Abstract
Varicella-zoster virus (VZV) is the etiological agent of chickenpox and shingles. Due to the virus's restricted host and cell type tropism and the lack of tools for VZV proteomics, it is one of the least-characterized human herpesviruses. We generated 251 monoclonal antibodies (MAbs) against 59 of the 71 (83%) currently known unique VZV proteins to characterize VZV protein expression in vitro and in situ. Using this new set of MAbs, 44 viral proteins were detected by Western blotting (WB) and indirect immunofluorescence (IF); 13 were detected by WB only, and 2 were detected by IF only. A large proportion of viral proteins was analyzed for the first time in the context of virus infection. Our study revealed the subcellular localization of 46 proteins, 14 of which were analyzed in detail by confocal microscopy. Seven viral proteins were analyzed in time course experiments and showed a cascade-like temporal gene expression pattern similar to those of other herpesviruses. Furthermore, selected MAbs tested positive on human skin lesions by using immunohistochemistry, demonstrating the wide applicability of the MAb collection. Finally, a significant portion of the VZV-specific antibodies reacted with orthologs of simian varicella virus (SVV), thus enabling the systematic analysis of varicella in a nonhuman primate model system. In summary, this study provides insight into the potential function of numerous VZV proteins and novel tools to systematically study VZV and SVV pathogenesis.
INTRODUCTION
Varicella-zoster virus (VZV) belongs to the alphaherpesvirus subfamily and is the only member of the genus Varicellovirus that can infect humans (1, 2). Primary infection causes chickenpox and typically occurs in early childhood with a prominent, highly contagious vesicular rash (3). During primary infection, VZV establishes latency in sensory trigeminal and dorsal root ganglia. Reactivation from latency results in a secondary disease called herpes zoster (HZ), or shingles, that is more common in elderly people (4). HZ most frequently occurs in the thoracic or lumbar nerve segments and the distribution area of the trigeminal nerve, causing a painful rash in the corresponding dermatome. While the molecular mechanism for reactivation from latency is not well characterized, it is more frequent in immunocompromised patients (5). The most common sequela of HZ is postherpetic neuralgia (PHN). In addition, VZV reactivation can lead to zoster ophthalmicus, acute retinal necrosis, meningitis, and vasculopathy (6).
The seroprevalence of VZV differs significantly between countries, but the majority of individuals are seropositive by the time of adolescence (7). While in otherwise healthy children and adolescents, primary VZV infection mostly resolves spontaneously without sequelae, severe symptoms may occur in immunocompromised people and during pregnancy (6). Vertical transmission of VZV during the first trimester causes congenital varicella syndrome (CVS), which is characterized by skin lesions, hypoplasia, low birth weight, and neurological disorders, while perinatal infection at the time of delivery leads to neonatal varicella, with high morbidity and mortality rates. Chickenpox and shingles vaccines based on the live-attenuated VZV v-Oka strain have been developed, and the former has been introduced into childhood immunization schemes in several countries (8). However, the increased incidence of HZ-related complications within the expanding elderly population calls for more effective ways to prevent primary infection with VZV and to limit reactivation from latency.
With a genome of approximately 125,000 bp containing 74 open reading frames (ORFs), 3 of which are duplicated (ORF62/71, ORF63/70, and ORF64/69), leaving 71 unique ORFs, VZV is the smallest human herpesvirus (9–11). A recent report in which the genome was systematically mutagenized determined that 44 of 71 VZV ORFs are essential for viral replication (12). VZV contains 5 unique genes (ORF1, ORF2, ORF13, ORF32, and ORF57) not present in herpes simplex virus 1 (HSV-1) and lacks 15 genes expressed by HSV-1 (9, 11, 13). Like all herpesviruses, the VZV virion consists of a nucleocapsid that harbors the double-stranded DNA genome surrounded by a tegument protein layer and a host-derived plasma membrane called the envelope, containing viral glycoproteins. Nucleocapsids formed in the infected nucleus are thought to gain access to the cytoplasm by budding through the nuclear envelope and receive their secondary envelope at the trans-Golgi network (TGN). Considering the high infectivity of VZV virions released from cutaneous lesions, it is surprising that in vitro VZV propagation is highly cell associated. This is caused at least in part by diversion of newly formed virions from the TGN to late endosomes (14). VZV infection is restricted to humans, and consequently, an appropriate animal model is lacking. Experimental inoculation of mice, rats, and nonhuman primates with VZV leads to seroconversion but not to disease resembling chickenpox and herpes zoster (15). This limitation is partially overcome by a SCID-humanized mouse model in which fetal human tissue is grafted and subsequently infected with VZV (16). More recent research revealed that infection of nonhuman primates with simian varicella virus (SVV) recapitulates most features of VZV infection in humans (3, 17–19). SVV is a member of the Varicellovirus genus along with VZV, equine herpesvirus 1 (EHV-1), EHV-4, pseudorabies virus (PRV), Marek's disease virus (MDV), and bovine herpesvirus 1 (BHV-1). Monkeys infected with SVV develop an exanthema mimicking chickenpox, which is spontaneously resolved, leading to latency in ganglia along the entire neuroaxis as well as the induction of SVV-specific B- and T-cell responses (17–19).
Due to its highly cell-associated character in vitro as well as the lack of appropriate animal models and virus-specific tools such as monoclonal antibodies (MAbs), many aspects of the VZV life cycle are currently still poorly understood. Previously, only 8 monoclonal and 29 polyclonal antibodies against 37 VZV proteins were described. To provide these tools and to be able to further investigate the molecular pathogenesis of VZV, we used a VZV ORF clone collection made recently (20–23) to generate a genome-scale MAb collection that was subsequently used to perform a comprehensive analysis of VZV proteins.
MATERIALS AND METHODS
Viruses and cells.
Uninfected and infected MeWo human melanoma cells (ATCC HTB-65) and the ARPE-19 human retinal pigment epithelial cell line (ATCC CRL-2302) were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS), l-glutamine, and antibiotics. To generate stable mouse hybridomas, SP2/O myeloma cells (ATCC CRL 1581) cultured in supplemented or plain RPMI 1640 medium were used. The VZV pOKA strain was used as the PCR template and for all infections (24). VZV was propagated by adding pOKA-infected MeWo cells to uninfected MeWo cells at a ratio of 1:50. Cell-free VZV strain EMC-1 was obtained as described previously (25, 26) and used for the infection of ARPE-19 cells. Telomerized rhesus fibroblasts (TRFs) were maintained in DMEM supplemented with 10% FCS and antibiotics. The recombinant SVV delta, in which enhanced green fluorescent protein (eGFP) was inserted between US2 and US3 through homologous recombination (27), was propagated on TRFs by inoculating uninfected cells with SVV-infected TRFs at a ratio of 1:10.
Generation of expression vectors.
An ORFeome-wide pOKA VZV entry library that was previously generated by using Gateway recombinational cloning (20) was subcloned into pETG-based vectors encoding N-terminally 6×His-, C-terminally 6×His-, or N-terminally maltose-binding protein (MBP)-tagged proteins. T7-driven expression of VZV proteins was performed in Escherichia coli BL21(DE3) and Rosetta DE3 cells. For baculovirus-based expression, VZV ORFs were subcloned into the DEST10 vector, providing an N-terminal 6×His tag (Invitrogen), by using the Gateway recombinational system.
Protein expression and purification.
BL21(DE3) and Rosetta DE3 cells were used for bacterial expression of 6×His- or MBP-tagged VZV proteins. Twenty milliliters of a culture of BL21(DE3) or Rosetta DE3 cells transformed with expression vectors encoding VZV ORFs and grown overnight was inoculated into 1 liter of selective LB medium (50 μg/ml of ampicillin, 50 μg/ml of chloramphenicol) and grown at 37°C until an optical density at 600 nm (OD600) of 0.6 was reached. Protein expression was induced by addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and further shaking of the culture for 4 to 6 h at 37°C. Alternatively, proteins were produced by using the baculovirus system essentially according to the manufacturer's protocol (Invitrogen). In brief, to reconstitute baculovirus, Sf9 cells (ATCC CRL-1711) were seeded into 6-well plates (900,000 cells/well), incubated at 27°C for 2 h, and transfected with DEST10 containing VZV ORF coding sequences (1 μg/well) and Cellfectin reagent (6 μl). Cells were maintained in Grace's insect cell culture medium either nonsupplemented for transfection or supplemented for cell growth. Three days later, the supernatant containing reconstituted virus was collected (P1 virus stock) and used to generate the P2 virus stock on Sf9 insect cells. The titration of the P2 stock was done by using serial dilutions, and baculovirus plaques were counted by using trypan blue. Protein production was monitored by Western blotting (WB) at 24 h, 48 h, and 72 h postinfection (hpi), using multiplicities of infection (MOIs) of 5 and 20.
To generate cell lysates, bacterial cells or baculovirus-infected cells were harvested, resuspended in 40 ml of lysis buffer (50 mM Tris-HCl [pH 6.8], 0.3 M NaCl, 6 M guanidinium hydrochloride, and 5 mM imidazole), and lysed by sonication. Soluble proteins were recovered by centrifugation and incubated with 1 ml of preequilibrated Ni-nitrilotriacetic acid (NTA) agarose beads. Following incubation of the protein solution for 30 min, the slurry was allowed to drain by gravity and washed extensively with urea buffer with stepwise increasing concentrations of imidazole (0, 10, and 50 mM imidazole). Finally, the protein was eluted 3 times with 500 μl of elution buffer (50 mM Tris-HCl [pH 6.8], 0.3 M NaCl, 4 M urea, and 450 mM imidazole). Eluted fractions and samples of solutions were analyzed by SDS-PAGE and WB.
