Abstract
The adenovirus (Ad) L4-33K protein has been linked to disparate functions during infection. L4-33K is a virus-encoded alternative RNA splicing factor which activates splicing of viral late gene transcripts that contain weak 3′ splice sites. Additionally, L4-33K has been indicated to play a role in adenovirus assembly. We generated and characterized an Ad5 L4-33K mutant virus to further explore its function(s) during infection. Infectivity, viral genome replication, and most viral gene expression of the L4-33K mutant virus are comparable to those of the wild-type virus, except for a prominent decrease in the levels of the late proteins IIIa and pVI. The L4-33K mutant virus produces only empty capsids, indicating a defect in viral DNA packaging. We demonstrate that L4-33K does not preferentially bind to viral packaging sequences in vivo, and mutation of L4-33K does not interfere with the binding of the known viral packaging proteins IVa2, L4-22K, L1-52/55K, and IIIa to the packaging sequences in vivo. Collectively, these results demonstrate that the phenotype of an Ad5 L4-33K mutant virus is complex. The L4-33K protein regulates the accumulation of selective Ad late gene mRNAs and is involved in the proper transition of gene expression during the late phase of infection. The L4-33K protein also plays a role in adenovirus morphogenesis by promoting the packaging of the viral genome into the empty capsid. These results demonstrate the multifunctional nature of the L4-33K protein and its involvement in several different and critical aspects of viral infection.
INTRODUCTION
Adenovirus (Ad) gene expression is complex and tightly regulated (1, 2). Defined by the onset of viral genome replication, the Ad life cycle is divided into early and late phases of infection. Following infection, the immediate early gene E1A is the first to be synthesized and activates expression of the other early genes (E1B, E2, E3, and E4). Ad early proteins counteract host responses to infection, including the induction of apoptosis, a DNA damage response, and aspects of the immune response, as well as set the stage for viral genome replication (1). The initiation of viral DNA replication activates late gene expression. Late regions L1 and L4 are the first late genes of the late gene family (L1 to L5) to be synthesized (3). The role of expression of the L1-52/55K protein during the early stage of the late phase of Ad infection is not understood, although there is a better understanding of the timing of such expression of the L4 gene. At this stage, the L4-22K protein is transcribed from a novel, internal L4 promoter (L4P) embedded in the open reading frame of L4-100K (4). L4-22K activates the expression of the full panel of Ad late genes at the transcriptional level by binding to the downstream element of the major late promoter (MLP) and at the posttranscriptional level by an unknown mechanism (5–7). At the same time, L4-22K also suppresses early gene expression, presumably to redistribute the cellular transcriptional/translational machinery from early genes to late genes in order to maximize progeny virus production (5). Following full activation of the MLP, another layer of regulation involves alternative RNA splicing (2). The Ad L4-33K protein is a virus-encoded alternative RNA splicing factor which activates splicing of Ad late gene transcripts that contain weak 3′ splice sites (8). The L4-33K protein may be phosphorylated by two cellular protein kinases: DNA-dependent protein kinase (DNA-PK) and protein kinase A (PKA) (9). L4-33K interacts with the catalytic subunit of DNA-PK, and phosphorylation by DNA-PK has an inhibitory role in L4-33K-mediated alternative RNA splicing. On the other hand, the phosphorylation of L4-33K by PKA stimulates L4-33K-mediated alternative RNA splicing (9). Through this process, L4-33K is believed to coordinately control late gene expression to optimize the conditions for maximal virus production. These roles of the L4-22K and L4-33K proteins are consistent with the work of Farley et al., who demonstrated that L4 is a key component in the early-to-late switch in Ad late gene expression by promoting the production of cytoplasmic mRNAs from the Ad5 major late transcription unit (10).
The mature Ad virion consists of major capsid proteins (hexon, penton, and fiber), minor capsid proteins (IIIa, VI, VIII, and IX), and core proteins (V, VII, Mu, and TP) that associate with the viral genome (11). The L4-100K protein is required for hexon to trimerize and likely for hexon transport into the nucleus (12, 13). In addition, the precursor form of VI (pVI) has also been shown to mediate hexon nuclear transport (14). From a number of studies, it is believed that Ad empty capsids (EC) are first assembled in the nucleus and that the viral genome is subsequently encapsidated (15). The EC band at a lighter density (1.29 to 1.30 g/cc) than the mature virions (MV) (1.34 g/cc) in a CsCl density equilibrium gradient. MV contain proteolytically processed forms of the proteins VI, VII, and VIII; precursor forms of pVI and pVIII are present in ECs. In contrast to the MV, EC also contain the L1-52/55K protein (16, 17). Core proteins V and VII, which are associated with the viral genome, are present in MV but are absent or present at lower levels in EC. The Ad genome is packaged into the capsid in a polar fashion (15). The packaging sequences (PS) are located at the left end of the Ad genome and are essential cis-acting elements for viral genome encapsidation (15). In addition, the viral proteins IVa2, L1-52/55K, L4-22K, and IIIa are also required for the viral genome packaging process through their interaction with the PS (5, 17–21). If any of these packaging proteins are deleted or mutated within the viral genome, only EC, but no MV, are produced. After the full-length genome is packaged into the capsid, adenovirus protease (AVP) becomes fully activated and cleaves the precursor forms of the capsid proteins IIIa, pVI, pVII, pVIII, pX, pTP, and L1-52/55K to make fully infectious mature virions (22). In order to gain full activity, AVP requires two cofactors: viral DNA and pVIc, an 11-amino-acid peptide from the C terminus of pVI (22).
Besides being a virus-encoded alternative RNA splicing factor, previous studies also suggested that L4-33K plays a role in virus assembly (23–25). However, when Fessler and Young introduced an amber mutation at amino acid position 20 of L4-33K, they also disrupted the L4-22K protein, which shares the N-terminal 105 amino acids with L4-33K (23). Since L4-22K is required for viral genome packaging (5), the assembly defect observed with this L4-33K mutant virus might be due to the deficiency in L4-22K instead of L4-33K. In an independent study, Finnen and colleagues made an L4-33K mutant full-length virus clone which truncated the C-terminal 47 amino acids of the L4-33K protein, without disrupting the adjacent E2 early (E2E) promoter and preserving the L4-22K reading frame; this L4-33K mutation was lethal for virus viability with a defect in virion production (24). In addition, Kulshreshtha and colleagues showed that L4-33K also plays a role in bovine Ad assembly (25). However, both of the latter studies did not conclusively examine viral late gene expression patterns. Given the known role of L4-33K as an alternative RNA splicing factor, the assembly defect observed in those two studies might reflect a deficiency in the expression of one or more late gene products whose expression is regulated by L4-33K, instead of a direct role for L4-33K in the virus assembly process. In order to determine the function(s) of the L4-33K protein in the context of viral infection, we characterized an Ad5 L4-33K mutant virus generated by using an L4-33K complementing cell line. These results showed that the L4-33K mutant virus has infectivity and viral genome replication comparable to those of wild-type Ad5. We confirmed the role of L4-33K as an alternative RNA splicing factor in the context of viral infection and identified the primary targets for L4-33K regulation during infection as the proteins IIIa and pVI, with a lesser effect on fiber. We observed only EC production, but no MV production, from L4-33K mutant virus-infected cells, demonstrating a role of L4-33K in packaging of the viral genome into the EC. However, the L4-33K protein neither preferentially binds to the PS nor influences the interaction of other packaging proteins with the PS in vivo. These results demonstrate the multifunctional nature of the L4-33K protein and its involvement in several different and critical aspects of Ad infection.
MATERIALS AND METHODS
Cells and viruses.
A549 (ATCC) and 293 cells were maintained in Dulbecco's modified Eagle medium (DMEM) containing penicillin and streptomycin and supplemented with 10% bovine calf serum (HyClone). TetC4 cells (20) that constitutively express the tetracycline (Tet)-controlled transactivator tTA in the background of N52.E6-Cre cells (26) (a kind gift from Gudrin Schiedner and Stefan Kochanek, University Ulm) were maintained in DMEM containing penicillin and streptomycin, 200 μg/ml Geneticin, and 200 μg/ml hygromycin B and supplemented with 10% FetalClone III serum (HyClone).
