Abstract
Productive replication of the Epstein-Barr virus (EBV) occurs in discrete sites in nuclei, called replication compartments, where viral genome DNA synthesis and transcription take place. The replication compartments include subnuclear domains, designated BMRF1 cores, which are highly enriched in the BMRF1 protein. During viral lytic replication, newly synthesized viral DNA genomes are organized around and then stored inside BMRF1 cores. Here, we examined spatial distribution of viral early and late gene mRNAs within replication compartments using confocal laser scanning microscopy and three-dimensional surface reconstruction imaging. EBV early mRNAs were mainly located outside the BMRF1 cores, while viral late mRNAs were identified inside, corresponding well with the fact that late gene transcription is dependent on viral DNA replication. From these results, we speculate that sites for viral early and late gene transcription are separated with reference to BMRF1 cores.
INTRODUCTION
The Epstein-Barr virus (EBV), a human lymphotropic herpesvirus that infects more than 95% of the human adult population (1), is the causative agent of infectious mononucleosis and is also associated with some B cell and epithelial cell malignancies (2, 3). After primary infection, EBV persists lifelong as a chronic asymptomatic infection by establishing latency in resting memory B cells (4, 5). Although the hallmark of latency is the absence of a complete viral productive cycle, EBV productive reactivation from latency, which occurs spontaneously in a subset of cells or can be induced artificially, leads to viral production through expression of a variety of lytic genes (6, 7). The EBV genome is amplified 100- to 1,000-fold by viral replication machinery composed of BALF5 DNA polymerase, BMRF1 polymerase processivity factor, BALF2 single-stranded DNA binding protein, and the BBLF4-BSLF1-BBLF2/3 helicase-primase complex in discrete sites in nuclei, called replication compartments (8, 9). With progression of lytic replication, the replication compartments become enlarged and fuse to form large globular structures that eventually fill the nucleus in late stages (8).
Analysis of the architecture of the EBV replication compartments has revealed that BMRF1 cores, where BMRF1 protein is enriched, constitute subnuclear domains inside the replication compartment (10). BMRF1 is a major phosphoprotein abundantly expressed during EBV productive infection (11, 12) which forms a heterodimer with the BALF5 polymerase catalytic subunit to enhance polymerase processivity (13). Furthermore, crystal structure analysis has demonstrated that BMRF1 by itself forms head-to-head homodimer or tetrameric ring structures (14, 15), presumably contributing to BMRF1 core structures. Thus, the BMRF1 protein may play dual roles during lytic replication, one as a polymerase processivity factor and the other in protecting the viral genome after synthesis. De novo synthesis of viral DNA occurs mainly outside the BMRF1 cores coupled with homologous recombination repair (HHR), which is followed by storage of replicated DNAs inside the cores where mismatch repair (MMR) factors are recruited to guarantee their integrity (10). The envisioned scenario is that viral DNA replication and viral genome maturation are assigned to outside and inside the BMRF1 cores, respectively.
During productive replication, EBV genes are expressed in a highly regulated temporal cascade. Three different classes of EBV genes have been characterized, the immediate-early (IE), early (E), and late (L) genes (16). The switch from latent to lytic infection is triggered by expression of the IE BZLF1 gene product, a b-Zip transcriptional factor (15, 17, 18). The BZLF1 protein binds to the promoters of E genes encoding proteins involved in viral DNA replication and metabolism (19, 20) and recruits RNA polymerase II on their promoters (21), initiating the lytic cascade program. Intriguingly, the transcription of L genes encoding virion structural components and DNA packaging proteins (20) is known to be tightly coupled to viral DNA replication (22). However, it remains unknown where E and L genes are transcribed within viral replication compartments.
Here, we examined spatial distribution of viral E and L gene mRNAs within the replication compartments using confocal laser scanning microscopy and three-dimensional (3D) surface reconstruction imaging. Viral early gene mRNAs were found to be located mainly outside the BMRF1 cores. In contrast, viral L gene mRNAs were identified inside, corresponding well with the fact that L gene transcription is dependent on viral DNA replication. These results indicate that the transcription sites for viral E and L genes are different, and that they are separated by the BMRF1 cores. This is the first report to describe a spatial relationship between the sites of viral gene transcription and replication compartments during EBV lytic replication.
MATERIALS AND METHODS
Cells and reagents.
Tet-BZLF1/B95-8 cells were maintained as described previously (10). The anti-EBV EA-D-p52/50 (BMRF1 gene product) mouse monoclonal antibody (clone R3) was purchased from Chemicon, and anti-RNA polymerase II mouse monoclonal antibodies were from Abcam. Fluorescein isothiocyanate (FITC)-conjugated anti-digoxigenin (DIG) sheep monoclonal antibodies were obtained from Roche Applied Science, and secondary goat anti-mouse IgG antibodies conjugated with Alexa 488 or 594, a Zenon mouse IgG labeling kit (Alexa 594), and a Zenon rabbit IgG labeling kit (Alexa 594) were from Molecular Probes. To block late gene transcription, phosphonoacetic acid (PAA), a herpesvirus DNA polymerase inhibitor, was added to the culture medium at a final concentration of 400 μg/ml.
Plasmid.
For pcDNA-HA-BcRF1, the BcRF1 sequence was amplified from the B95-8 genome using the following primers: 5′-CGCGGGTACCGCCACCATGTATCCATATGACGTTCCAGATTACGCTACACAAGGTAAGAGGGAGAT-3′ (forward) and 5′-CGCGGAATTCTTACACTTGAGCATCACGGC-3′ (reverse). Underlined nucleotides indicate the hemagglutinin (HA) epitope. The amplified DNA was digested with KpnI and EcoRI and then inserted into the KpnI/EcoRI sites of pcDNA3.