Generation of monoclonal antibodies.
BALB/c mice were injected subcutaneously with recombinantly expressed protein (50 μg) or a peptide conjugated to keyhole limpet hemocyanin (KLH) (50 μg) in complete Freund's adjuvant. Two weeks later, mice were boosted with the same protein in incomplete Freund's adjuvant by injecting a two-thirds volume subcutaneously and a one-third volume intraperitoneally (i.p.). After an additional 2-week period, the sera of immunized mice were screened for antibody titers against the immunogen by using an enzyme-linked immunosorbent assay (ELISA). The best responders were additionally boosted i.p. with the immunogen dissolved in phosphate-buffered saline (PBS). Three days later, spleen cells were collected and, after lysis of red blood cells, fused with SP2/0 myeloma cells at a ratio of 1:1. The cells were seeded onto 96-well tissue culture plates in 20% RPMI 1640 medium containing hypoxanthine, aminopterin, and thymidine for hybridoma selection. The cultures were screened for MAbs reactive against immunogens by using an ELISA. Positive mother wells were expanded and cloned.
Indirect immunofluorescence.
Uninfected and infected MeWo cells were cultured at a ratio of 50:1 on glass coverslips for 48 h and subsequently processed for immunofluorescence (IF) essentially as described previously (28). Nonspecific binding was blocked by incubating cells for 1 h with PBS containing 10% goat serum and 0.3% Triton X-100. In the case of sample preparation for confocal microscopy, 0.2 mg human IgG/ml (Sigma) and 0.3% Triton X-100 were used for blocking. Cells were incubated for 2 to 3 h at room temperature (RT) with MAb supplemented with 3% goat serum or human IgG and 0.3% Triton X-100. For confocal microscopy, antibodies recognizing lamin B (Santa Cruz) and TGN46 (AbD Serotec) were used in parallel to mark the nuclear envelope and trans-Golgi network (TGN), respectively. After several washes, the samples were incubated with Alexa 488- or Alexa 594-conjugated secondary reagents (Invitrogen). Nuclear chromatin was visualized by DAPI (4′,6-diamidino-2-phenylindole) staining. Finally, cells were embedded in mounting medium (Vectashield) and examined by using a Leica fluorescence microscope at a magnification of ×40. Alternatively, an SP5 confocal microscope was used.
Western blot analysis.
Total lysates of mock- or pOKA-infected MeWo cells were separated on 6 to 15% SDS-PAGE gels. Upon transfer onto nitrocellulose, membranes were incubated with MAb followed by goat anti-mouse antibodies conjugated to horseradish peroxidase (Dianova). Binding of antibodies was detected by enhanced chemiluminescence (ECL) (GE Healthcare). Mock- and SVV-infected TRFs were lysed at 48 hpi by adding 2× reducing Laemmli sample buffer and cleared by using QIAshredder columns (Qiagen). Proteins were resolved on 6 to 15% SDS-PAGE gels and transferred onto nitrocellulose membranes, which were incubated with the MAb followed by goat anti-mouse antibody conjugated to horseradish peroxidase (Santa Cruz). Specific staining was detected by ECL (Thermo Scientific). Cell-free VZV was obtained as described previously (25, 26). Briefly, ARPE-19 human retinal pigment epithelial cells were infected with a clinical VZV isolate (EMC-1) and scraped when the cells showed 30 to 50% cytopathic effect (25, 26). Infected cells were sonicated 3 times for 15 s, clarified for 15 min at 1,000 × g, and frozen at −80°C until use. For kinetic analysis, 5 × 105 ARPE-19 cells/well were seeded into 12-well plates and infected with cell-free VZV EMC-1 (passage 30) at an MOI of 0.12, followed by spin inoculation (20 min at 1,000 × g at RT). After incubation of cells at 37°C for 40 min, the virus inoculum was washed away, and the cells were harvested at the indicated time points. To prepare lysates, cells were washed with PBS, trypsinized, spun down, and, after another wash step, lysed in radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitors (Roche Applied Science). Western blots were probed with MAbs recognizing VZV ORF4, ORF8, ORF18, ORF24, ORF48, ORF63, and ORF68 or an actin-specific MAb to standardize the amount of protein loaded, followed by peroxidase-labeled secondary antibodies, and developed by using a Western blot imaging system (UVItec).
Immunohistology.
Punch biopsy specimens of suspected herpes zoster skin lesions were obtained for diagnostic purposes. Control human skin tissue included was breast skin from a healthy woman undergoing breast reconstruction. The tissue derived from herpes zoster skin lesions was confirmed to be VZV DNA positive by successive in-house PCR analyses (29). Punch biopsy specimens were snap-frozen in N2. Frozen skin sections (6 μm thick) were fixed in acetone containing 0.3% hydrogen peroxide for 10 min (30). Sections were stained for VZV proteins by using the newly developed MAbs directed to VZV ORF4, ORF21, ORF62, ORF63, and ORF66. Negative and positive controls included appropriate isotype controls and well-described anti-VZV MAbs directed to VZV ORF62 (clone 8616; Millipore) and ORF63 (clone 9D12; a generous gift from C. Sadzot-Delvaux, Liège, Belgium) (31), respectively. Antibody staining was visualized by using an avidin-biotin complex system (DakoCytomation) and 3-amino-9-ethylcarbazole (AEC; Sigma-Aldrich) as the substrate. Sections were counterstained with hematoxylin (Sigma-Aldrich). Study procedures to acquire and process the skin biopsy specimens were performed in compliance with Dutch laws and institutional guidelines and in accordance with the ethical standards of the Declaration of Helsinki.
RESULTS
Generation of monoclonal antibodies against VZV proteins.
To express VZV proteins for immunization, each of the 71 known and unique ORFs was expressed (i) as a full-length protein and/or fragment, (ii) with different N- and C-terminal, large and peptide tags, or (iii) with different expression systems. In summary, 160 plasmid constructs were generated for expression with an N- or C-terminal His tag (99 full-length constructs and 61 fragments), and 94 constructs were constructed for expression with an N-terminal MBP tag (65 full-length constructs and 29 fragments) in E. coli, along with 19 constructs for expression by baculovirus recombinants. With this approach, we were able to express 62 distinct viral proteins in amounts sufficient for immunization of mice. For an additional six ORFs, which could not be expressed by using any of the above-mentioned methods, peptides derived from their primary sequence were used as immunogens to generate MAbs, as described in Materials and Methods. In this study, we generated 251 novel MAbs against 59 of 71 (83%) unique VZV proteins, including 24 ORFs that are covered exclusively by this new MAb collection (Fig. 1). This enabled us to perform an extended analysis of VZV protein expression during lytic infection.
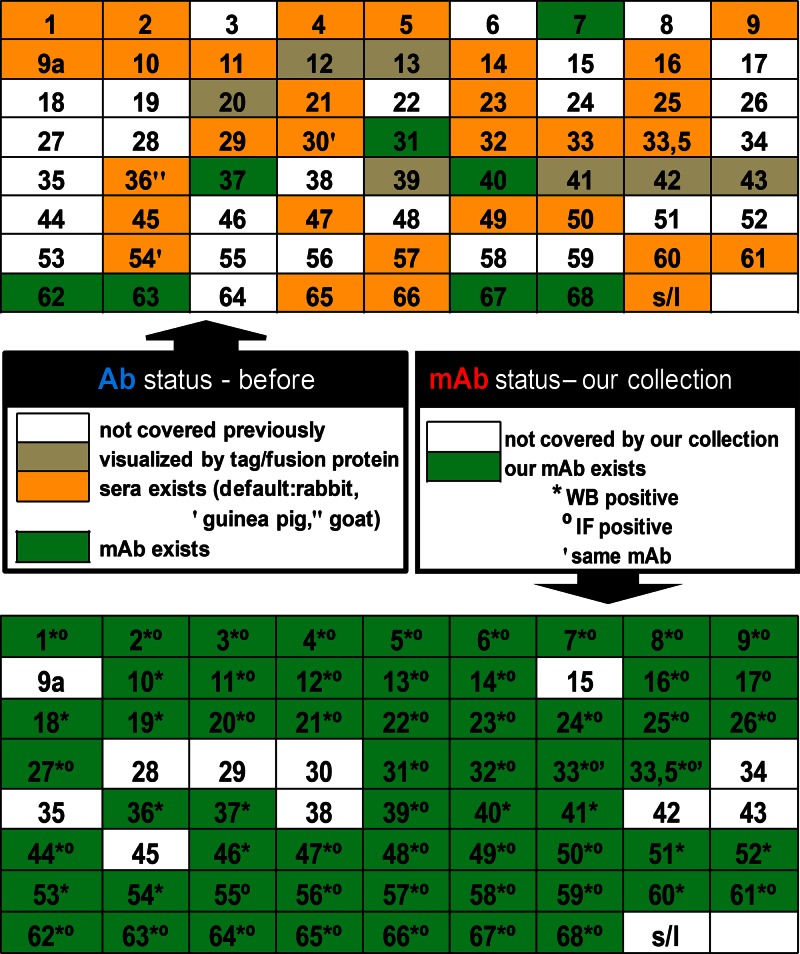
Fig 1.
Coverage of the novel VZV-specific monoclonal antibody collection. To summarize all available antibodies to VZV proteins, several search engines were applied. Coverage of 71 VZV ORFs by currently available antibodies (top) and monoclonal antibodies newly generated with our collection (bottom) is presented. In addition, in the top panel, ORFs previously visualized via tags or fusion proteins are noted.
