and lung tissue to study the tissue tropism, virus replication kinetics, apoptosis, and innate immune responses of HCoV-EMC infection in comparison with those of infections with highly pathogenic SARS-CoV and common cold-causing HCoV-229E.
As there are no specific antivirals for HCoV-EMC infection, we explore the antiviral effects of IFNs. IFN has previously been reported to inhibit SARS-CoV replication both in vitro and in animal models (23–28).
MATERIALS AND METHODS
Viruses.
Human betacoronavirus of lineage C virus (HCoV-EMC) was obtained from R. Fouchier, Erasmus MC, Rotterdam, the Netherlands, and the virus seed stock was prepared in Vero cell culture (ATCC) in minimal essential medium (MEM) containing 2% fetal bovine serum (FBS) and 100 units/ml penicillin–100 μg/ml streptomycin (PS). HCoV-EMC produced a cytopathic effect (CPE) within 1 to 2 days after infection of Vero cells. SARS-CoV (strain HK39849) and HCoV-229E were propagated in Vero cells and MRC-5 cells, respectively, as described previously (1). Highly pathogenic avian influenza (HPAI) viruses cause severe human respiratory disease, and stocks of A/Hong Kong/483/1997 (H5N1) virus were prepared in MDCK cells in MEM with no FBS and with 1% PS. All the experiments were performed in a biosafety level 3 biocontainment facility at The University of Hong Kong.
Virus titration by TCID50 assay.
Confluent 96-well tissue culture plates of the respective cells were used for the virus titration assay for HCoV-EMC and SARS-CoV (Vero cells), HCoV-229E (MRC-5 cells), or influenza A H5N1 virus (MDCK cells). The cells were washed once with phosphate-buffered saline (PBS), and the Vero and MRC-5 cells were replenished with MEM with 2% FBS and 1% PS, while MDCK cells were replenished with MEM with no FBS and 1% PS. Serial half-log10 dilutions (from 0.5 log unit to 7 log units) of virus-infected culture supernatants were added onto the wells in quadruplicate. The plates were observed for a CPE daily for 5 days. The viral dilution leading to a CPE in 50% of inoculated wells was estimated by using the Karber method, designated 1 50% tissue culture infectious dose (TCID50), and used to compute the viral titer in the test sample.
Ex vivo organ cultures and infection.
Fresh biopsy specimens of human bronchus and lung parenchyma that were obtained from patients undergoing surgical resection of lung tissue at Queen Mary Hospital as part of clinical care but that were surplus for routine diagnostic requirements were used in this study. This study was approved by the Institutional Review Board of The University of Hong Kong/Hospital Authority Hong Kong West Cluster. Ex vivo cultures of human bronchial and lung tissues were performed as previously described (21, 22, 29). The bronchial mucosae were placed on a surgical sponge with their apical epithelial surface facing upwards, while the lung parenchymal tissues were placed into a 24-well plate directly with 1 ml of F12K culture medium and with 1% PS at 37°C. Bronchial and lung tissues were infected with HCoV-229E, HCoV-EMC, and SARS-CoV at a viral titer of 106 TCID50s/ml for 1 h at 37°C and washed with 5 ml of warm PBS three times to remove unbound virus, as previously described (21, 22, 29). UV-inactivated virus and mock-infected cells were used as controls. Culture supernatants from the infected cultures were collected at 1, 24, 48, and 72 h postinfection (hpi) and titrated for infectious virus using the TCID50 assay for HCoV-229E, HCoV-EMC, and SARS-CoV, as described previously (1, 6). Increasing virus titers over time provided evidence of productive virus replication. Tissues were collected at 1, 24, 48, and 72 hpi for RNA extraction and at 24, 48, and 72 hpi for fixation in 10% formalin or 2.5% glutaraldehyde for immunohistochemistry and electron microscopy, respectively.
In vitro cell culture and infection.
Cells of a human alveolar epithelial cell line (A549) were cultured using Dulbecco modified Eagle medium with 10% FBS and 1% PS and were seeded at 1 × 105 cells per well in 24-well tissue culture plates. The cells were infected with HCoV-EMC, SARS-CoV, and HPAI H5N1 virus at a multiplicity of infection (MOI) of 1. After 1 h of virus adsorption at 37°C, the virus inoculum was removed, the cells were washed with PBS, and the culture was replenished with fresh culture medium without FBS. Evidence of viral infection and virus replication kinetics were determined by (i) assaying RNA of HCoV-EMC, SARS-CoV, and influenza type A virus at 1, 24, and 48 hpi by quantitative reverse transcription-PCR (RT-PCR) and (ii) titrating infectious virus in infected culture supernatants to demonstrate productive virus replication in Vero or MDCK cells, as appropriate.
Quantification of viral and host cytokine and chemokine mRNAs by quantitative RT-PCR.
Bronchial and lung tissue fragments were homogenized using a TissueRuptor device (Qiagen, Hilden, Germany) in 700 μl RNeasy lysis buffer with beta-mercaptoethanol on ice. A549 cell cultures were lysed in 350 μl RLT lysis buffer with beta-mercaptoethanol. RNA extraction was carried out using an RNeasy minikit (Qiagen, Hilden, Germany) following the manufacturer's instructions with the addition of DNase treatment, and the RNA was eluted in 50 μl RNase-free water.
A one-step RT-PCR assay specific for the region upstream of the E gene of HCoV-EMC was adapted from a recently described protocol (18). In brief, 5 μl of purified RNA was amplified in a 25-μl reaction mixture containing 1 μl SSIII-Taq enzyme mix (SuperScript III one-step RT-PCR system with Platinum Taq DNA polymerase [Invitrogen]), 12.5 μl of 2× reaction buffer, 0.8 mM MgSO4, 0.4 μM forward primer upE-Fwd (5′-GCAACGCGCGATTCAGTT-3′), 0.4 μM reverse primer upE-Rev (5′-GCCTCTACACGGGACCCATA-3′), and 0.2 μM probe upE-Prb (5′-FAM-CTCTTCACATAATCGCCCCGAGCTCG-TAMRA-3′, where FAM is 6-carboxyfluorescein and TAMRA is 6-carboxytetramethylrhodamine). The reaction mixtures were first incubated at 55°C for 20 min. After a 3-min denaturation at 95°C, the reaction mixtures were then thermal cycled for 40 cycles (94°C for 15 s, 58°C for 30 s). Total RNA harvested from HCoV-EMC-infected Vero cells and water were used as positive and negative controls, respectively. The expression of the HCoV-EMC gene was expressed as the fold change compared to its expression at 1 hpi, using the 2−ΔΔCT method, where CT is the threshold cycle, as described previously (30). SARS-CoV nucleocapsid (N) protein gene expression (31) and influenza virus matrix protein gene expression were also quantified as previously described (31) and expressed in terms of the fold change compared to the expression detected at 1 hpi.
The host gene expression profiles for proinflammatory cytokines (tumor necrosis factor alpha [TNF-α], IFN-β, interleukin-29 [IL-29]), chemokines (IFN-γ-induced protein 10 [IP-10], monocyte chemotactic protein-1 [MCP-1]), and the housekeeping (β-actin) gene were detected in absolute copy numbers determined from a standard curve generated from a standard plasmid with a known copy number which was simultaneously included in the quantitative PCR (qPCR) mixture, as previously described (21, 32). The mRNA levels of selected genes were quantified by real-time qPCR analysis with an ABI 7500 real-time PCR system (Applied Biosystems). Expression of these genes was normalized by using the product of the β-actin housekeeping gene mRNA.
