Abstract
Plant reoviruses are thought to replicate and assemble within cytoplasmic, nonmembranous structures called viroplasms. Here, we established continuous cell cultures of the white-backed planthopper (Sogatella furcifera Horváth) to investigate the mechanisms for the genesis and maturation of the viroplasm induced by Southern rice black-streaked dwarf virus (SRBSDV), a fijivirus in the family Reoviridae, during infection of its insect vector. Electron and confocal microscopy revealed that the viroplasm consisted of a granular region, where viral RNAs and nonstructural proteins P6 and P9-1 accumulated, and a filamentous region, where viral RNAs, progeny cores, viral particles, as well as nonstructural proteins P5 and P6 accumulated. Our results suggested that the filamentous viroplasm matrix was the site for the assembly of progeny virions. Because viral RNAs were produced by assembled core particles within the filamentous viroplasm matrix, we propose that these viral RNAs might be transported to the granular viroplasm matrix. P5 formed filamentous inclusions and P9-1 formed granular inclusions in the absence of viral infection, suggesting that the filamentous and granular viroplasm matrices were formed primarily by P5 and P9-1, respectively. P6 was apparently recruited in the whole viroplasm matrix by direct interaction with P9-1 and P5. Thus, the present results suggested that P5, P6, and P9-1 are collectively required for the genesis and maturation of the filamentous and granular viroplasm matrix induced by SRBSDV infection. Based on these results, we propose a new model to explain the genesis and maturation of the viroplasms induced by fijiviruses in insect vector cells.
INTRODUCTION
Plant viruses such as tospoviruses, rhabdoviruses, marafiviruses, tenuiviruses, and plant reoviruses are transmitted by their respective insect vectors in a persistent-propagative manner (1, 2). Plant reoviruses are found in the genera Phytoreovirus, Fijivirus, and Oryzavirus in the family Reoviridae (3). The insect species for transmission of persistent-propagative plant viruses includes thrips, aphids, leafhoppers, and planthoppers (2). Gaining insight into the mechanism(s) underlying viral replication in the insect vectors for these plant viruses may lead to new strategies to control viral transmission by the insect vectors. Continuous cell cultures of leafhopper have been extensively used to study the replication cycles of phytoreoviruses in their insect vectors, including viral entry, replication, and spread (4–9). Primary cell cultures of aphids, thrips, and white-backed planthopper (WBPH) (Sogatella furcifera Horváth) have been developed to investigate the localization of viral proteins from Sowthistle yellow vein virus, a rhabdovirus; Tomato spotted wilt virus, a tospovirus; and Southern rice black-streaked dwarf virus (SRBSDV), a tentatively identified species in the genus Fijivirus, in their respective insect vectors (3, 10–12). Because continuous insect cell cultures are uniquely able to develop a uniform and synchronous viral infection, we can use them to trace the replication process of plant virus (4–6). Here, we established continuous cell cultures of WBPH to study the replication cycle of SRBSDV in the vector insect.
SRBSDV, transmitted by WBPH vectors in a persistent-propagative manner, has spread rapidly throughout southern China, northern Vietnam, and western Japan (10, 13–18). SRBSDV virions are double shelled, spherical, and 65 to 70 nm in diameter (13, 17, 18). The core, approximately 50 nm in diameter, encapsidates 10 segments of double-stranded RNA (dsRNA) (13, 17, 18). Phylogenetic analyses showed that SRBSDV is most closely related to two other fijiviruses, Rice black-streaked dwarf virus (RBSDV) and Mal de Río Cuarto virus (MRCV) (17, 19). A comparison of the different genomic segments of SRBSDV with their counterparts in RBSDV and MRCV suggests that SRBSDV encodes at least six putative structural proteins (P1, P2, P3, P4, P8, and P10) and six putative nonstructural proteins (P5, P6, P7-1, P7-2, P9-1, and P9-2) (17, 19, 20). Among the putative structural proteins, P1 contains the characteristic sequence motifs of an RNA-dependent RNA polymerase; P2 and P4 are the putative capsid shell protein and spike protein, respectively; P3 is a putative capping enzyme; and P8 and P10 are the core and major outer capsid proteins, respectively (17, 21). However, the process of viral replication and assembly of progeny core and virions during viral infection is still poorly understood.
Replication and assembly of progeny virions of plant reoviruses are thought to occur within granular or filamentous inclusion bodies, namely, the viroplasm, during viral infection of the insect vectors (3). Currently, the mechanisms for the genesis and maturation of the viroplasm induced by fijiviruses remain largely unknown due to the lack of continuous cell lines derived from their insect vectors. Using a primary cell culture of WBPH, we recently clarified that newly synthesized viral RNAs, outer capsid structural proteins, and viral particles accumulated within the matrix of the viroplasms induced by SRBSDV infection and revealed that the nonstructural protein P9-1 plays an essential role in viroplasm formation and viral replication (10). However, whether other nonstructural proteins are involved in the genesis and maturation of the viroplasm induced by SRBSDV infection is still unknown. P6 of SRBSDV has about 63% amino acid identity with its counterpart, P6 of RBSDV, which forms viroplasm-like structures when expressed in nonhost plant cells through its interaction with P9-1, suggesting that P6 of SRBSDV might also be involved in viroplasm formation (17, 22). Of the other four putative nonstructural proteins of SRBSDV, P7-1 is a component of virus-containing tubular structures (23), P7-2 and P9-2 have not been detected in Western blots of SRBSDV-infected plant and insect tissues (our unpublished data), and the function of P5 is unknown (17). Thus, P7-1, P7-2, and P9-2 may not be involved in the formation of the viroplasm matrix.
In the present study, we successfully established continuous cell cultures of WBPH to investigate the mechanisms underlying the genesis and maturation of the viroplasm induced by SRBSDV infection in its insect vector. Our results suggest that the viral nonstructural proteins P5, P6, and P9-1 are collectively required for the genesis and maturation of the viroplasm induced by SRBSDV infection.
MATERIALS AND METHODS
Antibody preparations.
SRBSDV P9-1 and P10 antibodies and rabbit polyclonal antisera against P5, P6, and P8 of SRBSDV were prepared as described previously (10, 24). IgG was isolated from specific polyclonal antisera by using a protein A-Sepharose affinity column (Pierce). IgGs were conjugated directly to fluorescein isothiocyanate (FITC) or rhodamine according to the manufacturer's instructions (Invitrogen).
Establishment of continuous cell cultures derived from WBPH for viral infection.
The WBPH cell line was established by adapting the protocol of Kimura and Omura (5). The primary cell cultures of WBPH, originally established from embryonic fragments dissected from eggs of WBPH, were maintained in a monolayer culture at 25°C in Kimura's insect medium (10). When the cells grew almost confluently in the dish about 1 month after initiation, they were transferred into new dishes and again cultured to confluence (about 2 weeks after subculture). Such cells were then transferred into culture flasks for further subculturing. After 40 passages of subculturing at 10-day intervals, the dominant cell type in the established WBPH cell line was epithelial-like and approximately 25 to 35 μm in diameter (Fig. 1). Such vector cells in monolayers (VCMs) of WBPH were used for SRBSDV infection.
Fig 1.
Light micrograph of a monolayer culture of the WBPH cell line after 40 passages, showing the predominantly epithelial-like cell population. Bar, 30 μm (phase contrast).