Detection of VZV proteins in lytically infected cells in vitro.
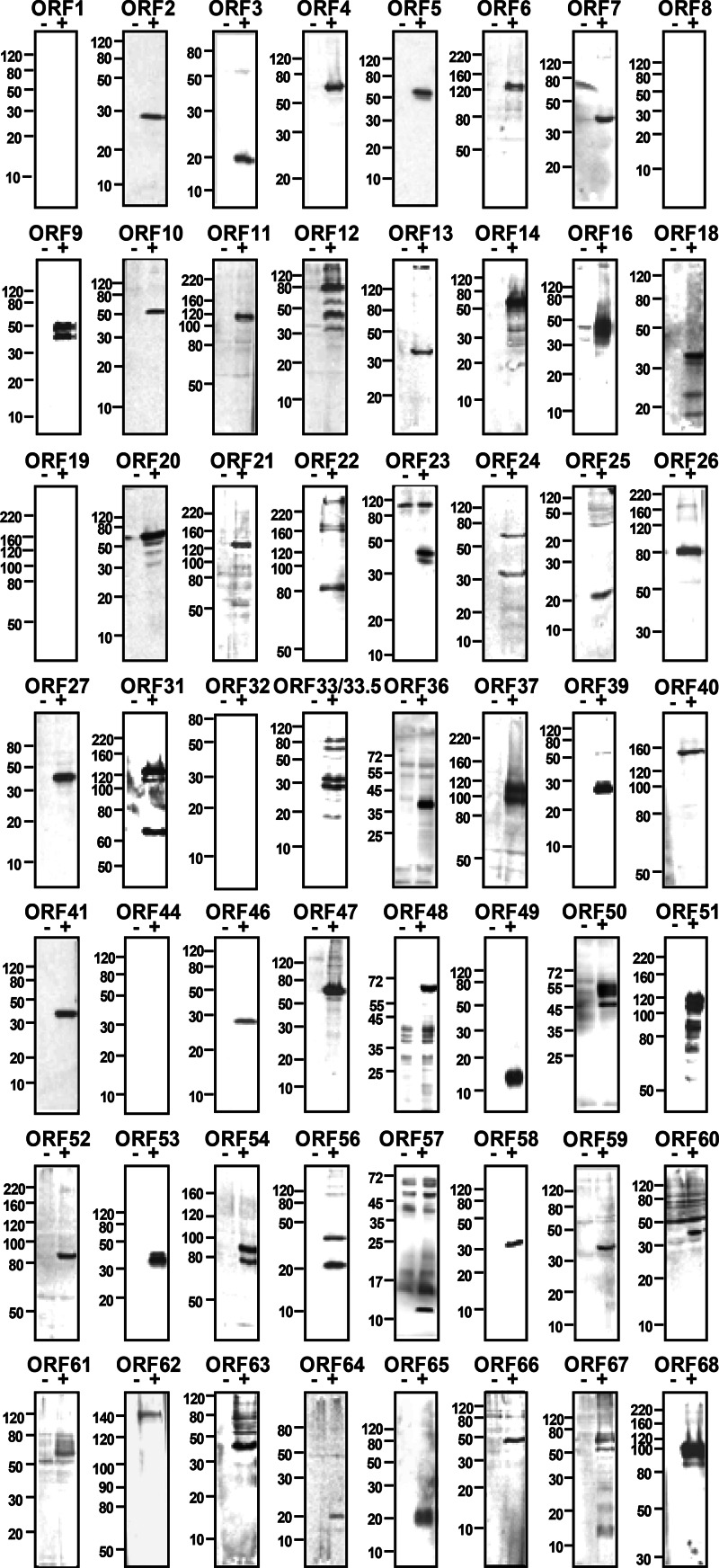
Due to the lack of specific antibodies, expression of numerous VZV proteins has remained obscure in the context of lytic infection. Using the new set of 251 VZV-specific MAbs, we validated the MAbs and determined the molecular weights of VZV proteins by WB analysis of infected MeWo cell lysates (Fig. 2). A large number of the 251 ELISA-positive hybridoma clones were reactive in WB experiments and detected 57 of 71 (80%) unique VZV proteins. All MAb-detected proteins were found exclusively in infected but not in uninfected cell lysates. The scaffold proteins ORF33 and ORF33.5 were recognized by the same MAb but could be distinguished by WB according to their molecular weight. In general, the migration of VZV proteins in SDS-PAGE gels correlated well with the predicted molecular weight, with only few exceptions. This is probably caused by proteolytic processing, a highly basic or acidic character, or posttranslational modification. This was particularly evident for the glycoproteins ORF5, ORF31, ORF37, and ORF60.
Fig 2.
Western blot analysis of VZV proteins expressed during lytic infection. Lysates of uninfected (−) or infected (+) cells were prepared at 2 days postinfection and analyzed by Western blotting using VZV-specific MAbs raised against the indicated ORFs. Molecular mass markers (in kDa) are shown on the left.
Kinetics of VZV protein expression during lytic infection in vitro.
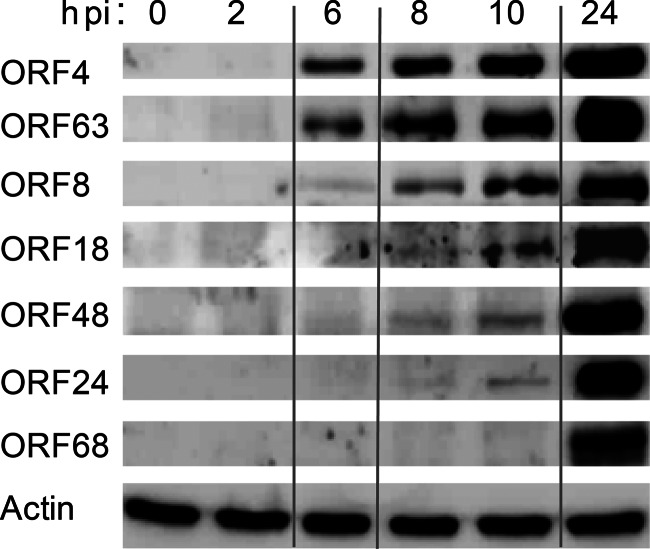
To determine the temporal gene expression level of VZV, an improved protocol was applied for the production of cell-free VZV that facilitated infectious virus titers of up to 5 × 105 PFU per ml (25, 26). Cell-free VZV was used to spin inoculate ARPE-19 cells, a human retinal pigment epithelial cell line, at an approximate MOI of 0.12. Cell lysates were prepared at 0 to 24 hpi and analyzed by WB (Fig. 3). Viral ORF4 and ORF63 products were detected at 6 hpi, as expected for transactivators generally expressed as immediate early (IE) proteins. Antibodies specific for proteins encoded by ORF8 (dUTPase), ORF18 (ribonucleotide reductase, small subunit), and ORF48 (DNase) were detected at between 8 and 10 hpi, typical for proteins involved in viral DNA replication at an early (E) phase of viral infection. The ORF24 product, the VZV ortholog of the HSV-1 nuclear egress protein UL34, was expressed with an early-late kinetic, while glycoprotein gE, encoded by ORF68, was detectable at 24 hpi, thereby exhibiting late kinetics. In conclusion, cascade-like VZV protein expression could be observed in in vitro lytic VZV infection experiments using cell-free virus.
Fig 3.
Kinetics of VZV protein expression during lytic infection. Shown is a time course analysis of VZV gene expression by Western blotting (WB) in in vitro infection assays with cell-free virus. To monitor infection with cell-free virus, ARPE-19 cells were spin inoculated at an MOI of 0.12. At the indicated time points, cell lysates were prepared and analyzed by WB using MAbs directed to VZV ORF4, ORF63, ORF8, ORF18, ORF48, ORF24, and ORF68. Equal amounts of lysates were controlled with an actin-specific antibody.
Subcellular distribution of VZV proteins during lytic in vitro infection.
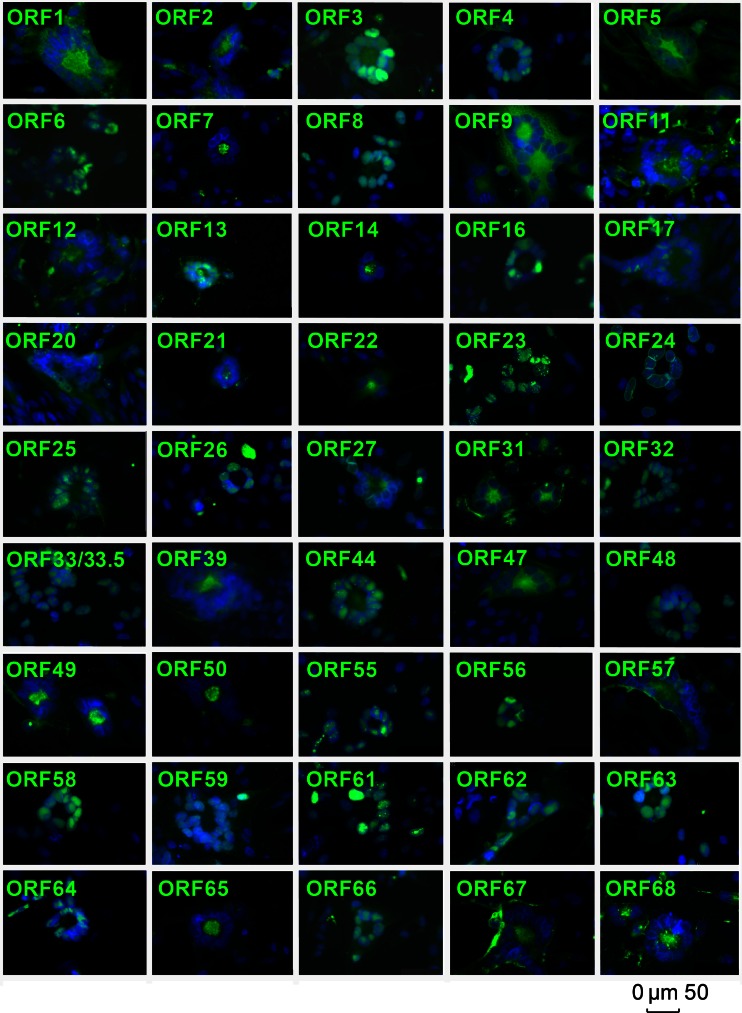
To determine the subcellular localization of viral proteins in VZV-infected cells, uninfected MeWo cells were cocultured with VZV-infected MeWo cells at a ratio of 50:1 for 48 h and subsequently analyzed by IF. In total, the subcellular localization of 46 of 71 (65%) unique VZV proteins could be determined. Screening of subcellular localizations of individual VZV proteins was analyzed in moderately sized syncytia at a rather late stage of infection (Fig. 4 and Table 1).