Immunofluorescence analyses.
Immunofluorescence (IF) analyses were performed based on our previous studies (8, 10). For immunofluorescence experiments, cells were extracted with 0.5% Triton X-100–mCSK buffer (8) and then fixed with 70% ethanol. After permeabilization, samples were blocked for 1 h in 10% normal goat serum in phosphate-buffered saline (PBS) and incubated overnight with the anti-RNA polymerase II mouse monoclonal antibody, followed by secondary goat anti-mouse IgG antibodies conjugated with Alexa Fluor 594 (1 h). Anti-BMRF1 antibodies were directly labeled with a Zenon tricolor mouse IgG1 labeling kit (Molecular Probes). Cells were incubated with Alexa Flour 488-labeled anti-BMRF1 antibodies for 45 min and washed three times with PBS, followed by a second fixation with 4% paraformaldehyde solution in PBS for 15 min at room temperature. All of the primary antibodies were employed at a 1:300 dilution, and the secondary antibodies were applied at a 1:500 dilution. All washes after antibody incubation were performed with PBS containing 0.1% normal goat serum and 0.01% Tween 20. The slides were mounted in ProLong Gold antifade reagent with 4′,6-diamidino-2-phenylindole (DAPI) (Molecular Probes) prior to microscopic analyses.
Quantitative RT-PCR.
Total cell RNA was purified using TriPure isolation reagent (Roche) and subjected to real-time reverse transcription-PCR (RT-PCR) using a one-step SYBR PrimeScript RT-PCR kit II (TaKaRa) and a real-time PCR system 7300 according to the manufacturer's instructions. PCR was performed in 10 μl of solution containing 0.2 μM primers, 0.2 μl ROX dye, and the sample RNA in 1× one-step SYBR RT-PCR buffer. The intensity of the ROX dye was used to compensate for volume fluctuations among the tubes. PCR included 5 min at 42°C, 10 s at 95°C, and 40 cycles at 95°C for 5 s, followed by 40 s at 60°C. Immediately after RT-PCR, the specificity of each PCR product was confirmed by dissociation curve analysis. An arbitrary RNA value was set to 1.0, and a standard curve was constructed using serial dilutions of RNA from the RNA set to 1.0. The amount of mRNA was quantitated based on the standard curve. Real-time PCR with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers was also performed to serve as an internal control for input RNA. Primers used for RT-PCR are listed in Table 1.
Table 1.
Oligonucleotide primer sequence
| Primer name | Sequence |
|---|---|
| BMRF1-forward | 5′-GCCGTTGAGGCCCACGTTGT-3′ |
| BMRF1-reverse | 5′-TGGGAATGGCAGGCGAGGGT-3′ |
| BALF5-forward | 5′-GCTGACGGACGGCAAGACCC-3′ |
| BALF5-reverse | 5′-GGGCAGTTCCTCGTTGCGCT-3′ |
| BALF4-forward | 5′-AACCTTTGACTCGACCATCG-3′ |
| BALF4-reverse | 5′-ACCTGCTCTTCGATGCACTT-3′ |
| BcLF1-forward | 5′-CATCCATGTTCATTGGGACC-3′ |
| BcLF1-reverse | 5′-CATTAGTCATACCTGCCAGG-3′ |
| BLLF1-forward | 5′-GTCAGTACACCATCCAGAGCC-3′ |
| BLLF1-reverse | 5′-TTGGTAGACAGCCTTCGTATG-3′ |
| GAPDH-forward | 5′-TGCACCACCAACTGCTAGC-3′ |
| GAPDH-reverse | 5′-GGCATGGACTGTGGTCATGAG-3′ |
Probe preparation for FISH.
The EBV E or L gene open reading frames (ORFs) coding for BMRF1, BALF5, BALF4, BcLF1, and BLLF1 were amplified by PCR from a bacterial artificial chromosome (BAC)-cloned copy of the EBV B95-8 genome. The PCR primer pairs used to amplify each ORF are given in Table 2. Fluorescence in situ hybridization (FISH) probes were prepared using a DIG-RNA labeling kit (SP6/T7) (Roche Applied Science).
Table 2.
Oligonucleotide primer sequencea
| Primer name | Sequence |
|---|---|
| BMRF1-antisense-f | 5′-GCCGTTGAGGCCCACGTTGT-3′ |
| BMRF1-antisense-r | 5′-TAATACGACTCACTATAGGGGTGGGAATGGCAGGCGAGGGT-3′ |
| BMRF1-sense-f | 5′-TAATACGACTCACTATAGGGGGCCGTTGAGGCCCACGTTGT-3′ |
| BMRF1-sense-r | 5′-TGGGAATGGCAGGCGAGGGT-3′ |
| BALF5-antisense-f | 5′-GCTGACGGACGGCAAGACCC-3′ |
| BALF5-antisense-r | 5′-TAATACGACTCACTATAGGGGGGGCAGTTCCTCGTTGCGCT-3′ |
| BALF5-sense-f | 5′-TAATACGACTCACTATAGGGGGCTGACGGACGGCAAGACCC-3′ |
| BALF5-sense-r | 5′-GGGCAGTTCCTCGTTGCGCT-3′ |
| BALF4-antisense-f | 5′-AACCTTTGACTCGACCATCG-3′ |
| BALF4-antisense-r | 5′-TAATACGACTCACTATAGGGGACCTGCTCTTCGATGCACTT-3′ |
| BALF4-sense-f | 5′-TAATACGACTCACTATAGGGGAACCTTTGACTCGACCATCG-3′ |
| BALF4-sense-r | 5′-ACCTGCTCTTCGATGCACTT-3′ |
| BcLF1-antisense-f | 5′-TTACCAGGGACGAGGACATC-3′ |
| BcLF1-antisense-r | 5′-TAATACGACTCACTATAGGGGCCCGTTCACCAAAACAGTCT-3′ |
| BcLF1-sense-f | 5′-TAATACGACTCACTATAGGGGTTACCAGGGACGAGGACATC-3′ |
| BcLF1-sense-r | 5′-CCCGTTCACCAAAACAGTCT-3′ |
| BLLF1-antisense-f | 5′-TGGAAACAGTTCCACATCCA-3′ |
| BLLF1-antisense-r | 5′-TAATACGACTCACTATAGGGGGAATCACCGCCGTAATCTGT-3′ |
| BLLF1-sense-f | 5′-TAATACGACTCACTATAGGGGTGGAAACAGTTCCACATCCA-3′ |
| BLLF1-sense-r | 5′-GAATCACCGCCGTAATCTGT-3′ |
f, forward; r, reverse. Underlining indicates the T7 promoter.