To make the L4-33K Tet-inducible cell line, the coding region of Ad5 L4-33K was amplified by PCR and cloned into pUHD 10-3 (27) (kindly provided by H. Bujard, Zentrum für Molekulare Biologie, Germany) under the control of the tetracycline response element and a minimal cytomegalovirus (CMV) promoter by using EcoRI and XbaI sites, respectively. The resulting plasmid, pUHD-33K, was cotransfected with pSuper-Blasticidin (Invitrogen) into TetC4 cells. An L4-33K-expressing cell line (termed TetC4-33K) was selected and maintained in DMEM containing penicillin and streptomycin, 200 μg/ml Geneticin, 200 μg/ml hygromycin B, 10 μg/ml blasticidin S, and 1 μg/ml doxycycline and supplemented with 10% fetal bovine serum (HyClone).
To make the Ad5 L4-33K mutant virus, an amber mutation (TAG) was introduced at amino acid position 138 of L4-33K in the background of Ad5 infectious clone pTG3602 (28), termed pTG3602-33K−. The PacI-linearized pTG3602-33K− plasmid was transfected by using NanoJuice (EMD Millipore) into TetC4-33K cells, which had been seeded without doxycycline for 24 h, and the transfected cells were overlaid for a plaque assay at 48 h posttransfection. At 10 days posttransfection, individual plaques were picked and amplified in TetC4-33K cells without doxycycline in the culture medium. The titer of the passage 2 lysate of the Ad5 L4-33K mutant virus was determined by a plaque assay using TetC4-33K cells and used to generate purified virions. The purity of the mutant virus stock was verified by restriction endonuclease digestion (the amber codon insertion generates an AvrII site) and Southern blot analysis.
A wild-type Ad5 (Ad5-WT) virion stock and an Ad5 containing loxP sites flanking the PS (termed Ad5-Ψ-loxP) were previously described (5). All virions were purified by using CsCl equilibrium density gradient centrifugation, as described previously (20). Throughout this study, A549 cells were infected at a multiplicity of infection (MOI) of 5 PFU/cell; 293, TetC4, and TetC4-33K cells were infected at an MOI of 1.5 PFU/cell.
L4-33K antibody.
The distinct C terminus (amino acid 106 to the end) of the Ad5 L4-33K coding region (termed L4-33K-C) was amplified by PCR and cloned into pET-28a using NheI and EcoRI sites. The resulting His-L4-33K-C fusion protein was expressed in Escherichia coli BL21(DE3) cells, purified by SDS-PAGE, and used to raise polyclonal rabbit antibodies (Lampire Biological Laboratories, Pipersville, PA).
Virus growth and infectivity.
For complementation assays, TetC4-33K cells were seeded in culture medium with or without doxycycline 24 h before infection by Ad-WT or 33K− virus particles. Infected-cell lysates were harvested at 48 h postinfection (hpi), and titers were determined by a plaque assay on TetC4-33K cells, as described above. For viral growth curves, A549 cells were infected with Ad5-WT or 33K− virus particles, and infected-cell lysates were harvested at 6, 12, 24, and 48 hpi for titration by a plaque assay on TetC4-33K cells, as described above.
For fluorescence focus assays, A549 cells grown on glass coverslips were infected with 106 Ad5-WT or 33K− virus particles in 24-well plates. At 18 hpi, cells were fixed and subjected to immunofluorescence, as previously described (5). For plaque assays to determine particle-to-PFU ratios, TetC4-33K cells were seeded in culture medium without doxycycline 24 h before infection with 1,000 Ad-WT or 33K− virus particles; cells were overlaid at 1 hpi. At 8 days postinfection, plaques from each set of infected plates were counted, and the particle-to-PFU ratio was determined. The purified WT and 33K− MVs from CsCl gradients were lysed in 0.1% SDS, and the absorbance at 260 nm was measured; particle numbers were calculated by using the following formula: 1 OD260 (optical density at 260 nm) unit equals 1 × 1012 particles/ml.
Viral genome replication.
A549 cells were infected with Ad5-WT or 33K− virus particles and harvested at 6, 12, 24, and 48 hpi. Cell pellets were subjected to genomic DNA extraction and quantitative PCR measurement, as previously described (5).
Northern blot analysis.
A549 cells infected with Ad5-WT or 33K− virus particles were harvested at 12, 24, and 48 hpi for preparation of total cytoplasmic RNA by using an RNeasy minikit (Qiagen) according to the manufacturer's instructions. A total of 4 or 12 μg RNA of each sample was separated on a 1% formaldehyde-agarose gel and transferred onto a positively charged nylon membrane (GE Healthcare). The probes for detecting L1 to L5 mRNAs correspond to Ad5 nucleotides (nt) 13026 to 13751, 16834 to 17452, 21573 to 22322, 26769 to 27590, and 31920 to 32465, respectively. The probe that specifically detects pVI mRNA corresponds to Ad5 nt 18003 to 18755. The probes were amplified by PCR, purified, and labeled with [32P]dATP by random primer labeling using Exo− Klenow DNA polymerase (NEB).
Reverse transcription-PCR (RT-PCR).
Two micrograms of cytoplasmic RNA from Ad5-WT- or 33K−-infected A549 cells was reverse transcribed by using the oligo(dT) primer (NEB) and SuperScript II reverse transcriptase (Invitrogen). Five percent of the reverse transcription reaction was used as the template for the following 20-cycle PCR. The primer sets for detecting L4-22K Ad5 nt 9700 to 9719 (located in the tripartite leader sequence) and Ad5 nt 26703 to 26682 (located in the L4-33K intron). The primer sets for detecting glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were GAPDH-1 (ACCCAGAAGACTGTGGATGG) and GAPDH-2 (TTCTAGACGGCAGGTCAGGT).
Western blot analysis.
Mock- or virus-infected cells were lysed in 2× Laemmli sample buffer (0.15 M Tris [pH 6.8], 4% SDS), and the protein concentration was determined by a bicinchoninic acid (BCA) protein assay kit (Pierce). Thirty micrograms of whole-cell extract was analyzed, as previously described (5). Primary antibodies included rabbit polyclonal E1A antibody (SC430; Santa Cruz Biotechnology) (1:500 dilution), mouse monoclonal DNA binding protein (DBP) antibody (Arnold Levine, Princeton University) (1:1,000 dilution), rabbit polyclonal IVa2 antibody (29) (1:1,000 dilution), rabbit polyclonal L1-52/55K antibody (29) (1:1,000 dilution), rabbit polyclonal IIIa antibody (30) (1:1,000 dilution), rabbit polyclonal penton antibody (Carl Anderson, Brookhaven National Laboratory) (1:1,000 dilution), rabbit polyclonal VII antibody (Daniel Engel, University of Virginia) (1:2,000 dilution), rabbit polyclonal V antibody (David Matthews, University of Bristol) (1:1,000 dilution), rabbit polyclonal VI antibody (Christopher Wiethoff, Loyola University Chicago) (1:5,000 dilution), mouse monoclonal hexon antibody (MAB0850; Abnova) (1:5,000 dilution), rabbit polyclonal L4-100K antibody (5) (1:1,000 dilution), rabbit polyclonal L4-22K antibody (against the distinct C terminus of L4-22K) (31) (1:2,000 dilution), rabbit polyclonal L4-33K antibody (against the distinct C terminus of L4-33K, as described above) (1:2,000 dilution), rabbit polyclonal VIII antibody (Ann Tollefson and William Wold, St. Louis University) (1:400 dilution), rabbit polyclonal fiber antibody (Carl Anderson) (1:1,000 dilution), rabbit polyclonal AVP antibody (Maxim Balakirev, CEA-Grenoble) (1:500 dilution), and mouse monoclonal α-tubulin antibody (T5192; Sigma) (1:10,000 dilution).