Fig 4.
Subcellular localization of VZV proteins in infected cells. VZV-infected cells were analyzed by indirect immunofluorescence at 2 days postinfection by using the indicated VZV MAbs and an Alexa 488-conjugated secondary antibody. Nuclei were counterstained with DAPI, and pictures were obtained by using a Leica fluorescence microscope. The size bar corresponds to 50 μm.
Table 1.
Varicella-zoster virus ORFsa
| VZV gene | Molecular mass (kDa) |
Function | Localization in infected cells | HSV-1 ortholog | |
|---|---|---|---|---|---|
| Calculated | Apparent | ||||
| Orf0(S/L) | 14.1 | Membrane-associated protein | NA | UL56 | |
| Orf1 | 12.1 | 16–20 | Tail-anchored membrane phosphoprotein | Golgi/M | None |
| Orf2 | 24.1 | 28 | Membrane-associated phosphoprotein | Golgi network | None |
| Orf3 | 19.1 | 19 | Potential role in gene expression | N≫C | UL55 |
| Orf4 | 51.5 | 60 | Transactivator | N | UL54 |
| Orf5 | 38.6 | 55 | Glycoprotein K | Golgi/M | UL53 |
| Orf6 | 122.6 | 125 | DNA helicase-primase complex | N | UL52 |
| Orf7 | 28.2 | 35 | Virion phosphoprotein | Golgi network | UL51 |
| Orf8 | 44.8 | 50 | dUTPase | N≫C | UL50 |
| Orf9 | 32.9 | 35 + 50 | Tegument protein | C | UL49 |
| Orf9a | 9.8 | Glycoprotein N | NA | UL49a | |
| Orf10 | 46.6 | 50 | Transactivator | NA | UL48 |
| Orf11 | 91.8 | 110 | Tegument protein | Golgi/M | UL47 |
| Orf12 | 74.3 | 80b | Tegument protein | Golgi/M | UL46 |
| Orf13 | 34.5 | 35 | Thymidylate synthetase | Golgi/M | None |
| Orf14 | 61.4 | 70 | Glycoprotein C | Golgi network | UL44 |
| Orf15 | 44.5 | Membrane protein | NA | UL43 | |
| Orf16 | 46.1 | 46 | DNA polymerase-associated protein | N≫C | UL42 |
| Orf17 | 51.4 | Host shutoff protein | N≫C | UL41 | |
| Orf18 | 35.4 | 36 | Ribonucleotide reductase (small SU) | NA | UL40 |
| Orf19 | 86.8 | 90 | Ribonucleotide reductase (large SU) | NA | UL39 |
| Orf20 | 54 | 55 | Capsid protein | N≫C | UL38 |
| Orf21 | 115.8 | 120 | Tegument protein | Golgi/M | UL37 |
| Orf22 | 306.3 | 310b | Tegument protein | Golgi/M | UL36 |
| Orf23 | 24.4 | 35 | Capsid protein | N | UL35 |
| Orf24 | 23.8 | 32 + 60 | Nuclear egress complex | N | UL34 |
| Orf25 | 17.5 | 20 | DNA packaging protein | N≫C | UL33 |
| Orf26 | 65.7 | 80 | Virion protein | N | UL32 |
| Orf27 | 35.7 | 40 | Nuclear egress complex | N | UL31 |
| Orf28 | 134.1 | DNA polymerase | NA | UL30 | |
| Orf29 | 132.1 | SS-DNA binding protein | NA | UL29 | |
| Orf30 | 87 | Virion protein | NA | UL28 | |
| Orf31 | 105.4 | 65 + 125 | Glycoprotein B | Golgi/M | UL27 |
| Orf32 | 16 | 18 + 20 | Potential regulatory role in replication | N | None |
| Orf33 | 66 | 30/34 + 66/80b | Protease, scaffold | N | UL26 |
| ORF33.5 | 32.8 | 30/34b | Scaffold | N | UL26.5 |
| Orf34 | 65.2 | Virion protein | NA | UL25 | |
| Orf35 | 29 | Virion protein | NA | UL24 | |
| Orf36 | 37.8 | 38 | TK | NA | UL23 |
| Orf37 | 93.7 | 100 + 110 | Glycoprotein H | NA | UL22 |
| Orf38 | 60.4 | Tegument protein | NA | UL21 | |
| Orf39 | 25.1 | 28 | Membrane protein | Golgi/M | UL20 |
| Orf40 | 155 | 155 | Major capsid protein | NA | UL19 |
| Orf41 | 34.4 | 35 | Capsid protein | NA | UL18 |
| Orf42 | 82.8 | Terminase | NA | UL15 | |
| Orf43 | 74 | Tegument protein | NA | UL17 | |
| Orf44 | 40.3 | 41b | Tegument protein | N≫C | UL16 |
| Orf45 | 40.2 | DNA packaging protein | NA | UL15 | |
| Orf46 | 22.6 | 28 | Minor tegument protein | NA | UL14 |
| Orf47 | 57.4 | 60 | S/T kinase | C | UL13 |
| Orf48 | 61.3 | 65 | DNase | N | UL12 |
| Orf49 | 8.9 | 15 | Myristylated virion protein | Golgi/M | UL11 |
| Orf50 | 48.7 | 50–55 | Glycoprotein M | Golgi network | UL10 |
| Orf51 | 94.4 | 120b | Origin binding protein | NA | UL9 |
| Orf52 | 86.4 | 86 | DNA helicase-primase complex | NA | UL8 |
| Orf53 | 37.4 | 38 | Potential role in mitochondrial function | NA | UL7 |
| Orf54 | 86.8 | 90b | Virion protein | NA | UL6 |
| Orf55 | 98.9 | DNA helicase-primase complex | N≫C | UL5 | |
| Orf56 | 21.8 | 22 + 35 | Virion protein | N | UL4 |
| Orf57 | 8.1 | 12 | Membrane-associated protein | Golgi/M | none |
| Orf58 | 25.1 | 30 | Nuclear phosphoprotein | N | UL3 |
| Orf59 | 34.4 | 35 | Uracil DNA glycosylase | N | UL2 |
| Orf60 | 17.6 | 35 | Glycoprotein L | NA | UL1 |
| Orf61 | 50.9 | 55c | Transactivator | N | RL2 |
| Orf62 | 140 | 140 | Major transactivator | N≫C | RS1 |
| Orf63 | 30.5 | 35c | Cotransactivator | N≫C | US1 |
| Orf64 | 19.9 | 20 | Virion protein | N | US10 |
| Orf65 | 11.4 | 20 | Tegument phosphoprotein | Golgi network | US9 |
| Orf66 | 43.7 | 48 | S/T kinase | N≫C | US3 |
| Orf67 | 39.4 | 70b | Glycoprotein I | Golgi/M | US7 |
| Orf68 | 70 | 100b | Glycoprotein E | Golgi/M | US8 |
Calculated and apparent molecular masses, function, and localization as determined by using our antibody collection and a herpes simplex virus 1 ortholog. N, nucleus (nuclear localization includes the nuclear egress complex); C, cytoplasm; N≫C, majority in the nucleus; Golgi/M, Golgi network/membranes; NA, not analyzed; SU, subunit; TK, thymidine kinase; SS-DNA, single-stranded DNA; S/T, serine/threonine.
Possible degradation products or posttranslational processing/alternative translational initiation.
Possible higher-molecular-mass products.