FISH.
For FISH experiments, cells were fixed in 4% paraformaldehyde in PBS containing 10% acetic acid for 15 min at room temperature and washed with PBS, followed by a second fixation with 70% ethanol for 24 h at −20°C. After washing in Tris-buffered saline, cells were treated with 0.01% pepsin–0.01N HCl for 5 min at 37°C and immediately rinsed with double-distilled water, followed by washing with PBS for 5 min and treatment with 1% formaldehyde in PBS for 10 min. For dehydration, fixed cells were treated with 70, 90, and 100% ethanol for 5 min each and then air dried. For immuno-RNA-FISH, postfixed cells were incubated with a prehybridization buffer containing 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 10% dextran sulfate, 0.2% bovine serum albumin (BSA), sheared salmon DNA (0.5 μg/μl), Escherichia coli tRNA (0.25 μg/μl), and RNase inhibitor and hybridized with RNA probes in prehybridization buffer overnight at 42°C in a moist chamber. After sequential washes 3 times with 50% formamide in 1× SSC at room temperature, with 50% formamide in 1× SSC at 42°C, and with 2× SSC at room temperature, samples were blocked for 30 min in 4% milk in PBS containing 0.05% Tween 20 and then exposed at room temperature to anti-BMRF1 mouse monoclonal antibodies (3 h), followed by Alexa 594-conjugated secondary antibodies (2 h) and FITC-labeled anti-DIG sheep monoclonal antibodies (1 h). Samples then were mounted in ProLong Gold antifade reagent with DAPI prior to microscopic analysis.
Microscopy and image analyses.
Images were captured and processed using an LSM510 Meta microscope (Carl Zeiss) with a Plan Apochromat 100×/1.4-numerical-aperture oil immersion objective. For 3D reconstruction with confocal laser scanning microscopy, images observed with an LSM510 Meta microscope were computerized to automatically make 50 to 100 serial optical sections at intervals of around 0.26 μm. The 3D reconstruction was performed with Imaris software (Carl Zeiss) based on the appropriate intensity threshold.
RESULTS AND DISCUSSION
RNA polymerase II migrates into BMRF1 cores with progression of lytic replication.
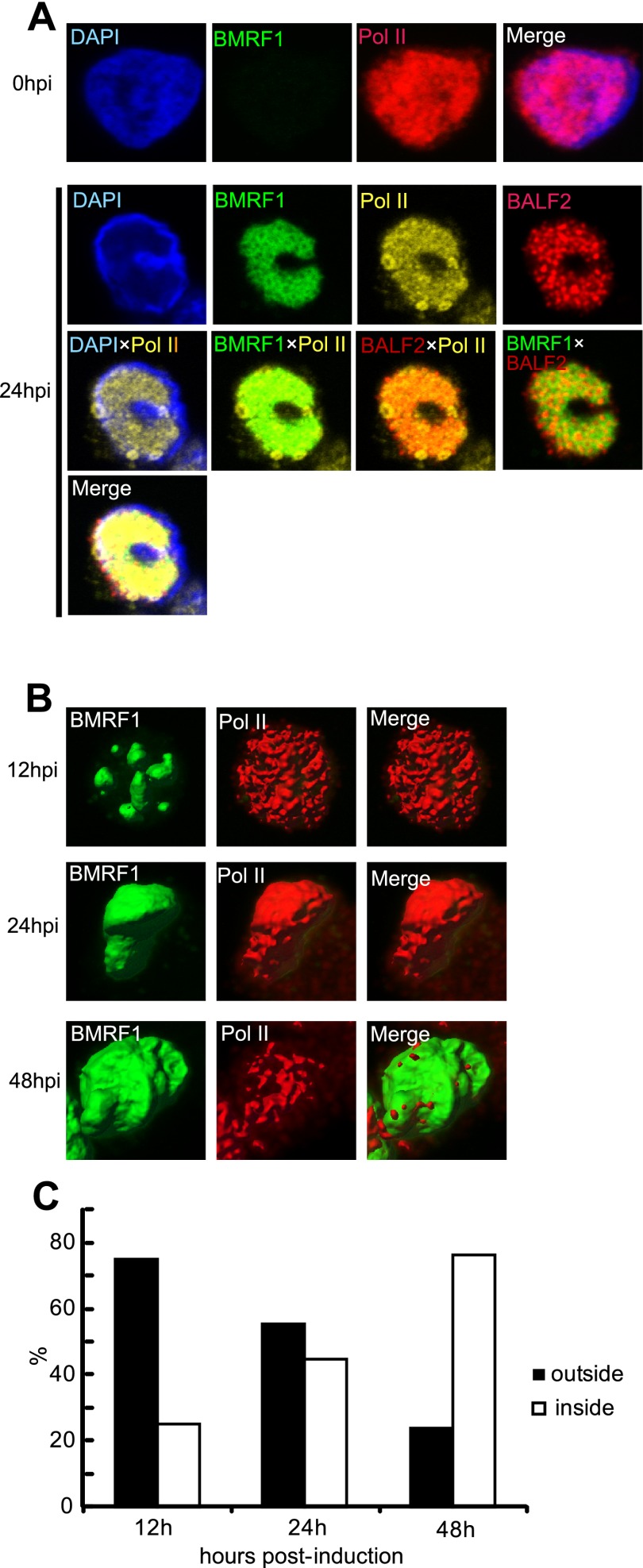
Viral IE, E, and L genes, like cellular genes, are transcribed by RNA polymerase II (RNAPII) into mRNAs. We first examined the localization of cellular RNAPII in latent or lytic replication-induced tet-BZLF1/B95-8 cells (Fig. 1A). In the latent phase, RNAPII was distributed throughout nuclei, colocalizing with chromosomal DNA as judged by DAPI staining. With lytic replication, RNAPII was recruited to viral replication compartments where BMRF1 and BALF2 replication proteins were localized (Fig. 1A). Since viral transcription dominates in lytically replicating cells, the majority of cellular RNAPII may be recruited to viral transcription sites. Spatial arrangement of RNAPII relative to the BMRF1 cores within replication compartments was examined by time course imaging utilizing a 3D surface reconstruction technique. The distribution of RNAPII relative to BMRF1 