Abstract
The recent identification of nonprimate hepaciviruses in dogs and then in horses prompted us to look for pegiviruses (GB virus-like viruses) in these species. Although none were detected in canines, we found widespread natural infection of horses by a novel pegivirus. Unique genomic features and phylogenetic analyses confirmed that the tentatively named equine pegivirus (EPgV) represents a novel species within the Pegivirus genus. We also determined that EPgV causes persistent viremia whereas its clinical significance is undetermined.
TEXT
Hepatitis C virus (HCV) and human pegiviruses (HPgV) infect an estimated 2 and 5% of the world's population, respectively. HCV, HPgV (formerly referred to as GB virus C or hepatitis G virus), and other genetically related viruses belong to two closely related genera of the family Flaviviridae, named Hepacivirus and Pegivirus (1). HCV is hepatotropic, and infection of humans can trigger liver damage characterized by fibrosis, cirrhosis, and hepatocellular carcinoma (2). HPgV is lymphotropic (3), but its pathogenicity in humans is unknown. Among people with other blood-borne or sexually transmitted infections, HPgV is more prevalent. Up to 40% of HIV-infected individuals have HPgV viremia (1, 4, 5). The viruses genetically most similar to human HCV include GB virus B (GBV-B) and the recently discovered nonprimate hepacivirus (NPHV) (6). The natural host of NPHV is the horse (6, 7), while GBV-B's origin and natural host remain elusive. The viruses genetically most similar to HPgV include simian (GBV-A) and bat (GBV-D) pegiviruses (SPgV and BPgV, respectively) (1, 8, 9). The recent identification of NPHV in horses (7) prompted us to look for the presence of pegivirus-like virus infections in dogs and horses. Here we report the identification, complete genome sequence, and initial characterization of a pegivirus that infects horses.
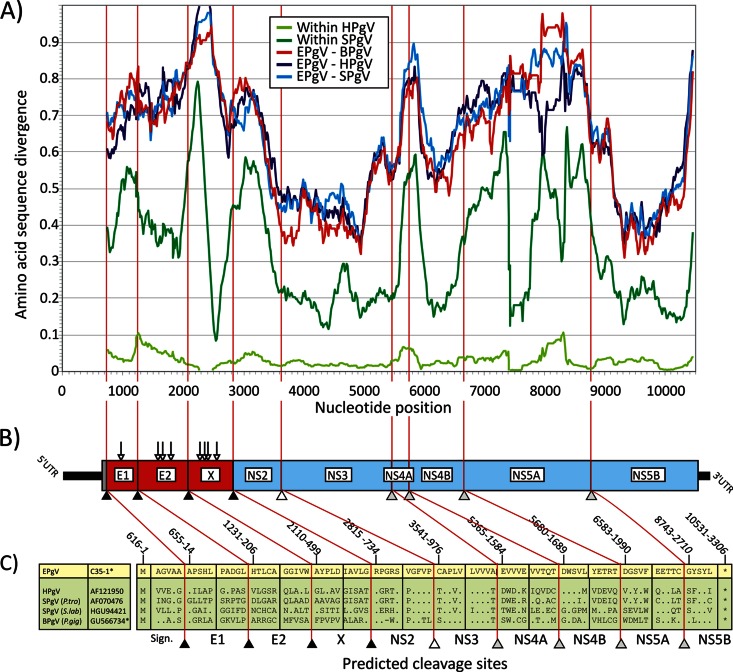
Identification, complete genome, and polyprotein of equine pegivirus (EPgV).
Degenerate primers targeting the conserved helicase motifs of known pegivirus species were used to screen serum samples from 12 horses with elevated liver enzyme levels. The degenerate PCR assay used primers AK4340F1 (5′-GTACTTGCTACTGCNACNCC-3′) and AK4630R1 (5′-TACCCTGTCATAAGGGCRTC-3′) for the first round of PCR. Primers AK4340F2 (5′-CTTGCTACTGCNACNCCWCC-3′) and AK4630R2 (5′-TACCCTGTCATAAGGGCRTCNGT-3′) were used for the second round. For the first round, the PCR cycle included 8 min of denaturation at 95°C; 10 cycles of 95°C for 40 s, 60°C for 1 min, and 72°C for 40 s; 30 cycles of 95°C for 30 s, 56°C for 45 s, and 72°C for 40 s; and a final extension at 72°C for 10 min. For the second round, the PCR cycle included 8 min of denaturation at 95°C; 10 cycles of 95°C for 40 s, 64°C for 1 min, and 72°C for 40 s; 30 cycles of 95°C for 30 s, 58°C for 45 s, and 72°C for 40 s; and a final extension at 72°C for 10 min. All PCR products approximately 300 bp in length were sequenced to confirm the results.