Generation of antisera against BtCoV HKU5 N protein.
The gene for BtCoV HKU5 N protein (GenBank accession no. EF065512) was inserted into the Venezuelan equine encephalitis (VEE) virus replicon plasmid pVR21 by overlap PCR, the replicon particles (VRP-HKU5 N protein) were packaged, and titers were determined on baby hamster kidney cells as previously described (33, 34). Five-week-old BALB/c mice (Harlan Laboratories) were immunized with 105 infectious units (IU) of VRP-HKU5 N protein in a 10-μl volume, and 21 days later, the mice were again boosted with the same dose of antigen. At 21 days after the boost, serum was collected by tail nick and used in immunohistochemistry analysis. The BtCoV HKU5 N protein shares 68% identity to the N protein of HCoV-EMC (35), and the antiserum against the N protein was found to cross-react with the HCoV-EMC nucleocapsid protein.
Immunohistochemistry.
The 10% formalin-fixed tissues were embedded in paraffin and stained using a polyclonal mouse antibody raised against coronavirus HKU5 N glycoprotein (1:200), which cross-reacts with HCoV-EMC by Western blotting and immunofluorescence assays; mouse monoclonal antibody against SARS-CoV nucleoprotein, which is reactive with SARS-CoV at a 1:50 dilution, as previously described (8); mouse monoclonal antibody 1E7, which is reactive with HCoV-229E at a 1:100 dilution (provided by Lia van der Hoek); and cleaved caspase 3 (CST-9661S; Cell Signaling) as a marker of apoptotic cells. The reaction of the primary antibody was revealed by the use of biotinylated goat anti-mouse antibody (1:500; 115-065-146; Jackson) and developed using a NovaRed substrate kit (SK-4800; Vector Labs). Antibodies to CD68 for macrophages, AE1/AE3 for epithelial cells, β-tubulin (F-2043; Sigma) for ciliated bronchial epithelial cells, MUC5AC (18-2261; Life Technology) for goblet cells, and podoplanin for type I pneumocytes were used for double labeling using immunofluorescence with secondary antibody conjugated with fluorescein isothiocyanate (FITC). HCoV-EMC was detected using HKU5 N protein antiserum and Vector Red substrate in these double-labeling experiments, as this substrate is also fluorescent when using a tetramethyl rhodamine isocyanate filter. The stained preparations were examined using a Nikon Ni immunofluorescence microscope, and images were captured using a SPOT Slider 2-megapixel camera. Double-chromogen immunohistochemistry for caspase 3 was performed by first detecting the viral antigen using an alkaline phosphatase-conjugated streptavidin technique, followed by the use of ImmPRESS horseradish peroxidase (MP-7401; Vector Labs) for detection of the cleaved caspase 3 (9661S; Cell Signaling).
Transmission electron microscopy.
Bronchial and lung tissues were fixed in 2.5% glutaraldehyde, washed three times in PBS, and serially dehydrated. The tissues were postfixed in 1% osmium tetroxide and embedded in Araldite resin (Polysciences, Inc., Warrington, PA). Sections were examined with a transmission electron microscope (CM100; Philips).
Antiviral effect of interferon treatment in ex vivo human lung tissue.
Recombinant IFN-α (PHC4014; Invitrogen) and IFN-β (PHC4244; Invitrogen) were reconstituted into 1,000 U/ml of F12K medium with 1% PS. Preinfection and postinfection administration regimes were applied. For the preinfection IFN treatment, lung tissue fragments were pretreated with either IFN-α or IFN-β 24 h prior to infection. The lung tissue fragments preincubated with IFN were then infected with HCoV-EMC or SARS-CoV as described above, and the infected culture was replenished with culture medium with the corresponding IFN after infection. For the postinfection treatment, untreated lung tissue fragments were infected, and at 1 hpi, the infected culture was replenished with medium containing 1,000 U/ml of IFN. Control lung tissue cultures without any IFN were used for comparison. Supernatants from the infected lung tissue explant cultures were collected at 1, 24, 48, and 72 hpi and titrated for infectious virus using the TCID50 assay.
Statistical analysis.
Experiments were performed independently with specimens from at least three different donors in duplicate. Results shows in the figures are the calculated mean and standard error of the mean. Mock-infected tissue served as a negative control. The differences in the log10-transformed viral titers among different viruses at different time points postinfection and differences in the amounts of cytokine and chemokine mRNAs of coronavirus-infected cells were compared by using one-way analysis of variance, followed by a Bonferroni multiple-comparison test. Differences were considered significant at a P value of <0.05. The statistical analysis was performed using GraphPad Prism (version 5) software.
RESULTS
Infection and replication of HCoV-EMC, SARS-CoV, and HCoV 229E in ex vivo cultures of human bronchial and lung tissue.
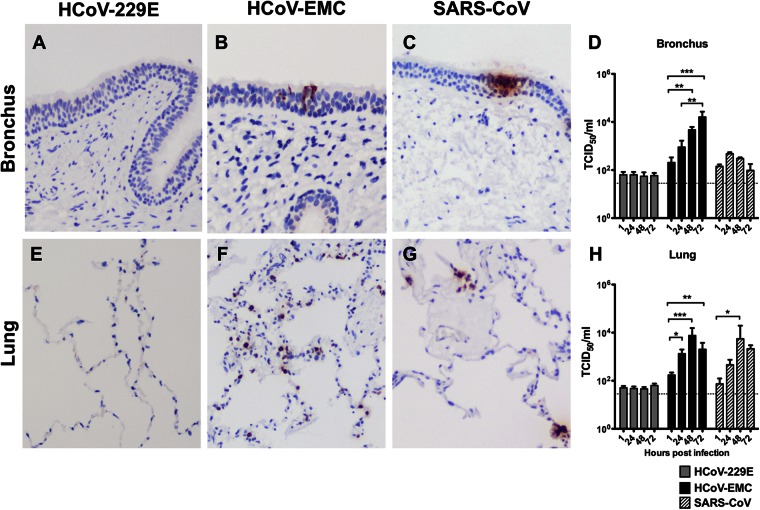
In ex vivo cultures of human bronchial tissue, HCoV-EMC infected the tissue and productively replicated, with a 2-log-unit increase in viral titer from 1 hpi to 72 hpi (P < 0.005) (Fig. 1B and D). SARS-CoV infected bronchial tissue (Fig. 1C) and showed a less than 1-log-unit increase in titer from 1 hpi to 24 hpi, but the increase did not reach statistical significance (Fig. 1D). HCoV-229E did not detectably infect or replicate in bronchial tissues (Fig. 1A and D). In ex vivo cultures of human lung tissue, viral antigen was not detected in HCoV-229E-inoculated tissues (Fig. 1E), and no productive replication was observed (Fig. 1H), indicating that HCoV-229E does not replicate in the human lung. Both HCoV-EMC (Fig. 1F) and SARS-CoV (Fig. 1G) extensively infected and replicated in (Fig. 1H) the lung parenchymal tissues, as shown by the presence of viral antigen in these tissues along with an approximately 2-log-unit increase in HCoV-EMC and SARS-CoV loads from 1 hpi to 48 hpi.