cores was examined in cells at 12, 24, and 48 h postinduction, and the frequencies of cells with RNAPII outside and inside BMRF1 cores were determined. At 12 h postinduction, corresponding to the early stages of productive replication, RNAPII was distributed throughout nuclei and surrounded the BMRF1 cores in more than 70% of cells (Fig. 1C; a representative image is shown in panel B). At this time point, a subset of cells having RNAPII inside the BMRF1 core may reflect spontaneous reactivation without induction. At 24 h postinduction, RNAPII localization tended to become more restricted within nuclei (a representative image is shown in Fig. 1B), and the frequency of cells with RNAPII outside the BMRF1 cores decreased to approximately 50% (Fig. 1C). At 48 h postinduction, corresponding to late stages of infection, the cells with RNAPII inside the BMRF1 cores accounted for more than 70% (Fig. 1C; a representative image is shown in panel B). Thus, the localization of RNAPII shifted from outside to inside the BMRF1 cores in concordance with progression of EBV lytic replication. These observations let us hypothesize that viral E gene transcription and L gene transcription take place at different loci within nuclei.
Fig 1.
RNAPII becomes localized inside BMRF1 cores at late stages of productive replication. Tet-BZLF1/B95-8 cells were treated with doxycycline to induce lytic replication and harvested at the indicated postinduction times. After treatment with mCSK buffer, they were fixed. (A) Harvested cells were stained with anti-BMRF1 (green), anti-BALF2 (red), and anti-RNAPII (yellow) antibodies and observed by laser scanning confocal microscopy. The 2D images show brightest-point projections. Lower panels are merged images of the indicated combinations of the proteins. Pol II, RNA polymerase II. (B) Lytic replication-induced cells were stained with anti-BMRF1 (green) and anti-RNAPII (red). Projections of 60 images collected at 0.33-μm steps in the z axis are displayed as 3D topographical reconstruction images of BMRF1 and RNAPII (left and middle panels, respectively). Representative 3D surface reconstruction images are presented. Right panels are merged 3D surface reconstruction images. (C) The ratio of cells in which RNAPII was located outside the BMRF1 core to total BMRF1-positive cells was determined (black bar). White bars show the ratios of cells in which RNAPII was located inside the BMRF core to total BMRF1-positive cells. We regard cells as inside when 50% or more of RNAPII signal was detected inside the core. More than 100 cells were counted at each of the indicated time points.
Spatial distributions of viral E and L gene transcripts.
We designed experiments to examine the intracellular distribution of viral E and L transcripts with mRNA FISH, expecting that abundantly expressed viral mRNAs should transiently accumulate in loci of transcription. mRNA FISH was combined with immunostaining of BMRF1 protein, and the results were analyzed using 3D surface reconstruction imaging.