Synchronous infection of VCMs by SRBSDV was initiated as described previously (6, 25). VCMs used for an infectivity assay of the virus were grown on coverslips with a 15-mm diameter. To prepare monolayers, we harvested cells from 2- to 3-day-old cultures and seeded them over the entire coverslip. When the monolayer on the coverslip reached 90% confluence, cells were washed with a solution of 0.1 M histidine that contained 0.01 M MgCl2 (pH 6.2) (His-Mg) and then inoculated with the viral inoculum from infected plants. After 2 h, cells were washed with His-Mg and then covered with growth medium prior to fixation.
Immunofluorescence microscopy.
VCMs grown on glass coverslips were fixed for 30 min in 4% paraformaldehyde and processed for immunofluorescence microscopy, as described previously (6, 9). Cells were incubated with a 100-fold-diluted solution of the directly conjugated IgG. Cells were visualized with a Leica TCS SP5 confocal microscope as described previously (10, 14).
Immunofluorescence detection of newly synthesized viral RNAs in vivo.
VCMs on coverslips were inoculated with SRBSDV. Infection was allowed to proceed for 34.5, 58.5, or 82.5 h, and cells were then treated for 30 min with actinomycin D (Sigma) to inhibit cellular RNA polymerase II. Cells were transfected with bromouridine 5′-triphosphate (BrUTP) in the presence of Cellfectin (Invitrogen), according to the manufacturer's instructions, and were incubated for 60 min before fixation and immunostaining. BrUTP-labeled viral RNAs were stained with anti-bromodeoxyuridine (BrdU) from mouse (Sigma), followed by anti-mouse IgG conjugated to rhodamine (Sigma).
Electron microscopy.
VCMs on coverslips were fixed, dehydrated, and embedded as described previously (6, 7). Cell sections were then incubated with IgGs and immunogold labeled with goat antibodies against rabbit IgG that had been conjugated with 15-nm gold particles (Sigma) (6, 7).
Baculovirus expression of nonstructural proteins of SRBSDV.
The baculovirus system was used to express three nonstructural proteins, P5, P6, and P9-1, as described previously (6, 7, 10, 24). Recombinant baculovirus vectors containing P5, P6, and P9-1 were introduced into DH10Bac (Invitrogen) for transposition into the bacmid. Sf9 cells were transfected with recombinant bacmids in the presence of Cellfectin (Invitrogen), incubated for 72 h, and then fixed in 4% paraformaldehyde and processed for immunofluorescence microscopy as described previously (10, 14, 24).
Yeast two-hybrid assay.
Yeast cells were transformed by using a lithium acetate-based protocol for preparing and transforming yeast competent cells in the user manual with the DUALmembrane pairwise interaction kit (Dualsystems Biotech).
RESULTS
Granular and filamentous morphology of viroplasm induced by SRBSDV infection.
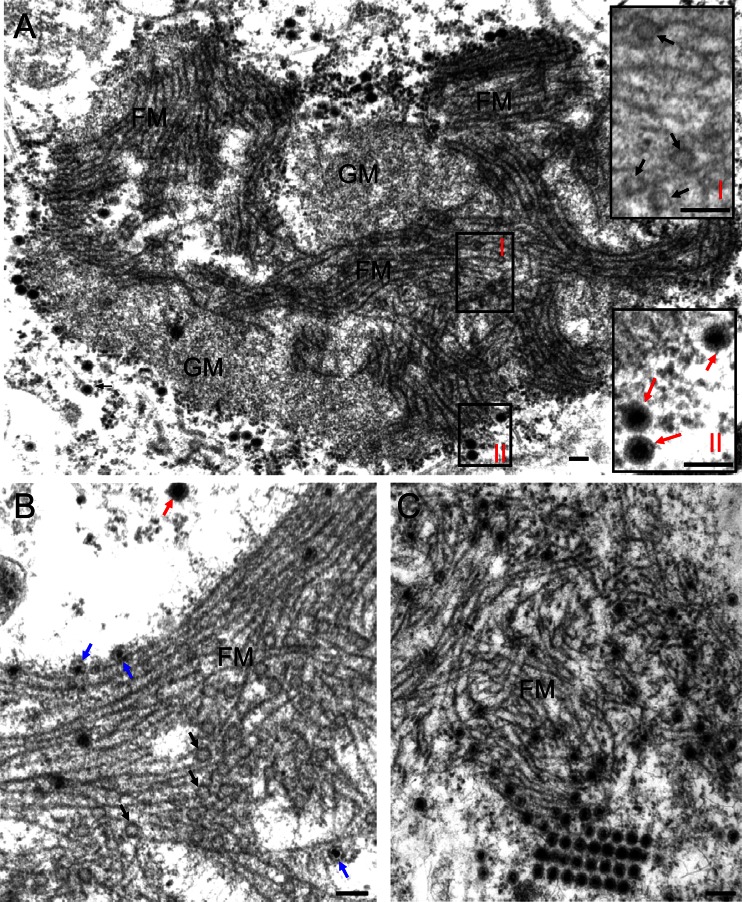
In electron micrographs of thin sections of SRBSDV-infected VCMs at 84 h postinoculation (hpi), the viroplasm matrix contained a granular, electron-dense region mostly devoid of virus particles and a filamentous region where subviral or viral particles had accumulated (Fig. 2). A careful analysis of the micrographs suggested a possible sequential process of the assembly of progeny virions during viral infection. Initially, a large mass of semiopaque (empty) particles, ∼50 nm in diameter, possibly inner capsids, was closely associated with the edges of filaments (∼7 nm in diameter) within the filamentous region (Fig. 2A). Apparently, when charged with viral nucleic acid, these inner capsids became densely stained and were still confined to the filamentous region of the viroplasm (Fig. 2B). Finally, the filled single-layer core particles apparently matured into intact, double-layered viral particles with a diameter of ∼70 nm, which accumulated within or at the periphery of the filamentous matrix of the viroplasm (Fig. 2C). SRBSDV particles at the margin of the filamentous matrix appeared to aggregate to form paracrystalline arrays (Fig. 2C). The filamentous matrix of the viroplasm thus appeared to be the site for the assembly of progeny viral particles during viral replication.
Fig 2.
Transmission electron micrographs of the viroplasms induced by SRBSDV infection in VCMs. (A) The viroplasm matrix consists of granular and filamentous regions. Insets I and II are enlarged images of boxed areas I and II, respectively. (B) Within the filamentous viroplasm matrix, the core of the single-layer particles was densely stained, indicating that viral RNAs are packaged into empty core particles. (C) Within or at the periphery of the filamentous viroplasm matrix, double-layer viral particles accumulated. Black arrows mark single-layer empty particles (∼50 nm in diameter) on the edges of filaments. Blue arrows show single-layer core particles (∼50 nm in diameter). Red arrows mark intact double-layer viral particles (∼70 nm in diameter). GM, granular matrix; FM, filamentous matrix. Bars, 100 nm.
Viral nonstructural proteins P5 and P6 were the components of the filamentous matrix; P6 and P9-1 were the components of the granular matrix of the viroplasm.