For virus particle analyses, 1 × 108 TetC4 cells, 2 × 108 293 cells, and 5 × 107 A549 cells were infected with Ad5-WT, Ad5-Ψ-loxP, or 33K− virus particles. At 48 to 72 hpi, infected cells were harvested for preparation of virus particles by one-step and two-equilibrium CsCl density gradients. Two micrograms of isolated MV or EC from the CsCl gradient was subsequently analyzed by silver staining and Western blot analysis, as described previously (20).
ChIP.
A549 cells were infected with Ad5-WT or 33K− virus particles and cross-linked with formaldehyde at 18 hpi. The samples were then sonicated and subjected to chromatin immunoprecipitation (ChIP) procedures, as previously described (5). The final DNA sample was dissolved in 50 μl Tris-EDTA (TE) and subjected to quantitative PCR analysis using two sets of primers: the PACK-4/5 primer set to assess the copy number of the PS that had been specifically immunoprecipitated by the antibodies and the E4-ORF6 primer set to assess the copy number of an Ad5 right-end DNA fragment that had been nonspecifically immunoprecipitated or reflected incomplete sonication products. The capacity of each viral protein (L4-22K, L4-33K, IVa2, L1-52/55K, or IIIa) to bind the PS in virus-infected cells was calculated as the value of the PS copy number divided by the E4-ORF6 copy number. For comparison, the PACK/E4-ORF6 ratio of each antibody in a particular virus infection was normalized to ChIP values using preimmune serum.
Electron microscopy.
Ten microliters of virus particles purified from one-step and two-equilibrium CsCl gradients was applied to the shiny side of Formvar/carbon film on 300-square-mesh nickel grids (FCF300-Ni; Electron Microscopy Sciences) and fixed with 5% electron microscopy-grade glutaraldehyde (catalog number 16316-10; Electron Microscopy Sciences). After washes with TNE buffer (10 mM Tris [pH 7.5], 250 mM NaCl, 0.5 mM EDTA) and water, the grids were stained with NanoVan (catalog number 2011; Nanoprobes) for 15 s. The grids with negatively stained virus particles were examined on an FEI Tecnai 12 BioTwin G02 microscope at 80 kV, and digital images were acquired by the use of an AMT XR-60 charge-coupled-device digital camera system.
RESULTS
Isolation of an Ad5 L4-33K mutant virus, infectivity, and viral genome replication.
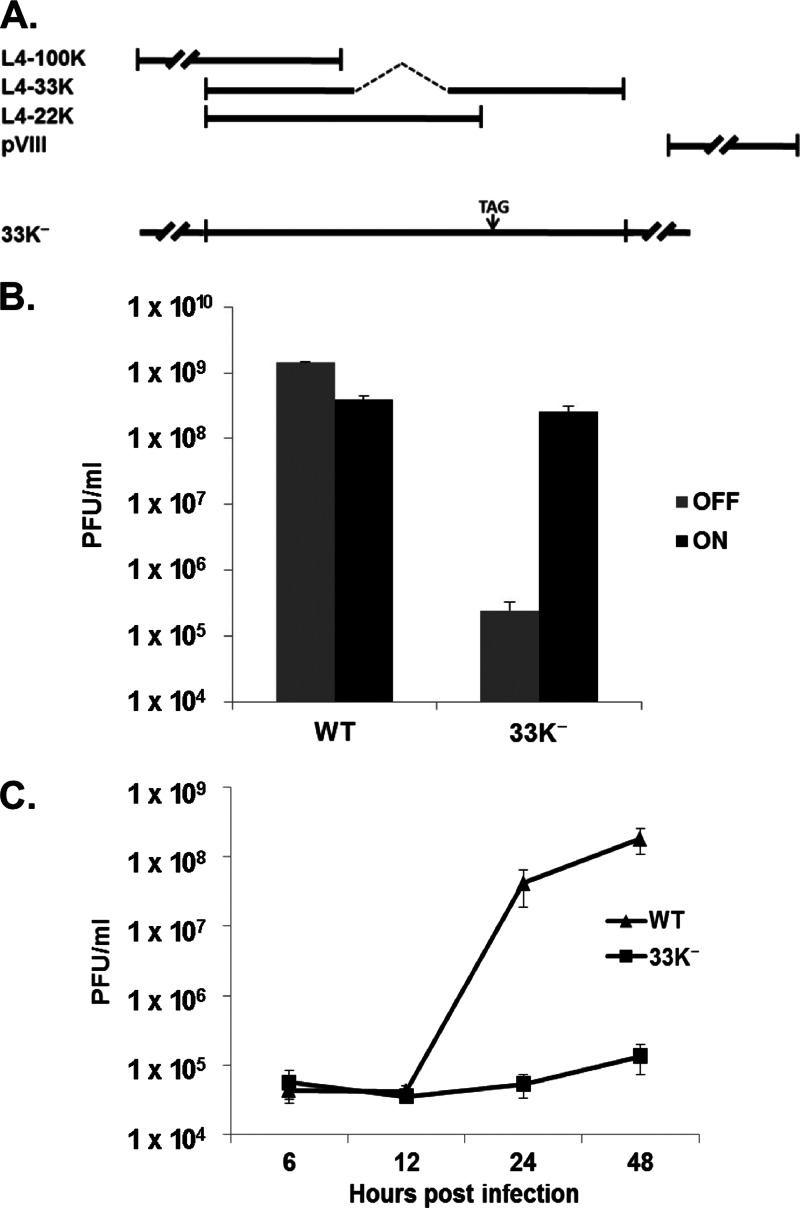
The Ad5 L4 region encodes four proteins: L4-100K, L4-33K, L4-22K, and pVIII (Fig. 1A). L4-33K and L4-22K share a 105-amino-acid N terminus; the L4-33K mRNA is spliced to generate a unique L4-33K C terminus. The E2 early (E2E) promoter is located on the opposite strand of the L4 region and overlaps part of the L4-33K C terminus (24). In order to make an Ad5 L4-33K mutant virus, an amber mutation (TAG) was introduced at amino acid position 138 of L4-33K in the background of the Ad5 infectious clone pTG3602 (termed pTG3602-33K−) in which both the L4-22K reading frame and E2E promoter were kept intact (Fig. 1A). Consistent with previous reports that mutation of L4-33K is lethal (24, 25), pTG3602-33K− was not viable in noncomplementing cells (data not shown). A tetracycline-inducible cell line expressing full-length Ad5 L4-33K, termed TetC4-33K, was isolated to complement the growth of the L4-33K mutant virus. Plaque-purified 33K− virus was amplified in TetC4-33K cells, and mutant virus particles were purified by CsCl equilibrium centrifugation for use in all of the following experiments. To confirm the complementation of the L4-33K mutant in TetC4-33K cells, the titers of Ad5-WT and 33K− viruses produced in TetC4-33K cells with (off) or without (on) doxycycline in the culture medium were measured. These results showed that the TetC4-33K cell line complemented the growth of the 33K− mutant virus to a level comparable to that of Ad5-WT when L4-33K expression was turned on (Fig. 1B). In addition, a one-step growth curve assay confirmed that mutation of L4-33K was lethal to virus viability in noncomplementing A549 cells (Fig. 1C). We note that turning on L4-33K expression slightly decreased the yield of Ad5-WT (Fig. 1B), which may reflect premature changes in normal major late transcription unit (MLTU) splicing patterns that alter the balance of late gene products and reduce virus yield in light of the role of L4-33K in regulating alternative RNA splicing (8). We also note that extensive cell death was observed after turning on L4-33K expression for 72 h (data not shown). Thus, L4-33K expression has a toxic effect on cells that may relate to the disruption of normal splicing patterns of cellular transcripts that may influence virus yield.
Fig 1.