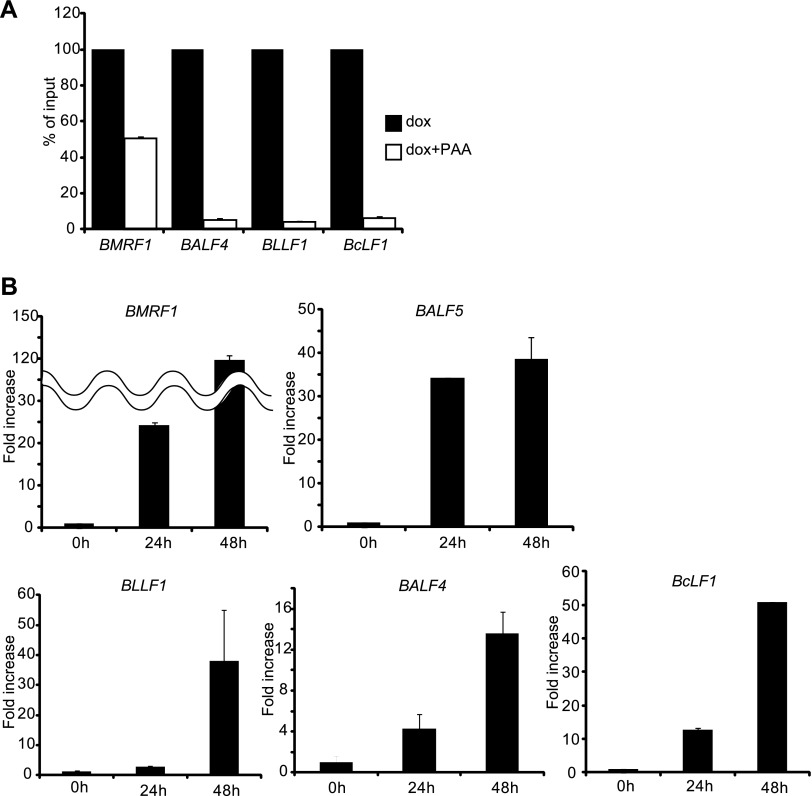
BMRF1 and BALF5 (DNA polymerase) were selected as viral E genes, while BALF4 (gB, gp110), BcLF1 (major capsid protein), and BLLF1 (gp350/220) were chosen as L genes. Levels of BALF4, BcLF1, and BLLF1 mRNA signals were very low compared to that of BMRF1 mRNA signal in the presence of PAA treatment after 48 h postinduction (Fig. 2A), confirming that BALF4, BcLF1, and BLLF1 are L genes. We performed quantitative RT-PCR analysis at 0, 24, and 48 h postinduction to establish the expression time courses of E and L genes in our system. As shown in Fig. 2B, upper, mRNA levels of the E genes BMRF1 and BALF5 became high at 24 h postinduction. In contrast, as shown in Fig. 2B, lower, mRNA levels of L genes such as BALF4, BcLF1, and BLLF1 were still low at 24 h postinduction, and their expression levels became high at 48 h postinduction. Sense and antisense RNA probes were synthesized for each transcript in order to make sure that only the antisense probes, but not the sense probes, generated signals. As expected, cells undergoing lytic replication, verified by BMRF1 protein expression, did not exhibit any mRNA FISH signals when sense probes of BMRF1, BALF5, BALF4, and BcLF1 were used (Fig. 3B′, C′, D′, E′, and F′).
Fig 2.
BALF4, BLLF1, and BcLF1 are late (L) genes. (A) Lytic replication-induced tet-BZLF1/B95-8 cells were cultured in the absence or presence of PAA (phosphonoacetic acid) and harvested at 48 h postinduction. Total RNAs were purified using TriPure isolation reagent and applied for real-time RT-PCR as described in Materials and Methods. Levels of BMRF1, BALF4, BcLF1, and BLLF1 mRNAs were normalized to GAPDH mRNA levels. The value obtained in the absence of PAA was set to 100%, and data are expressed as means ± standard deviations (SD) from 3 biological replicates. dox, doxycycline. (B) Expression profiles for mRNA levels of EBV early (E) and L genes. Lytic replication-induced tet-BZLF1/B95-8 cells were harvested at 0, 24, and 48 h postinduction. Total RNAs were purified using TriPure isolation reagent and applied for real-time RT-PCR as described in Materials and Methods. Levels of BMRF1, BALF5, BALF4, BcLF1, and BLLF1 mRNAs were normalized to GAPDH mRNA levels. The value obtained at 0 h postinduction was set to 1, and data are expressed as means ± SD from 3 biological replicates.
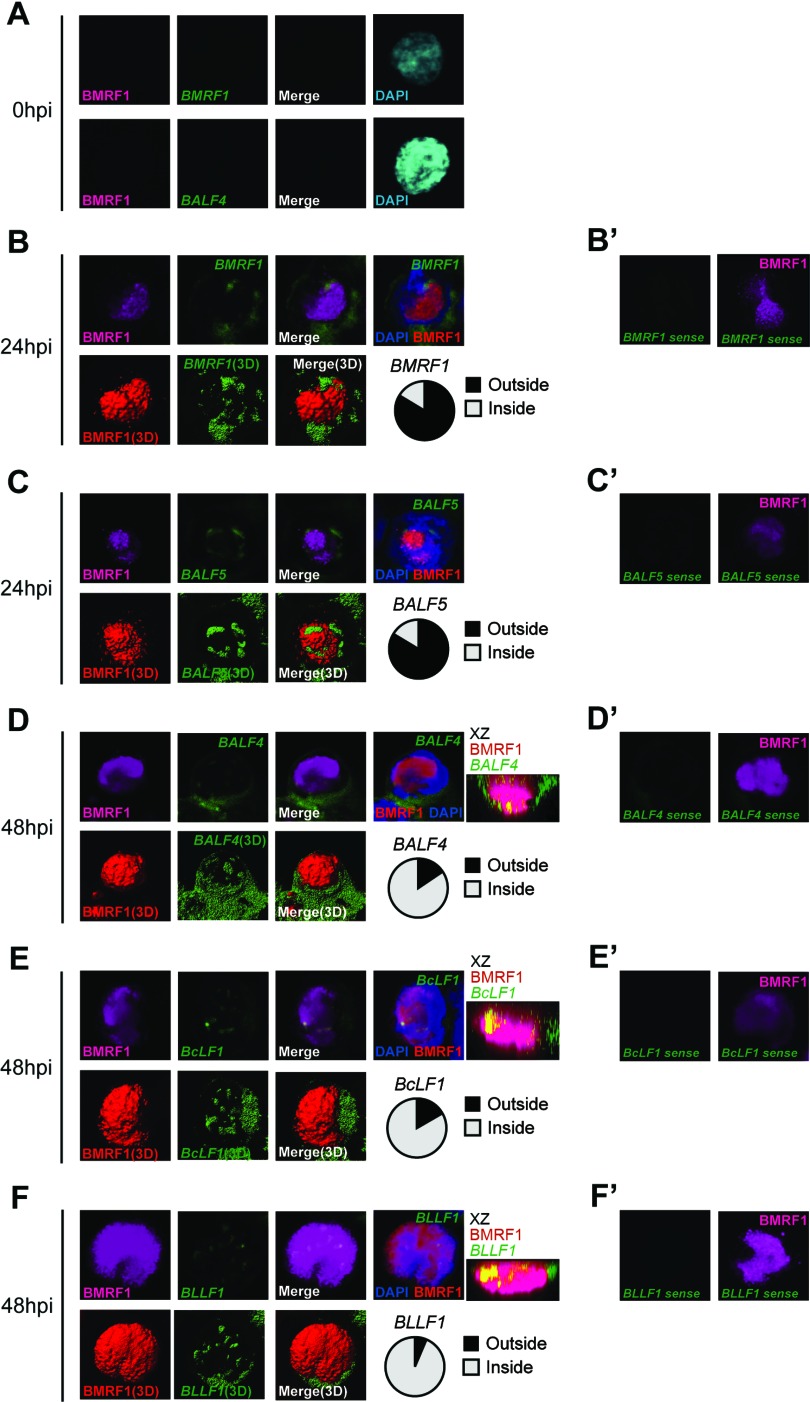
Fig 3.