Two samples were PCR positive, and amplicon sequencing revealed viral sequences highly divergent from all known pegiviruses. Unbiased high-throughput sequencing using the Ion Torrent Personal Genome Machine was done to acquire more sequences of the novel virus. A complete genome was acquired by primer walking, gap-filling PCRs, 5′ random amplification of cDNA ends (RACE), and 3′ poly(A/G/U) tailing (6, 7). The complete genome sequence of the new virus (isolate C35) contains 11,197 nucleotides (nt) and is the longest genome of any known Hepacivirus or Pegivirus described to date. Following the use of host names to describe pegiviruses (1), we have therefore tentatively named this EPgV. Deduced translation of the EPgV genome indicated the presence of three possible initiation codons at nt 499, 592, and 616. However, the presence of a polypyrimidine tract (PPT) at nt 573 to 589, of single nucleotide insertions/deletions at nt 570 and 581 in different EPgV isolates, and of a stable stem-loop formed by nt 589 to 607 indicated that the AUG codon at position 616 is most likely to be the initiation codon. Assuming this initiation site, the EPgV genome includes a 5′ untranslated region (UTR) of 615 nt, a single open reading frame (ORF) encoding a putative polyprotein 3,305 amino acid (aa) residues in length, and a 3′ UTR of up to 664 nt. Comparative genetic analysis indicated that the EPgV polyprotein contains three structural (E1, E2, X) and six nonstructural (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) proteins (Fig. 1A). In common with a previous analysis of BPgV (8), the EPgV polyprotein sequence shows additional predicted signalase sites at positions 13 and 498 (Fig. 1C). The first represents a signal peptide for the translocation of E1 on the endoplasmic reticulum membrane. The second, in conjunction with the downstream signalase site at the start of NS2 (position 734), creates a potential, moderately glycosylated potential homolog of the X protein reported in BPgV. The NS3 protein of EPgV contains a sequence motif for serine proteases (histidine 57, aspartic acid 81, serine 139) in the N-terminal portion and for nucleoside triphosphatase and RNA helicase (GSGKS at positions 208 to 212) in the C-terminal portion. The NS5B protein of EPgV is 596 aa long and contains functional RNA-dependent RNA polymerase motifs in palm, fingers, and thumb subdomains. The palm region contains five different motifs, A to E, that play a major role in the polymerization ability of viral polymerase. EPgV motif A contains the D-X4-D region (DATCFD), motif B contains Gx3TTx4N (GVYTTSSAN), and motif C contains the highly conserved GDD active site (HGDD) (10).
Fig 1.
(A) Amino acid sequence divergence between EPgV and HPgV, SPgV, and BPgV based on 201-nt fragments in 6-nt increments across the genome alignment (midpoint plotted on the y axis). Within-species distances for HPgV and SPgV are included for comparison. The divergence scan commenced at the predicted signalase cleavage site at the start of the E1-encoding genes. (B, C) Genome diagram of EPgV showing predicted N-linked glycosylation sites (Nx[S/T]; vertical arrows) and proposed cleavage sites (sequence positions were numbered on the basis of the EPgV sequence). Cleavage sites in the EPgV polyprotein and in other pegiviruses were predicted by alignment and homology to sites previously identified in SPgV (sites NS3/NS4A, NS4A/NS4B, NS4B/NS5A, and NS5A/NS5B) and by comparison with predicted signalase sites between structural proteins and the NS2/NS3 cleavage site of BPgV. Potential signalase sites in EPgV were evaluated by using the SignalP 4.1 server, which identified homologous cleavage sites that aligned with those in BPgV, including the boundaries of the novel proposed X protein.
Sequence relationships with known viruses.