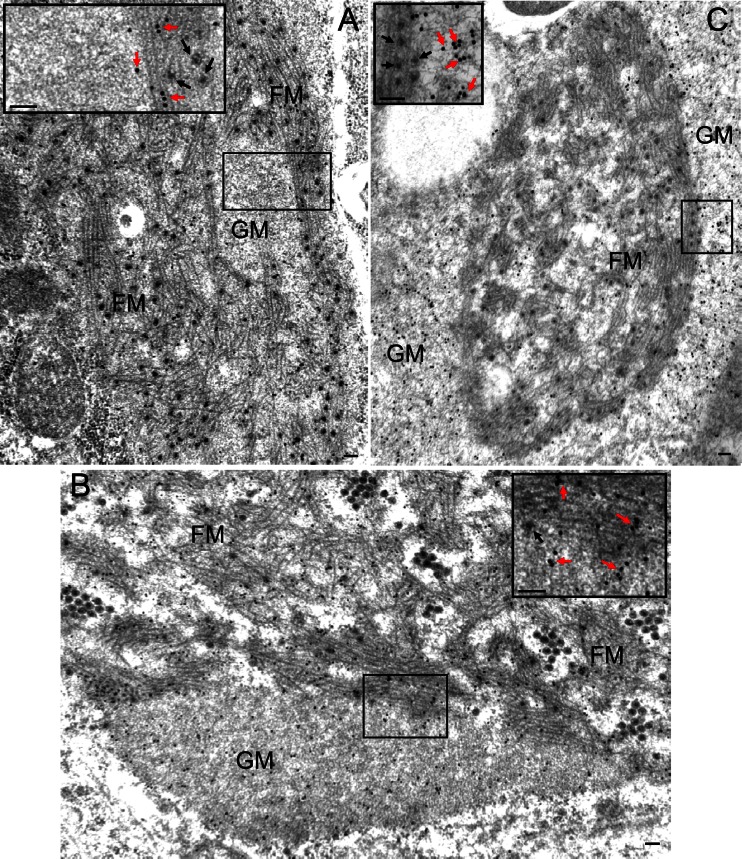
We used immunogold electron microscopy to localize nonstructural proteins P5, P6, and P9-1 of SRBSDV in infected VCMs at 84 hpi and to determine their role in the formation of the matrix of viroplasm. Our results showed that P5 antibodies reacted specifically with the filamentous matrix of the viroplasm (Fig. 3A), whereas P6 antibodies reacted evenly with both the granular and filamentous regions (Fig. 3B). These results suggested that P5 was associated only with the filamentous matrix, whereas P6 was involved in the formation of both the filamentous and granular matrices of the viroplasm. On the other hand, P9-1 antibodies reacted strongly with the granular matrix and reacted poorly with the filamentous matrix (Fig. 3C), suggesting that P9-1 was the major component of the granular matrix of viroplasm. None of these three nonstructural proteins were associated with viral particles (Fig. 3). Taken together, our results suggested that the viral nonstructural proteins P5, P6, and P9-1 were involved in the formation of the viroplasm induced by SRBSDV infection.
Fig 3.
Viral nonstructural proteins P5, P6, and P9-1 are the constituents of the matrix of the viroplasm induced by SRBSDV infection in VCMs. Shown is immunogold labeling of P5 (A), P6 (B), and P9-1 (C) in the matrix of the viroplasm in SRBSDV-infected VCMs at 84 hpi. Insets are the enlarged images of the boxed areas. Cells were immunostained for P5, P6, and P9-1 with P5-, P6-, and P9-1-specific antibodies in panels A, B, and C, respectively, as primary antibodies, followed by treatment with goat antibodies against rabbit IgG that had been conjugated with 15-nm-diameter gold particles as secondary antibodies. Black arrows, viral particles; red arrows, gold particles; GM, granular matrix; FM, filamentous matrix. Bars, 100 nm.
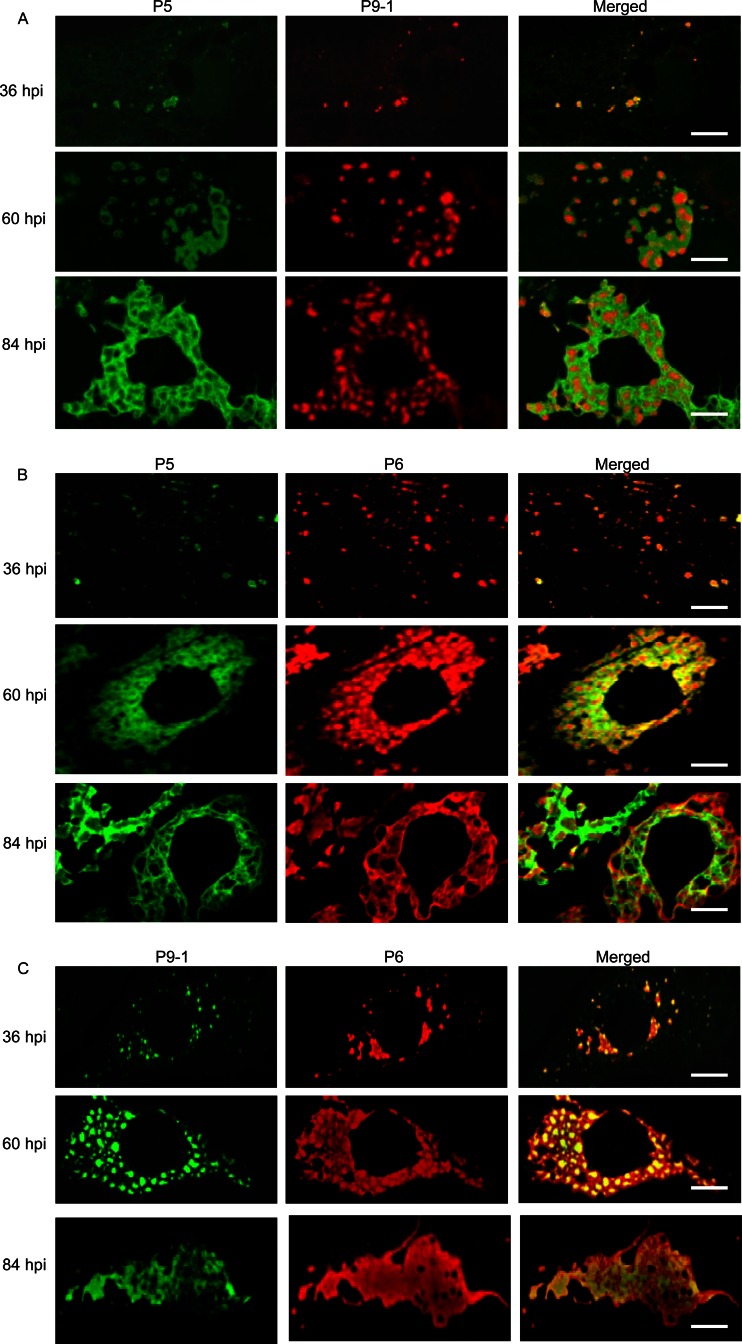
To confirm our observations, we used immunofluorescence microscopy to examine the subcellular distribution of P5, P6, and P9-1 over a time course of viral infection. We immunostained SRBSDV-infected VCM with P5-specific IgG conjugated to FITC (P5-FITC) and P9-1-specific IgG conjugated to rhodamine (P9-1–rhodamine), with P5-FITC and P6-specific IgG conjugated to rhodamine (P6-rhodamine), or with P9-1-specific IgG conjugated to FITC (P9-1–FITC) and P6-rhodamine. When the cells were examined by immunofluorescence microscopy as described above, P5, P6, and P9-1 were first detectable at 36 hpi, when they were scattered throughout the cytoplasm in discrete, punctate inclusions (Fig. 4A to C). These proteins had obvious regions of colocalization throughout the cytoplasm (36 hpi) (Fig. 4A to C). By 60 hpi, P9-1 had formed granular inclusions throughout the cytoplasm, whereas P5 was localized to ringlike structures at the margins of the granular inclusions of P9-1 (Fig. 4A). P6 was distributed throughout the central granular region of P9-1 and the peripheral region of P5 (60 hpi) (Fig. 4B and C). By 84 hpi, granular inclusions of P9-1 were irregularly grouped to form a large, amorphous mass (Fig. 4A and C), and the earlier ringlike structures of P5 had merged to ultimately form filamentous and reticular structures, which were evident at the periphery of each granular inclusion of P9-1 in the aggregates (Fig. 4A). P6 was distributed throughout the granular region of P9-1 and the filamentous region of P5 (84 hpi) (Fig. 4B and C). The granular inclusion of P9-1 and the reticular structures of P5 obviously corresponded to the granular and fibrillar matrices of the viroplasm, respectively, as viewed by electron microscopy (Fig. 2 and 3). Taken together, our results indicated that the viral nonstructural proteins P5, P6, and P9-1 had different roles in the formation of viroplasm during viral infection and were apparently required for the genesis and maturation of viroplasm in virus-infected cells.