Alteration of viral gene transcription foci at early and late stages of EBV lytic replication. Lytic replication-induced tet-BZLF1/B95-8 cells were hybridized with DIG-labeled RNA FISH probes and then exposed to anti-BMRF1 antibodies (magenta), anti-DIG antibodies (green), and DAPI (blue). The data are presented as 3D reconstruction images (lower panels; merged images are at the right) and corresponding 2D images (upper panels). The 2D images show brightest-point projections of 60 images collected at 0.33-μm steps on the z axis. (A) Lytic replication-induced tet-BZLF1/B95-8 cells were harvested at 0 h postinduction and hybridized with antisense mRNA probes for BMRF1 (upper panels) and BALF4 (lower panels), followed by exposure to anti-BMRF1 monoclonal antibodies (magenta). (B and C) Lytic replication-induced tet-BZLF1/B95-8 cells were harvested at 24 h postinduction and hybridized with antisense mRNA probes for BMRF1 (B) and BALF5 (C), followed by exposure to anti-BMRF1 monoclonal antibodies (magenta). More than 30 cells were counted, and the ratio of the cells in which FISH signals were located outside the BMRF1 core to total BMRF1-positive cells was determined (circle chart, black). (D to F) Lytic replication-induced tet-BZLF1/B95-8 cells were harvested at 48 h postinduction and hybridized with antisense mRNA probes for BALF4 (D), BcLF1 (E), and BLLF1 (F), followed by exposure to anti-BMRF1 monoclonal antibodies (magenta). x-z 2D merged images, which show brightest-point sections in the y axis, are also shown. More than 30 cells were counted, and the ratio of the cells in which FISH signals were located inside the BMRF1 core to total BMRF1-positive cells was determined (circle chart, gray). (B′ to F′) As negative controls, cells were hybridized with a sense RNA probe for BMRF1 (B′), BALF5 (C′), BALF4 (D′), BcLF1 (E′), and BLLF1 (F′), followed by exposure to anti-BMRF1 monoclonal antibodies (magenta).
On the other hand, when antisense probes of viral E (BMRF1 and BALF5) and L transcripts (BALF4, BcLF1, and BLLF1) were used as probes, nearly all of the cells undergoing lytic replication (with BMRF1 protein expression) exhibited mRNA FISH signals (Fig. 3B, C, D, E, and F). Importantly, the same antisense probes for BMRF1, BALF4, and BALF5 did not raise any signals in cells prior to the induction of lytic replication (Fig. 3A and data not shown). The results indicated that the FISH signals obtained with the antisense probes actually represent the localization of viral E and L mRNAs.
The distribution of viral E transcripts (BMRF1 and BALF5) relative to BMRF1 protein in lytically replicating cells was examined at 24 h postinduction. The mRNAs were found to form small globular bodies at the nuclear periphery, partly concealing the BMRF1 cores (Fig. 3B and C). Signals were also detected in the cytoplasm, presumably representing mRNA that had been exported from nuclei. The frequencies of cells with viral E transcripts at the periphery or inside BMRF1 cores were determined. BMRF1 mRNA signals were observed outside BMRF1 cores in more than 80% of cells that were positive for BMRF1 protein expression (Fig. 3B). Similarly, BALF5 mRNA signals were observed outside BMRF1 cores in more than 80% of BMRF1-positive cells (Fig. 3C). The same results were obtained at 48 h postinduction (data not shown).
We then examined the spatial distribution of viral L transcripts (BALF4, BcLF1, and BLLF1) in cells at 48 h postinduction, which was expected to be in the phase of maximal L gene expression (20). In contrast to the localization of E transcripts, viral L transcripts were frequently found to form globular structures in the inner space of BMRF1 cores. The frequencies of cells with viral L transcripts being inside BMRF1 cores were 85% for BALF4, 83% for BcLF1, and 93% for BLLF1 (Fig. 3D, E, and F). L gene signals were hardly detected at 24 h postinduction (data not shown) because mRNA levels of BALF4, BcLF1, and BLLF1 remained low at 24 h postinduction (Fig. 2B). Thus, these results support the idea that EBV L gene transcription takes place inside the BMRF1 cores, while E genes are predominantly transcribed at the periphery.