To assess the relatedness of EPgV to other pegiviruses, their coding sequences were aligned and pairwise distances between structural and nonstructural genome regions were computed (Table 1). Proteins were aligned by using MUSCLE with default settings in the SSE sequence editor (11–13) (http://www.virus-evolution.org/Software). EPgV was substantially divergent from all other sequences, with amino acid divergences ranging from 62 to 77% in structural genes (E1, E2, and X) and from 49 to 59% in the nonstructural region. The 5′ UTR showed short regions of homology to HPgV and SPgV, while the 3′ UTRs matched no other pegivirus or other sequences found in GenBank by BLAST searching. With a scanning window of 67 aa, the sequence divergence between EPgV and other pegiviruses was analyzed across the genome (Fig. 1B). By comparison of BPgV and primate pegivirus (4), the most conserved regions within the Pegivirus genus were the NS3 and NS5B genes, with high sequence divergence in the E1 and E2 glycoproteins and NS4B and no homology to other sequences in GenBank in extended regions of NS4A and NS5A.
Table 1.
Nucleotide and translated amino acid sequence divergences of structural and nonstructural genes of EPgV and other pegiviruses
| Gene category and virus | % Divergence |
|||
|---|---|---|---|---|
| EPgV | HPgV | SPgV | BPgV | |
| Structural (E1, E2, X) | ||||
| EPgV | 59.2a | 63.0a | 62.8a | |
| HPgV | 69.7b | 52.2a | 54.2a | |
| SPgV | 76.6b | 61.8b | 56.4a | |
| BPgV | 77.3b | 62.6b | 67.0b | |
| Nonstructural (NS2-NS5B) | ||||
| EPgV | 51.7a | 52.3a | 50.4a | |
| HPgV | 57.7b | 46.4a | 50.9a | |
| SPgV | 59.1b | 48.8b | 51.4a | |
| BPgV | 56.3b | 56.1b | 57.6b | |
Nucleotide level.
Amino acid level.
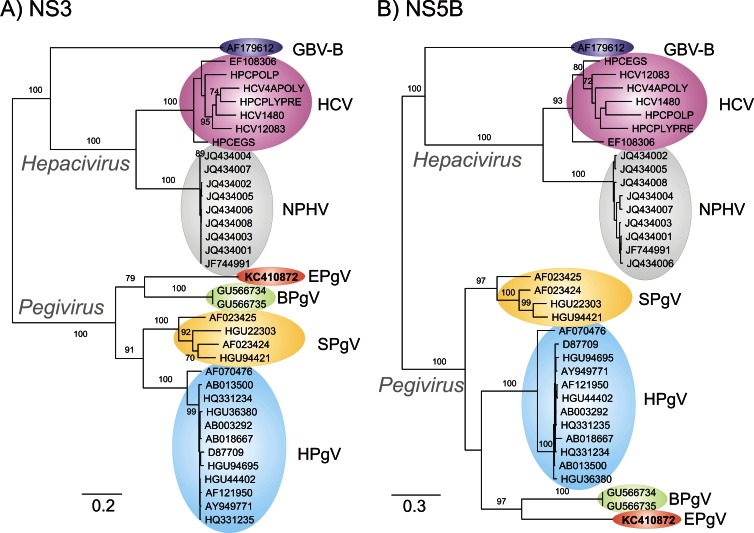
Sequence comparisons were extended to include homologous sequences from the Hepacivirus genus identified in the NS3 and NS5B regions that could be aligned (Fig. 2). Bootstrapped maximum-likelihood trees of the NS3 and NS5B regions of pegivirus and homologous regions of HCV and other hepaciviruses were generated by using RAxML with the PROTGAMMA model (gamma distribution for rates over sites, Dayhoff amino acid similarity matrix with all model parameters estimated by RAxML) and 100 bootstraps (14). Phylogenetic trees of the two genome regions were topologically equivalent, although with some differences in relative branch length. The analysis confirmed the separate grouping of EPgV from all other pegiviruses (Fig. 2).
Fig 2.