L4-33K mutant virus. (A) Schematic diagram of the Ad5 L4 region. Coding regions for L4-100K, L4-33K, L4-22K, and pVIII are shown. L4-33K mutant virus 33K− is depicted with a stop codon introduced at amino acid position 138 of L4-33K. (B) Complementation of Ad5-WT and 33K− virus in TetC4-33K cells with (OFF) or without (ON) doxycycline (1 μg/ml) in the culture medium. Infected-cell lysates were harvest at 48 hpi and subsequently titrated on TetC4-33K cells cultured without doxycycline by a plaque assay. (C) Growth curves of Ad5-WT and 33K− virus in A549 cells. Infected A549 cell lysates were harvest at 6, 12, 24, and 48 hpi and subsequently titrated on TetC4-33K cells by a plaque assay.
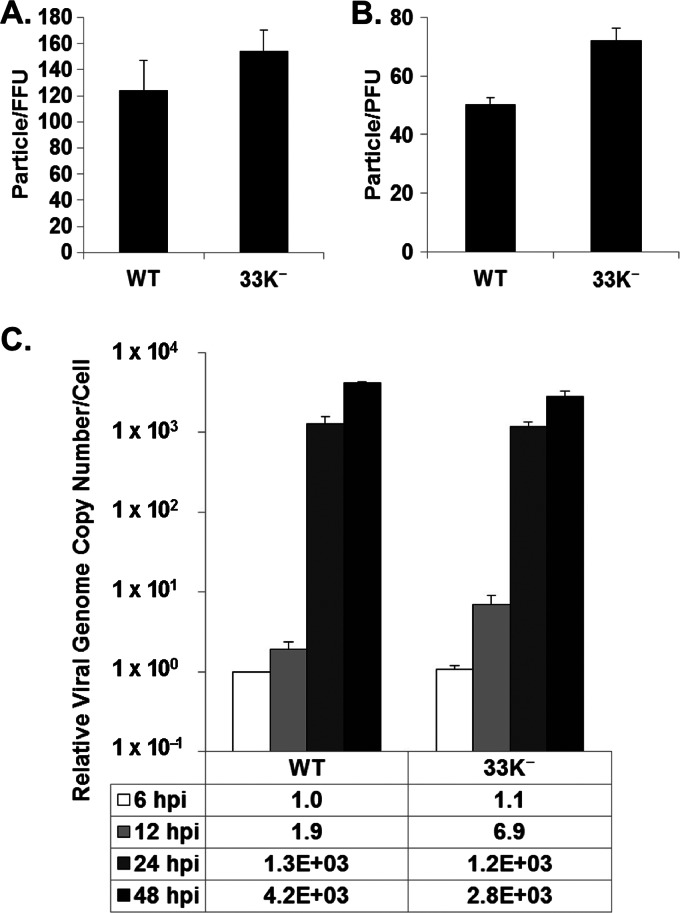
In order to investigate the step(s) of the Ad life cycle in which L4-33K plays an essential role(s), the infectivity of 33K− virus particles was determined by a fluorescence focus assay in A549 cells by using antibody against Ad5 DNA binding protein (DBP) and a plaque assay in TetC4-33K cells in comparison to Ad5-WT. The results from both assays demonstrated that the 33K− mutant virus had ratios of particles to focus-forming units (FFU) and PFU similar to those of Ad5-WT (Fig. 2A and B), suggesting that the 33K− virus behaved comparably to Ad5-WT from the onset of infection to viral genome replication. Next, viral genome replication of Ad5-WT and 33K− in infected A549 cells was measured (Fig. 2C). These results exhibited comparable replication kinetics between the two viruses, although 33K− had slightly lower replication levels than Ad5-WT at 48 hpi. From these data, we conclude that the L4-33K− virus has infectivity and viral genome replication similar to those of Ad5-WT and that the growth defect of the 33K− virus is likely due to later events after viral genome replication.
Fig 2.
L4-33K mutant virus infectivity and genome replication. (A and B) Ratios of physical particles to infectious particles of Ad5-WT and 33K− virus were determined in A549 cells (particle/FFU ratio) (A) and TetC4-33K cells (particle/PFU ratio) (B). (C) Viral genome replication in Ad5-WT-, or 33K−-infected A549 cells. The genome copy number was determined by quantitative PCR and normalized to the GAPDH value; this value was relative to that of Ad5-WT at 6 hpi.
L4-33K mutant virus gene expression.
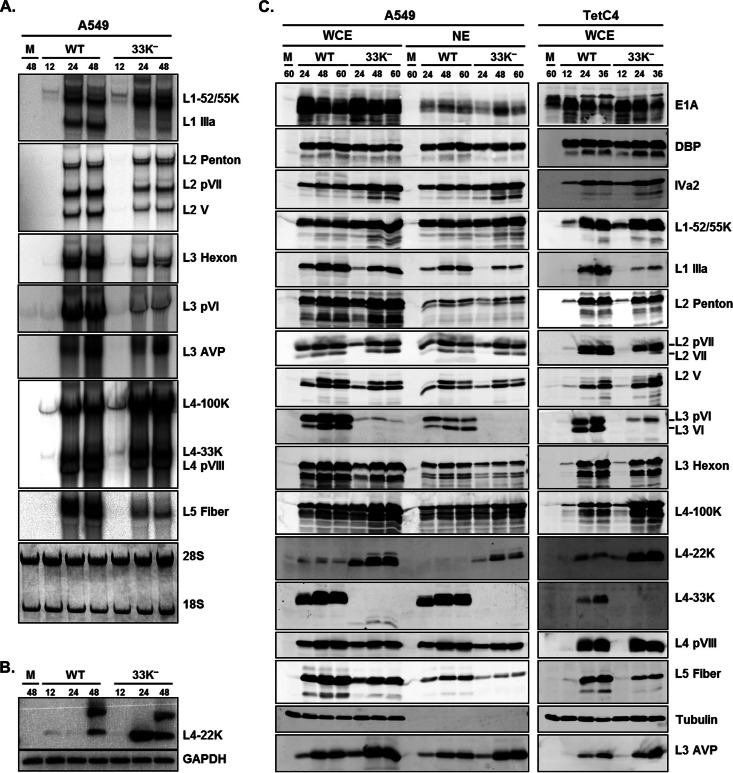
Given the facts that the 33K− virus has infectivity and genome replication comparable to those of Ad5-WT and that the L4-33K protein is known as an alternative RNA splicing factor (8), we performed Northern blot and Western blot analyses to evaluate viral gene expression patterns. Consistent with previous results using an in vitro splicing assay (8), Northern blot results confirmed that IIIa mRNA levels were dramatically reduced with the 33K− virus (Fig. 3A). L3 pVI mRNA levels were also dramatically reduced in 33K−-infected cells, and L2 V, L2 pVII, L3 hexon, and L5 fiber mRNA levels were also diminished but to a lesser extent (Fig. 3A). In contrast, notable increases in levels of L1-52/55K, L4-100K, and L4-22K mRNAs were observed in 33K−-infected A549 cells (Fig. 3A and B). L4-22K mRNAs were not detectable by Northern blotting, so they were evaluated by semiquantitative RT-PCR (Fig. 3B).
Fig 3.
L4-33K mutant virus gene expression. (A) Northern blot analysis of Ad late gene mRNAs in Ad5-WT- or 33K−-infected A549 cells. Total cytoplasmic mRNAs isolated from the infected cells at 12, 24, and 48 hpi were analyzed by Northern blotting using 32P-labeled probes for each late region. (B) RT-PCR analysis of L4-22K mRNA levels in Ad5-WT- or 33K−-infected A549 cells. (C) Western blot analyses of Ad early and late proteins in A549 and TetC4 cells. Representative early (E1A and DBP), intermediate (IVa2), and late (L1 to L5) gene products were analyzed by using whole-cell extracts (WCE) and nuclear extracts (NE) isolated from Ad5-WT- and 33K−-infected A549 cells (left) or WCE isolated from Ad5-WT- and 33K−-infected TetC4 cells (right) at different times (hours) postinfection (indicated at the top). Protein designations are indicated on the right; tubulin was analyzed as a loading control.