Fig 1.
Tissue tropism of HCoV-EMC in human bronchus and lung. Bronchial (A to C) and lung (E to G) tissues were infected with HCoV-229E, HCoV-EMC, and SARS-CoV. At 24 hpi, the tissues were fixed and tissue sections were stained for human coronavirus N protein (reddish brown), as described in Materials and Methods. Viral replication kinetics in ex vivo cultures of bronchial (D) and lung (H) biopsy tissue specimens infected with 106 TCID50s/ml of coronaviruses were determined by virus titration at 37°C. The bar charts show the mean and the standard error of the mean of the virus titer pooled from at least three independent experiments. Asterisks indicate a statistically significant increase in viral yield compared to that at 1 hpi: *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
HCoV-EMC cell tropism in the human respiratory tract.
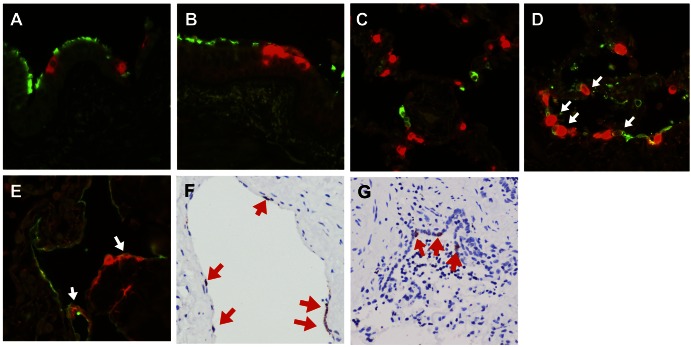
In order to identify the target cells of HCoV-EMC in the human respiratory tract, we performed costaining of HCoV-EMC antigen (stained in red) with specific cell markers (stained in green) in HCoV-EMC-infected human bronchial and lung tissues in ex vivo culture (Fig. 2). In bronchial tissues, we stained the ciliated cells (Fig. 2A) and goblet cells (Fig. 2B) using β-tubulin and MUC5AC, respectively. The infected cells (in red) were not colocalized with these two major cell types in the bronchus, and viral antigen was mainly found in nonciliated bronchial epithelial cells.
Fig 2.
Cellular localization of HCoV-EMC in lung tissue. HCoV-EMC was stained with Vector Red (red) and cell markers conjugated with FITC (green) for detection of β-tubulin (ciliated cell marker) (A) and MUC5AC (goblet cell marker) (B) in bronchial tissue and CD68 (macrophage marker) (C), AE1/AE3 (epithelial cell marker) (D), and podoplanin (type I pneumocyte marker) (E) in lung tissue at 24 hpi. White arrows in panels D and E denote cells with costaining. (F) Cellular tropism of HCoV-EMC in lung tissue. Human coronavirus N protein (stained in reddish brown and marked with red arrows) was identified in endothelial cells at 24 hpi. (G) Bronchiolar epithelial cells at 48 hpi.
In lung, macrophages, epithelial cells, and type I pneumocytes were stained with the specific markers CD68 (Fig. 2C), AE1/AE3 (Fig. 2D), and podoplanin (Fig. 2E), respectively. HCoV-EMC did not costain with macrophages (Fig. 2C), but there was overlapping of staining with the AE1/AE3 marker (Fig. 2D), which suggested that the principal target cells of HCoV-EMC infection were of epithelial origin, and focal colocalization was found in type I pneumocytes (Fig. 2E).
Cellular morphology and immunohistochemistry for viral antigen (nucleoprotein) in the HCoV-EMC-infected lung tissues (Fig. 1F) gave additional information on cell types targeted by HCoV-EMC at 48 hpi. Endothelial cells within the medium-size interstitial vessels of the lung (Fig. 2F) and the bronchiolar epithelial cells (Fig. 2G) were found to stain positive for the nucleoprotein of HCoV-EMC. The tropism of HCoV-EMC in lung endothelial cells may suggest the possibility of extrapulmonary dissemination.
Electron microscopy shows budding HCoV-EMC virions in bronchial and alveolar epithelial cells.
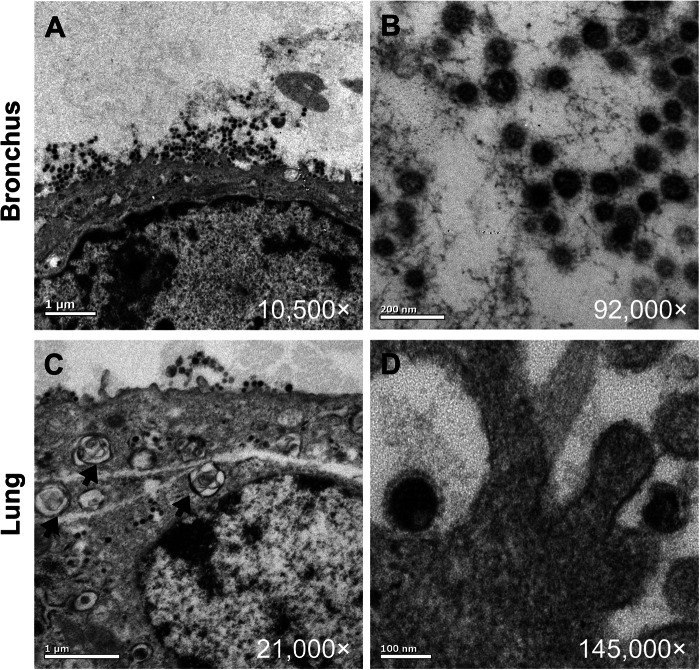
To further confirm the sites of HCoV-EMC replication within the human lung, transmission electron microscopy of the ex vivo-infected bronchial and lung parenchymal tissues was performed. Virion-containing cells in the bronchus were flattened nonciliated bronchiolar-type epithelial cells (Fig. 3A), and those in the lung were type II pneumocytes with visible lamellar bodies (Fig. 3C). While virions were identified intracellularly, large aggregates of virions in intracytoplasmic secretory vesicles were not conspicuous. Ultrastructural examination of the bronchus and lung showed enveloped viral particles from 75 to 85 nm in diameter (Fig. 3B and D). These were often associated with a rim of spikes or corona measuring approximately 8 nm in length.
Fig 3.
Transmission electron microscopy locating the budding site of HCoV-EMC in the human bronchus and lung. Budding of virions from bronchial epithelial cells (A, B) and alveolar epithelial cells (C, D) at 48 hpi. Black arrows, type II alveolar epithelial cells with lamellar bodies.
Extensive apoptosis in HCoV-EMC-infected human lung tissue ex vivo cultures.
