Abstract
The HIV-1 spike is composed of three protomeric units, each containing a peripheral gp120 and a transmembrane gp41 subunit. Binding to the CD4 and the chemokine receptors triggers them to mediate virus entry into cells by membrane fusion. The spikes also represent the major target for neutralizing antibodies (Abs) against the virus. We have studied how two related broadly neutralizing Abs, PG9 and PG16, react with the spike. Unexpectedly, this also suggested how the functions of the individual protomers in the spike depend on each other. The Abs have been shown to bind the V1/V2 loops of gp120, located at the top of the spike. Using blue native-polyacrylamide gel electrophoresis (BN-PAGE), we show that only single Abs or antigen-binding fragments could bind to the spikes of HIV-1 virus-like particles. Apparently, binding to one gp120 sterically interferes with binding to the other two subunits in the spike top. Despite this constraint, all of the protomers of the spike became resistant to CD4 binding and subsequent formation of the coreceptor binding site. These activities were measured by monitoring the sequential complex formation of the spike first with Abs and then with soluble 2d- or 4d-CD4 or with soluble CD4 and the CD4 inducible coreceptor binding site Ab 17b in BN-PAGE. The inhibition of the spike by single-Ab binding suggested that the activation reactions of the individual protomeric units are linked to each other in a coordinated activation process.
INTRODUCTION
The HIV-1 spike is a trimer where the protomeric unit is composed of the noncovalently linked peripheral (gp120) and transmembrane (gp41) subunits (1–3). The spike mediates entry of the virus into the cell through membrane fusion. In this process, gp120 binds successively to the primary, CD4, and the secondary, chemokine receptors on the cell surface. This activates gp41 to interact with the lipid bilayer of the cell via its fusion peptide (4, 5). The back-folding of the gp41 subunits from a trimeric prehairpin to a hairpin structure then approaches the viral and the cell membranes so they can merge and allow the viral capsid with the viral genome to enter into the cell (6–8).
It has been shown that binding of CD4 induces changes in the gp120 structure that result in formation of the binding site for the secondary receptor and that changes induced by both receptors release the gp120 constraints on gp41 activation (1, 3, 9–11). However, an important question concerns the possible coordination of the activating processes in the individual protomers of the trimeric spike. Are the activating changes in the protomers linked to each other and therefore have to occur simultaneously? A coordinated process would ensure the formation of symmetrical intermediate forms of the trimeric spike, which should be important for their stability and function.
The spike is also the target for neutralizing antibodies (NAbs). Several broadly neutralizing antibodies (bNAbs) against HIV-1 have been characterized (12–19). The elucidation of their neutralization mechanisms should be useful for the development of an HIV-1 vaccine and also to explain the spike activation mechanism. Some bNAbs like 2G12 and VRC01stabilize the native unliganded conformation of the spike and thereby inhibit spike activation (10, 20). Most interestingly, single Ab binding seems to be sufficient for neutralization (21). This can be explained assuming a coordinated activation mechanism of the three protomers of the spike. In this case, binding of a single stabilizing bNAb to one protomer will also prevent the activation of the unliganded protomers.
In the present study we have studied the neutralization mechanism of the bNAbs PG9 and PG16 (17, 22). These are somatically related Abs that neutralize the majority of the HIV-1 strains. They are sensitive to changes in the gp120 variable (V) loops V1/V2 and also V3, require the Asn 160-linked sugar unit in V1/V2, and bind preferentially to native spikes. PG9 has also been crystallized in complex with a recombinant glycopeptide mimic of V1/V2 and the atomic structure determined (23). This showed Ab binding to the peptide proximal sugar residue of N160 and to the main chain of one of the four strands of the V1/V2 sheet. Further mapping of the V1/V2 binding sites in PG9 by Arg mutations indicated that these defined much of the epitope (22, 23).
We show here that only single PG9 and PG16 Abs can bind to the trimeric spike, apparently for steric reasons. Nevertheless, they stabilize the oligomeric structure of the spike and inhibit CD4 binding to and activation of all the protomers. This suggests that the spike activation is indeed a coordinated process occurring simultaneously in all the protomers of the trimer. The Ab-induced inhibition of activation was found to be reversible. Soluble CD4 at a high concentration was apparently able to catch spikes from dissociated spike-Ab complexes and activate them.
MATERIALS AND METHODS
PG9 and PG16 Abs directed against the V1/V2 loops were obtained from Dennis R. Burton, the Scripps Research Institute, La Jolla, CA. The CD4 binding site Ab b12 was purchased from Polymun Scientific (Klosterneuburg, Austria). The 17b antibody, with a CD4 inducible binding site, and soluble 2d- and 4d-CD4 were obtained from the NIH AIDS Research and Reference Reagent Program (17b from James E. Robinson (24), sCD4-183, i.e., 2d-CD4, was obtained from Pharmacia, Inc. (25), and soluble human CD4, i.e., 4d-CD4, was obtained from Progenics. VLPs carrying HIV-1 JR-FL spikes with a deleted cytoplasmic tail (truncated at residue 708), disulfide linked gp120 and gp41 (Cys501-Cys605), and E168K and N189A mutations were produced in 293T cells in a 150-cm2 culture flask by calcium phosphate precipitation-mediated cotransfection using 10 μg of pCAGGS JRFL gp160SOS,ΔCT,E168K,N189A and 10 μg of pNL4-3.Luc.R-E DNA (3, 26, 27). The 293T cells were maintained as described previously (28). [35S]Cys was incorporated into the VLPs through metabolic labeling in Cys-free Dulbecco modified Eagle medium (National Veterinary Institute, Uppsala, Sweden) supplemented with 10 μM unlabeled Cys and 100 μCi of [35S]Cys/ml (Fisher Scientific, Gothenburg, Sweden) from 24 to 48 h after the transfection. The [35S]Cys-labeled VLPs were isolated by ultracentrifugation in a sucrose step gradient (3). Fabs were generated by digesting 18 μg of monoclonal Ab in 25 μl of phosphate-buffered saline (PBS) containing 10 mM EDTA, 5 mM Cys, and 0.5 μg of papain for 4 h at 37°C. The digestion was terminated by addition of iodoacetamide to 5 mM. Samples were diluted with 175 μl of HNC buffer (50 mM HEPES, 100 mM NaCl, 1.8 mM CaCl2 [pH 7.4]) and uncleaved Abs and Fc fragments were removed by binding to protein A-Sepharose (GE Healthcare, Uppsala, Sweden) overnight at 4°C. VLPs were incubated with ligands in HNC buffer for 2 h at room temperature. The formation of liganded spike complexes was followed by BN-PAGE and phosphorimaging. For this, the samples were solubilized by incubation for 10 min at 37°C in HNC buffer containing 0.15 to 0.3% Triton X-100. One volume of two times-concentrated BN-PAGE sample buffer was added, and the samples were incubated for 10 min at room temperature and analyzed by gradient BN-PAGE containing 4.5 to 8% total acrylamide as described previously (29). The dried gels were exposed to phosphorimage screens (BAS MS2025; Fujifilm, Science Imaging, Nacka, Sweden) and the radiolabeled bands visualized and quantified using a Molecular Imager FX and the Quantity One software (Bio-Rad, Hercules, CA).
RESULTS
A single PG9 or PG16 Fab binds to the HIV-1 spike.