To explore viral gene expression at the protein level, we performed Western blot analyses of viral early and late proteins using Ad5-WT- or 33K−-infected A549 and TetC4 whole-cell extracts (WCE) and nuclear extracts (NE) (Fig. 3C). These results largely, but not completely, recapitulated the results of the Northern blot assays and demonstrated that mutation of L4-33K led to decreased levels of IIIa, fiber, and pVI and increased levels of the L1-52/55K, L4-100K, and L4-22K proteins. The levels of V, pVII, and hexon remained almost the same as with Ad5-WT (Fig. 3C). The most notable effects in these analyses with the 33K− virus was a dramatic reduction in pVI levels and increase in L4-22K levels. It is notable that the reduction in IIIa protein levels was much less that the reduction in IIIa mRNA levels, and conversely, the reduction in pVI protein levels was greater than the reduction in pVI mRNA levels. Comparable results were obtained by using nuclear extracts from infected A549 cells, indicating no apparent changes in nuclear import of Ad proteins with the 33K− virus. Notably, the early and intermediate gene products E1A, DBP, and IVa2 were not affected in 33K−-infected cells; this also confirmed that mutation of L4-33K did not disrupt E2E transcription. In addition, there was no full-length L4-33K expression with the 33K− virus, but trace amounts of an apparently truncated version were evident, which ensured the integrity of the L4-33K mutant virus stock. The reduction in the levels of most viral late proteins (Fig. 3C) was not as evident as the corresponding reduction of mRNA levels (Fig. 3A) in 33K−-infected cells. This discrepancy might be due to the increased level of L4-100K, which is known to increase translation of viral late mRNAs that contain the 5′ tripartite leader sequence (31). In conclusion, comprehensive Northern blot and Western blot analyses demonstrate that the L4-33K protein regulates different aspects of viral late gene expression.
The L4-33K mutant virus produces only empty capsids.
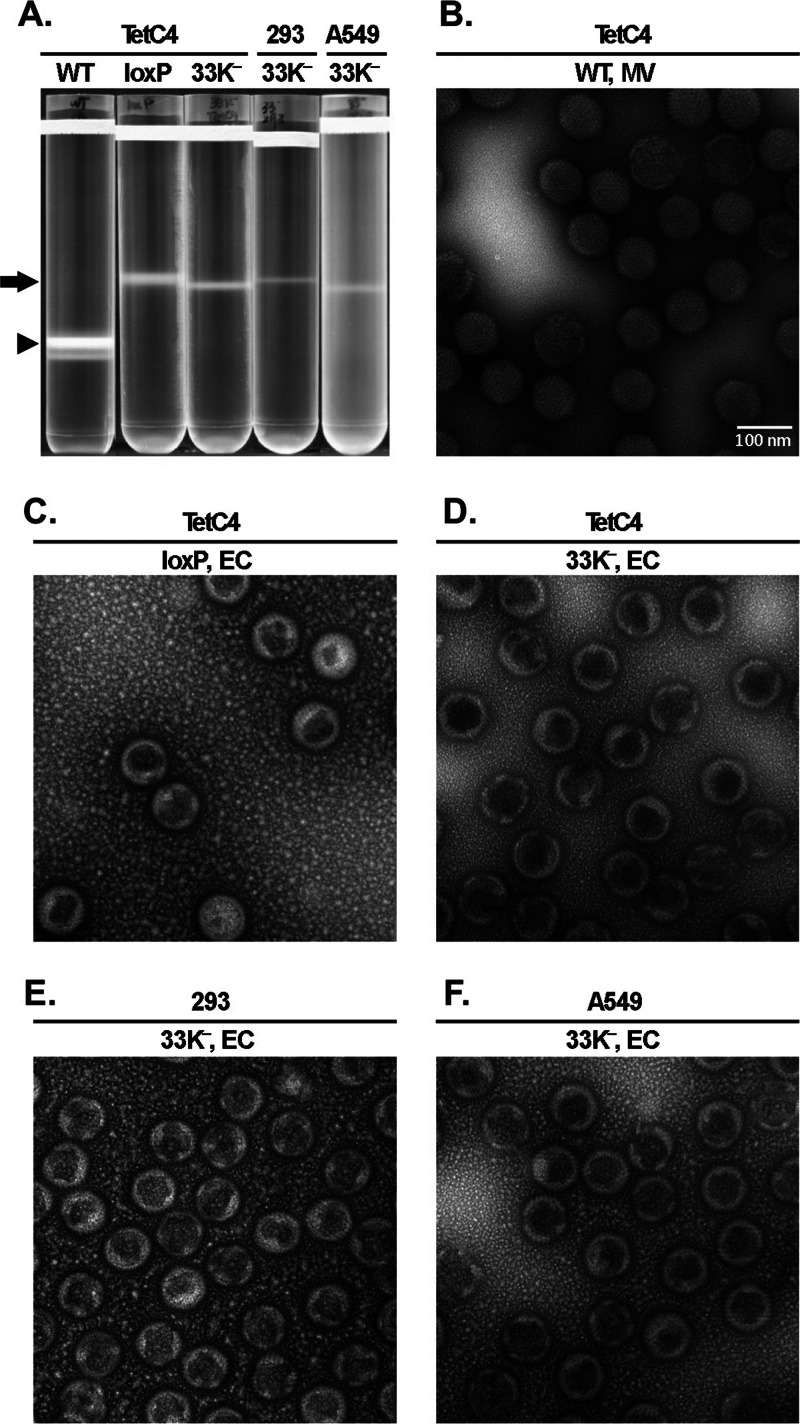
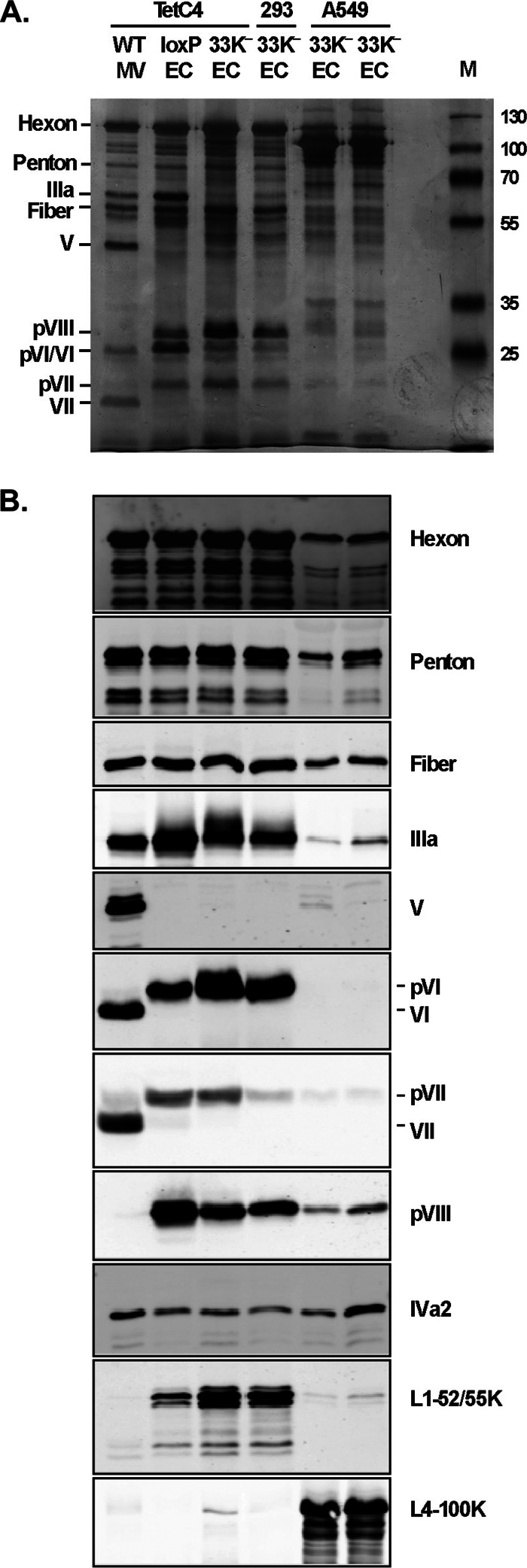
We analyzed virus production in 33K−-infected cells to determine whether mutation of L4-33K has any effect on virus assembly, viral genome packaging, or other late events of the Ad life cycle. Only EC, but no MV, were produced in 33K−-infected A549, 293, and TetC4 cells (Fig. 4A). This result demonstrates that instead of having a virion assembly defect, mutation of L4-33K impaired the viral genome packaging process. Note that the yield of EC from 33K−-infected cells is lower than that from a virus lacking the PS (Ad5-Ψ-loxP grown in Cre-expressing cells), which is likely due to decreased late gene expression with the 33K− mutant virus (Fig. 3). The morphology of EC produced from 33K−-infected cells was assessed by electron microscopy (Fig. 4D to F). The 33K− EC exhibited higher electron density than MV produced by Ad5-WT (Fig. 4B), which was very similar to that of the EC produced from a virus lacking the PS (Ad5-Ψ-loxP grown in Cre-expressing cells) (Fig. 4C). Next, we used silver staining and Western blotting to analyze the EC generated from 33K−-infected cells in comparison to Ad5-WT MV as well as the control EC generated by using the Ad5-Ψ-loxP virus. These results revealed that EC generated from 33K− virus infection in TetC4 and 293 cells had a protein composition very similar to that of bona fide EC produced by using Ad5-Ψ-loxP (Fig. 5A and B). These EC lacked core protein V and had reduced levels of core protein VII, both characteristic of MV, and contained precursor forms of pVI and pVIII as well as readily evident levels of L1-52/55K, which were absent or present in a very low abundance in MV. These EC contained similar amounts of IIIa compared to those of Ad5-WT MV and the loxP EC (Fig. 5B), despite the finding that the total pool of IIIa in 33K−-infected TetC4 cells was reduced (Fig. 3C). Interestingly, the EC generated from 33K− virus-infected A549 cells presented a different profile of virion protein composition (Fig. 5A and B). Although these EC presented a morphology similar to that of the EC generated from 33K−-infected TetC4 or 293 cells (Fig. 4D to F), they had lower protein levels for most