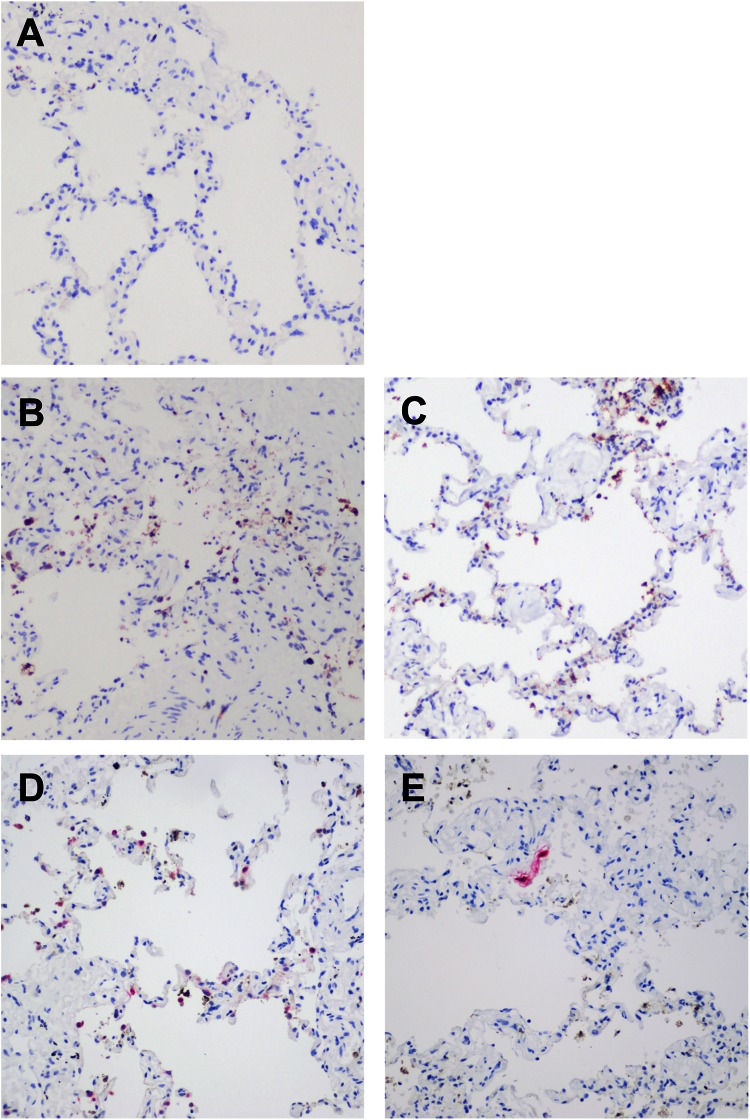
By immunohistochemistry, we found extensive expression of cleaved caspase 3, an apoptosis marker, in ex vivo lung tissue infected with HCoV-EMC (Fig. 4B) and SARS-CoV (Fig. 4C) but not in mock-infected (Fig. 4A) human lung tissue. In order to investigate if the apoptosis was induced directly by coronavirus infection in the human lung, we performed costaining by immunohistochemistry of HCoV-EMC antigen (stained in pink) with cleaved caspase 3 (stained in reddish brown) using HCoV-EMC-infected (Fig. 4D) and SARS-CoV-infected (Fig. 4E) human lung tissues in ex vivo culture. Examination of both HCoV-EMC- and SARS-CoV-infected lung tissue revealed that the apoptotic cells were not the viral protein-expressing cells (Fig. 4D and E), suggesting that paracrine mechanisms may contribute to induction of apoptosis.
Fig 4.
Apoptotic cells identified in human lung tissue ex vivo culture upon HCoV-EMC and SARS-CoV infection. (A to C) Ex vivo culture of lung tissue mock infected (A) or infected with HCoV-EMC (B) or SARS-CoV (C) at 48 hpi. The reddish brown stain identifies the presence of cleaved caspase 3. (D and E) Costaining of HCoV-EMC (D) and SARS-CoV (E) antigen (pink stain) with cleaved caspase 3 (reddish brown stain).
Viral and host gene expression upon HCoV-EMC infection in ex vivo cultures of bronchial and lung tissue and in vitro culture of A549 cells.
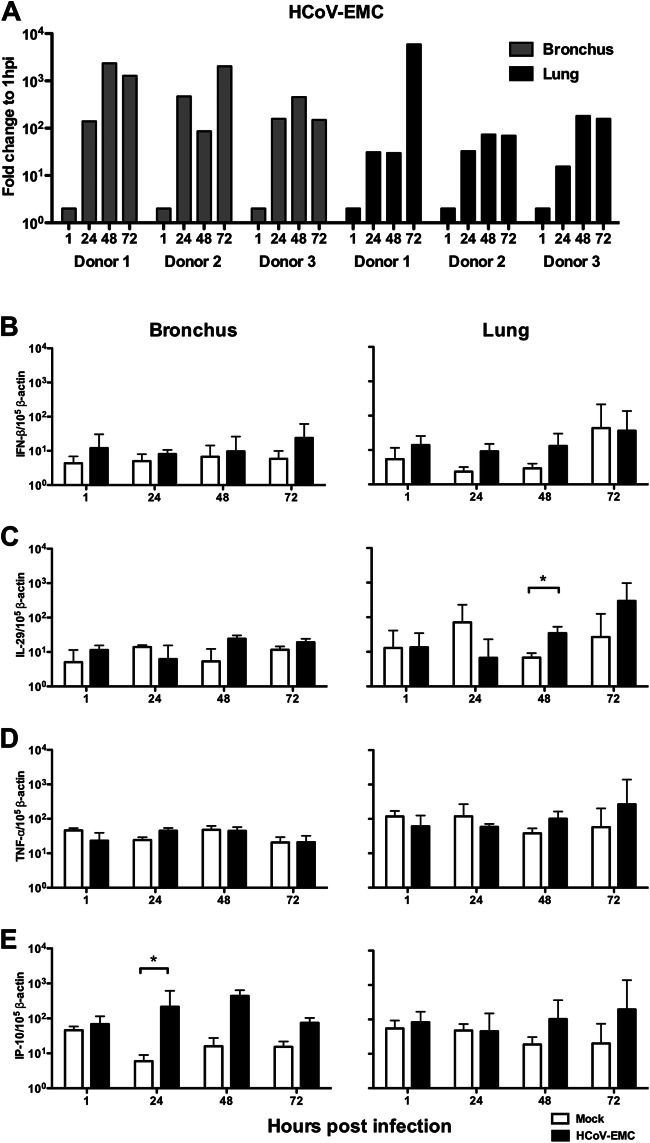
Ex vivo bronchial and lung tissues from three donors were infected with HCoV-EMC and RNA was extracted from infected cells at 1, 24, 48, and 72 hpi. Viral RNA was quantitated by RT-PCR. Host expression of mRNA for type I (IFN-β) and type III (IL-29) interferons and proinflammatory cytokines and chemokines TNF-α and IP-10 was quantitated in HCoV-EMC- or mock-infected bronchial and lung tissues. Viral gene expression increased by more than 2,000-fold in bronchial tissue cultures and more than 180-fold in lung tissue cultures, with one donor showing an exceptional 6,000-fold increase in a lung tissue culture (Fig. 5A). However, compared to mock-infected cultures, HCoV-EMC infection in bronchial and lung tissues failed to induce IFN-β or TNF-α (Fig. 5B and D). There was marginal induction of IL-29 in virus-infected lung tissue cultures at 48 hpi (P < 0.05) compared with that for mock-infected tissue (Fig. 5C) and higher IP-10 mRNA expression at 24 hpi compared with that for mock-infected bronchial tissue (P < 0.05) (Fig. 5E). IL-1β, MCP-1, and RANTES mRNAs were also similarly quantified, with no upregulation of these genes detected in bronchial or lung tissues infected with HCoV-EMC (data not shown). Inactivation of the HCoV-EMC by UV irradiation prior to infection of ex vivo bronchial and lung tissue cultures completely abolished viral replication and any cytokine induction (data not shown).