In the present study, we used an HIV-1 JR-FL mutant spike (gp160SOS,ΔCT,E168K,N189A) incorporated into VLPs (26). This contains an engineered disulfide bond between the gp120 C-terminal region and the small disulfide region of gp41, which increases spike stability. It also contains a deletion of the cytoplasmic domain. This increases spike incorporation into VLPs but have a negligible effect on the neutralization sensitivity (30). A Glu-to-Lys substitution at position 168 of the V2 loop and an Asn-to-Ala substitution at position 189 are introduced to allow binding of the PG9 and PG16 Abs to the JR-FL spike (17, 27, 31). The mutant spike can bind CD4 and coreceptors and, after dithiothreitol-mediated reduction of the engineered disulfide, infect cells like wild-type (wt) virus (3, 32, 33).
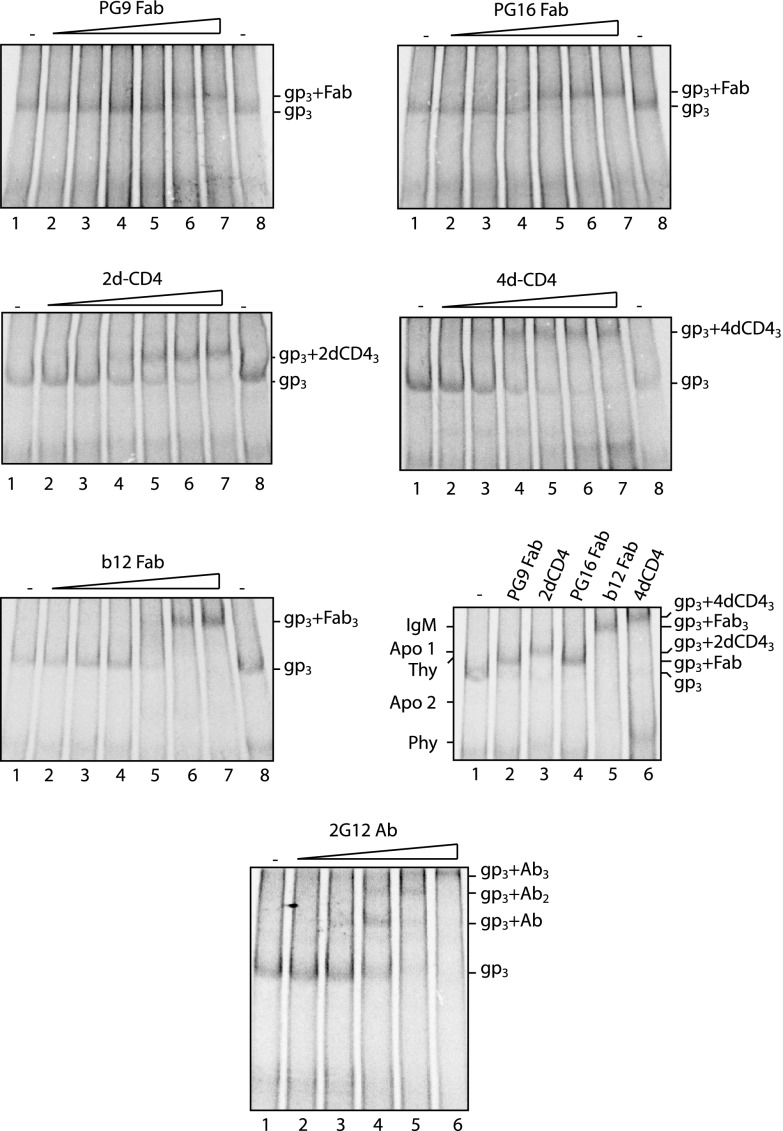
We prepared fragments (Fabs) of PG9 and PG16 Abs by papain digestion and analyzed their binding stoichiometry to the trimeric HIV-1 mutant spike by 4.5 to 8% BN-PAGE. The Fab of the b12 Ab and soluble 2d- and 4d-CD4 were used as controls. Spikes of [35S]Cys-labeled VLPs were reacted with increasing concentrations of each ligand at room temperature for 2 h and then solubilized and analyzed by BN-PAGE (Fig. 1). A side-by-side comparison of the migrations of the different ligand complexes in BN-PAGE was also performed (Fig. 1, lower right panel). We found that both PG9 and PG16 Fabs formed a complex with the spike that migrated clearly slower than the unliganded spike (gp3) (Fig. 1, upper panels). The 2d-CD4 generated a complex that migrated even slower (Fig. 1, middle left panel and lower right panel, lanes 2 to 4), and the 4d-CD4 and the b12 Fab formed complexes that migrated much slower than the spike-PG9/16 Fab complexes (Fig. 1, middle right panel, lower left panel and lower right panel, lane 2 and lanes 4 to 6). The unliganded spike band and the CD4 liganded spike bands have earlier been shown to represent gp120-gp41 trimers, i.e., spikes, and fully saturated spike-CD4 complexes, respectively, by three-dimensional-structure analyses of corresponding material using cryo-electron microscopy (cryo-EM) (3). Considering the approximate molecular masses of an Fab (∼50 kDa), the 2d-CD4 (∼26 kDa), and the 4d-CD4 (∼46 kDa), the b12 liganded spike band should represent a spike in complex with three b12 Fabs. Consequently, the characteristic migrations of the spike-PG9/16 bands slightly faster than the spike-2d-CD4 band indicate that no more than one PG9 or PG16 Fab can bind to the spike.
Fig 1.
One PG9 or PG16 Fab binds to the spike. [35S]Cys-labeled VLPs were incubated with PG9 Fab (upper left panel), PG16 Fab (upper right panel), 2d-CD4 (middle left panel), 4d-CD4 (middle right panel), b12 Fab (lower left panel), or 2G12 Ab (IgG) (bottom panel) at increasing concentrations for 2 h at room temperature and then solubilized for analysis of liganded spikes by 4.5 to 8% BN-PAGE. The Fab concentrations were 0.25, 0.75, 2.5, 7.5, 25, and 75 μg/ml, the 2G12 Ab concentrations were 0.3, 1, 3, 10, and 30 μg/ml, and the sCD4 concentrations were 1.25, 2.5, 7.5, 10, and 12.5 μg/ml. The spike and the various spike-sCD4 or Fab complexes were also analyzed next to each other for close-by comparisons of band migrations (lower right panel). Unliganded (gp3) and liganded spike bands are indicated to the right of each panel. The migrations of molecular mass standards (IgM, IgM pentamer, 1,048 kDa; Apo 1, apoferritin band 1, 720 kDa; Thy, thyroglobulin, 669 kDa; Apo 2, apoferritin band 2, 480 kDa; Phy, B-phycoerythrin, 242 kDa) are indicated to the left of the lower right panel. The panels represent phosphorimages of the gels. Note that the bottom BN-PAGE with spike-2G12 Ab complexes has been run longer than the other ones in order to resolve complexes with two and three Abs.