Fig 4.
Confocal immunofluorescence micrographs of colocalization of viral nonstructural proteins P5, P6, and P9-1 within the matrix of the viroplasm induced by SRBSDV infection in VCMs. SRBSDV-infected VCMs were immunostained with P5-FITC and P9-1–rhodamine (A), with P5-FITC and P6-rhodamine (B), or with P9-1–FITC and P6-rhodamine (C) at 36, 60, and 84 hpi. Bars, 5 μm.
P6 was recruited to the granular inclusion formed by P9-1 or to the filamentous inclusion formed by P5 through interaction with P9-1 or P5.
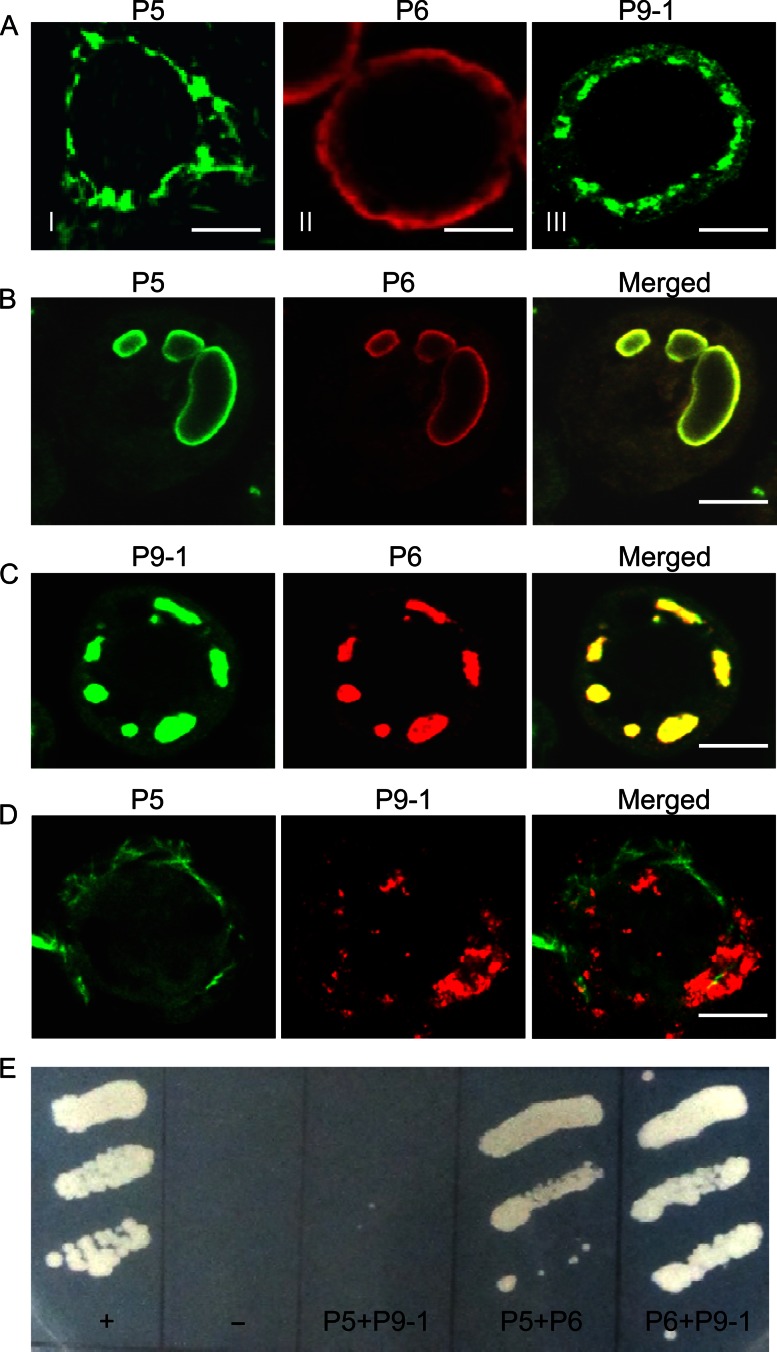
To identify which nonstructural protein encoded by SRBSDV had an inherent ability to form the viroplasm matrix, we used a baculovirus system to express one of the three nonstructural proteins P5, P6, and P9-1 in nonhost Sf9 cells. As seen by immunofluorescence microscopy, P5 was aggregated to form filament-like structures in the cytoplasm (Fig. 5AI), P6 was distributed diffusely throughout the cytoplasm (Fig. 5AII), and P9-1 was aggregated to form granular inclusions in the cytoplasm (Fig. 5AIII). Thus, our results confirmed that P9-1 can form granular matrix-like structures in the absence of viral proliferation, as described previously (10). The filamentous distribution of P5 in Sf9 cells suggested that the filamentous viroplasm matrix in SRBSDV-infected cells was composed primarily of P5. Furthermore, P6 did not form viroplasm-like inclusions when expressed alone. This finding, combined with our observation that P6 could colocalize with P5 and P9-1 in the different regions of the viroplasm throughout infection, led us to hypothesize that P6 might be recruited to inclusions of P5 or P9-1 when they were coexpressed in Sf9 cells.
Fig 5.
SRBSDV P6 was associated with inclusions formed by P5 or P9-1 through protein-protein interactions. (A to D) Subcellular localization of nonstructural proteins P5, P6, and P9-1 in the absence of viral infection. Sf9 cells infected with recombinant baculoviruses containing P5, P6, or P9-1 were fixed 3 days after infection and prepared for immunofluorescence microscopy. Cells were immunostained with P5-FITC, P6-rhodamine, P9-1–FITC, or P9-1-*rhodamine. (A) P5 formed filament-like structures (frame I), P6 was distributed diffusely in the cytoplasm (frame II), and P9-1 formed punctate structures (frame III). Bars, 5 μm. (B) P6 and P5 were colocalized on large circular structures when coinfected with recombinant baculoviruses containing either P6 or P5. Bar, 5 μm. (C) P6 and P9-1 colocalized to punctate structures when coinfected with recombinant baculoviruses containing either P6 or P9-1. Bar, 5 μm. (D) The filament-like structures of P5 were not associated with the punctate structures of P9-1 when coinfected with recombinant baculoviruses containing either P5 or P9-1. Bar, 5 μm. (E) Yeast two-hybrid assay of protein-protein interactions among the SRBSDV P5, P6, and P9-1 proteins. Transformants were plated onto synthetic defined minimal medium minus leucine, tryptophan, histidine, and adenine (SD/−Leu/−Trp/−His/−Ade) for 4 days. +, positive control, i.e., pTSU2-APP/pNubG-Fe65; −, negative control, i.e., pTSU2-APP+pRR3-N; P5+P9-1, pBT3-STE-P5+pPR3-N-P9-1; P5+P6, pBT3-STE-P5+pPR3-N-P6; P6+P9-1, pBT3-STE-P6+pPR3-N-P9-1.