of the virion proteins (despite loading of equal amounts of total protein onto the gel), dramatically higher levels of L4-100K protein, IVa2 levels similar to those of the EC generated from 33K−-infected TetC4 or 293 cells, and reduced levels of the L1-52/55K protein compared to the other EC (Fig. 5B). Additional proteins were detected in EC from 33K−-infected A549 cells that were unique to these particles; some of the bands in the ∼50-kDa range were L4-100K breakdown products, while the origin of other bands is not known. The discrepancy in protein composition of the 33K− EC in TetC4, 293, and A549 cells is unknown but might be due to the different natures of these cell lines (see Discussion).
Fig 4.
L4-33K mutant virus particle production. (A) CsCl density equilibrium gradient profiles of virus particles produced from Ad5-WT-, Ad5-Ψ-loxP-, or 33K−-infected TetC4 (Cre recombinase-positive), 293, or A549 cells. The arrow and arrowhead indicate empty capsids (EC) and mature virions (MV), respectively. Ad5-Ψ-loxP contains loxP sites flanking the PS and thus produces only EC in TetC4 cells due to cleavage of the PS from the genome by Cre recombinase. (B to F) Electron microscopy examination of the MV and EC isolated in panel A.
Fig 5.
Protein composition of the empty capsids generated by 33K− virus. (A) Silver staining of protein components of 2 μg of MV and EC isolated from virus particles indicated in Fig. 4A. Protein designations are indicated on the left. M, molecular weight markers. (B) Western blot analysis of protein components of 2 μg of MV and EC isolated from virus particles indicated in panel A. Protein designations are indicated on the right. Virus particles used in these analyses were banded twice by CsCl equilibrium centrifugation. Two independent batches of 33K− EC samples from A549 cells were prepared and included in the analysis (adjacent lanes).
The L4-33K protein does not preferentially bind to the PS in vivo.
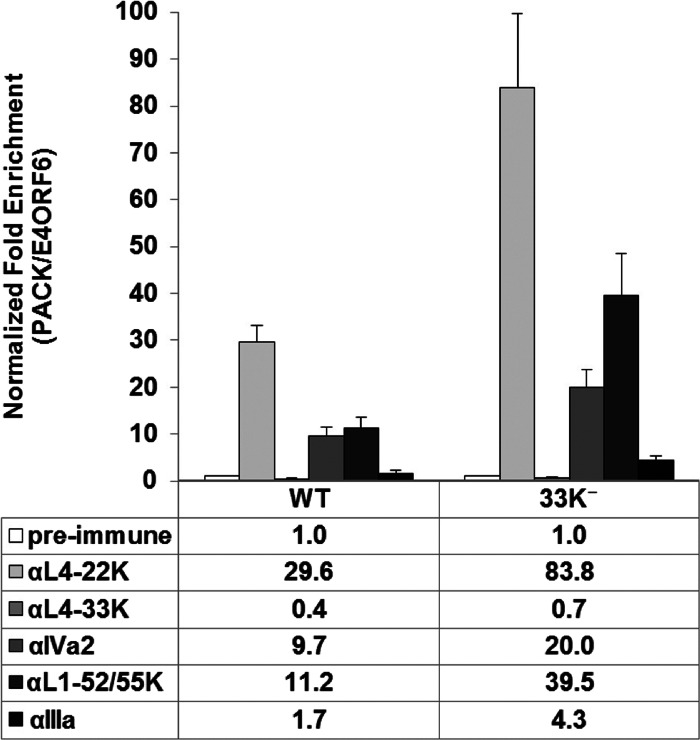
Given the viral genome packaging defect observed in 33K−-infected cells, we performed chromatin immunoprecipitation (ChIP) assays to determine if the L4-33K protein is part of the genome packaging machinery that interacts with the PS during infection. The results from Ad5-WT-infected A549 cells showed that L4-33K did not preferentially bind to the PS in vivo (Fig. 6, αL4-33K versus preimmune and WT versus 33K−). Next, we examined the binding of other viral packaging proteins to the PS in Ad5-WT- or 33K−-infected A549 cells. These results showed that mutation of L4-33K did not interfere with the binding of the viral packaging proteins IVa2, L4-22K, L1-52/55K, and IIIa to the PS in vivo (Fig. 6, WT versus 33K−). Based on these results, we conclude that L4-33K is not likely to be involved in the viral genome packaging process via direct interaction with the packaging domain.
Fig 6.
ChIP assays for packaging protein binding to the PS in vivo. A549 cells were infected with Ad5-WT or 33K−, and L4-22K-, L4-33K-, IVa2-, L1-52/55K-, and IIIa-specific antibodies were used for ChIP to quantify binding to the PS. The results are presented as normalized fold enrichment by dividing the copy number of PS pulled down specifically by the antibody-antigen complex to the copy number of the E4-ORF6 fragment that was pulled down nonspecifically by the antibody. The fold enrichment value of each antibody was normalized to that of the preimmune serum negative control.
DISCUSSION
The Ad L4-33K protein has been linked with two disparate functions: one as a virus-encoded alternative RNA splicing factor that regulates Ad late mRNA splicing patterns and a second as a protein involved in virus assembly (8, 10, 22–24). To further explore the function(s) of L4-33K in the context of natural Ad infection, we made and characterized an Ad5 L4-33K mutant virus using a newly developed complementing cell line. Characterization of this mutant virus demonstrated that mutation of L4-33K is lethal to Ad5 viability. Our results demonstrate that mutation of L4-33K does not block virus assembly but impairs viral genome packaging. Furthermore, our results confirm in vivo the essential role of L4-33K as an Ad late mRNA splicing regulator in the context of virus infection.