A puzzling feature in these experiments was that neither the soluble CD4 nor the b12 Fab formed complexes with the spike containing one or two ligands. Only fully saturated spike complexes were observed when the virus was incubated with increasing ligand concentrations (Fig. 1, middle left, middle right, and lower left panels). To exclude the possibility that incompletely saturated ligand complexes were inherently unstable in our BN-PAGE, we also tested the bNAb 2G12. This bNAb contains a domain switch, which allows it to recognize single spikes, but not cross-link separate ones (13). With this Ab, we observed the formation of spike complexes containing one and two ligands, respectively (Fig. 1, bottom panel). Similar results have been shown before with 2G12 Fab using a comparable BN-PAGE (34). This suggests that incompletely saturated complexes of spikes with soluble CD4 and b12 Fab complexes are not dissociated in the analysis system but are simply not formed.
One PG9 or PG16 Ab binds to one solubilized spike but cross-links two spikes in the viral membrane.
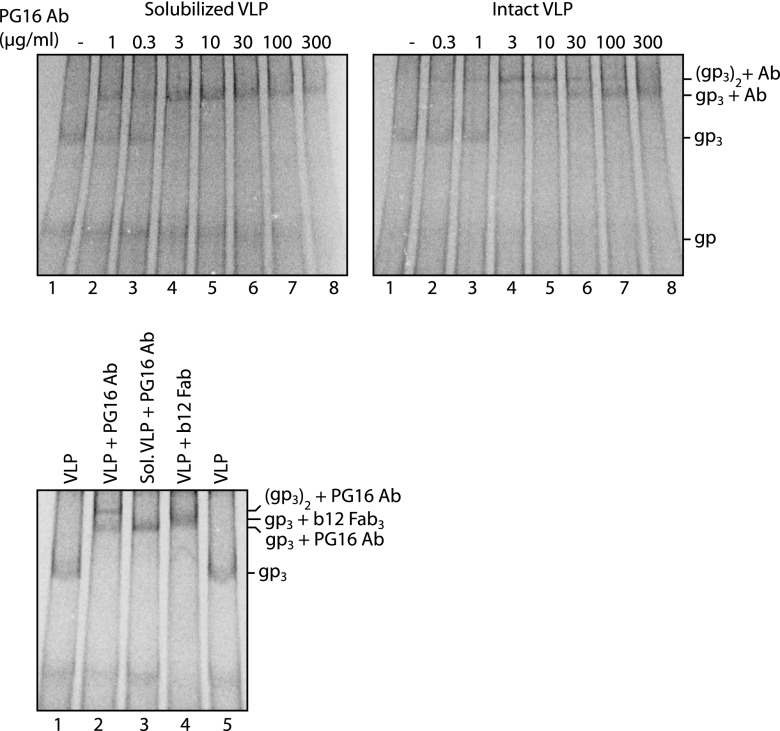
The binding of complete divalent PG9 and PG16 Abs (IgG) to solubilized spikes and to spikes in intact particles was also studied. When spikes solubilized from labeled VLPs were treated with PG16 Abs at increasing concentrations for 2 h at room temperature and the complexes were analyzed by BN-PAGE, we observed a shift of the spike band into one much slower-migrating band that moved somewhat faster than the spike in complex with three b12 Fabs (Fig. 2, upper left panel and lower panel). This is consistent with the binding of a single PG16 Ab to one spike. However, if the spikes on intact VLPs were treated with a low concentration of PG16 Abs for 2 h at room temperature and then solubilized, we found that the spike band shifted into a still slower-migrating band (Fig. 2, upper right panel, lanes 1 to 4 and lower panel). At higher Ab concentrations, this slower band shifted gradually into the faster-moving band observed after treating the solubilized spikes with the Abs (Fig. 2, compare upper left and right panels, lanes 4 to 8, and the lower panel). Apparently, at low concentrations, single Abs first bind to a fraction of the spikes with one arm and then, during the 2 h of incubation, probably after diffusion in the membrane, the bound Ab cross-links another spike with its free arm. At higher concentrations, single Abs most likely bind directly to all spikes and thereby prevent cross-linking. If the slower-migrating band had represented a complex between one spike and several Abs, it would had been present most prevalently at higher Ab concentrations. Thus, these results confirm that only a single Ab binding site can be used in the trimeric spike. Similar results were obtained with the PG9 Ab.
Fig 2.
One PG16 Ab binds to one spike in solution but cross-links two spikes in the particle. Solubilized [35S]Cys-labeled VLPs were incubated with PG16 Ab at increasing concentrations for 2 h at room temperature and then analyzed by 4.5 to 8% BN-PAGE (upper left panel). Intact labeled particles were similarly incubated with the Ab and then solubilized for BN-PAGE (upper right panel). PG16 Ab complexes formed with spikes of solubilized and intact VLP and the b12 Fab complex formed with spikes of intact VLP were also analyzed next to each other for comparisons of band migrations (bottom panel). Unliganded (gp3) and liganded spike bands are indicated as is also the single disulfide-linked gp120-gp41 complex band (gp). The panels represent phosphorimages of the gels.
The spike oligomer is stabilized by a single PG9 or PG16 Ab binding.
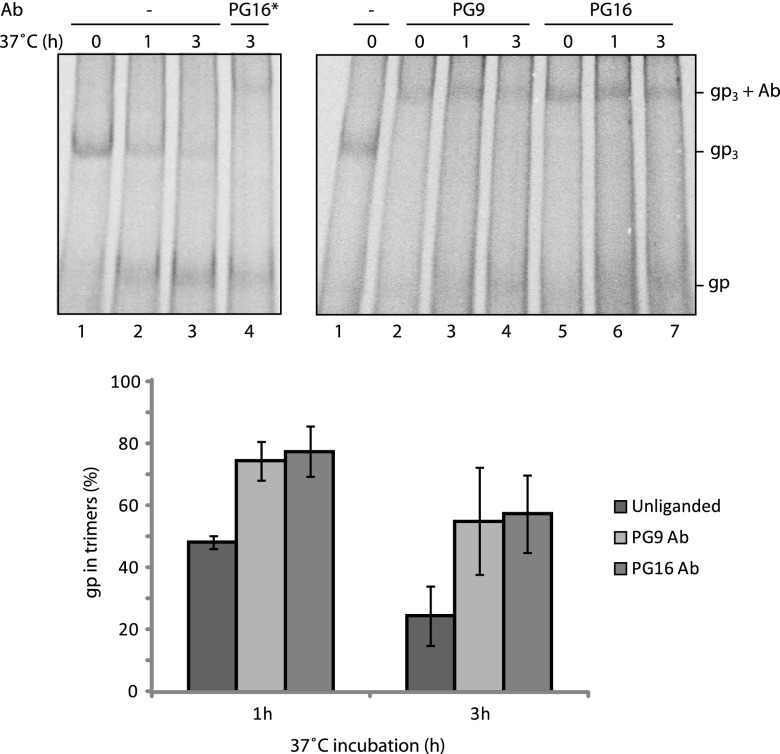
The stability of the spike solubilized from labeled VLPs was studied in unliganded form and as a complex with PG9 or PG16 Ab by BN-PAGE after incubation at 37°C for 1 and 3 h. The unliganded oligomer was found to progressively dissociate into single disulfide-linked gp120-gp41 complexes (gp) after 1 and 3 h of incubation (Fig. 3, upper left panel, lanes 1 to 3). However, when the spike had been complexed with one PG9 or PG16 antibody, most of it still resisted as Ab complexed trimers after 3 h of incubation at 37°C (Fig. 3, upper right panel). The corresponding quantification is shown in Fig. 3, bottom panel. If PG9/16 Abs were added to the solubilized spikes after the 37°C incubation, no detectable Ab binding was found to the dissociated gp120-gp41 complexes, only to the remaining trimers, thus confirming that trimerization is important for PG9/16 Ab binding (Fig. 3, upper left panel, lane 4). These data showed that the binding of PG9 or PG16 Ab to a single binding site on the spike inhibits trimer dissociation into separate gp120-gp41 complexes.