To confirm this hypothesis, we coinfected Sf9 cells with recombinant baculoviruses encoding P5, P6, or P9-1 and examined the localization by immunostaining. Coexpression experiments revealed that P6 and P5 were associated with the large circular structures (Fig. 5B). Furthermore, P6 and P9-1 were colocalized on apparent granular inclusions in the cytoplasm (Fig. 5C). In contrast, P5 was not observed in association with the inclusions formed by P9-1 (Fig. 5D). Thus, our findings suggested that P6 might be recruited to the granular or filamentous matrices of the viroplasm through the direct interaction of P6 with P9-1 and P5 during viral infection.
The DUALmembrane system, a variant of the yeast two-hybrid assay, was used to determine whether there were any protein-protein interactions among the three proteins P5, P6, and P9-1. Our results showed that P6 interacted directly with P5 and P9-1 (Fig. 5E). However, P5 did not interact with P9-1 (Fig. 5E). Taken together, these results provided convincing evidence that P6 localized to the granular matrix of the viroplasm via its direct interaction with P9-1 and to the filamentous matrix of the viroplasm by direct interaction with P5.
Assembly of progeny virions within the filamentous viroplasm matrix.
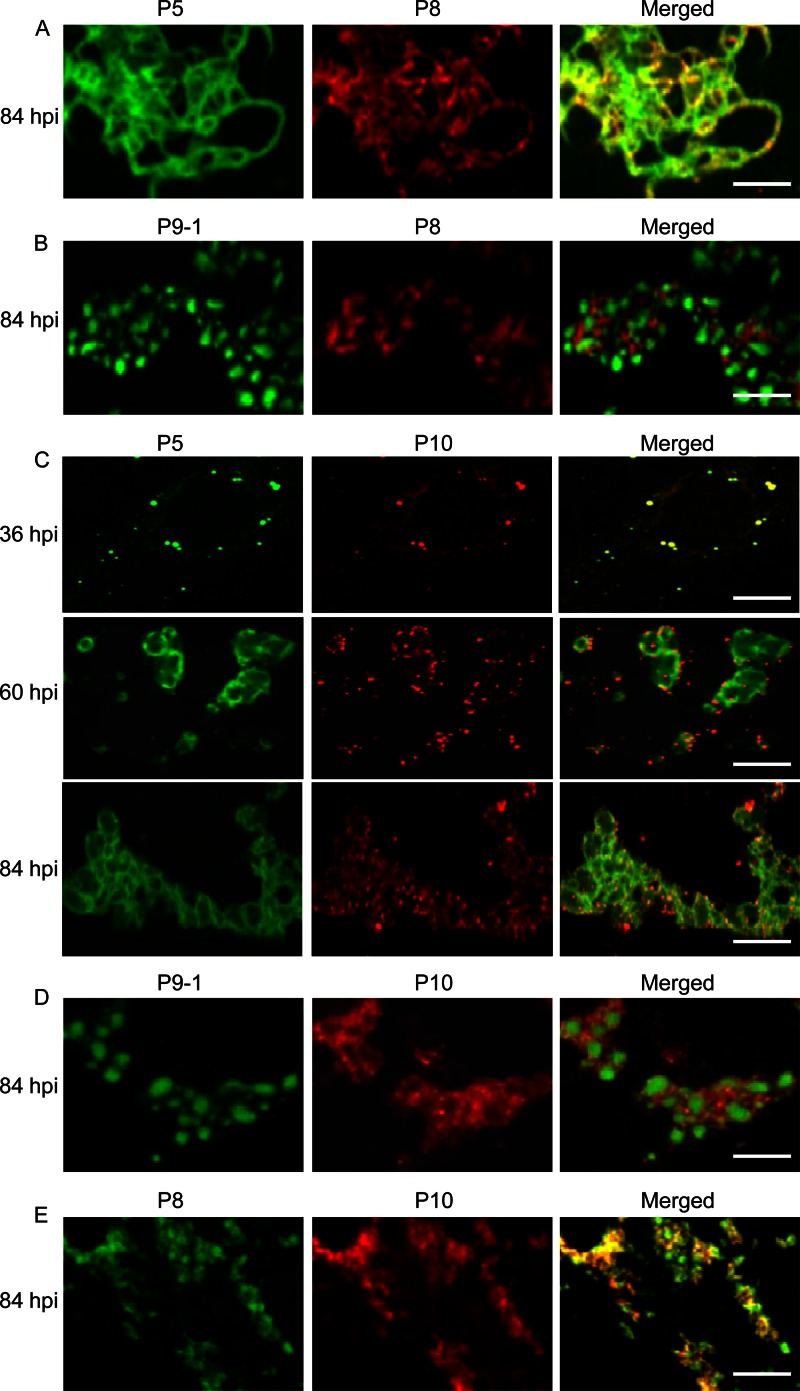
To determine whether the filamentous viroplasm matrix was required for assembly of core particles during virus replication, infected cells were fixed at 84 hpi, immunostained with P5-FITC and viral core protein P8-specific IgG conjugated to rhodamine (P8-rhodamine) or with P9-1–FITC and P8-rhodamine, and then imaged by immunofluorescence microscopy. During viral infection, core protein P8 became associated with P5 on the filamentous materials in the reticular structures formed by P5 (Fig. 6A). However, P8 had accumulated in the space between adjacent granular inclusions of P9-1 in the aggregates (Fig. 6B). Thus, our observations suggested that the filamentous viroplasm matrix was the probable site of core particle assembly.
Fig 6.
Confocal immunofluorescence micrographs of the colocalization of SRBSDV core protein P8 and outer capsid protein P10 with nonstructural proteins P5 and P9-1 within the matrix of the viroplasm induced by SRBSDV infection in VCMs. SRBSDV-infected VCMs were immunostained with P5-FITC and P8-rhodamine (A), with P9-1–FITC and P8-rhodamine (B), with P5-FITC and P10-rhodamine (C), with P9-1–FITC and P10-rhodamine (D), or with P8-FITC and P10-rhodamine (E). Bars, 5 μm.
To determine whether the filamentous viroplasm matrix was required for assembly of complete virions during virus replication, we used immunofluorescence microscopy to examine the colocalization of viral outer capsid protein P10 and P5 over a time course of viral infection. Infected cells were fixed at 36, 60, and 84 hpi; immunostained with P5-FITC and P10-specific IgG conjugated to rhodamine (P10-rhodamine); and then imaged by immunofluorescence microscopy. At an early stage after viral infection, by 36 hpi, the outer capsid protein P10 was visualized as small, discrete punctate inclusions, which were attached to the punctate inclusions of P5 (Fig. 6C). By 60 hpi, P10 was then associated with the ringlike structures formed by P5 (Fig. 6C). By 84 hpi, P10 was attached to the filamentous materials of the reticular structures formed by P5 (Fig. 6C). Thus, our observations confirmed that the filamentous viroplasm matrix was the site for the accumulation of viral particles. To further observe the colocalization of P10 with P9-1 and P8, infected cells were fixed at 84 hpi, immunostained with P9-1–FITC and P10-rhodamine or with P8-specific IgG conjugated to FITC (P8-FITC) and P10-rhodamine, and then imaged by immunofluorescence microscopy. As expected, punctate inclusions of P10 were present in the vicinity of granular inclusions of P9-1 (Fig. 6D). Moreover, outer capsid protein P10 and core protein P8 were colocalized on the punctate inclusions (Fig. 6E). Taken together, the above-described observations suggested that the filamentous viroplasm matrix, rather than the granular viroplasm matrix, was the probable site of progeny virion assembly in which the outer capsid proteins became attached to core particles.