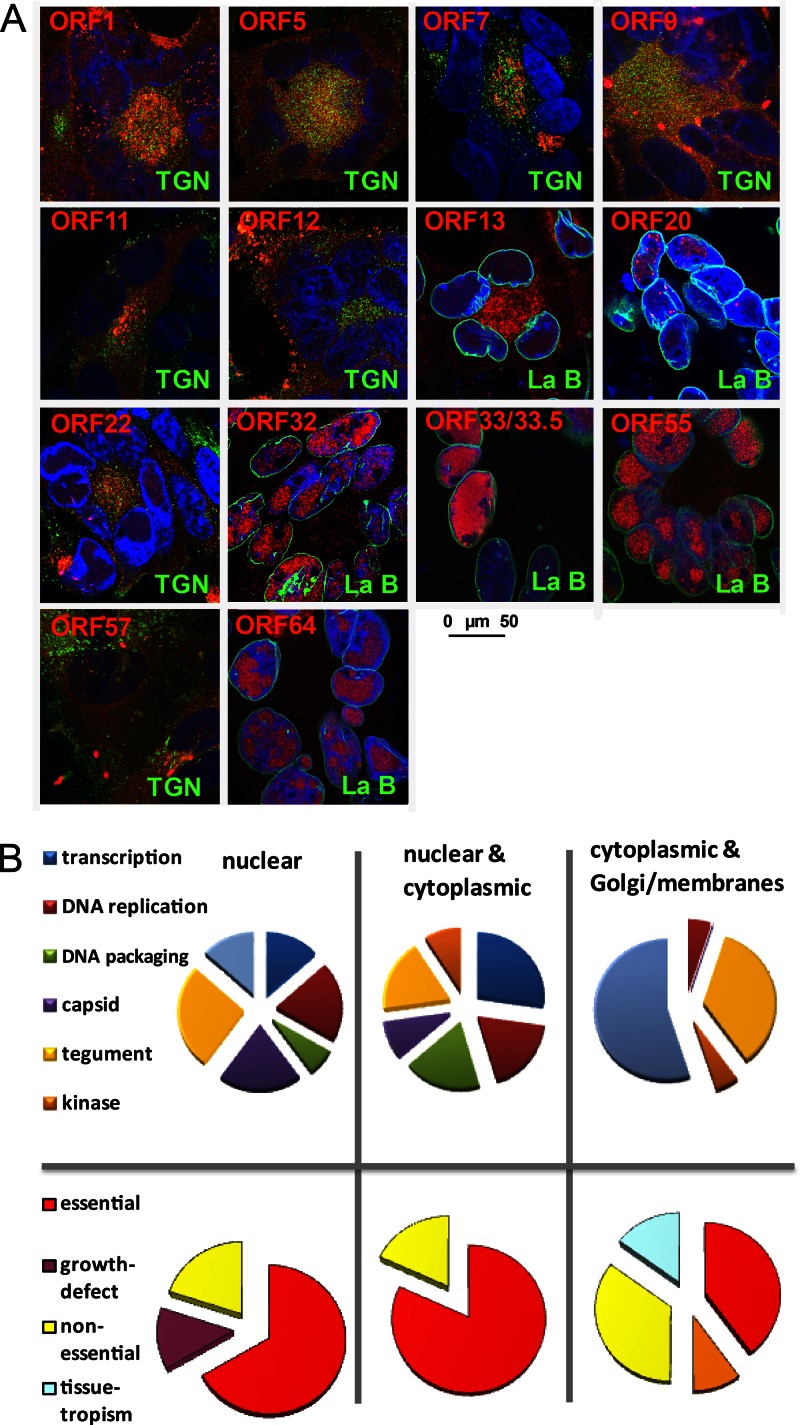
Several representative VZV ORFs were further investigated by high-resolution confocal microscopy (Fig. 5A). Several until now poorly characterized ORFs, four of which are unique to VZV (ORF1, ORF13, ORF32, and ORF57), were analyzed together with ORFs belonging to different localization and functional classes. Capsid proteins (e.g., ORF20), proteins involved in capsid packaging and maturation (e.g., ORF33, ORF33.5, and ORF55), and proteins involved in DNA replication and metabolism, as was proposed for ORF32 (32), were found primarily in the nucleus (Fig. 5A and B). Several representatives of tegument proteins were localized to either the nucleus (e.g., ORF64), both the cytoplasm and center of syncytia/Golgi networks (e.g., ORF7, ORF9, ORF11, ORF12, and ORF22), or the nucleus and cytoplasm (e.g., ORF13), probably depending on their role in primary or secondary envelopment. Glycoproteins, including ORF5 (gK) and the membrane protein ORF1 (33), localized to the Golgi network and other membranes. Similarly, a Golgi network- and membrane-associated localization was observed for ORF57, a poorly characterized VZV protein (34).
Fig 5.
Detailed analysis of localization and function of selected VZV proteins. (A) At 2 days postinfection, VZV-infected cells were stained with MAbs against the VZV ORFs indicated in red and in parallel with antibodies specific to either TGN46 (TGN) or lamin B (La B), indicated in green. VZV MAbs were detected by Alexa 594 (red)-conjugated secondary antibodies, and TGN/lamin B MAbs were detected by Alexa 488 (green)-conjugated secondary antibodies. Nuclei were counterstained with DAPI (blue), and pictures were obtained by using a Leica SP5 confocal microscope. The size bar corresponds to 50 μm. (B) Analysis of VZV ORFs based on their subcellular localization (panel A and Fig. 4) and their function (top) or essentiality (bottom). Nuclear localization includes the nuclear egress complex.
The results concur with knowledge of VZV biology reported in the literature; for example, in comparison to cytoplasmic VZV proteins, a higher percentage of nuclear VZV proteins was found to be essential (P < 0.031 by Fisher's test) (Fig. 5B) (12). The data suggest that VZV proteins involved in DNA replication and morphogenesis are localized primarily in the nucleus of a lytically infected cell.
VZV protein expression in herpes zoster lesions.
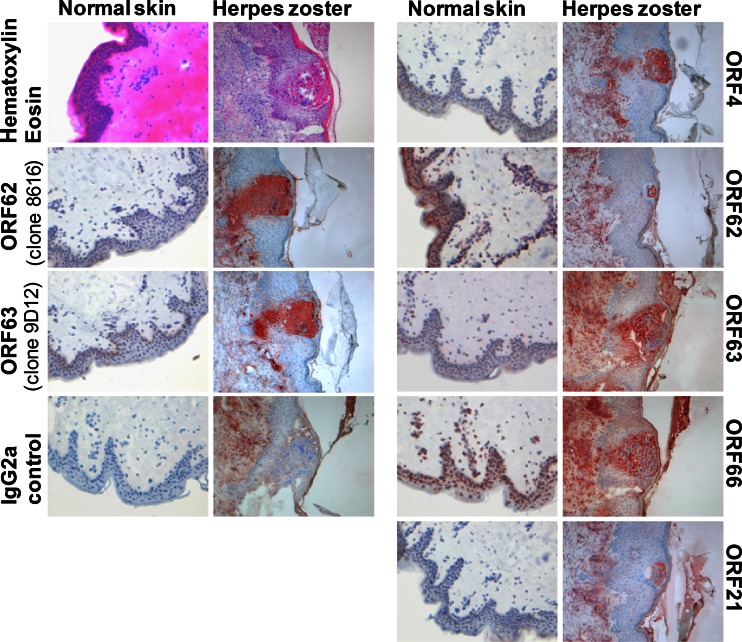
Herpes zoster is a VZV-induced disease characterized by a painful skin rash, generally limited to one side of the body in a dermatomal pattern (4). Detection of VZV DNA and antigens in zoster skin lesions by PCR and immunohistochemistry (IHC), respectively, is of diagnostic value (35, 36). The availability of VZV ORF-specific MAbs for IHC purposes is limited. A selection of the newly developed VZV-specific MAbs was used to study the expression of the respective VZV proteins in snap-frozen tissue sections of a herpes zoster lesion (Fig. 6). Compared to the IgG2a isotype control, all MAbs tested showed specific staining mainly at the border and within the vesicular zoster lesion. The IHC staining pattern of the new ORF62- and ORF63-specific MAbs resembled that of the previously established ORF62 (clone 8616)- and ORF63 (clone 9D12)-specific MAbs (31, 37). As reported previously, positive staining of ORF4, ORF62, and ORF63 occurred in both the nuclei and cytoplasm of epidermal and infundibular keratinocytes (38, 39). Overall, the data demonstrate the applicability of these MAbs for IHC purposes.
Fig 6.
VZV protein expression in herpes zoster skin lesions. Frozen sections of suspected herpes zoster skin lesions and healthy control skin were processed for immunohistochemistry using VZV MAbs directed to ORF4, ORF21, ORF62, ORF63, or ORF66. The previously established anti-VZV MAbs directed to ORF62 (clone 8616) and ORF63 (clone 9D12) were included as positive controls. A MAb (IgG2a subclass) directed to an irrelevant antigen was used as a negative control. To present the overall histology of the herpes zoster and control skin, staining with hematoxylin and eosin is shown. Antibody binding was visualized by using the avidin-biotin complex system and 3-amino-9-ethylcarbazole substrate (red).
Cross-reactivity of anti-VZV MAbs with simian varicella virus proteins.