Fig 3.
Single PG9 or PG16 binding stabilizes the spike. Solubilized [35S]Cys-labeled VLPs were treated with saturating amounts of PG9 or PG16 Ab (75 μg/ml) for 2 h at room temperature (right panel) or mock treated (left panel, lanes 1 to 3) and then incubated for 1 or 3 h at 37°C before analyses by 4.5 to 8% BN-PAGE. Solubilized spikes were also treated with the PG16 Ab after the 37°C incubation (left panel, lane 4, PG16*). Bands corresponding to spikes (gp3) and its liganded forms and single disulfide linked gp120-gp41complexes (gp) are indicated. The fraction of gp that were still in trimeric form, i.e., as spikes or liganded spikes, after the incubations was quantified from the phosphorimages of the gels and expressed as a percentage of the total trimeric gp, unliganded or liganded, at the start of incubation. The means ± the standard deviations (SD) are shown (n = 3). In the left panel the background (BG) for the spike band was taken from the corresponding position in lane 4 (saturating amounts of PG16 Ab added after solubilization) and for the gp band from lane 1(no incubation). Note that this BG will include some gp that has dissociated already before the incubations. In right panel, the BG for the spike-Ab band was taken from lane 1 (no Ab and no incubation) and the BGs for the gp band in the 1-h and the 3-h incubations from lanes 2 and 5 (no incubation), respectively.
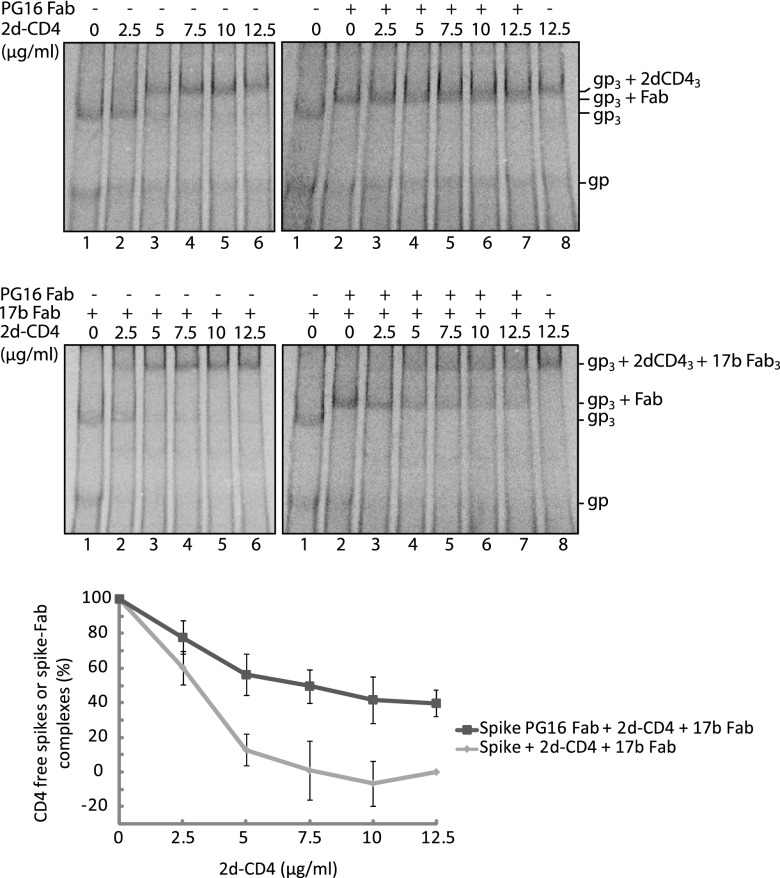
The binding of one PG9 or PG16 Ab or Fab to the spike reversibly inhibits 2d- or 4d-CD4 binding and spike activation.
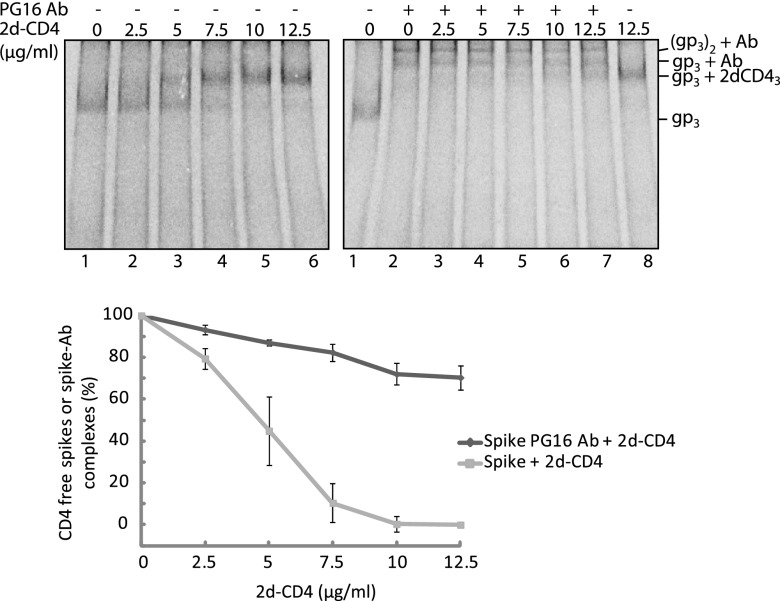
To study the functional effect of binding a PG9 or PG16 Ab or Fab to the trimeric HIV-1 spike, we followed the ability of the complex to bind CD4 and, with the Fab, also to form the coreceptor binding site. The formation of the latter was monitored by binding of the 17b Fab, which recognizes a CD4 induced epitope that overlaps the binding site of the coreceptor (4, 24). Spikes in VLPs were first treated with the PG9/16 Abs or Fabs and then with soluble 2d- or 4d-CD4, alone or together with 17b Fab, and the resulting spike-ligand complexes were identified by BN-PAGE after solubilization. When PG16 Ab was reacted with the VLP spikes, most of the spike band shifted to the slower-migrating bands corresponding to the spike-Ab-spike and the spike-Ab complexes (Fig. 4, upper right panel, lane 2). After a second incubation with increasing concentrations of 2d-CD4, these bands shifted gradually into a faster-migrating one corresponding to the saturated spike-2d-CD4 complex (Fig. 4, upper right panel, lanes 3 to 8). These band shifts can be explained by a reversible binding of the PG16 Ab to the spikes and the formation of an Ab free spike complex with three molecules of 2d-CD4. Although the PG16 Ab was unable to prevent 2d-CD4 binding at high concentrations, it is possible that it increases spike resistance toward 2d-CD4 binding. To find out, we compared the binding of 2d-CD4 to unliganded spikes at corresponding 2d-CD4 concentrations. We found that most unliganded spikes bound 2d-CD4 at a concentration of 7.5 μg/ml (Fig. 4, upper left panel, lane 4), a concentration that had no significant effect on spike-Ab-spike and spike-Ab complexes (Fig. 4, upper right panel, lane 5). The resistance of the spike-Ab complexes to 2d-CD4 binding was quantified by measuring the relative number of spikes in such complexes in phosphorimages. The results showed a significantly increased resistance of the spikes toward 2d-CD4 binding when the spike was bound to the Ab (Fig. 4, bottom panel). We conclude that PG16 Ab binding to the spike inhibits 2d-CD4 binding but, due to the dynamic kinetics of spike-Ab association and dissociation the 2d-CD4 can, at higher concentrations, bind Ab-dissociated spikes.