Fig 5.
Human HCoV-EMC gene expression and the major cytokine and chemokine expression in bronchial and lung tissues in response to infection. (A) Expression of HCoV-EMC genes expressed as the fold change at 24, 48, and 72 hpi compared with that at 1 hpi. Gray bars, data from ex vivo bronchial tissue from three donors; black bars, data from ex vivo lung tissue from three donors. (B to E) IFN-β (B), IL-29 (C), TNF-α (D), and IP-10 (E) expression from mock-infected and infected ex vivo cultures at 1, 24, 48, and 72 hpi. The graph shows the mean and standard error of mean numbers of copies of mRNA per 105 β-actin copies from three representative experiments. *, P < 0.05.
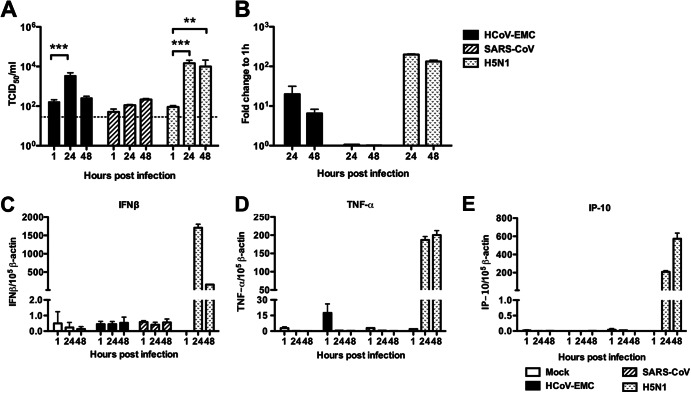
In order to confirm the apparent lack of host IFN responses elicited by HCoV-EMC in the ex vivo bronchial and lung tissue cultures, we carried out further experiments quantitating viral RNA and host type I and III IFN, TNF-α, and IP-10 mRNA in cells of the alveolar epithelial cell line A549 infected with HCoV-EMC, SARS-CoV (nonreplicating negative control), and HPAI H5N1 virus (high cytokine-inducing positive control) or uninfected A549 cells (control). We demonstrated by TCID50 assay and viral gene expression that HCoV-EMC and HPAI H5N1 virus replicated in A549 cells, while SARS-CoV showed no evidence of replication (Fig. 6A and B). While influenza A H5N1 virus strongly induced IFN-β (Fig. 6C), TNF-α (Fig. 6D), and IP-10 (Fig. 6E) gene expression, none of these cytokines were induced by HCoV-EMC infection of A549 cells.
Fig 6.
Viral replication and cytokine and chemokine expression of HCoV-EMC in A549 cells. (A) A549 cells infected with HCoV-EMC, SARS-CoV, and influenza H5N1 virus (MOI, 1). Viral replication was determined by TCID50 assay. The bar chart shows the mean and the standard error of mean titer of virus pooled from three independent experiments. Asterisks indicate a statistically significant increase in viral yield compared to that at 1 hpi: **, P < 0.05; ***, P < 0.0005. (B) Expression of HCoV-EMC, SARS-CoV, and influenza H5N1 virus genes expressed as the fold change compared with that at 1 hpi. (C to E) IFN-β (C), TNF-α (D), and IP-10 (E) gene expression from mock-infected and HCoV-EMC-, SARS-CoV-, and influenza H5N1 virus-infected A549 cells at 1, 24, and 48 hpi. The graph shows the mean and standard error of the mean numbers of copies of mRNA per 105 β-actin copies from three representative experiments.
IFNs inhibit HCoV-EMC replication in ex vivo lung tissue cultures.
HCoV-EMC and SARS-CoV productively replicated in ex vivo lung tissue cultures (Fig. 1H). Using this ex vivo lung tissue culture model, we investigated the effect of IFN-α or IFN-β treatment commencing 1 h after infection on viral replication. A significant decrease in the replication kinetics of HCoV-EMC was observed, with an approximately 2-log-unit decrease in infectious viral titers being observed at 48 hpi and 72 hpi (Fig. 7A). This decrease paralleled a reduction in viral gene copy load assayed by quantitative RT-PCR assays (data not shown). IFN-α or IFN-β treatment also inhibited SARS-CoV replication in ex vivo lung tissue cultures, but the effect was less pronounced than that with HCoV-EMC infection (Fig. 7B). In addition, the effect of IFN-α or IFN-β treatment 24 h prior to infection continued into the postinfection period was also examined. This treatment did not further enhance the antiviral effect on HCoV-EMC replication (data not shown).
Fig 7.
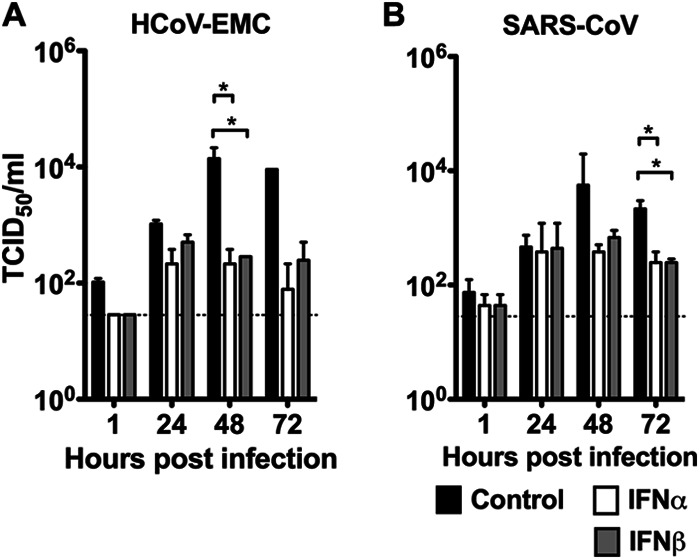
Interferon treatments suppressed HCoV-EMC and SARS CoV replication in human lung ex vivo culture. The human lung ex vivo cultures were infected with 106 TCID50s/ml of HCoV-EMC or SARS-CoV for 1 h at 37°C. A control culture consisting of F12K medium plus 1% PS (black bars) or culture medium with IFN-α (white bars) or IFN-β (gray bars) at a concentration of 1,000 U/ml in F12K medium plus 1% PS was used to replenish the medium at 1 hpi. Supernatants were collected from infected cultures at 1, 24, 48, and 72 hpi and titrated in Vero cells. Replication kinetics of HCoV-EMC (A) and SARS-CoV (B) were plotted to show the inhibitory effect of IFNs on virus replication posttreatment. *, a statistically significant difference (P < 0.05) at the same time point between control and IFN treatments.