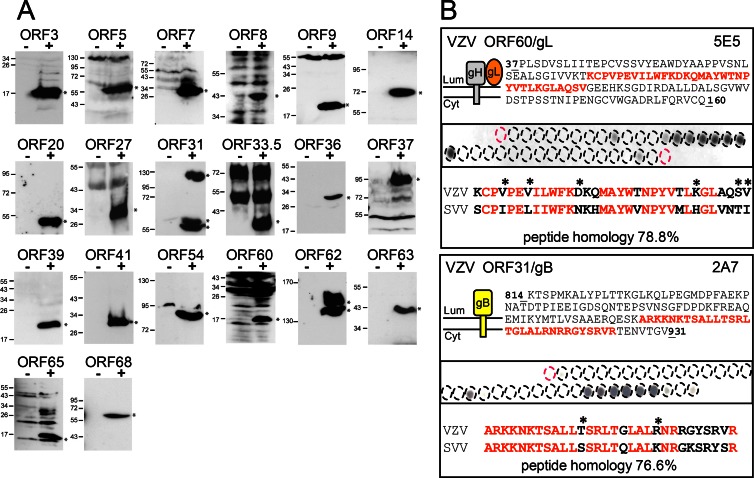
SVV infection of rhesus macaques represents a valuable nonhuman primate model to analyze the immunobiology and pathogenesis of VZV infection (3, 17–19). However, a minority of SVV proteins have been studied until now. The newly developed collection of VZV-specific MAbs may provide novel tools for the biochemical characterization of SVV. Because VZV and SVV exhibit considerable sequence similarity at the protein level (Table 2), suggesting antigenic relatedness (17), we determined the potential cross-reactivity of the VZV MAbs to the orthologous SVV proteins. We analyzed all WB-reactive VZV MAbs using cell lysates prepared from SVV-infected TRFs, and here, we show the cross-reactivity of 20 anti-VZV MAbs with orthologous SVV proteins (Fig. 7). The migration of most of the SVV proteins correlated with the predicted molecular weight based on SVV genome sequence data (Table 2) (17). However, a significant proportion of detected SVV proteins appeared with a different size. The discrepant SVV protein size detected by WB may be caused by posttranslational modification or proteolytic processing of the respective viral proteins. The SVV proteins recognized by cross-reactive VZV were on average not more similar than undetected SVV proteins (17), but the binding sites were highly conserved. The epitopes of the MAbs directed against the ORF31 (gB) and ORF60 (gL) glycoproteins were mapped by using 15-mer synthetic peptides with an offset of 3 residues spotted onto nitrocellulose membranes. In both viral proteins, the MAb-reactive epitopes were linear peptides of 30 and 33 residues in length located within the proteins' cytoplasmic and extracellular domains, respectively. A comparison of these VZV epitopes with the corresponding SVV regions revealed striking sequence similarities of 76.6% (gB) and 78.8% (gL) between both varicelloviruses, which explains the MAbs' cross-reactivity. Moreover, the epitope recognized by the anti-ORF60 MAb seems to be well conserved within all herpesviral gL orthologs (40). In summary, our data show that many VZV-specific MAbs are cross-reactive with SVV, thereby representing new valuable tools for future research on both VZV and SVV pathogenesis.
Table 2.
Hybridoma clones positive in SVV-infected cells by Western blottingb
| SVV clone | Molecular mass (kDa) |
% sequence similarity to VZV | |
|---|---|---|---|
| Calculated | Apparent | ||
| ORF3 | 19.7 | 20 | 63.7 |
| ORF5 | 42 | 55 | 59.3 |
| ORF7 | 25.3 | 30 | 72.9 |
| ORF8 | 44.9 | 45 | 38.2 |
| ORF9 | 33.2 | 20 | 59.4 |
| ORF14 | 60.5 | 72 | 43.8 |
| ORF20 | 52.9 | 53 | 60.5 |
| ORF27 | 35.4 | 35 | 74.4 |
| ORF31 | 104 | 55 + 60 + 125 | 75.4 |
| ORF33/33.5 | 65.1 | 30a | 64.4 |
| ORF36 | 37.9 | 30 | 52.3 |
| ORF37 | 96.8 | 97 | 55.5 |
| ORF39 | 25.4 | 20 | 53.1 |
| ORF41 | 34.3 | 32 | 70.5 |
| ORF54 | 83.5 | 85 | 59.5 |
| ORF60 | 20.2 | 17 + 26a | 43.5 |
| ORF62 | 136.8 | 140 + 160 | 58.0 |
| ORF63 | 29.3 | 40 | 52.0 |
| ORF65 | 9.0 | 17 + 30a | 49.0 |
| ORF68 | 67.6 | 65 | 47.0 |
Possible higher-molecular-mass products.
The sequence similarity was previously described by Gray (17).
Fig 7.
Cross-reactivity of VZV-specific antibodies with SVV proteins in Western blots. (A) To determine if the VZV-specific MAbs recognized SVV proteins, protein lysates were generated from mock- and SVV-infected monkey cells at 48 h postinfection and analyzed by Western blotting using MAbs raised against the indicated VZV ORFs. Molecular mass markers (in kDa) are shown on the left. (B) Synthetic peptides covering the domain of ORF31/gB (residues 814 to 931) and ORF60/gL (residues 37 to 160, shown together with its interacting glycoprotein gH), previously used as immunogens, were synthesized as 15-mers with an offset of 3 residues and coupled to cellulose membranes. Membranes were incubated with ORF-specific antibodies. The topology of these membrane proteins is depicted graphically; the sequence of reactive peptides is shown in red. The epitope region identified in VZV ORF31/gB and ORF60/gL (top) is aligned with the amino acid sequence of the respective ORF31 and ORF60 of SVV (bottom). Asterisks represent conserved but not identical amino acids.
DISCUSSION
In this study, we present the first comprehensive characterization of VZV and, to some extent, SVV proteins expressed in vitro and in situ. Our collection of VZV-specific MAbs covers 83% of the VZV ORFeome. The fact that 40% of the VZV proteins were recognized by using the same MAb clone by both WB and IF, and on occasion by IHC, provides an internal quality control of the collection. However, the true power of the collection lies in the availability of different MAb clones for each VZV ORF, expressing different IgG isotypes, recognizing different epitopes, and supporting different methods, including biochemical, cell-biological, as well as immunohistological studies. We were able to analyze the subcellular distribution of 46 and the expression pattern of 57 VZV proteins, with almost half of them being detected for the first time.
Previous approaches to systematically localize herpesviral proteins in infected cells used transfection-based expression of tagged recombinant viral proteins (41, 42). Since these proteins were expressed in a modified form and individually, which can influence folding and binding to other viral proteins, localization may be altered in the context of a natural lytic infection in vitro. In fact, localization of several VZV proteins differed from that of HSV-1 orthologs, as reported by Salsman and colleagues, which is unlikely to represent dissimilarities between these two alphaherpesvirus species (41). A prominent example is the DNA helicase-primase complex, consisting of ORF6, ORF52, and ORF55 in VZV, and its corresponding orthologous HSV-1 complex, which includes UL52, UL8, and UL5. In our study, the VZV ORF6 component was found to be nuclear, consistent with its function. In contrast, its HSV-1 ortholog UL52 was documented to be pancytoplasmic under isolated expression conditions (41). Similarly, the VZV DNase ORF48 was detected in the nucleus of infected cells, whereas its HSV-1 ortholog UL12 was reported to be cytoplasmic. Thus, our antibody collection enables the direct detection of native and functional viral proteins, which represents the best existing tool for the analysis of their expression, distribution, and dynamics in the course of a lytic VZV infection.
Kinetics of VZV gene expression.
VZV propagates in vitro in a cell-associated manner, forming multinucleated syncytia. Recent advances in the production of cell-free VZV enabled the synchronous infection of cells in vitro (25, 26). Using these reagents, we showed that infection with cell-free VZV causes a lytic infection cycle with a cascade-like protein expression profile of presumed IE, E, and L viral genes. While in cell tracker experiments, expression of VZV ORF4 and ORF63 could be observed as early as 2 hpi (43), gene expression using cell-free virus was delayed, with ORF4 and ORF63 being detected at around 6 hpi. ORF68 (gE), considered an L protein in other herpesviruses, was expressed rather early in cell tracker experiments, as reported by Reichelt and colleagues (43), while it exhibited late kinetics in our study. Thus, infection with cell-free VZV confirms cascade-like protein expression with immediate early, early, and late kinetics. Possibly, formation of syncytia between the naive and inoculum cell leads to untimely flooding of the naive cell with massive amounts of viral proteins, leading to altered expression patterns. Alternatively, the otherwise regulated kinetic expression is overruled by the high virus load provided by the inoculum cells. Thus, cell-based propagation of VZV may more closely reflect the in vivo situation during chickenpox and HZ.
Functional implications.
VZV encodes 5 viral proteins (ORF1, ORF2, ORF13, ORF32, and ORF57) that are missing in the HSV-1 genome. Notably, these VZV ORFs are covered by our VZV-specific MAb collection, which enabled us to study the localization of these viral proteins during lytic infection in vitro. Like ORF1, a tail-anchored membrane protein (33), VZV ORF2 is recruited to the Golgi network and potentially other membranes, consistent with biochemical fractionation experiments (44). Similarly, ORF13, a thymidylate synthetase (TS), was predominantly detected in the center of syncytia and the Golgi network. Interestingly, the interaction of ORF13 with ORF6 (20, 23), a component of the DNA helicase-primase complex, suggests a regulatory interaction between precursor synthesis and DNA encapsidation and implies the partial localization of ORF13 to the nucleus. This observation was confirmed by confocal analysis. ORF32, however, was found primarily in the nucleus, while a fraction may also be found in the cytosol (32). The ORF32-specific MAb detected two bands of 16 to 18 kDa, consistent with phosphorylation by the VZV kinase ORF47 (32). Finally, ORF57 is found in the center of syncytia, where the Golgi network resides. Since biochemical analysis classified ORF57 as a cytosolic protein (34), ORF57 may be part of the VZV tegument recruited for secondary envelopment. In yeast two-hybrid studies, ORF57 interacted with ORF39, a membrane protein orthologous to HSV-1 UL20, and ORF9A, the VZV glycoprotein gN (20, 23). Furthermore, ORF57 interacts with additional proteins that are expected to be recruited to the VZV virion based on HSV orthologs detected in extracellular viral particles (45). These proteins include ORF10 (HSV-1 UL48), ORF18 (UL40), ORF38 (UL21), ORF42 (UL15 terminase), ORF46 (UL14), and ORF62 (RS1) (20–23). Furthermore, with the exception of ORF32, all viral proteins unique to VZV (ORF1, ORF2, ORF13, and ORF57) were either exclusively or predominantly located to the Golgi network. We thus hypothesize that in the course of secondary envelopment, most if not all of the unique VZV proteins are incorporated into the maturing particle, thereby determining critical steps of VZV pathogenesis in vivo.