Fig 4.
Binding of the PG16 Ab to the spike reversibly inhibits 2d-CD4 binding. Spikes in [35S]Cys-labeled VLPs, first incubated with PG16 Ab and then with 2d-CD4 at increasing concentrations (right panel) or directly with 2d-CD4 (left panel), were solubilized and analyzed by 4.5 to 8% BN-PAGE. Bands corresponding to the spike (gp3) and its liganded forms are indicated to the right. The resistance of the unliganded and the PG16 Ab bound form of the spike toward 2d-CD4 binding were quantified from the phosphorimages and expressed as percentage of total spikes. The means ± the SD are shown (n= 3; lower panel). For quantitations, the BG levels of the different bands were defined as follows: in the left panel for the spike band, we used the gp3 position of lane 6 (saturating 2d-CD4) and for the 2d-CD4 liganded spike band the gp3+2dCD43 position of lane 1 (no 2d-CD4). In the right panel for the spike-Ab and the spike-Ab-spike bands we used the gp3+Ab and (gp3)2+Ab positions of lane 8 (no Ab), and for the 2d-CD4 liganded spike band we used the gp3+2dCD4 position of lane 2 (no 2d-CD4). The top panels represent phosphorimages of the gels.
When the spikes of labeled VLPs were treated with the PG16 Fab and then analyzed by BN-PAGE, we observed the formation of the spike-Fab band (Fig. 5, upper right panel, lane 2). Subsequent incubation with 2d-CD4 at increasing concentrations caused a gradual shift of the spike-Fab band into a somewhat slower-migrating band, which corresponded to the spike saturated with 2d-CD4 (Fig. 5, upper right panel, lanes 3 to 8). Together, the two bands appeared as one broad band. Most likely the 2d-CD4 is, at higher concentrations, able to bind to spikes from which Fab dissociates, as described above in the case of the Ab liganded spikes (Fig. 4). When the Fab reacted VLPs were incubated with both the 2d-CD4 and the 17b Fab, we found that the spike-Fab band shifted into a much slower one corresponding to a spike-2d-CD4-17b Fab complex (Fig. 5, middle right panel). Apparently, 2d-CD4 binding to spikes from which PG16 Fab has dissociated can activate the spikes to form the coreceptor binding site. The formation of the spike-2d-CD4-17b Fab complex facilitated a quantitative comparison of the resistance of the spike-PG16 Fab complex toward 2d-CD4 binding to that of unliganded spikes. This comparison was not possible using the spike-Fab and the spike-2d-CD4 complexes due to their similar migration in BN-PAGE (Fig. 5, upper panels). When the fraction of remaining spike-Fab complexes was measured after reaction with 2d-CD4 at increasing concentrations, and with 17b Fab, we found that this was significantly higher than the fraction of unliganded spikes that remained after direct incubations of the spike with 2d-CD4, at corresponding concentrations, and with 17b Fab (Fig. 5, middle left and bottom panels).
Fig 5.
Binding of the PG16 Fab to the spike reversibly inhibits 2d-CD4 binding. Spikes in [35S]Cys labeled VLPs incubated first with PG16 Fab and then with 2d-CD4 at increasing concentrations (upper right panel), with PG16 Fab and then with 2d-CD4 and 17b Fab (middle right panel), with 2d-CD4 only (upper left panel) or with 2d-CD4 and 17b Fab (middle left panel) were solubilized and analyzed by 4.5–8% BN-PAGE. Bands corresponding to the spike (gp3), its liganded forms and single disulfide linked gp120-gp41complexes (gp) are indicated to the right. The resistance of the unliganded and the PG16 Fab bound form of the spike toward 2d-CD4 + 17b Fab binding at the different 2d-CD4 concentrations were quantified from the phosphorimages and expressed as percentage of total spikes. The means ± the SD are shown (n= 3; lower panel). The BG levels were defined as follows. In the middle left panel for the spike band we used gp3 position of lane 6 (saturating 2d-CD4 and 17b Fab), and for the 2d-CD4 and 17b Fab liganded spike band we used the gp3+2dCD43+17b Fab3 position of lane 1 (no PG16 Fab, no 2d-CD4); in the middle right panel for the PG16 Fab liganded spike band we used the gp3+Fab position of lane 8 (no PG16 Fab), and for the 2d-CD4 and 17b Fab liganded spike band we used the gp3+2dCD43+17b Fab3 position of lane 2 (no 2d-CD4). The top and middle panels represent phosphorimages of the gels.
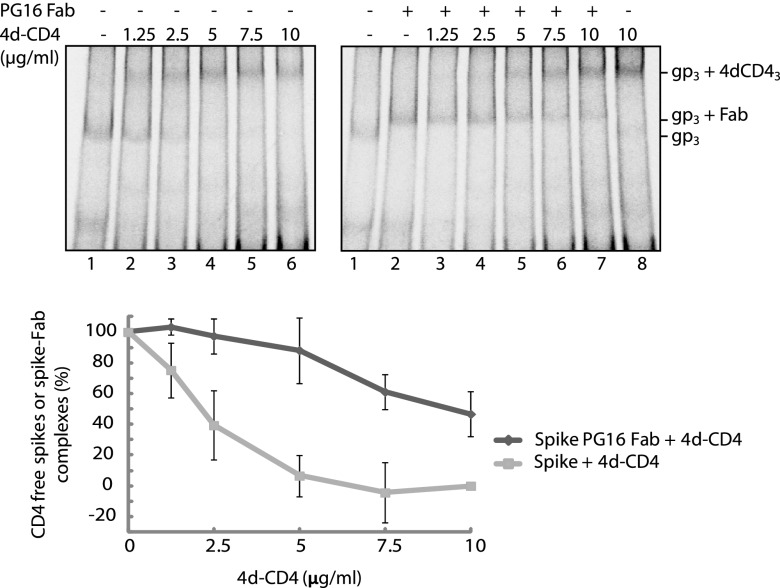
Although the experiments described above suggested that the binding of PG16 Ab or Fab to the spike prevented simultaneous 2d-CD4 binding, the formation of some ternary spike-Fab(Ab)-2d-CD4 complexes could not completely be ruled out. For instance, the size of the spike in complex with three 2d-CD4 is expected to be very similar to that of a complex formed between one spike, one Fab and one 2d-CD4, thus weakening our interpretation of the band shift shown in Fig. 5, upper right panel. To settle this important question, we analyzed the binding of the larger soluble 4d-CD4 to the PG16 Fab liganded spike by BN-PAGE. Incubation of the spike in labeled VLPs with the Fab resulted in formation of the corresponding band (Fig. 6, right panel, lane 2). Further incubation in the presence of 4d-CD4 at increasing concentrations caused a shift of the spike-Fab band into a much larger one corresponding to the fully saturated spike-4d-CD4 complex (Fig. 6, right panel, lanes 3 to 8). We could not detect any bands corresponding to spike-Fab-4d-CD4 ternary complexes, e.g., the binding of one 4d-CD4 to a spike-Fab complex, which should result in a clear band shift. We also noted that the shift of the spike-Fab band into the spike-4d-CD4 band occurred without any intermediates, suggesting immediate full occupancy of the Fab dissociated spike with 4d-CD4 molecules. Additional analyses in which the 4d-CD4 was bound directly to the spike without Fab at corresponding concentrations and subsequent band quantifications showed that the spike-Fab complex was more resistant also toward 4d-CD4 binding than the unliganded spike (Fig. 6, upper left, upper right, and bottom panels). Altogether, we conclude that the binding of one PG16 Ab or Fab to the trimeric spike is able to inhibit the binding of 2d- or 4d-CD4 to all of its protomers. However, the Ab or Fab binding is reversible and at higher concentrations the 2d- or 4d-CD4 can catch a dissociated spike and transform it into the coreceptor active form, which is not any more able to bind the PG16 Ab (16). Similar results were obtained with PG9 Fab and Ab.