As mutation of L4-33K is lethal to virus growth (Fig. 1C), we determined the step(s) at which L4-33K may play an essential role(s). The L4-33K mutant virus had infectivity and viral genome replication comparable to those of Ad5-WT (Fig. 2), which demonstrates that there are no apparent differences between Ad5-WT and the 33K− mutant virus from the onset of infection through viral genome replication. These results suggested that the growth defect arose from a late event during infection. Northern blot analysis of cytoplasmic viral mRNAs (Fig. 3A) confirmed previous reports that L4-33K functions as a virus-encoded alternative RNA splicing factor (8, 10). Consistent with those reports, we found that IIIa, V, pVII, and hexon mRNA levels were regulated by L4-33K, although the magnitude of the effect on V and pVII mRNA levels in vivo was modest compared to that observed in vitro. Multiple factors may influence cytoplasmic mRNA levels in vivo, including nuclear transport and mRNA stability, which may be related to these differences. In addition, we also found that pVI and fiber mRNA levels were reduced in L4-33K mutant virus-infected cells, whereas increased levels of L1-52/55K, L4-100K, and L4-22K mRNAs were observed in the absence of L4-33K (Fig. 3A and B). A previous report showed that IIIa mRNA accumulation does not occur at the expense of L1-52/55K mRNA accumulation by the use of an in vivo splicing assay by cotransfecting an L4-33K expression vector with another vector expressing an L1/L2 region splicing substrate (8). These subtle differences likely reflect the different natures of the assays utilized but do not affect the clear interpretation that L4-33K is highly important for the appropriate accumulation of certain Ad late mRNAs.
Our analysis of the full panel of Ad late gene transcripts in the context of 33K− mutant virus infection demonstrates that there is a clear switch of viral late gene mRNA accumulation mediated by L4-33K: the early set of late gene mRNAs (L1-52/55K, L4-100K, and L4-22K) first accumulate at relatively early points of the late phase (12 hpi) (Fig. 3A and B), consistent with a prior report (3). Subsequently, Ad late mRNAs corresponding to all five late gene families (L1 to L5) are produced, some of whose expression depends completely (IIIa) or in part (V, pVII, hexon, pVI, and fiber) upon the expression of L4-33K. The observed increased mRNA levels of L1-52/55K, L4-100K, and L4-22K strongly suggest that the switch in mRNA expression patterns did not occur efficiently in the absence of L4-33K. It is possible that other viral proteins whose levels are altered in 33K−-infected cells (IIIa, V, pVII, pVI, hexon, fiber, L1-52/55K, or L4-100K) might contribute to the switch of viral late gene mRNA accumulation, but this possibility is discounted because those gene products have no known function in gene transcription, RNA processing, or mRNA export. However, since the L4-22K protein activates Ad late gene expression at the transcriptional and posttranscriptional levels (5–7), we cannot rule out the possibility that increased L4-22K levels (Fig. 3C) may contribute to the increased mRNA levels of L1-52/55K and L4-100K. Interestingly, viral mRNA levels that are increased in 33K−-infected cells (L1-52/55K, L4-100K, and L4-22K) are located in the L1 and L4 regions, which corresponds to a previous report that L1 and L4 transcripts accumulate during the early time points of the late phase of Ad infection (3). These results lead us to conclude that L4-33K temporally controls the switch from the early set of viral late mRNAs to the late ones during progression of the late phase of Ad infection. Finally, we note that the decrease in L4-33K protein levels and the corresponding increase in L4-22K mRNA and protein levels (Fig. 3A and B) suggest a feedback loop of splicing regulation, whereby L4-33K promotes splicing of its own precursor RNA. When L4-33K protein is eliminated, the level of the unspliced version of this RNA precursor that gives rise to the L4-22K protein (Fig. 1A) is increased. However, this possibility is discounted by the observation that the total level of L4-33K mRNA was not detectably reduced in 33K−-infected cells. Since levels of L1-52/55K and L4-100K mRNAs were also elevated in 33K−-infected cells, we believe that these results likely reflect increased L1 and L4 gene transcription due to the delay in the late-phase switch in late gene transcription patterns.
Western blot analyses largely, but not completely, recapitulated changes in Ad late mRNA levels observed by Northern blot analyses (Fig. 3). For example, the dramatic decrease in IIIa cytoplasmic mRNA levels observed in 33K−-infected cells resulted in only a moderate decrease in IIIa protein levels. One explanation for this discrepancy is that the increased level of L4-100K observed in 33K−-infected cells might compensate for reduced viral mRNA levels, since an important function of L4-100K is to enhance translation of viral late mRNAs through its interaction with both the 5′ tripartite leader sequence and the translation initiation complex (32). However, in contrast to IIIa, the decrease in pVI protein levels was greater than the decrease in pVI mRNA levels, making this interpretation uncertain, since all of the Ad late gene transcripts contain a tripartite leader. The exact basis for the differences in viral late mRNA levels versus proteins levels is unknown at this time. We note that the significant decrease in pVI expression when L4-33K was missing matches what Morris et al. observed using an L4P-negative Ad genome with L4-22K protein expressed in trans (4), a situation that resembles infection with an L4-33K mutant virus. Since pVI has been reported to augment hexon protein transport into the nucleus (14), the reduction in pVI levels in 33K−-infected cells could explain the previously reported assembly defect of L4-33K mutants (23–25). However, our results showed a similar nuclear accumulation of hexon in Ad5-WT- and 33K−-infected cells (Fig. 3C), discounting this interpretation of the previously reported results.
Three previous reports concluded that L4-33K plays a role in virus assembly (23–25). Our results clarify the interpretations of results obtained with different L4-33K mutants. Only EC, and no MV, were produced in L4-33K mutant virus-infected A549, 293, and TetC4 cells (Fig. 4). This demonstrates that there is no virus assembly defect but rather a viral genome packaging defect in the absence of the L4-33K protein. The accumulation of EC also confirms that most viral late gene products accumulate properly in the nucleus, as shown by Western blotting (Fig. 3C), except for decreased levels of IIIa, pVI, and fiber. The lack of any capsid production following transfection of a full-length Ad clone containing an L4-33K mutation in previous reports may reflect the inefficient nature of the transfection approach. Interestingly, the EC generated from 33K−-infected A549 cells had a protein composition distinct from that of EC produced in TetC4 or 293 cells (Fig. 5). As TetC4 and 293 cells express the Ad5 E1 region, the alteration of cellular components in these highly transformed cells, compared to A549 cells, might contribute to the different phenotypes observed. Alternatively, since the Ad5 E1 proteins are already expressed in TetC4 and 293 cells, one might anticipate that the kinetics of virus infection in these cells, compared to A549 cells, may be augmented and influence the results. We also suggest that the EC observed in 33K−-infected A549 cells may represent a novel Ad assembly intermediate not previously described. Since the EC produced in A549 cells contain large amounts of L4-100K and smaller amounts of other viral proteins evident in bona fide EC, the A549 EC may represent a precursor to EC found with the other cells. L4-100K and pVI both serve as chaperones for hexon trimerization and hexon nuclear import, so perhaps the reduced levels of pVI in 33K−-infected A549 cells prevent the displacement of L4-100K from EC, although the level of pVI was also greatly reduced in TetC4 cells infected with this mutant, where elevated L4-100K levels in EC were not observed. Alternatively, a component(s) specific to A549 cells may influence the displacement of L4-100K from hexon and, thus, affect hexon biogenesis and virion assembly. These speculations will be the subject of future studies.
The viral genome packaging defect observed with the 33K− mutant virus led us to investigate how L4-33K might be involved in this process. ChIP analyses (Fig. 6) showed that the L4-33K protein itself does not preferentially bind to the PS in vivo, and its mutation does not impair the interaction of other viral packaging proteins (IVa2, L4-22K, L1-52/55K, and IIIa) with the PS. In fact, we observed stronger binding of these packaging proteins with the PS in L4-33K mutant virus-infected cells (Fig. 6). The reason for this stronger binding may be due to the increased levels of L4-22K and L1-52/55K in 33K−-infected cells (Fig. 3C). As L4-22K and IVa2 are dependent on each other and are essential to recruit L1-52/55K and IIIa to bind to the PS in vivo (5), the increased levels of L4-22K and L1-52/55K in 33K−-infected cells may enhance the formation of the packaging complex, even though the total pool of IIIa is reduced. However, the discrepancy of the strong binding of the known viral packaging proteins with the PS without effective genome packaging suggests either that another important component(s) of the viral genome packaging machinery is reduced or missing in 33K−-infected cells or that the ChIP approach does not reveal the complete nature of the packaging process.