Accumulation of newly synthesized viral RNAs within the granular and filamentous viroplasm matrices.
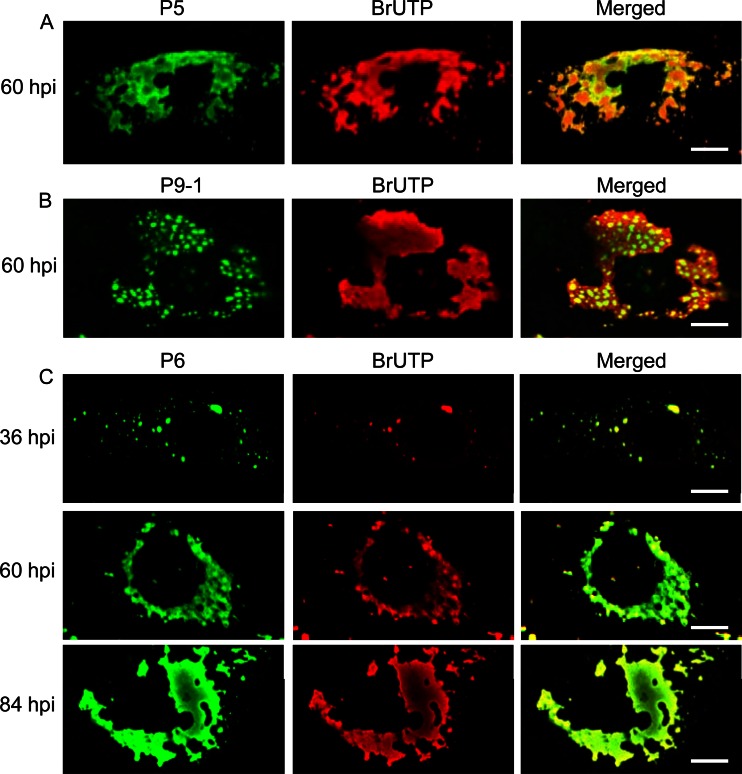
Using immunofluorescence microscopy, by 60 hpi, we observed that the newly synthesized SRBSDV RNAs were distributed throughout the whole viroplasm matrix, including the granular region formed by P9-1 and the filamentous region formed by P5 (Fig. 7A and B). Because P6 was distributed throughout the granular region of P9-1 and the filamentous region of P5, we then investigated the distribution of newly synthesized viral RNAs within the whole matrix of viroplasm by colocalization of BrUTP and P6 over a time course of viral infection. Our results showed that SRBSDV RNAs were accumulated within the whole matrix of viroplasm at 36, 60, and 84 hpi (Fig. 7C). Thus, both the granular and filamentous matrices of the viroplasm were apparently the sites for the accumulation of SRBSDV RNAs during viral replication.
Fig 7.
Intracellular sites of RNA synthesis in SRBSDV-infected VCMs. BrUTP-labeled viral RNA was immunostained with anti-BrdU from mouse, followed by anti-mouse IgG conjugated to rhodamine. (A) Filamentous viroplasm matrix immunostained with P5-FITC. (B) Granular viroplasm matrix immunostained with P9-1–FITC. (C) Whole viroplasm matrix immunostained with P6-FITC. Bars, 5 μm.
DISCUSSION
The greater uniformity of continuous cell cultures of insect vectors than of primary cell cultures of insect vectors enabled us to develop a consistent and synchronous viral infection in insect vector cells (4–6). In this study, using a continuous cell culture of the insect vector in a monolayer (VCM) derived from WBPH, we were able to trace the molecular entities responsible for biological events during SRBSDV replication in its insect vector cells. We showed that the viroplasm induced by SRBSDV infection in VCMs consisted of a granular region, where viral RNAs and nonstructural proteins P6 and P9-1 accumulated, and a filamentous region, where viral RNAs, progeny cores, viral particles, as well as nonstructural proteins P5 and P6 accumulated (Fig. 2 to 4, 6, and 7). P5 formed filamentous inclusions and P9-1 formed granular inclusions in the absence of viral infection (Fig. 5), suggesting that the filamentous and granular viroplasm matrices were formed primarily by P5 and P9-1, respectively. P6 was apparently recruited in the whole viroplasm matrix by direct interaction with P9-1 and P5 (Fig. 5). The direct interaction of P6 with P5 and P9-1 may be the reason why the filamentous matrix formed by P5 was distributed in close proximity to the granular matrix formed by P9-1 during SRBSDV infection. Taken together, our cytopathological analyses strongly suggest that the viral nonstructural proteins P5, P6, and P9-1 are collectively required for the genesis and maturation of the granular and filamentous matrices of the viroplasm in virus-infected cells.
The continuous cell culture of WBPHs allowed us to define the process of progeny virion assembly during viral infection. Our results demonstrated that empty core particles initially were assembled and aligned along the filaments within the viroplasm filamentous matrix formed by P5 and that viral RNAs apparently were then packaged into these preformed, empty core particles to assemble viral RNA-containing core particles (Fig. 2 to 4, 6, and 7). These observations are consistent with those of dsRNA bacteriophage φ6 of the Cystoviridae family, in which viral plus-strand RNAs are packaged into empty core particles (26, 27). The packaging of viral plus-strand RNAs would be followed by minus-strand RNA synthesis, yielding dsRNAs that are protected in core particles (Fig. 2), as in the case of viruses in the family Reoviridae (3, 28). Our observations also showed that the virions matured within the filamentous viroplasm matrix, where the outer capsid proteins were assembled on newly formed core particles to generate mature virions (Fig. 2 and 6). On the basis of these results, we propose a model in which filamentous viroplasm matrix formed by nonstructural protein P5 might recruit and concentrate nonstructural protein P6, viral structural proteins, viral RNAs, and, possibly, host factors, thus building the blocks needed for viral assembly.
Our present study suggests that the granular viroplasm matrix formed by nonstructural protein P9-1 can recruit nonstructural protein P6 and viral RNAs. P9-1 of SRBSDV has about 77% amino acid identity with its counterpart, P9-1 of RBSDV (17). In RBSDV, P9-1 has a nonspecific RNA-binding ability and forms an octameric structure resembling the structures formed by NSP2 of a rotavirus and Pns9 of Rice gall dwarf virus, a phytoreovirus (20). Thus, P9-1 of SRBSDV may have a structure and RNA-binding activities similar to those of its counterpart, P9-1 of RBSDV. Furthermore, P6 of SRBSDV is a viral RNA-silencing suppressor in the plant host and contains the amino acid sequence motifs typical of an RNA-binding protein (17, 22, 29), suggesting that this protein might have RNA-binding activities. For viruses in the family Reoviridae, the core serves as the transcriptase particle for the production of virus plus-strand RNAs (3, 28); thus, we hypothesized that the localization of the cores to the filamentous viroplasm matrix of P5 might lead to the production of viral plus-strand RNAs during infection. This hypothesis is supported by our finding that the newly synthesized BrUTP-containing viral RNAs were localized in both the filamentous and granular viroplasm matrices (Fig. 7). On the basis of these results, we propose one mechanism for the generation, accumulation, and trafficking of viral RNAs within the matrix of the viroplasm. SRBSDV RNAs initially are synthesized by core particles localized within the filamentous viroplasm matrix and then migrate to the granular viroplasm matrix via nonstructural protein P6, which has possible RNA-binding activities and is distributed throughout the viroplasm matrix. The granular viroplasm matrix acts as a reservoir for retaining a large amount of viral RNAs. The viral RNAs within the granular viroplasm matrix might migrate to the adjacent cytoplasm to direct the synthesis of viral proteins that promote further maturation of viroplasms or migrate back to the filamentous viroplasm matrix for core assembly. Thus, the recruitment or retention of viral RNA within the granular viroplasm matrix should benefit viral replication by maximizing viral RNA and protein production and by supporting the maturation of filamentous viroplasm matrix, confirming that P9-1 is essential for viroplasm formation and viral replication (10).