Fig 6.
Binding of the PG16 Fab to the spike reversibly inhibits 4d-CD4 binding. Spikes in [35S]Cys-labeled VLPs incubated first with PG16 Fab and then with 4d-CD4 at increasing concentrations (right panel) or directly with 4d-CD4 (left panel) were solubilized and analyzed by 4.5 to 8% BN-PAGE. Bands corresponding to the spike (gp3) and its liganded forms are indicated to the right. The resistance of the unliganded and the PG16 Ab bound form of the spike toward 4d-CD4 binding were quantified from the phosphorimages and expressed as a percentage of the total spikes. The means ± the SD are shown (n= 3; lower panel). The BG levels used were as follows. In the left panel for the spike band we used the gp3 position of lane 6 (saturating 4d-CD4), and for the 4d-CD4 liganded spike we used the gp3+4dCD43 position of lane 1 (no ligand); in the right panel for the PG16 Fab liganded spike band we used the gp3+Fab position of lane 8 (no Fab), and for the 4d-CD4 liganded spike band we used the gp3+4dCD43 position of lane 2 (no 4d-CD43). The top panels represent phosphorimages of the gels.
DISCUSSION
Cryo-EM has revealed the HIV-1 spike as a cage-like structure, where the three protomeric units form separated lobes on the sides, a common cage roof and separated spike legs or a common spike stem, depending on the study (1–3). The atomic gp120 core structure from crystal analyses has been fitted into the side lobe in such a position that the missing V1/V2 and V3 loops could accommodate the cage roof. The location of the V1/V2 in the roof close to the 3-fold axis has been proven by comparing the density maps of the wt spike and the spike with a V1/V2 deletion (35, 36). Thus, the PG9 and PG16 antibodies should bind at the very top of the spike. In this position, close to the 3-fold axis of the spike, steric reasons most likely will limit the binding to involve only a single Ab molecule as found in the present study. This is consistent with the earlier finding that HIV-1 pseudovirus with spikes containing only one optimally PG9/16 Ab binding protomer (JR-FL, E168K and N189A) shows full neutralization sensitivity (31). Several other V1/V2 specific monoclonal Abs, when incubated with HIV-1 VLPs, have been shown to generate BN-PAGE bands like the spike-Ab and the spike-Ab-spike bands we show in Fig. 2, suggesting that they all are sterically restricted to bind to only one V1/V2 loop complex (27). After submission of this report, Julien et al. published the cryo-EM structure of an HIV-1 spike ectodomain trimer, BG505 SOSIP.664 gp140, in complex with PG9 Fab and confirmed the binding of a single Fab to the trimeric spike ectodomain (37).
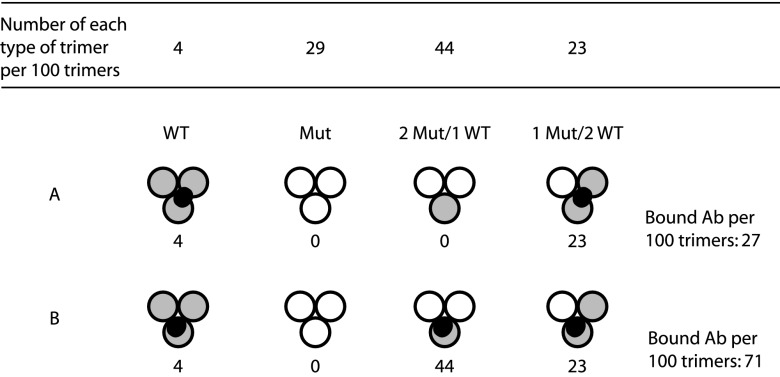
CD4 binding to the HIV-1 spike alters the structure of the gp120, in particular its inner domain, resulting in the formation of the bridging sheet between the inner and outer domains, reorganization of the V1/V2 and V3 loops and weakening of the gp120-gp41 association (4, 11, 38–40). Cryo-EM analyses have shown that the cage roof opens up by an apparent lateral displacement of the V1/V2 loops and rising up of the V3 loops (1, 3, 10). This exposes the binding site for the coreceptor in V3 and adjacent structures and opens a path for the gp41 subunits to reach the target membrane with their fusion peptides. We show here that when one PG9 or PG16 Ab or Fab binds to the spike, then the whole spike becomes stabilized and all its protomers becomes resistant to CD4 binding. If the Ab epitope is limited to one protomer or is shared between two protomers, then a complete inhibition of CD4 binding to the spike would imply that the Ab inhibits a coordinated spike activation process. In such a process, the binding of one Ab to one protomer will not only inhibit this one but also prevent the activation of the other two protomers even if they do not bind an Ab. This would also be the case for the single unliganded protomer if the epitope is shared between two protomers in the spike. However, if the Ab would cross-link all protomers via a shared epitope, then the stabilization and inhibition of the spike could be explained by a direct Ab binding effect. An earlier measurements of PG9 and PG16 Ab binding to cell surface spikes containing mixtures of wt and binding negative mutant protomers were interpreted in favor of protomer limited epitopes (17). However, in the present study, it was assumed that the spike bound three Abs. If the theoretical calculation of Ab binding to spikes generated by the synthesis of wt and mutant protomers in a 1:2 ratio described in Walker et al. is repeated, assuming a single Ab binding to one or two protomers, then a model wherein the epitope is shared between two protomers fits the measured values (Fig. 7). This was confirmed by the recent cryo-EM study of Julien et al. (37). Thus, it is possible that the V1/V2 and the V3 loops of neighboring protomers interact and form the complete PG9/16 epitope. Such an interaction has been suggested by the shielding of V3 of one protomer by the V1/V2 of an adjacent protomer (41).
Fig 7.