DISCUSSION
We compared the tropism of the novel human coronavirus EMC, SARS-CoV, and HCoV-229E on ex vivo cultures of human lung and bronchial tissue using immunohistochemistry to indicate infection and by quantitating infectious viral titers from 1 to 72 hpi to indicate productive viral replication. HCoV-EMC and SARS-CoV infected and productively replicated in ex vivo cultures of human lung. While both viruses also infected bronchial epithelial cells, as assessed by immunohistochemistry, there was no significant increase in infectious viral titers of SARS-CoV in the bronchial tissue ex vivo cultures, while HCoV-EMC demonstrated evidence of productive replication.
Human infections associated with HCoV-EMC have so far appeared to be relatively severe clinically, but it is unclear whether this severity reflects an ascertainment bias where milder disease goes unrecognized. Future studies, including seroepidemiological investigations, are needed to establish the true severity of this infection in human populations. The tropism and replication competence of HCoV-EMC in human lung and bronchus demonstrated here suggest that HCoV-EMC replicates at least as well as or even better than SARS-CoV in human lung and bronchial tissues and targets type I and type II alveolar epithelial cells, highlighting the potential threat posed by this novel virus. Type II alveolar epithelial cells are crucial in the regeneration of the alveolar epithelium following injury by infection, and a virus that targets this cell type is likely to lead to significant lung pathology. Previous studies have shown that HPAI H5N1 virus, one that is also known to cause severe primary viral pneumonia and acute respiratory distress syndrome, also replicates efficiently in the alveolar epithelium of ex vivo lung tissue cultures (29). In agreement with the milder clinical disease caused by HCoV-229E, HCoV-229E failed to infect or replicate in either bronchial or lung tissues. However, it is relevant to note that not all viruses that can replicate in the alveolar epithelium of the ex vivo cultures of lung tissue are consistently associated with severe respiratory disease (e.g., the 2009 pandemic H1N1 virus) (21). Thus, the capacity for replication in alveolar epithelium in ex vivo cultures of lung tissue appears to be a necessary but not sufficient correlate of disease severity.
In previous studies with swine influenza viruses, we found that a lack of tropism for ex vivo cultures of bronchial and nasopharyngeal tissue correlated with a lack of transmission in humans (22). HCoV-229E, which is known to transmit efficiently in humans, also failed to replicate efficiently in ex vivo bronchial tissue cultures, although HCoV-229E infection and replication were demonstrated in in vitro cultures of a pseudostratified human airway epithelial cell model (36) and well-differentiated normal human bronchial epithelial cells (6). Coronaviruses causing the common cold have been shown to infect the nasal mucosa of humans (37) and would presumably replicate in ex vivo cultures of human nasopharyngeal or tonsil tissue. Since human nasopharyngeal tissue for ex vivo culture is less readily available, we have not so far obtained data from cultures infected with HCoV-229E, SARS-CoV, or HCoV-EMC. More systematic investigations of HCoV-EMC in ex vivo nasopharyngeal tissue cultures would provide further information on the tropism of this virus for the upper respiratory tract, providing insights into potential human transmissibility or a lack thereof.
Immunohistochemical analysis of virus-infected ex vivo lung tissue cultures also demonstrated that endothelial cells within medium-size interstitial blood vessels of the lung were also targets for HCoV-EMC infection. This may imply that the virus, as with SARS-CoV, may spread systemically to affect distant organs. Thus, the renal dysfunction that was repeatedly seen in patients infected with HCoV-EMC may possibly be due to virus dissemination to the kidney, although there is no direct evidence of this at present. Further clinical studies are needed to address whether viral dissemination does in fact occur and whether the renal dysfunction is due to viral invasion of the kidneys.
Immunofluorescence study and transmission electron microscopy of infected ex vivo cultures of lung and bronchial tissue allowed us to further define the cell types targeted by HCoV-EMC. Nonciliated bronchial epithelial cells, bronchiolar epithelial cells, and type I and type II pneumocytes appear to be the major targets for HCoV-EMC infection. We did not observe virus-infected alveolar macrophages in ex vivo lung tissue cultures under these experimental conditions. A preliminary study of human peripheral blood monocyte-derived macrophages also showed that these cells did not support the replication of HCoV-EMC (unpublished data).
Mechanisms that contribute to the pathogenesis of respiratory viruses such as SARS-CoV and avian influenza A H5N1 virus include direct virus replication-induced cell apoptosis, necrosis, or autophagy; bystander apoptosis; or dysregulation of host innate inflammatory responses. We found extensive apoptosis in SARS-CoV- and HCoV-EMC-infected ex vivo lung tissues which did not colocalize with viral antigen, suggesting that both viruses can induce apoptosis via paracrine mechanisms. Unlike influenza A H5N1 virus, which induces a prominent proinflammatory cytokine response, and SARS-CoV, which induces a dysregulated cytokine responses (i.e., poor IFN response but potent proinflammatory chemokine responses), HCoV-EMC infection elicited poor proinflammatory chemokine and cytokine responses, including poor type I and III IFN responses both in ex vivo lung tissue cultures and in the alveolar epithelial cell line A549. Thus, immunomodulatory therapies that have been investigated in experimental models of H5N1 disease (38–40) may have no clinical utility in HCoV-EMC infection. On the other hand, IFN therapy may be of potential benefit. SARS-CoV did not appear to induce IFN responses, although other proinflammatory cytokines were potently induced (1), and it was found that a number of SARS-CoV proteins functioned as IFN antagonists in vitro (41). IFN therapy has been shown to have therapeutic potential in SARS-CoV infections using in vitro (42) and nonhuman primate (24) models, and some trend toward clinical benefit was observed in human studies that were based on retrospective controls rather than in randomized controlled clinical trials (43). Ex vivo cultures of human lung tissue have been used to demonstrate the therapeutic benefits of IFNs in HPAI H5N1 virus infections (44). Therefore, we explored the antiviral effects of IFNs on HCoV-EMC infection in a human ex vivo lung tissue culture model.
Both IFN-α and IFN-β significantly suppressed viral replication when added to ex vivo cultures of human lung tissue 1 h after HCoV-EMC infection. IFN treatment of the cells that began 24 h prior to infection and continued into the postinfection period did not enhance the antiviral effect on HCoV-EMC replication. As there are currently no antivirals for treatment of HCoV-EMC, these findings provide a therapeutic option for this serious disease. Further studies using relevant animal models would be a priority to confirm the utility of IFNs as therapeutic and/or prophylactic interventions for HCoV-EMC infection. The biological basis for innate immune evasion by HCoV-EMC deserves investigation.
A recent study (published online after the submission of our manuscript) has reported that HCoV-EMC replicates more efficiently than SARS-CoV in a pseudostratified culture of human airway epithelium (HAE) that morphologically and functionally resembles human upper conducting airways in vitro (36). Their data pertained only to the bronchial region of the respiratory tract, and furthermore, the extent of HCoV-EMC replication in that in vitro HAE model was more limited than what we observed in the ex vivo bronchial or lung tissue cultures. As with our results, they showed that HCoV-EMC is a weak inducer of IFNs and that IFNs can inhibit replication of HCoV-EMC in HAE cultures (36). Our results support and extend those observations using human bronchial tissue cultured ex vivo, add information on the tropism of HCoV-EMC in ex vivo lung and bronchial tissue infection, and demonstrate the effect of interferon on human tissues infected with this virus.