In the case of phytoreoviruses and oryzaviruses, the electron-dense inclusion bodies, called viroplasms, are globular structures identified as the machinery of viral replication and assembly of progeny virions (6, 7, 24). However, in the case of SRBSDV, a fijivirus, the granular viroplasm matrix formed by P9-1 was the site for the accumulation of newly synthesized viral RNA rather than for the assembly of viral particles. These analyses suggested that the granular viroplasm matrix formed by P9-1 may confer a selective disadvantage or, reciprocally, that the filamentous viroplasm matrix formed by P5 may confer a selective advantage for the assembly of progeny viral particles during persistent infection of SRBSDV in its WBPH vectors. One consequence of a filamentous matrix versus a globular matrix morphology is that the surface-area-to-volume ratio is much larger. The larger surface area of filamentous matrix may allow for more efficient viral replication and assembly through better access to newly synthesized viral proteins or RNAs. Thus, the filamentous matrix would provide a larger surface area for assembly of the outer capsid proteins onto newly forming core particles to generate a large amount of mature virions, as shown in Fig. 2. Furthermore, the filamentous matrix may also allow more efficient release of newly assembled viral particles from the edge of the viroplasm. Thus, we hypothesize that the viroplasm induced by SRBSDV in its WBPH vectors can produce many more progeny virions than the oryzaviruses or phytoreoviruses can in their respective insect vectors. To confirm this hypothesis, we compared the concentrations of SRBSDV and Rice dwarf virus (RDV), a phytoreovirus, in their respective infected VCMs and found that SRBSDV was about 10 times more concentrated than RDV in the VCMs (data not shown). Although the data obtained here are not direct evidence, the greater ability of SRBSDV to multiply in insect vector cells than of RDV may be attributed to the assembly of progeny virions within the filamentous viroplasm matrix. Because a persistent-propagative plant virus must encounter multiple tissues barriers in the path from the alimentary canal to the salivary glands in the vector insect (1, 2, 14), the highly efficient propagation of SRBSDV in WBPH vectors may confer a greater ability for the virus to overcome these barriers, consistent with the fact that SRBSDV can be transmitted by WBPH vectors with high efficiency (14, 16). These analyses suggest that SRBSDV, the first WBPH-borne reovirus, has evolved to be well adapted for persistent infection and maintenance in its WBPH vectors.
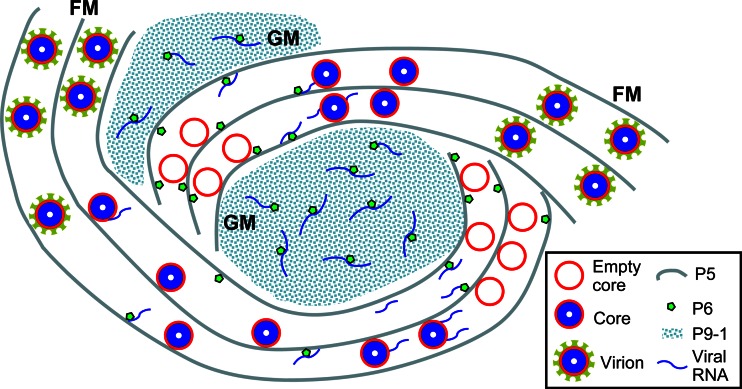
In light of the present data, we have developed a model to explain the assembly of viroplasm induced by SRBSDV and probably other fijiviruses in cells of the insect vectors (Fig. 8). In this model, following the entry and release of the virus into the cytoplasm, the SRBSDV core particle begins transcribing viral plus-strand RNAs, as in the case of other viruses in the family Reoviridae (3, 28). SRBSDV proteins, including the nonstructural proteins P5, P6, and P9-1, are synthesized by the host's cellular translational machinery. Following translation, P5 and P9-1 may associate to form small filamentous and granular viroplasm matrices, respectively. Proteins required for the assembly of progeny core particles, as well as P6, associate with filamentous matrix of P5 by either a direct or indirect association with P5. SRBSDV RNAs are bound by viral proteins to form replication and assembly complexes for the production of progeny core particles and virions within the filamentous matrix. P9-1 acts as a cytoplasmic scaffolding protein for the recruitment of P6 to the granular matrix. The filamentous matrix-localized core particle continues to transcribe the SRBSDV RNAs, some of which migrate to the granular matrix of P9-1, possibly by association with P6. The small viroplasms move and merge with other viroplasms during viral infection, ultimately forming the large viroplasms that are characteristic of SRBSDV-infected cells. Further molecular identification and characterization of the vector components involved in the formation of the viroplasm matrix induced by SRBSDV infection will certainly shed light on the replication mechanism of fijiviruses and related viruses.
Fig 8.
Proposed model for the assembly of viroplasm induced by SRBSDV in cells of its insect vector. The viroplasm consists of a granular matrix formed by nonstructural protein P9-1 for the accumulation of viral RNA and a filamentous matrix formed by nonstructural protein P5 for the assembly of progeny core and virions. Viral nonstructural protein P6 was concentrated in the granular or filamentous matrices by direct interaction with P9-1 or P5. Viral RNAs are produced by assembled core particles within the filamentous matrix, which might then be transported to the granular matrix by association with P6. GM, granular matrix; FM, filamentous matrix.
ACKNOWLEDGMENTS
This work was supported by grants from the Natural Science Foundation of China (31200118, 31130044, and 31070130), the Natural Science Foundation of Fujian Province (2012J01087, 2011J06008, and 2010J01075), and the Fund of the Ministry of Education of China (20113515130002, 20103515120007, and 211082).