Maximum-likelihood trees of amino acid sequences from the NS3 (nt 3545 to 5523 in the C35 EPgV genome) and NS5B (nt 8743 to 10231) regions of EPgV and other pegiviruses. Pegiviruses were compared with homologous regions of hepaciviruses (HCV genotypes 1a to 7a, NPHV, and GBV-B). Sequences used in phylogenetic analysis: SPgV and GBV-Ccpz, HGU22303, AF023424, AF023425, HGU94421, GVU84961, and AF070476; HPgV (all complete genomes showing >2% divergence from each other), AB008342, D87709, D87711, AB003290, D87714, D90601, D87712, D87708, D87715, D87263, D87262, AB003293, AB003288, D87710, D87713, HGU94695, HGU75356, AF006500, AB013501, HGU63715, AF309966, AF121950, AF031827, AB003289, AY196904, D87255, HGU44402, AF104403, D90600, HGU45966, AY949771, AB008336, AB003291, HGU36380, AB013500, AB021287, AB018667, AB003292, HQ331234, HQ331235, HQ331233, AB010193, AF017560, AB008335, AF081782, AF031828, and AF031829; BPgV, GU566735 and GU566734.
RNA secondary structure in the UTRs and coding regions of the EPgV genome.
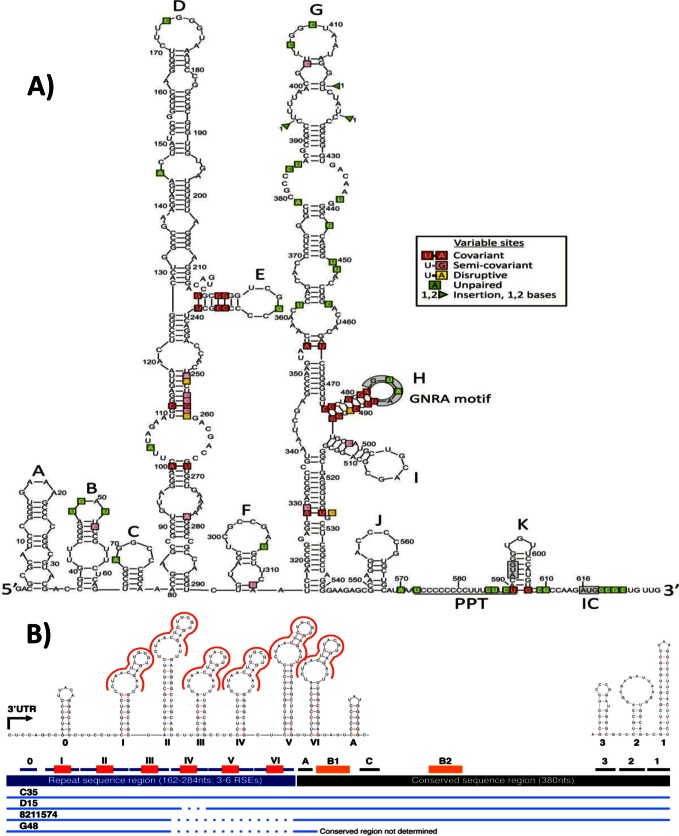
The 5′ UTR of EPgV is predicted to be 615 nt long, similar in length to those of other pegiviruses. To improve the accuracy of RNA structure predictions, complete or partial 5′ UTR sequences from 18 other infected horses in addition to C35 were included in the analysis. Structure prediction used PFOLD, a stochastic context-free grammar method, to identify phylogenetically conserved covariant sites supporting an RNA structure model (15), STRUCTUREDIST in the SSE package, which identifies conserved paired and unpaired bases in minimum-energy folds (16); and a combined minimum-energy/covariant site detection method implemented in ALIFOLD (16). Secondary-structure predictions were moderately consistent among all three methods, although PFOLD predicted no consistent structure of the 5′ UTR beyond position 290 and ALIFOLD between the start and position 200 and between 520 and the end of the 5′ UTR. Combining predictions, however, allowed a preliminary consensus structure model to be generated (Fig. 3A). It should be noted that none of the structure prediction methods can predict tertiary-structure elements, such as pseudoknots, that are present in some viral 5′ UTR structures. Furthermore, predictions could not be guided by comparison with other pegiviruses, as the regions of homology were highly restricted, and the HPgV and SPgV predicted structures differed from each other and from that of EPgV (17).
Fig 3.