In conclusion, this study illustrates the clinical utility of using ex vivo cultures of the human respiratory system to investigate newly emerging respiratory viruses. There have so far been no autopsy reports describing the virus-induced pathology in the lung, and such studies will complement and confirm the data that we report here. It must be noted that autopsy data, even when available, often reflect the late-stage disease in patients who may have been kept alive on mechanical ventilation for long periods of time. Thus, studies with ex vivo experimental infection of the human respiratory tract are invaluable to understand virus tropism and pathogenesis as well as to evaluate potential therapeutic options.
ACKNOWLEDGMENTS
We than Lia van der Hoek of the University of Amsterdam in the Netherlands for sharing the mouse anti-HCoV-229E antibody to carry out the immunohistochemistry analysis in this study. We also thank Kevin Fung of the Department of Pathology, The University of Hong Kong, Sara S. R. Kang, S. F. Sia, Icarus W. W. Chan, Christine B. H. Trang, Iris H. Y. Ng, Chloe K. S. Wong, Kenrie P. Y. Hui, and Francois Kien of the Centre of Influenza Research, School of Public Health, The University of Hong Kong, and W. S. Lee of the HKU Electron Microscope Unit for technical assistance with the experiment.
This study was supported in part by research grants to J.S.M.P. from the European Community Seventh Framework Program (FP7/200-2013) under the project European Management Platform for Emerging and Re-Emerging Disease Entities (EMPERIE; grant agreement no. 223498), the National Institute of Allergy and Infectious Diseases (NIAID; contract HHSN266200700005C), and the Research Fund for Control of Infectious Diseases of the Health & Welfare Bureau of the Government of the Hong Kong special administrative region grant A3-BSL3-P7. R.S.B. is supported by a Southeastern Regional Center of Excellence Grant in Biodefense and grants from the National Institutes of Health (U54-AI057157, R01AI085524, R01AI075297).
Footnotes
Published ahead of print 3 April 2013
REFERENCES
- 1. Cheung CY, Poon LL, Ng IH, Luk W, Sia SF, Wu MH, Chan KH, Yuen KY, Gordon S, Guan Y, Peiris JS. 2005. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J. Virol. 79:7819–7826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Law HK, Cheung CY, Ng HY, Sia SF, Chan YO, Luk W, Nicholls JM, Peiris JS, Lau YL. 2005. Chemokine up-regulation in SARS-coronavirus-infected, monocyte-derived human dendritic cells. Blood 106:2366–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. de Lang A, Baas T, Smits SL, Katze MG, Osterhaus AD, Haagmans BL. 2009. Unraveling the complexities of the interferon response during SARS-CoV infection. Future Virol. 4:71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Totura AL, Baric RS. 2012. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr. Opin. Virol. 2:264–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sims AC, Baric RS, Yount B, Burkett SE, Collins PL, Pickles RJ. 2005. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J. Virol. 79:15511–15524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chan MC, Chan RW, Tsao GS, Peiris JS. 2011. Pathogenesis of SARS coronavirus infection using human lung epithelial cells: an in vitro model. Hong Kong Med. J. 17(Suppl 6):31–35 [PubMed] [Google Scholar]
- 7. Mossel EC, Wang J, Jeffers S, Edeen KE, Wang S, Cosgrove GP, Funk CJ, Manzer R, Miura TA, Pearson LD, Holmes KV, Mason RJ. 2008. SARS-CoV replicates in primary human alveolar type II cell cultures but not in type I-like cells. Virology 372:127–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nicholls JM, Butany J, Poon LL, Chan KH, Beh SL, Poutanen S, Peiris JS, Wong M. 2006. Time course and cellular localization of SARS-CoV nucleoprotein and RNA in lungs from fatal cases of SARS. PLoS Med. 3:e27 doi: 10.1371/journal.pmed.0030027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Qian Z, Travanty EA, Oko L, Edeen K, Berglund A, Wang J, Ito Y, Holmes KV, Mason R. 15 February 2013. Innate immune response of human alveolar type II cells infected with SARS-coronavirus. Am. J. Respir. Cell Mol. Biol. [Epub ahead of print.] doi: 10.1165/rcmb.2012-0339OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bermingham A, Chand MA, Brown CS, Aarons E, Tong C, Langrish C, Hoschler K, Brown K, Galiano M, Myers R, Pebody RG, Green HK, Boddington NL, Gopal R, Price N, Newsholme W, Drosten C, Fouchier RA, Zambon M. 2012. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveill. 17(40):20290 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20290 [PubMed] [Google Scholar]
- 11. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367:1814–1820 [DOI] [PubMed] [Google Scholar]
- 12. WHO 2012. Background and summary of novel coronavirus infection. WHO, Geneva, Switzerland [Google Scholar]
- 13. Danielsson N, ECDC Internal Response Team C, Catchpole M 2012. Novel coronavirus associated with severe respiratory disease: case definition and public health measures. Euro Surveill. 17(39):pii=20282. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20282 [DOI] [PubMed] [Google Scholar]
- 14. European Centre for Disease Prevention and Control 2012. Communicable disease threats report. European Centre for Disease Prevention and Control, Stockholm, Sweden: http://ecdc.europa.eu/en/publications/Publications/CDTR online version 4 May 2012.pdf [Google Scholar]
- 15. Khan G. 2013. A novel coronavirus capable of lethal human infections: an emerging picture. Virol. J. 10:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Annan A, Baldwin HJ, Coman VM, Klose SM, Owuse M, Nkrumah EE, Badu EK, Anti P, Agbenyega O, Meyer B, Oppong S, Sarkodi YA, Kalko EKV, Lina PHC, Godlevska EV, Reusken C, Seebens A, Gloza-Rausch F, Vallo P, Tschapka M, Drosten C, Drexler JF. 2013. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg. Infect. Dis. 19:456–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chan JF, Li KS, To KK, Cheng VC, Chen H, Yuen KY. 2012. Is the discovery of the novel human betacoronavirus 2c EMC/2012 (HCoV-EMC) the beginning of another SARS-like pandemic? J. Infect. 65:477–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Corman V, Eckerle I, Bleicker T, Zaki A, Landt O, Eschbach-Bludau M, van Boheemen S, Gopal R, Ballhause M, Bestebroer T, Muth D, Muller M, Drexler J, Zambon M, Osterhaus A, Fouchier R, Drosten C. 2012. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 17(39):pii=20285. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20285 [DOI] [PubMed] [Google Scholar]