Footnotes
Published ahead of print 10 April 2013
REFERENCES
- 1. Ammar ED, Tsai CW, Whitfield AE, Redinbaugh MG, Hogenhout SA. 2009. Cellular and molecular aspects of rhabdovirus interactions with insect and plant hosts. Annu. Rev. Entomol. 54:447–468 [DOI] [PubMed] [Google Scholar]
- 2. Hogenhout SA, Ammar ED, Whitfield AE, Redinbaugh MG. 2008. Insect vector interactions with persistently transmitted viruses. Annu. Rev. Phytopathol. 46:327–359 [DOI] [PubMed] [Google Scholar]
- 3. Attoui H, Mertens PPC, Becnel J, Belaganahalli S, Bergoin M, Brussaard CP, Chappell JD, Ciarlet M, del Vas M, Dermody TS, Dormitzer PR, Duncan R, Fcang Q, Graham R, Guglielmi KM, Harding RM, Hillman B, Makkay A, Marzachì C, Matthijnssens J, Milne RG, Mohd JF, Mori H, Noordeloos AA, Omura T, Patton JT, Rao S, Maan M, Stoltz D, Suzuki N, Upadhyaya NM, Wei C, Zhou H. 2012. Family Reoviridae, p 541–637 In King AMQ, Adams MJ, Carstens EB, Lefkowits EJ. (ed), Virus taxonomy. Ninth report of the International Committee for the Taxonomy of Viruses Elsevier Academic Press, New York, NY [Google Scholar]
- 4. Creamer R. 1993. Invertebrate tissue culture as a tool to study insect transmission of plant viruses. In Vitro Cell Dev. Biol. 29A:284–288 [DOI] [PubMed] [Google Scholar]
- 5. Kimura I, Omura T. 1988. Leafhopper cell cultures as a means for phytoreovirus research. Adv. Dis. Vector Res. 5:111–135 [Google Scholar]
- 6. Wei T, Shimizu T, Hagiwara K, Kikuchi A, Moriyasu Y, Suzuki N, Chen H, Omura T. 2006. Pns12 protein of Rice dwarf virus is essential for formation of viroplasms and nucleation of viral-assembly complexes. J. Gen. Virol. 87:429–438 [DOI] [PubMed] [Google Scholar]
- 7. Wei T, Uehara-Ichiki T, Miyazaki N, Hibino H, Iwasaki K, Omura T. 2009. Association of Rice gall dwarf virus with microtubules is necessary for viral release from cultured insect vector cells. J. Virol. 83:10830–10835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wei T, Chen H, Ichiki-Uehara T, Hibino H, Omura T. 2007. Entry of Rice dwarf virus into cultured cells of its insect vector involves clathrin-mediated endocytosis. J. Virol. 81:7811–7815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wei T, Kikuchi A, Moriyasu Y, Suzuki N, Shimizu T, Hagiwara K, Chen H, Takahashi M, Ichiki-Uehara T, Omura T. 2006. The spread of Rice dwarf virus among cells of its insect vector exploits virus-induced tubular structures. J. Virol. 80:8593–8602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jia D, Chen H, Zheng A, Chen Q, Liu Q, Xie L, Wu Z, Wei T. 2012. Development of an insect vector cell culture and RNA interference system to investigate the functional role of fijivirus replication protein. J. Virol. 86:5800–5807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nagata T, Storms MM, Goldbach R, Peters D. 1997. Multiplication of Tomato spotted wilt virus in primary cell cultures derived from two thrips species. Virus Res. 49:59–66 [DOI] [PubMed] [Google Scholar]
- 12. Peters D, Black LM. 1970. Infection of primary cultures of aphid cells with a plant virus. Virology 40:847–853 [DOI] [PubMed] [Google Scholar]
- 13. Hoang AT, Zhang HM, Yang J, Chen JP, Hebrard E, Zhou GH, Vinh VN, Cheng JA. 2011. Identification, characterization, and distribution of southern rice black-streaked dwarf virus in Vietnam. Plant Dis. 95:1063–1069 [DOI] [PubMed] [Google Scholar]
- 14. Jia D, Chen H, Mao Q, Liu Q, Wei T. 2012. Restriction of viral dissemination from the midgut determines incompetence of small brown planthopper as a vector of Southern rice black-streaked dwarf virus. Virus Res. 167:404–408 [DOI] [PubMed] [Google Scholar]
- 15. Matsukura K, Towata T, Sakai JI, Onuki M, Okuda M, Matsumura M. 9 January 2013. Dynamics of Southern rice black-streaked dwarf virus in rice and implication for virus acquisition. Phytopathology [Epub ahead of print.] doi: 10.1094/PHYTO-10-12-0261-R [DOI] [PubMed] [Google Scholar]
- 16. Pu LL, Xie GH, Ji CY, Ling B, Zhang MX, Xu DL, Zhou GH. 2012. Transmission characteristics of Southern rice black-streaked dwarf virus by rice planthoppers. Crop Prot. 41:71–76 [Google Scholar]
- 17. Wang Q, Yang J, Zhou GH, Zhang HM, Chen JP, Adams MJ. 2010. The complete genome sequence of two isolates of Southern rice black-streaked dwarf virus, a new member of the genus Fijivirus. J. Phytopathol. 158:733–737 [Google Scholar]
- 18. Zhou GH, Wen JJ, Cai DJ, Li P, Xu DL, Zhang SG. 2008. Southern rice black-streaked dwarf virus: a new proposed Fijivirus species in the family Reoviridae. Chin. Sci. Bull. 53:3677–3685 [Google Scholar]
- 19. Maroniche GA, Mongelli VC, Llauger G, Alfonso V, Taboga O, del Vas M. 2012. In vivo subcellular localization of Mal de Río Cuarto virus (MRCV) non-structural proteins in insect cells reveals their putative functions. Virology 430:81–89 [DOI] [PubMed] [Google Scholar]
- 20. Akita F, Higashiura A, Shimizu T, Pu Y, Suzuki M, Uehara-Ichiki T, Sasaya T, Kanamaru S, Arisaka F, Tsukihara T, Nakagawa A, Omura T. 2012. Crystallographic analysis reveals octamerization of viroplasm matrix protein P9-1 of Rice black streaked dwarf virus. J. Virol. 86:746–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang H, Chen J, Adams M. 2001. Molecular characterisation of segments 1 to 6 of rice black-streaked dwarf virus from China provides the complete genome. Arch. Virol. 146:2331–2339 [DOI] [PubMed] [Google Scholar]
- 22. Wang Q, Tao T, Zhang Y, Wu W, Li D, Yu J, Han C. 2011. Rice black-streaked dwarf virus P6 self-interacts to form punctate, viroplasm-like structures in the cytoplasm and recruits viroplasm-associated protein P9-1. Virol. J. 8:24 doi: 10.1186/1743-422X-8-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu Y, Jia D, Chen H, Chen Q, Xie L, Wu Z, Wei T. 2011. The P7-1 protein of Southern rice black-streaked dwarf virus, a fijivirus, induces the formation of tubular structures in insect cells. Arch. Virol. 156:1729–1736 [DOI] [PubMed] [Google Scholar]
- 24. Jia D, Guo N, Chen H, Akita F, Xie L, Omura T, Wei T. 2012. Assembly of the viroplasm by viral non-structural protein Pns10 is essential for persistent infection of Rice ragged stunt virus in its insect vector. J. Gen. Virol. 93:2299–2309 [DOI] [PubMed] [Google Scholar]
- 25. Kimura I. 1986. A study of Rice dwarf virus in vector cell monolayers by fluorescent antibody focus counting. J. Gen. Virol. 67:2119–2124 [Google Scholar]
- 26. Kainov DE, Pirttimaa M, Tuma R, Butcher SJ, Thomas GJ, Jr, Bamford DH, Makeyev EV. 2003. RNA packaging device of double-stranded RNA bacteriophages, possibly as simple as hexamer of P4 protein. J. Biol. Chem. 278:48084–48091 [DOI] [PubMed] [Google Scholar]
- 27. Pirttimaa MJ, Paatero AO, Frilander MJ, Bamford DH. 2002. Nonspecific nucleoside triphosphatase P4 of double-stranded RNA bacteriophage φ6 is required for single-stranded RNA packaging and transcription. J. Virol. 76:10122–10127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ahlquist P. 2006. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat. Rev. Microbiol. 4:371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lu YH, Zhang JF, Xiong RY, Xu QF, Zhou YJ. 2011. Identification of an RNA silencing suppressor encoded by southern rice black-streaked dwarf virus S6. Sci. Agric. Sin. 14:2909–2917 (In Chinese.) [Google Scholar]