(A) Preliminary RNA secondary structure of the EPgV 5′ UTR based on consensus predictions from PFLOD, STRUCTUREDIST, and ALIFOLD. IRES-associated sequences (GNRA motif and PPT) are labeled. Bases polymorphic between different variants of EPgV are shown in boxes and color coded according to their effects on base pairing (see key). IC, initiation codon. (B) Schematic of elements in the 3′ UTR and prediction of strong stem-loop structures. The repeat sequence region is drawn in blue, the conserved sequence region is in black, and the RSE (CTAACTCTNCGTGAGAT) is in red. A different RSE found in the conserved region is in orange (B1, B2). Strong stem-loop structures are indicated with horizontal lines and numbered with roman numerals in the repeat sequence region (the first stem-loop is given a zero since it does not have the RSE), with letters in the first part of the conserved region, and with numbers at the genome terminus. Additional structures in the conserved sequence could not be accurately determined because of a lack of variation among the isolates analyzed and a lack of homologous sequences in other viruses. Sequence alignments of isolates C35, D15, 8211574, and G48 are indicated at the bottom, with dotted lines representing internal deletions.
The consensus structure of the EPgV 5′ UTR comprised two large stem-loops (labeled D and G), several possible smaller ones (A, B, C, J, and K) positioned 5′ and 3′ to them, and an intermediate stem-loop, F. These structures are larger than, although positioned similarly to, stem-loops II and IV described in HPgV and SPgV (17), whereas stem-loop H is positioned similarly to SL-IVB. Comparative sequence data from different variants of EPgV were strongly supportive of the model, with most variable sites and insertions/deletions occurring in predicted unpaired regions (shaded green) and with two exceptions, variable sites in paired regions showing covariant or semicovariant compensatory substitutions to maintain pairing (shaded red and pink). Several features of the 3′-terminal half of the EPgV 5′ UTR are consistent with an internal ribosome entry site (IRES) structure. It contains a predicted unpaired PPT between positions 574 and 590. Additionally, stem-loop H positioned upstream contains a terminal GNRA tetraloop found in most virus IRES structures. Indeed, the arrangement of stem-loops G and H, followed by the PPT, a cryptic base-paired start codon in stem-loop K, an additional unpaired region, and finally the likely authentic start codon, is remarkably similar to the structure of type I IRESs of enteroviruses (in the virus family Picornaviridae). Nonetheless, physical mapping of the 5′ UTR and functional characterization are required to confirm this putative structural homology.
Analysis of the 3′ UTRs from four different isolates revealed a 5′ 162- to 284-nt repeat element region, including three to six repeat sequence elements (RSEs), followed by a highly conserved 380-nt sequence. The conserved region contained additional but different RSEs. The RSEs in the 5′ region were predicted to fold into conserved stem-loop structures (Fig. 3B). The existence of multiple repeat elements and their incorporation into a repetitive array of stem-loops have not been described in other mammalian viruses previously, and their functional role remains to be determined. Notably, 3′ UTR sequences from other pegiviruses show no sequence homology with the EPgV 3′ UTR or comparable repeated elements.
Thermodynamic folding analysis of the EPgV genome revealed a 10.7% free-energy difference between its minimum folding energy and that of sequence order-randomized controls (MFED), an observation consistent with the presence of genome scale ordered RNA structure in the EPgV genome (7). This MFED was similar to those of HPgV (mean, 12.8% [range, 11.7 to 13.3%]), SPgV (mean, 13.3% [range, 12.7 to 13.8%]), and BPgV (9.7 and 10.7%) (6).
Prevalence and persistence of EPgV.
To estimate the prevalence of EPgV, we studied two sets of available serum/plasma samples collected in the United States by reverse transcription (RT)-PCR with conserved primers in NS3. For the one U.S. sample set, the viremia frequencies were 3/12 (25%) among horses with elevated liver enzymes and 4/62 (6.4%) among healthy animals (P = 0.08 by Fisher's exact test). In an Alabama herd of healthy horses that were tested at three time points between 2008 and 2012, 6/19 (32%), 6/29 (21%), and 4/27 (15%) were positive. Two of these horses remained infected over a period of at least 3.5 years, whereas four animals apparently cleared the infection and new infections were observed in other animals. Though our study was not designed to draw conclusions about tissue tropism, EPgV sequences were detected in liver and lymph node biopsy samples, as well as in peripheral blood mononuclear cells, from the two chronically infected Alabama horses. Semiquantitative PCR did not reveal major quantitative differences in the presence of EPgV RNA between these tissues. Using quantitative RT-PCR, the viral RNA concentration in serum was determined to range from 104.5 to 106.5 genome equivalents per ml. Viral RNA was not detected in tracheal wash samples from these animals. Among the liver autopsy samples from nine horses (in California) with a range of causes of death, one sample was positive (from a horse that died of emaciation and a collapsed trachea).