In summary, our results demonstrate that the phenotype of an Ad5 L4-33K mutant virus is complex. The L4-33K protein regulates the accumulation of selective Ad late gene mRNAs and is involved in the proper transition of gene expression during the late phase of infection. The L4-33K protein also plays a role in adenovirus morphogenesis by regulating the packaging of viral DNA into the EC by an unknown mechanism. Finally, the analysis of the L4-33K mutant virus in infected A549 cells may have revealed the existence of a new virus assembly intermediate not previously identified, since a novel composition of viral proteins was observed in EC produced in these cells.
ACKNOWLEDGMENTS
We thank our colleagues, including Gudrin Schiedner, Stefan Kochanek, Arnold Levine, Carl Anderson, Daniel Engel, David Matthews, Christopher Wiethoff, Maxim Balakirev, Ann Tollefson, and William Wold, for providing important reagents used in this study. We thank laboratory members for informative discussions throughout the project. We thank Mary Anderson and Ilana Shoshani for excellent technical assistance and Sharanya Chandrasekhar for constructing pTG3602-33K−.
This work was supported by NIH grant AI041636. D.G. was supported by a SUNY doctoral diversity fellowship, a W. Burghardt Turner fellowship, and NIH training grant 5T32AI007539.
Footnotes
Published ahead of print 3 April 2013
REFERENCES
- 1. Berk AJ. 2005. Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene 24:7673–7685 [DOI] [PubMed] [Google Scholar]
- 2. Akusjarvi G. 2008. Temporal regulation of adenovirus major late alternative RNA splicing. Front. Biosci. 13:5006–5015 [DOI] [PubMed] [Google Scholar]
- 3. Larsson S, Svensson C, Akusjarvi G. 1992. Control of adenovirus major late gene expression at multiple levels. J. Mol. Biol. 225:287–298 [DOI] [PubMed] [Google Scholar]
- 4. Morris SJ, Scott GE, Leppard KN. 2010. Adenovirus late-phase infection is controlled by a novel L4 promoter. J. Virol. 84:7096–7104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wu K, Orozco D, Hearing P. 2012. The adenovirus L4-22K protein is multifunctional and is an integral component of crucial aspects of infection. J. Virol. 86:10474–10483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Morris SJ, Leppard KN. 2009. Adenovirus serotype 5 L4-22K and L4-33K proteins have distinct functions in regulating late gene expression. J. Virol. 83:3049–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Backstrom E, Kaufmann KB, Lan X, Akusjarvi G. 2010. Adenovirus L4-22K stimulates major late transcription by a mechanism requiring the intragenic late-specific transcription factor-binding site. Virus Res. 151:220–228 [DOI] [PubMed] [Google Scholar]
- 8. Tormanen H, Backstrom E, Carlsson A, Akusjarvi G. 2006. L4-33K, an adenovirus-encoded alternative RNA splicing factor. J. Biol. Chem. 281:36510–36517 [DOI] [PubMed] [Google Scholar]
- 9. Tormanen Persson H, Aksaas AK, Kvissel AK, Punga T, Engstrom A, Skalhegg BS, Akusjarvi G. 2012. Two cellular protein kinases, DNA-PK and PKA, phosphorylate the adenoviral L4-33K protein and have opposite effects on L1 alternative RNA splicing. PLoS One 7:e31871 doi: 10.1371/journal.pone.0031871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Farley DC, Brown JL, Leppard KN. 2004. Activation of the early-late switch in adenovirus type 5 major late transcription unit expression by L4 gene products. J. Virol. 78:1782–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smith JG, Wiethoff CM, Stewart PL, Nemerow GR. 2010. Adenovirus. Curr. Top. Microbiol. Immunol. 343:195–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kauffman RS, Ginsberg HS. 1976. Characterization of a temperature-sensitive, hexon transport mutant of type 5 adenovirus. J. Virol. 19:643–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cepko CL, Sharp PA. 1982. Assembly of adenovirus major capsid protein is mediated by a nonvirion protein. Cell 31:407–415 [DOI] [PubMed] [Google Scholar]
- 14. Wodrich H, Guan T, Cingolani G, Von Seggern D, Nemerow G, Gerace L. 2003. Switch from capsid protein import to adenovirus assembly by cleavage of nuclear transport signals. EMBO J. 22:6245–6255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ostapchuk P, Hearing P. 2003. Regulation of adenovirus packaging. Curr. Top. Microbiol. Immunol. 272:165–185 [DOI] [PubMed] [Google Scholar]
- 16. Hasson TB, Ornelles DA, Shenk T. 1992. Adenovirus L1 52- and 55-kilodalton proteins are present within assembling virions and colocalize with nuclear structures distinct from replication centers. J. Virol. 66:6133–6142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gustin KE, Imperiale MJ. 1998. Encapsidation of viral DNA requires the adenovirus L1 52/55-kilodalton protein. J. Virol. 72:7860–7870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. D'Halluin JC, Milleville M, Boulanger PA, Martin GR. 1978. Temperature-sensitive mutant of adenovirus type 2 blocked in virion assembly: accumulation of light intermediate particles. J. Virol. 26:344–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hasson TB, Soloway PD, Ornelles DA, Doerfler W, Shenk T. 1989. Adenovirus L1 52- and 55-kilodalton proteins are required for assembly of virions. J. Virol. 63:3612–3621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ostapchuk P, Almond M, Hearing P. 2011. Characterization of empty adenovirus particles assembled in the absence of a functional adenovirus IVa2 protein. J. Virol. 85:5524–5531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang W, Imperiale MJ. 2003. Requirement of the adenovirus IVa2 protein for virus assembly. J. Virol. 77:3586–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mangel WF, Toledo DL, Ding J, Sweet RM, McGrath WJ. 1997. Temporal and spatial control of the adenovirus proteinase by both a peptide and the viral DNA. Trends Biochem. Sci. 22:393–398 [DOI] [PubMed] [Google Scholar]
- 23. Fessler SP, Young CS. 1999. The role of the L4 33K gene in adenovirus infection. Virology 263:507–516 [DOI] [PubMed] [Google Scholar]
- 24. Finnen RL, Biddle JF, Flint J. 2001. Truncation of the human adenovirus type 5 L4 33-kDa protein: evidence for an essential role of the carboxy-terminus in the viral infectious cycle. Virology 289:388–399 [DOI] [PubMed] [Google Scholar]
- 25. Kulshreshtha V, Babiuk LA, Tikoo SK. 2004. Role of bovine adenovirus-3 33K protein in viral replication. Virology 323:59–69 [DOI] [PubMed] [Google Scholar]
- 26. Schiedner G, Hertel S, Kochanek S. 2000. Efficient transformation of primary human amniocytes by E1 functions of Ad5: generation of new cell lines for adenoviral vector production. Hum. Gene Ther. 11:2105–2116 [DOI] [PubMed] [Google Scholar]
- 27. Gossen M, Bujard H. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. U. S. A. 89:5547–5551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chartier C, Degryse E, Gantzer M, Dieterle A, Pavirani A, Mehtali M. 1996. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J. Virol. 70:4805–4810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ostapchuk P, Yang J, Auffarth E, Hearing P. 2005. Functional interaction of the adenovirus IVa2 protein with adenovirus type 5 packaging sequences. J. Virol. 79:2831–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ma HC, Hearing P. 2011. Adenovirus structural protein IIIa is involved in the serotype specificity of viral DNA packaging. J. Virol. 85:7849–7855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ostapchuk P, Anderson ME, Chandrasekhar S, Hearing P. 2006. The L4 22-kilodalton protein plays a role in packaging of the adenovirus genome. J. Virol. 80:6973–6981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xi Q, Cuesta R, Schneider RJ. 2004. Tethering of eIF4G to adenoviral mRNAs by viral 100k protein drives ribosome shunting. Genes Dev. 18:1997–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]