The majority of the commercially available VZV-specific MAbs (4 of 6) are directed toward the VZV glycoproteins gB, gE, gH, and gI. In addition, polyclonal antibodies were generated for VZV gC, gM, and gL. In contrast, no antibodies were available for the type III transmembrane protein ORF39, the putative homolog of HSV-1 UL20, or gK (ORF5). However, both VZV proteins have been studied by using protein tagging (46, 47). Here we showed that they localized to the center of syncytia/Golgi networks and other membranes. ORF39 migrated according to its calculated molecular weight, consistent with the absence of glycosylation. In contrast to previous reports (48), migration of gK was much more delayed, consistent with the addition of fully matured glycosyl moieties at two acceptor sites. The discrepancy between these studies could be explained by the use of different antibody reagents and their ability to gain access to their epitopes. While our MAb may recognize exclusively mature forms, immunoprecipitation of tagged gK may have preselected an underglycosylated version. Interestingly, the molecular weights of both VZV and SVV gK observed by WB were similar, suggesting that our VZV/SVV cross-reactive MAb recognizes the glycosylated mature form of ORF5 in both VZV and SVV.
Two novel MAbs reactive to VZV gB (ORF31) and gL (ORF60) were chosen for epitope analysis. The gB-specific MAb recognized a single linear stretch of 30 residues enriched in positively charged and hydrophobic residues. Similar to gB, gL and its interacting partner gH are highly conserved among herpesviruses. VZV gH and gL closely resemble their HSV orthologs (49). In general, both glycoproteins are major targets of virus-neutralizing antibodies, involved in inhibiting virus entry and infectivity (50). The epitope analysis revealed that the VZV gL-specific MAb decorated a region highly conserved between gL orthologs (40), suggesting that this MAb will likely recognize gL orthologs of other members of the herpesvirus family. However, the same region is particularly important for the interaction of gL with gH, the transport of the complex to the cell surface, and its function during entry of HSV (51). It may be expected that none of the tested MAbs will show a neutralizing capacity. In the case of gB, it recognizes the cytoplasmic tail, while in the case of gL, the epitope is most likely hidden when the gH/gL complex has formed and is exposed on the cell surface. In line with this, a recently described recombinant MAb with a high capacity to neutralize VZV recognized the VZV gH/gL protein complex but not individual gH or gL proteins (52). The remaining MAbs of the collection that are still incompletely characterized may harbor the potential to neutralize VZV infection.
Cross-reactivity of anti-VZV MAbs with SVV.
Due to the highly restricted host range of VZV compared to that of HSV, detailed analyses of the pathogenesis of VZV in an appropriate experimental animal model are lacking. Recently, the nonhuman primate varicellovirus SVV has received great attention in studies of varicella and herpes zoster pathology in monkeys (53). However, due to the lack of specific reagents, SVV protein characteristics remain ill defined. Western blot analysis revealed that 20 SVV proteins were recognized by the MAbs developed for VZV orthologs. This reflects the sequence similarity between VZV and SVV ORFs, which varies between 27 and 75% amino acid identity (17). The predicted size for SVV proteins is based on published amino acid sequences. However, while attempting to clone certain open reading frames of the virus, we came across sequences that differ from the published SVV sequence. In the case of ORF65, the DNA sequence of this gene contains mistakes leading to a frameshift and, thus, a premature stop codon in the published sequence. The molecular mass of SVV ORF65 was 7 kDa higher than expected based on SVV genome sequence data (54). While, in general, the published sequence is correct, the SVV genomic sequences may need correction in certain regions. In summary, since very few SVV-specific antibodies have been available so far (18, 55, 56), our MAbs will facilitate the analysis of SVV in nonhuman primate models and provide novel insights into SVV pathogenesis.
In summary, the current study presents the generation of a comprehensive set of VZV-specific MAbs that were used here for detailed analyses of the majority of viral proteins expressed during VZV infection in vitro and in situ. The data provide novel insights into the potential function of many uncharacterized VZV proteins and tools to systematically investigate varicellovirus pathogenesis in future studies, which are mandatory to develop new intervention strategies to prevent and treat varicellovirus infections.
ACKNOWLEDGMENTS
This work was supported by the EU FP6 INCO-CT-2006-026278 grant, Establishment of High-Throughput Monoclonal Antibody Production and Hybridoma Bank (CAPRI) (S.J. and J.H.); the EU FP7 REGPOT 229585 grant, Center for Antibody Production Rijeka: Upgrading the Central Research and Service Infrastructure for the South Eastern Region of Europe (CAPRI2010) (S.J.); and the Bayerisches Staatsministerium für Wissenschaft, Forschung und Kunst (Baygene) (J.H.).
We thank Dietlind Rose, Manuel Endesfelder, Filip Marković, and Danijela Baždarić for technical assistance and R. M. Verdijk and E. P. Prens (Erasmus MC) for providing the herpes zoster and human control skin specimens, respectively.
Footnotes
Published ahead of print 17 April 2013
REFERENCES
- 1. Arvin AM. 1996. Varicella-zoster virus. Clin. Microbiol. Rev. 9:361–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arvin AM. 2001. Varicella-zoster virus: molecular virology and virus-host interactions. Curr. Opin. Microbiol. 4:442–449 [DOI] [PubMed] [Google Scholar]
- 3. Moffat J, Ku CC, Zerboni L, Sommer M, Arvin A. 2007. VZV: pathogenesis and the disease consequences of primary infection, p 675–688 In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K. (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom: [PubMed] [Google Scholar]
- 4. Cohen JI. 2007. VZV: molecular basis of persistence (latency and reactivation), p 689–699 In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K. (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom: [PubMed] [Google Scholar]
- 5. Arvin A, Abendroth A. 2007. VZV: immunobiology and host response, p 700–712 In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K. (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom: [PubMed] [Google Scholar]
- 6. Gilden D, Mahalingam R, Nagel MA, Pugazhenthi S, Cohrs RJ. 2011. The neurobiology of varicella zoster virus infection. Neuropathol. Appl. Neurobiol 37:441–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nardone A, de Ory F, Carton M, Cohen D, van Damme P, Davidkin I, Rota MC, de Melker H, Mossong J, Slacikova M, Tischer A, Andrews N, Berbers G, Gabutti G, Gay N, Jones L, Jokinen S, Kafatos G, de Aragon MV, Schneider F, Smetana Z, Vargova B, Vranckx R, Miller E. 2007. The comparative sero-epidemiology of varicella zoster virus in 11 countries in the European region. Vaccine 25:7866–7872 [DOI] [PubMed] [Google Scholar]
- 8. Gershon AA. 2007. Varicella-zoster vaccine, p 1262–1273 In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K. (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom: [PubMed] [Google Scholar]
- 9. Roizman B, Campadelli-Fiume G. 2007. Alphaherpes viral genes and their functions, p 70–92 In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K. (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom: [PubMed] [Google Scholar]
- 10. Cohen JI. 2010. The varicella-zoster virus genome. Curr. Top. Microbiol. Immunol. 342:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baines JD, Pellett PE. 2007. Genetic comparison of human alphaherpesvirus genomes, p 61–69 In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K. (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom: [PubMed] [Google Scholar]
- 12. Zhang Z, Selariu A, Warden C, Huang G, Huang Y, Zaccheus O, Cheng T, Xia N, Zhu H. 2010. Genome-wide mutagenesis reveals that ORF7 is a novel VZV skin-tropic factor. PLoS Pathog. 6:e1000971 doi: 10.1371/journal.ppat.1000971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kaufer BB, Smejkal B, Osterrieder N. 2010. The varicella-zoster virus ORFS/L (ORF0) gene is required for efficient viral replication and contains an element involved in DNA cleavage. J. Virol. 84:11661–11669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen JJ, Zhu Z, Gershon AA, Gershon MD. 2004. Mannose 6-phosphate receptor dependence of varicella zoster virus infection in vitro and in the epidermis during varicella and zoster. Cell 119:915–926 [DOI] [PubMed] [Google Scholar]
- 15. Myers MG, Connelly BL. 1992. Animal models of varicella. J. Infect. Dis. 166(Suppl 1):S48–S50 [DOI] [PubMed] [Google Scholar]
- 16. Ku CC, Besser J, Abendroth A, Grose C, Arvin AM. 2005. Varicella-zoster virus pathogenesis and immunobiology: new concepts emerging from investigations with the SCIDhu mouse model. J. Virol. 79:2651–2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gray WL. 2010. Simian varicella virus: molecular virology. Curr. Top. Microbiol. Immunol. 342:291–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gray WL, Mullis LB, Soike KF. 2001. Expression of the simian varicella virus glycoprotein E. Virus Res. 79:27–37 [DOI] [PubMed] [Google Scholar]