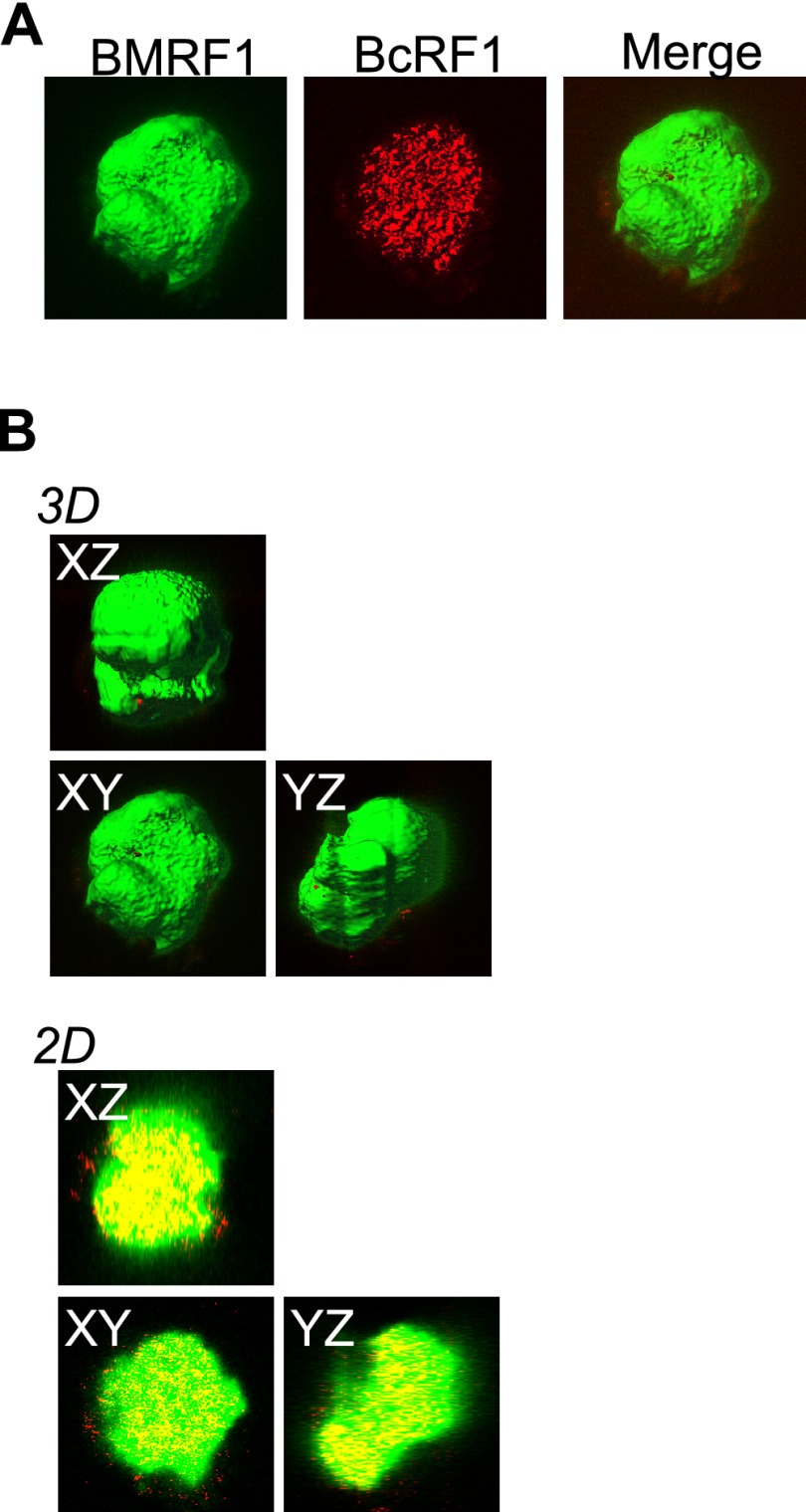
Recently, several studies have shed light on the regulation of herpesvirus L gene expression (22–25). It appears to depend not only on viral DNA replication but also on the presence of virally encoded factors. In the case of EBV, Gruffat et al. reported that the BcRF1 gene product binds to a TATT motif within the promoters of various L genes, such as BALF4, BcLF1, and BLLF1, and it works as an L gene-specific TATA binding protein (25). Supporting this idea, we observed that BcRF1 protein is localized inside BMRF1 cores in late stages of lytic replication (Fig. 4A and B).
Fig 4.
BcRF1, the EBV late gene transactivator, is located inside the BMRF1 core. Tet-BZLF1/B95-8 cells were transfected with HA-BcRF1 expression vector and then treated with doxycycline to induce lytic replication. Cells were harvested at 48 h postinduction, treated with mCSK buffer, fixed, and stained with anti-BMRF1 (green) and anti-HA (red) antibodies. (A) The data are presented as 3D surface reconstruction images of BMRF1 and HA-BcRF1 (left and middle panels). A merged 3D surface reconstruction image is also presented (right panel). (B) The data are presented as serial x-y, x-z, and y-z 3D surface reconstruction images (3D) and corresponding 2D images (2D). The 2D images show brightest-point sections in the x, y, or z axis.
We previously identified BMRF1 core structures within replication compartments and proposed a model for viral genome DNA synthesis and maturation (10). First, viral DNA genomes are synthesized by viral replication machinery outside the BMRF1 cores, coupled with homologous recombination by host HHR factors. The synthesized viral genomes are then stored inside the core, where host MMR factors are loaded on the synthesized DNA to repair mismatched base pairs for maturing intact viral genomes. The BMRF1 cores therefore partition the replication compartments into inside and outside subdomains, specifying viral genome synthesis and maturation, respectively. Consistent with this model, our present study showed that E gene transcripts were detected outside the BMRF1 cores, while L gene transcripts were found inside (Fig. 2). It was previously demonstrated that viral L gene transcription is dependent on viral DNA synthesis (23), and newly synthesized viral genomes, which serve as the templates of L gene transcription, are stored inside the BMRF1 cores (10). Thus, it is reasonable that viral L gene transcription occurs inside BMRF1 cores. Taking the available information together, we propose an advanced model of viral transcription loci in concordance with inbound transition of viral genomes while they are replicated by virally encoded replication machinery.