Calculated binding of PG9 and PG16 Abs to mixed trimers. In this example, spikes are assembled from wt (gray) and binding negative mutant (white) protomers in a 1:2 ratio (17). This should yield 4% wt homotrimeric spikes, 29% mutant homotrimers, 44% mixed trimers containing two mutant protomers, and 23% mixed trimers containing one mutant protomer using the formula described in reference 21. (A) If epitopes extend over two protomers (dark gray area) and only a single Ab can bind to each trimer, then the binding would be 27% of the binding to a homogeneous population of wt spikes. (B) However, if epitopes are confined to a single protomer the binding would be as high as 71%. The binding model with an epitope involving two protomers fits the measured binding described in reference 17.
A trivial explanation for the PG9 and PG16 Ab-mediated inhibition of CD4 binding we observed (Fig. 4 to 6) is that their epitopes overlap with the binding site for CD4. However, this should not be the case because the PG9 and PG16 epitopes have, as discussed above, been mapped to the top of the spike roof and the CD4 binding site to one phase of the side lobe of the spike (1, 3). In the spike roof only one PG9 or PG16 Ab has space to react, whereas three CD4 molecules can bind. Finally, the CD4 binding site Ab b12 has been shown not to compete with PG9 or PG16 binding to the spike (17).
In the present study we used a spike variant with disulfide-linked gp120 and gp41 subunits and with deleted cytoplasmic tail. We cannot exclude the possibility that the effect of PG9 and PG16 Ab binding would be different in the wt spike than in the variant. However, it has been shown that PG9 and PG16 Ab incubation with VLPs containing full-length spikes lacking the intersubunit disulfide (but still containing the mutations for PG9/16 Ab reactivity) generates a BN-PAGE band doublet similar to the spike-Ab and spike-Ab-spike bands we see in Fig. 2 (27). This suggests that only one binding site is used in the complete spike lacking the disulfide as in the variant we studied.
A coordinated spike activation could be triggered by a single spike protomer-CD4 interaction, or it might require a stoichiometric amount of receptors. Consistent with the latter model CD4 receptors have been suggested to bind with low affinity to the CD4 binding loop of the outer domain in the three gp120 subunits of the spike and together create an avidity-enhanced interaction with subsequent simultaneous structural changes, which will lock the CD4 molecules in place (19). A single receptor could trigger a coordinated activation of the spike if the activation processes of the individual protomers are cooperating. This model has been used to explain the finding that trimers with two different activation inhibiting mutations in separate protomers can rescue spike activation by complementation (42). For instance, complementation was observed between a CD4 binding site mutant and a fusion peptide mutant. It was suggested that the structural changes induced by CD4 binding to the protomer with the fusion peptide mutation was transmitted to the other protomer(s) with the CD4 binding site mutation causing gp41 activation. This model has also been used to explain how the apparent binding of a single soluble CD4 molecule can activate a simian immunodeficiency virus spike with a cytoplasmic tail deletion (34). The signal for the cooperativity of the CD4-induced changes in gp120 between the individual protomers of the spike could possibly be conducted via its roof structure, where the V1/V2 and V3 loops of the three gp120 units might interact.
Our present finding that the soluble 2-d and 4-dCD4 and the b12 Fab could, opposite to the 2G12 Ab, not form intermediate spike complexes containing one or two ligands, only fully saturated complexes (Fig. 1), can be interpreted in favor of a coordinated activation of the spike protomers. The b12 Fab binding has been shown to result in a partial CD4-like change of the spike structure, whereas the 2G12 Ab stabilizes the native spike structure (1, 20). Thus, it is possible that the spike protomers undergo reversible changes into intermediate structures along the activation pathway in a dynamic but coordinated fashion and that CD4 and b12 bind to these. Because of the coordination the binding of one ligand will lock all of the protomers into the ligand binding stage. This will facilitate the binding of the second and third ligand after the first has bound and in our binding experiments with increasing ligand concentrations show up as a saturation of ligand binding without intermediate complexes. In contrast, Abs like 2G12, do not have to wait for a dynamic change of the spike structure for binding and therefore, the binding of one Ab will not facilitate the binding of additional ones and spikes containing one and two Abs can be seen.
The reversible spike stabilization and inhibition of CD4 binding of PG9 and PG16 Abs are features shared with the 2G12 Ab (13, 20). The latter bNAb stabilized the native form of the HIV-1 spike and inhibited CD4 association by binding to several Asn linked sugar units on gp120 by virtue of its unique heavy chain variable domain switch. Binding was reversible and a high concentration of sCD4 could displace 2G12 and activate the spike. It was suggested that CD4 activation-induced changes in gp120 resulted in a relocation of the 2G12 Ab target sugar units and that prebound 2G12 caused an allosteric constraint that reciprocally reduced CD4 binding. However, several 2G12 Abs or Fabs can bind to the spike, and therefore the 2G12 Ab-induced spike inactivation cannot be used to support the coordinated activation model as in the case of the PG9 and PG16 Abs (27). Nevertheless, it has been shown in general that single Ab binding to the HIV-1 spike is enough for neutralization (21). This suggests that the fundamental mechanism underlying stabilization-mediated spike inactivation by neutralizing Abs is based on a coordinated activation process of the individual spike protomers.
ACKNOWLEDGMENTS
We are grateful to Dennis R. Burton for providing the PG9 and PG16 Abs.
Swedish Science Foundation grant 4964, Swedish Cancer Foundation grant 110405, EU FP7-People-ITN-2008 Marie Curie actions Project Virus Entry grants 235649 (to H. G.) and AI93278 (to J. M. B.), and Stiftelsen Läkare mot AIDS Forskningsfond to R.L. supported this work.