- 19. Lau SK, Woo PC, Yip CC, Tse H, Tsoi HW, Cheng VC, Lee P, Tang BS, Cheung CH, Lee RA, So LY, Lau YL, Chan KH, Yuen KY. 2006. Coronavirus HKU1 and other coronavirus infections in Hong Kong. J. Clin. Microbiol. 44:2063–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van der Hoek L, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJ, Wolthers KC, Wertheim-van Dillen PM, Kaandorp J, Spaargaren J, Berkhout B. 2004. Identification of a new human coronavirus. Nat. Med. 10:368–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chan MC, Chan RW, Yu WC, Ho CC, Yuen KM, Fong JH, Tang LL, Lai WW, Lo AC, Chui WH, Sihoe AD, Kwong DL, Wong DS, Tsao GS, Poon LL, Guan Y, Nicholls JM, Peiris JS. 2010. Tropism and innate host responses of the 2009 pandemic H1N1 influenza virus in ex vivo and in vitro cultures of human conjunctiva and respiratory tract. Am. J. Pathol. 176:1828–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chan RW, Kang SS, Yen HL, Li AC, Tang LL, Yu WC, Yuen KM, Chan IW, Wong DD, Lai WW, Kwong DL, Sihoe AD, Poon LL, Guan Y, Nicholls JM, Peiris JS, Chan MC. 2011. Tissue tropism of swine influenza viruses and reassortants in ex vivo cultures of the human respiratory tract and conjunctiva. J. Virol. 85:11581–11587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dahl H, Linde A, Strannegard O. 2004. In vitro inhibition of SARS virus replication by human interferons. Scand. J. Infect. Dis. 36:829–831 [DOI] [PubMed] [Google Scholar]
- 24. Haagmans BL, Kuiken T, Martina BE, Fouchier RA, Rimmelzwaan GF, van Amerongen G, van Riel D, de Jong T, Itamura S, Chan KH, Tashiro M, Osterhaus AD. 2004. Pegylated interferon-alpha protects type 1 pneumocytes against SARS coronavirus infection in macaques. Nat. Med. 10:290–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kumaki Y, Ennis J, Rahbar R, Turner JD, Wandersee MK, Smith AJ, Bailey KW, Vest ZG, Madsen JR, Li JK, Barnard DL. 2011. Single-dose intranasal administration with mDEF201 (adenovirus vectored mouse interferon-alpha) confers protection from mortality in a lethal SARS-CoV BALB/c mouse model. Antiviral Res. 89:75–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sainz B, Jr, Mossel EC, Peters CJ, Garry RF. 2004. Interferon-beta and interferon-gamma synergistically inhibit the replication of severe acute respiratory syndrome-associated coronavirus (SARS-CoV). Virology 329:11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Smits SL, de Lang A, van den Brand JM, Leijten LM, van IJcken WF, Eijkemans MJ, van Amerongen G, Kuiken T, Andeweg AC, Osterhaus AD, Haagmans BL. 2010. Exacerbated innate host response to SARS-CoV in aged non-human primates. PLoS Pathog. 6:e1000756 doi: 10.1371/journal.ppat.1000756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stroher U, DiCaro A, Li Y, Strong JE, Aoki F, Plummer F, Jones SM, Feldmann H. 2004. Severe acute respiratory syndrome-related coronavirus is inhibited by interferon-alpha. J. Infect. Dis. 189:1164–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nicholls JM, Chan MC, Chan WY, Wong HK, Cheung CY, Kwong DL, Wong MP, Chui WH, Poon LL, Tsao SW, Guan Y, Peiris JS. 2007. Tropism of avian influenza A (H5N1) in the upper and lower respiratory tract. Nat. Med. 13:147–149 [DOI] [PubMed] [Google Scholar]
- 30. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 31. Poon LL, Chan KH, Wong OK, Cheung TK, Ng I, Zheng B, Seto WH, Yuen KY, Guan Y, Peiris JS. 2004. Detection of SARS coronavirus in patients with severe acute respiratory syndrome by conventional and real-time quantitative reverse transcription-PCR assays. Clin. Chem. 50:67–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cheung CY, Poon LL, Lau AS, Luk W, Lau YL, Shortridge KF, Gordon S, Guan Y, Peiris JS. 2002. Induction of proinflammatory cytokines in human macrophages by influenza A (H5N1) viruses: a mechanism for the unusual severity of human disease? Lancet 360:1831–1837 [DOI] [PubMed] [Google Scholar]
- 33. Debbink K, Donaldson EF, Lindesmith LC, Baric RS. 2012. Genetic mapping of a highly variable norovirus GII.4 blockade epitope: potential role in escape from human herd immunity. J. Virol. 86:1214–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lindesmith LC, Beltramello M, Donaldson EF, Corti D, Swanstrom J, Debbink K, Lanzavecchia A, Baric RS. 2012. Immunogenetic mechanisms driving norovirus GII.4 antigenic variation. PLoS Pathog. 8:e1002705 doi: 10.1371/journal.ppat.1002705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van Boheemen S, de Graaf M, Lauber C, Bestebroer TM, Raj VS, Zaki AM, Osterhaus AD, Haagmans BL, Gorbalenya AE, Snijder EJ, Fouchier RA. 2012. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio 3(6):e00473–12 doi: 10.1128/mBio.00473-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kindler E, Jonsdottir HR, Muth D, Hamming OJ, Hartmann R, Rodriguez R, Geffers R, Fouchier RA, Drosten C, Muller MA, Dijkman R, Thiel V. 2013. Efficient replication of the novel human betacoronavirus EMC on primary human epithelium highlights its zoonotic potential. mBio 4(1):e00611–12 doi: 10.1128/mBio.00611-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Afzelius BA. 1994. Ultrastructure of human nasal epithelium during an episode of coronavirus infection. Virchows Arch. 424:295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee SM, Gai WW, Cheung TK, Peiris JS. 2011. Antiviral effect of a selective COX-2 inhibitor on H5N1 infection in vitro. Antiviral Res. 91:330–334 [DOI] [PubMed] [Google Scholar]
- 39. Moseley CE, Webster RG, Aldridge JR. 2010. Peroxisome proliferator-activated receptor and AMP-activated protein kinase agonists protect against lethal influenza virus challenge in mice. Influenza Other Respi. Viruses 4:307–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zheng BJ, Chan KW, Lin YP, Zhao GY, Chan C, Zhang HJ, Chen HL, Wong SS, Lau SK, Woo PC, Chan KH, Jin DY, Yuen KY. 2008. Delayed antiviral plus immunomodulator treatment still reduces mortality in mice infected by high inoculum of influenza A/H5N1 virus. Proc. Natl. Acad. Sci. U. S. A. 105:8091–8096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kopecky-Bromberg SA, Martinez-Sobrido L, Frieman M, Baric RA, Palese P. 2007. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 81:548–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cinatl J, Morgenstern B, Bauer G, Chandra P, Rabenau H, Doerr HW. 2003. Treatment of SARS with human interferons. Lancet 362:293–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Loutfy MR, Blatt LM, Siminovitch KA, Ward S, Wolff B, Lho H, Pham DH, Deif H, LaMere EA, Chang M, Kain KC, Farcas GA, Ferguson P, Latchford M, Levy G, Dennis JW, Lai EK, Fish EN. 2003. Interferon alfacon-1 plus corticosteroids in severe acute respiratory syndrome: a preliminary study. JAMA 290:3222–3228 [DOI] [PubMed] [Google Scholar]
- 44. Jia D, Rahbar R, Chan RW, Lee SM, Chan MC, Wang BX, Baker DP, Sun B, Peiris JS, Nicholls JM, Fish EN. 2010. Influenza virus non-structural protein 1 (NS1) disrupts interferon signaling. PLoS One 5:e13927 doi: 10.1371/journal.pone.0013927 [DOI] [PMC free article] [PubMed] [Google Scholar]