ACKNOWLEDGMENTS
This work was supported by grants in aid for Scientific Research from the Ministry of Education, Science, Sports, Culture and Technology of Japan (no. 23114512, 3390118, and 24659213 to T.T.). A.S. is supported by a Research Fellowship of the Japanese Society for the Promotion of Science for Young Scientists.
Footnotes
Published ahead of print 3 April 2013
REFERENCES
- 1. Baer R, Bankier AT, Biggin MD, Deininger PL, Farrell PJ, Gibson TJ, Hatfull G, Hudson GS, Satchwell SC, Seguin C, et al. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207–211 [DOI] [PubMed] [Google Scholar]
- 2. Imai S, Nishikawa J, Takada K. 1998. Cell-to-cell contact as an efficient mode of Epstein-Barr virus infection of diverse human epithelial cells. J. Virol. 72:4371–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yoshiyama H, Imai S, Shimizu N, Takada K. 1997. Epstein-Barr virus infection of human gastric carcinoma cells: implication of the existence of a new virus receptor different from CD21. J. Virol. 71:5688–5691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Babcock GJ, Decker LL, Volk M, Thorley-Lawson DA. 1998. EBV persistence in memory B cells in vivo. Immunity 9:395–404 [DOI] [PubMed] [Google Scholar]
- 5. Henle W. 1972. Role of Epstein-Barr virus in infectious mononucleosis and malignant lymphomas in man. Fed. Proc. 31:1674. [PubMed] [Google Scholar]
- 6. Ben-Sasson SA, Klein G. 1981. Activation of the Epstein-Barr virus genome by 5-aza-cytidine in latently infected human lymphoid lines. Int. J. Cancer 28:131–135 [DOI] [PubMed] [Google Scholar]
- 7. Ragona G, Ernberg I, Klein G. 1980. Induction and biological characterization of the Epstein-Barr virus (EBV) carried by the Jijoye lymphoma line. Virology 101:553–557 [DOI] [PubMed] [Google Scholar]
- 8. Daikoku T, Kudoh A, Fujita M, Sugaya Y, Isomura H, Shirata N, Tsurumi T. 2005. Architecture of replication compartments formed during Epstein-Barr virus lytic replication. J. Virol. 79:3409–3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fixman ED, Hayward GS, Hayward SD. 1995. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J. Virol. 69:2998–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sugimoto A, Kanda T, Yamashita Y, Murata T, Saito S, Kawashima D, Isomura H, Nishiyama Y, Tsurumi T. 2011. Spatiotemporally different DNA repair systems participate in Epstein-Barr virus genome maturation. J. Virol. 85:6127–6135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cho MS, Milman G, Hayward SD. 1985. A second Epstein-Barr virus early antigen gene in BamHI fragment M encodes a 48- to 50-kilodalton nuclear protein. J. Virol. 56:860–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li JS, Zhou BS, Dutschman GE, Grill SP, Tan RS, Cheng YC. 1987. Association of Epstein-Barr virus early antigen diffuse component and virus-specified DNA polymerase activity. J. Virol. 61:2947–2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tsurumi T, Daikoku T, Kurachi R, Nishiyama Y. 1993. Functional interaction between Epstein-Barr virus DNA polymerase catalytic subunit and its accessory subunit in vitro. J. Virol. 67:7648–7653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Murayama K, Nakayama S, Kato-Murayama M, Akasaka R, Ohbayashi N, Kamewari-Hayami Y, Terada T, Shirouzu M, Tsurumi T, Yokoyama S. 2009. Crystal structure of Epstein-Barr virus DNA polymerase processivity factor BMRF1. J. Biol. Chem. 284:35896–35905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nakayama S, Murata T, Yasui Y, Murayama K, Isomura H, Kanda T, Tsurumi T. 2010. Tetrameric ring formation of Epstein-Barr virus polymerase processivity factor is crucial for viral replication. J. Virol. 84:12589–12598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. 2007. Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 17. Amon W, Farrell PJ. 2005. Reactivation of Epstein-Barr virus from latency. Rev. Med. Virol. 15:149–156 [DOI] [PubMed] [Google Scholar]
- 18. Tsurumi T, Fujita M, Kudoh A. 2005. Latent and lytic Epstein-Barr virus replication strategies. Rev. Med. Virol. 15:3–15 [DOI] [PubMed] [Google Scholar]
- 19. Furnari FB, Adams MD, Pagano JS. 1992. Regulation of the Epstein-Barr virus DNA polymerase gene. J. Virol. 66:2837–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kieff E, Richinson A. 2001. Epstein-Barr virus and its replication, p 2512–2573 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 4th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 21. Lehman AM, Ellwood KB, Middleton BE, Carey M. 1998. Compensatory energetic relationships between upstream activators and the RNA polymerase II general transcription machinery. J. Biol. Chem. 273:932–939 [DOI] [PubMed] [Google Scholar]
- 22. Serio TR, Kolman JL, Miller G. 1997. Late gene expression from the Epstein-Barr virus BcLF1 and BFRF3 promoters does not require DNA replication in cis. J. Virol. 71:8726–8734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Amon W, Binne UK, Bryant H, Jenkins PJ, Karstegl CE, Farrell PJ. 2004. Lytic cycle gene regulation of Epstein-Barr virus. J. Virol. 78:13460–13469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chang PJ, Chang YS, Liu ST. 1998. Characterization of the BcLF1 promoter in Epstein-Barr virus. J. Gen. Virol. 79(Pt 8):2003–2006 [DOI] [PubMed] [Google Scholar]
- 25. Gruffat H, Kadjouf F, Mariame B, Manet E. 2012. The Epstein-Barr virus BcRF1 gene product is a TBP-like protein with an essential role in late gene expression. J. Virol. 86:6023–6032 [DOI] [PMC free article] [PubMed] [Google Scholar]