Footnotes
Published ahead of print 17 April 2013
REFERENCES
- 1. Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mao Y, Wang L, Gu C, Herschhorn A, Xiang SH, Haim H, Yang X, Sodroski J. 2012. Subunit organization of the membrane-bound HIV-1 envelope glycoprotein trimer. Nat. Struct. Mol. Biol. 19:893–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wu SR, Loving R, Lindqvist B, Hebert H, Koeck PJ, Sjoberg M, Garoff H. 2010. Single-particle cryoelectron microscopy analysis reveals the HIV-1 spike as a tripod structure. Proc. Natl. Acad. Sci. U. S. A. 107:18844–18849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wyatt R, Sodroski J. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884–1888 [DOI] [PubMed] [Google Scholar]
- 6. Chan DC, Fass D, Berger JM, Kim PS. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263–273 [DOI] [PubMed] [Google Scholar]
- 7. Furuta RA, Wild CT, Weng Y, Weiss CD. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat. Struct. Biol. 5:276–279 [DOI] [PubMed] [Google Scholar]
- 8. Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426–430 [DOI] [PubMed] [Google Scholar]
- 9. Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, Katinger H, Parren PW, Robinson J, Van Ryk D, Wang L, Burton DR, Freire E, Wyatt R, Sodroski J, Hendrickson WA, Arthos J. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678–682 [DOI] [PubMed] [Google Scholar]
- 10. Tran EE, Borgnia MJ, Kuybeda O, Schauder DM, Bartesaghi A, Frank GA, Sapiro G, Milne JL, Subramaniam S. 2012. Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog. 8:e1002797 doi: 10.1371/journal.ppat.1002797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xiang SH, Finzi A, Pacheco B, Alexander K, Yuan W, Rizzuto C, Huang CC, Kwong PD, Sodroski J. 2010. A V3 loop-dependent gp120 element disrupted by CD4 binding stabilizes the human immunodeficiency virus envelope glycoprotein trimer. J. Virol. 84:3147–3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, Gruber G, Tauer C, Steindl F, Jungbauer A, et al. 1994. Generation of human monoclonal antibodies against HIV-1 proteins: electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retrovir. 10:359–369 [DOI] [PubMed] [Google Scholar]
- 13. Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, Zhu P, Wormald MR, Stanfield RL, Roux KH, Kelly JW, Rudd PM, Dwek RA, Katinger H, Burton DR, Wilson IA. 2003. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300:2065–2071 [DOI] [PubMed] [Google Scholar]
- 14. Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, Ruker F, Katinger H. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67:6642–6647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Scanlan CN, Pantophlet R, Wormald MR, Ollmann Saphire E, Stanfield R, Wilson IA, Katinger H, Dwek RA, Rudd PM, Burton DR. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1→2 mannose residues on the outer face of gp120. J. Virol. 76:7306–7321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Koff WC, Wilson IA, Burton DR, Poignard P. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, O'Dell S, Perfetto S, Schmidt SD, Shi W, Wu L, Yang Y, Yang ZY, Yang Z, Zhang Z, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Simek M, Burton DR, Koff WC, Doria-Rose NA, Connors M, Mullikin JC, Nabel GJ, Roederer M, Shapiro L, Kwong PD, Mascola JR. 2011. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333:1593–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445:732–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Platt EJ, Gomes MM, Kabat D. 2012. Kinetic mechanism for HIV-1 neutralization by antibody 2G12 entails reversible glycan binding that slows cell entry. Proc. Natl. Acad. Sci. U. S. A. 109:7829–7834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang X, Kurteva S, Lee S, Sodroski J. 2005. Stoichiometry of antibody neutralization of human immunodeficiency virus type 1. J. Virol. 79:3500–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Doria-Rose NA, Georgiev I, O'Dell S, Chuang GY, Staupe RP, McLellan JS, Gorman J, Pancera M, Bonsignori M, Haynes BF, Burton DR, Koff WC, Kwong PD, Mascola JR. 2012. A short segment of the HIV-1 gp120 V1/V2 region is a major determinant of resistance to V1/V2 neutralizing antibodies. J. Virol. 86:8319–8323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, O'Dell S, Patel N, SShahzad-ul-Hussan Yang Y, Zhang B, Zhou T, Zhu J, Boyington JC, Chuang GY, Diwanji D, Georgiev I, Kwon YD, Lee D, Louder MK, Moquin S, Schmidt SD, Yang ZY, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Burton DR, Koff WC, Walker LM, Phogat S, Wyatt R, Orwenyo J, Wang LX, Arthos J, Bewley CA, Mascola JR, Nabel GJ, Schief WR, Ward AB, Wilson IA, Kwong PD. 2011. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480:336–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Garlick RL, Kirschner RJ, Eckenrode FM, Tarpley WG, Tomich CS. 1990. Escherichia coli expression, purification, and biological activity of a truncated soluble CD4. AIDS Res. Hum. Retrovir. 6:465–479 [DOI] [PubMed] [Google Scholar]
- 26. Moore PL, Crooks ET, Porter L, Zhu P, Cayanan CS, Grise H, Corcoran P, Zwick MB, Franti M, Morris L, Roux KH, Burton DR, Binley JM. 2006. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J. Virol. 80:2515–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tong T, Crooks ET, Osawa K, Binley JM. 2012. HIV-1 virus-like particles bearing pure env trimers expose neutralizing epitopes but occlude nonneutralizing epitopes. J. Virol. 86:3574–3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Löving R, Kronqvist M, Sjöberg M, Garoff H. 2011. Cooperative cleavage of the R peptide in the Env trimer of Moloney murine leukemia virus facilitates its maturation for fusion competence. J. Virol. 85:3262–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu SR, Sjoberg M, Wallin M, Lindqvist B, Ekstrom M, Hebert H, Koeck PJ, Garoff H. 2008. Turning of the receptor-binding domains opens up the murine leukaemia virus Env for membrane fusion. EMBO J. 27:2799–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Crooks ET, Moore PL, Richman D, Robinson J, Crooks JA, Franti M, Schulke N, Binley JM. 2005. Characterizing anti-HIV monoclonal antibodies and immune sera by defining the mechanism of neutralization. Hum. Antibodies 14:101–113 [PMC free article] [PubMed] [Google Scholar]
- 31. Doores KJ, Burton DR. 2010. Variable loop glycan dependency of the broad and potent HIV-1-neutralizing antibodies PG9 and PG16. J. Virol. 84:10510–10521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Abrahamyan LG, Markosyan RM, Moore JP, Cohen FS, Melikyan GB. 2003. Human immunodeficiency virus type 1 Env with an intersubunit disulfide bond engages coreceptors but requires bond reduction after engagement to induce fusion. J. Virol. 77:5829–5836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Binley JM, Cayanan CS, Wiley C, Schulke N, Olson WC, Burton DR. 2003. Redox-triggered infection by disulfide-shackled human immunodeficiency virus type 1 pseudovirions. J. Virol. 77:5678–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Crooks ET, Jiang P, Franti M, Wong S, Zwick MB, Hoxie JA, Robinson JE, Moore PL, Binley JM. 2008. Relationship of HIV-1 and SIV envelope glycoprotein trimer occupation and neutralization. Virology 377:364–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hu G, Liu J, Taylor KA, Roux KH. 2011. Structural comparison of HIV-1 envelope spikes with and without the V1/V2 loop. J. Virol. 85:2741–2750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. White TA, Bartesaghi A, Borgnia MJ, Meyerson JR, de la Cruz MJ, Bess JW, Nandwani R, Hoxie JA, Lifson JD, Milne JL, Subramaniam S. 2010. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure. PLoS Pathog. 6:e1001249 doi: 10.1371/journal.ppat.1001249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Julien JP, Lee JH, Cupo A, Murin CD, Derking R, Hoffenberg S, Caulfield MJ, King CR, Marozsan AJ, Klasse PJ, Sanders RW, Moore JP, Wilson IA, Ward AB. 2013. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc. Natl. Acad. Sci. U. S. A. 110:4351–4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Finzi A, Xiang SH, Pacheco B, Wang L, Haight J, Kassa A, Danek B, Pancera M, Kwong PD, Sodroski J. 2010. Topological layers in the HIV-1 gp120 inner domain regulate gp41 interaction and CD4-triggered conformational transitions. Mol. Cell 37:656–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sattentau QJ, Moore JP. 1991. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J. Exp. Med. 174:407–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wyatt R, Moore J, Accola M, Desjardin E, Robinson J, Sodroski J. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 69:5723–5733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rusert P, Krarup A, Magnus C, Brandenberg OF, Weber J, Ehlert AK, Regoes RR, Gunthard HF, Trkola A. 2011. Interaction of the gp120 V1V2 loop with a neighboring gp120 unit shields the HIV envelope trimer against cross-neutralizing antibodies. J. Exp. Med. 208:1419–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Salzwedel K, Berger EA. 2000. Cooperative subunit interactions within the oligomeric envelope glycoprotein of HIV-1: functional complementation of specific defects in gp120 and gp41. Proc. Natl. Acad. Sci. U. S. A. 97:12794–12799 [DOI] [PMC free article] [PubMed] [Google Scholar]