- 19. Messaoudi I, Barron A, Wellish M, Engelmann F, Legasse A, Planer S, Gilden D, Nikolich-Zugich J, Mahalingam R. 2009. Simian varicella virus infection of rhesus macaques recapitulates essential features of varicella zoster virus infection in humans. PLoS Pathog. 5:e1000657 doi: 10.1371/journal.ppat.1000657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Uetz P, Dong YA, Zeretzke C, Atzler C, Baiker A, Berger B, Rajagopala SV, Roupelieva M, Rose D, Fossum E, Haas J. 2006. Herpesviral protein networks and their interaction with the human proteome. Science 311:239–242 [DOI] [PubMed] [Google Scholar]
- 21. Cagney G, Uetz P, Fields S. 2000. High-throughput screening for protein-protein interactions using two-hybrid assay. Methods Enzymol. 328:3–14 [DOI] [PubMed] [Google Scholar]
- 22. Fossum E, Friedel CC, Rajagopala SV, Titz B, Baiker A, Schmidt T, Kraus T, Stellberger T, Rutenberg C, Suthram S, Bandyopadhyay S, Rose D, von Brunn A, Uhlmann M, Zeretzke C, Dong YA, Boulet H, Koegl M, Bailer SM, Koszinowski U, Ideker T, Uetz P, Zimmer R, Haas J. 2009. Evolutionarily conserved herpesviral protein interaction networks. PLoS Pathog. 5:e1000570 doi: 10.1371/journal.ppat.1000570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stellberger T, Hauser R, Baiker A, Pothineni VR, Haas J, Uetz P. 2010. Improving the yeast two-hybrid system with permutated fusions proteins: the varicella zoster virus interactome. Proteome Sci. 8:8 doi: 10.1186/1477-5956-8-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Takahashi M, Otsuka T, Okuno Y, Asano Y, Yazaki T. 1974. Live vaccine used to prevent the spread of varicella in children in hospital. Lancet ii:1288. [DOI] [PubMed] [Google Scholar]
- 25. Schmidt-Chanasit J, Bleymehl K, Rabenau HF, Ulrich RG, Cinatl J, Jr, Doerr HW. 2008. In vitro replication of varicella-zoster virus in human retinal pigment epithelial cells. J. Clin. Microbiol. 46:2122–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harper DR, Mathieu N, Mullarkey J. 1998. High-titre, cryostable cell-free varicella zoster virus. Arch. Virol. 143:1163–1170 [DOI] [PubMed] [Google Scholar]
- 27. Mahalingam R, Wellish M, White T, Soike K, Cohrs R, Kleinschmidt-DeMasters BK, Gilden DH. 1998. Infectious simian varicella virus expressing the green fluorescent protein. J. Neurovirol. 4:438–444 [DOI] [PubMed] [Google Scholar]
- 28. Ott M, Tascher G, Hassdenteufel S, Zimmermann R, Haas J, Bailer SM. 2011. Functional characterization of the essential tail anchor of the herpes simplex virus type 1 nuclear egress protein pUL34. J. Gen. Virol. 92:2734–2745 [DOI] [PubMed] [Google Scholar]
- 29. Remeijer L, Duan R, van Dun JM, Wefers Bettink MA, Osterhaus AD, Verjans GM. 2009. Prevalence and clinical consequences of herpes simplex virus type 1 DNA in human cornea tissues. J. Infect. Dis. 200:11–19 [DOI] [PubMed] [Google Scholar]
- 30. Verjans GM, Hintzen RQ, van Dun JM, Poot A, Milikan JC, Laman JD, Langerak AW, Kinchington PR, Osterhaus AD. 2007. Selective retention of herpes simplex virus-specific T cells in latently infected human trigeminal ganglia. Proc. Natl. Acad. Sci. U. S. A. 104:3496–3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kennedy PG, Grinfeld E, Bontems S, Sadzot-Delvaux C. 2001. Varicella-zoster virus gene expression in latently infected rat dorsal root ganglia. Virology 289:218–223 [DOI] [PubMed] [Google Scholar]
- 32. Reddy SM, Cox E, Iofin I, Soong W, Cohen JI. 1998. Varicella-zoster virus (VZV) ORF32 encodes a phosphoprotein that is posttranslationally modified by the VZV ORF47 protein kinase. J. Virol. 72:8083–8088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Koshizuka T, Sadaoka T, Yoshii H, Yamanishi K, Mori Y. 2008. Varicella-zoster virus ORF1 gene product is a tail-anchored membrane protein localized to plasma membrane and trans-Golgi network in infected cells. Virology 377:289–295 [DOI] [PubMed] [Google Scholar]
- 34. Cox E, Reddy S, Iofin I, Cohen JI. 1998. Varicella-zoster virus ORF57, unlike its pseudorabies virus UL3.5 homolog, is dispensable for viral replication in cell culture. Virology 250:205–209 [DOI] [PubMed] [Google Scholar]
- 35. Nuovo GJ. 2006. The surgical and cytopathology of viral infections: utility of immunohistochemistry, in situ hybridization, and in situ polymerase chain reaction amplification. Ann. Diagn. Pathol. 10:117–131 [DOI] [PubMed] [Google Scholar]
- 36. Muraki R, Baba T, Iwasaki T, Sata T, Kurata T. 1992. Immunohistochemical study of skin lesions in herpes zoster. Virchows Arch. A Pathol. Anat. Histopathol. 420:71–76 [DOI] [PubMed] [Google Scholar]
- 37. Ouwendijk WJ, Flowerdew SE, Wick D, Horn AK, Sinicina I, Strupp M, Osterhaus AD, Verjans GM, Hufner K. 2012. Immunohistochemical detection of intra-neuronal VZV proteins in snap-frozen human ganglia is confounded by antibodies directed against blood group A1-associated antigens. J. Neurovirol. 18:172–180 [DOI] [PubMed] [Google Scholar]
- 38. Nikkels AF, Debrus S, Sadzot-Delvaux C, Piette J, Rentier B, Pierard GE. 1995. Immunohistochemical identification of varicella-zoster virus gene 63-encoded protein (IE63) and late (gE) protein on smears and cutaneous biopsies: implications for diagnostic use. J. Med. Virol. 47:342–347 [DOI] [PubMed] [Google Scholar]
- 39. Huch JH, Cunningham AL, Arvin AM, Nasr N, Santegoets SJ, Slobedman E, Slobedman B, Abendroth A. 2010. Impact of varicella-zoster virus on dendritic cell subsets in human skin during natural infection. J. Virol. 84:4060–4072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fan Q, Lin E, Spear PG. 2009. Insertional mutations in herpes simplex virus type 1 gL identify functional domains for association with gH and for membrane fusion. J. Virol. 83:11607–11615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Salsman J, Zimmerman N, Chen T, Domagala M, Frappier L. 2008. Genome-wide screen of three herpesviruses for protein subcellular localization and alteration of PML nuclear bodies. PLoS Pathog. 4:e1000100 doi: 10.1371/journal.ppat.1000100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sander G, Konrad A, Thurau M, Wies E, Leubert R, Kremmer E, Dinkel H, Schulz T, Neipel F, Sturzl M. 2008. Intracellular localization map of human herpesvirus 8 proteins. J. Virol. 82:1908–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Reichelt M, Brady J, Arvin AM. 2009. The replication cycle of varicella-zoster virus: analysis of the kinetics of viral protein expression, genome synthesis, and virion assembly at the single-cell level. J. Virol. 83:3904–3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sato H, Pesnicak L, Cohen JI. 2002. Varicella-zoster virus open reading frame 2 encodes a membrane phosphoprotein that is dispensable for viral replication and for establishment of latency. J. Virol. 76:3575–3578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Loret S, Guay G, Lippe R. 2008. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J. Virol. 82:8605–8618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Govero J, Hall S, Heineman TC. 2007. Intracellular localization of varicella-zoster virus ORF39 protein and its functional relationship to glycoprotein K. Virology 358:291–302 [DOI] [PubMed] [Google Scholar]
- 47. Hall SL, Govero JL, Heineman TC. 2007. Intracellular transport and stability of varicella-zoster virus glycoprotein K. Virology 358:283–290 [DOI] [PubMed] [Google Scholar]
- 48. Mo C, Suen J, Sommer M, Arvin A. 1999. Characterization of varicella-zoster virus glycoprotein K (open reading frame 5) and its role in virus growth. J. Virol. 73:4197–4207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Grose C, Carpenter JE, Jackson W, Duus KM. 2010. Overview of varicella-zoster virus glycoproteins gC, gH and gL. Curr. Top. Microbiol. Immunol. 342:113–128 [DOI] [PubMed] [Google Scholar]
- 50. Vleck SE, Oliver SL, Reichelt M, Rajamani J, Zerboni L, Jones C, Zehnder J, Grose C, Arvin AM. 2010. Anti-glycoprotein H antibody impairs the pathogenicity of varicella-zoster virus in skin xenografts in the SCID mouse model. J. Virol. 84:141–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chowdary TK, Cairns TM, Atanasiu D, Cohen GH, Eisenberg RJ, Heldwein EE. 2010. Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nat. Struct. Mol. Biol. 17:882–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Birlea M, Owens GP, Eshleman EM, Ritchie A, Traktinskiy I, Bos N, Seitz S, Yevgeniy A, Mahalingam R, Gilden D, Cohrs RJ. 2013. Human anti-varicella-zoster virus (VZV) recombinant monoclonal antibody produced after Zostavax immunization recognizes gH/gL complex and neutralizes VZV infection. J. Virol. 87:415–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mahalingam R, Gilden DH. 2007. Simian varicella virus, p 1043–1050 In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K. (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom: [PubMed] [Google Scholar]
- 54. Gray WL, Starnes B, White MW, Mahalingam R. 2001. The DNA sequence of the simian varicella virus genome. Virology 284:123–130 [DOI] [PubMed] [Google Scholar]
- 55. Ashburn CV, Gray WL. 2002. Expression of the simian varicella virus glycoprotein L and H. Arch. Virol. 147:335–348 [DOI] [PubMed] [Google Scholar]
- 56. Mueller NH, Bos NL, Seitz S, Wellish M, Mahalingam R, Gilden D, Cohrs RJ. 2012. Recombinant monoclonal antibody recognizes a unique epitope on varicella-zoster virus immediate-early 63 protein. J. Virol. 86:6345–6349 [DOI] [PMC free article] [PubMed] [Google Scholar]