Conclusions.
The recently designated genus Pegivirus of the family Flaviviridae currently includes three virus species that infect humans, primates, and bats. Recently, we identified hepaciviruses in horses (NPHV) that represent the closest known animal homolog of HCV (7). In the present study, we report the identification of the first pegivirus that infects horses. While our paper was in revision, a new Pegivirus species, named Theiler's disease-associated virus (TDAV), was identified in horses (18). Our sequence analysis confirmed that EPgV and TDAV are two genetically distinct Pegivirus species. These studies suggest that both hepaciviruses and pegiviruses may be widely distributed among different mammalian species. Our study and available samples were not sufficient to determine the health relevance of EPgV infection in horses; however, the limited biopsy data suggest that this virus may not be strictly hepatotropic. The complete genome sequence of EPgV will enable studies of EPgV sequence diversity and the development of effective diagnostic reagents to conduct studies of transmission, tissue tropism, and disease association.
Nucleotide sequence accession number.
The nucleotide sequence of the new virus (isolate C35) has been submitted to GenBank and assigned accession no. KC410872.
ACKNOWLEDGMENTS
This work was supported by awards AI090196, AI079231, AI57158, AI070411, AI090055, AI072613, CA057973, and EY017404 from the National Institutes of Health, by the Division of Intramural Research, National Institute of Dental and Craniofacial Research, by the Greenberg Medical Research Institute, by the Starr Foundation, and by the Danish Council for Independent Research.
Footnotes
Published ahead of print 17 April 2013
REFERENCES
- 1. Stapleton JT, Foung S, Muerhoff AS, Bukh J, Simmonds P. 2011. The GB viruses: a review and proposed classification of GBV-A, GBV-C (HGV), and GBV-D in genus Pegivirus within the family Flaviviridae. J. Gen. Virol. 92:233–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beames B, Chavez D, Guerra B, Notvall L, Brasky KM, Lanford RE. 2000. Development of a primary tamarin hepatocyte culture system for GB virus-B: a surrogate model for hepatitis C virus. J. Virol. 74:11764–11772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Berg T, Muller AR, Platz KP, Hohne M, Bechstein WO, Hopf U, Wiedenmann B, Neuhaus P, Schreier E. 1999. Dynamics of GB virus C viremia early after orthotopic liver transplantation indicates extrahepatic tissues as the predominant site of GB virus C replication. Hepatology 29:245–249 [DOI] [PubMed] [Google Scholar]
- 4. Blair CS, Davidson F, Lycett C, McDonald DM, Haydon GH, Yap PL, Hayes PC, Simmonds P, Gillon J. 1998. Prevalence, incidence, and clinical characteristics of hepatitis G virus/GB virus C infection in Scottish blood donors. J. Infect. Dis. 178:1779–1782 [DOI] [PubMed] [Google Scholar]
- 5. Williams CF, Klinzman D, Yamashita TE, Xiang J, Polgreen PM, Rinaldo C, Liu C, Phair J, Margolick JB, Zdunek D, Hess G, Stapleton JT. 2004. Persistent GB virus C infection and survival in HIV-infected men. N. Engl. J. Med. 350:981–990 [DOI] [PubMed] [Google Scholar]
- 6. Kapoor A, Simmonds P, Gerold G, Qaisar N, Jain K, Henriquez JA, Firth C, Hirschberg DL, Rice CM, Shields S, Lipkin WI. 2011. Characterization of a canine homolog of hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 108:11608–11613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burbelo PD, Dubovi EJ, Simmonds P, Medina JL, Henriquez JA, Mishra N, Wagner J, Tokarz R, Cullen JM, Iadarola MJ, Rice CM, Lipkin WI, Kapoor A. 2012. Serology-enabled discovery of genetically diverse hepaciviruses in a new host. J. Virol. 86:6171–6178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Epstein JH, Quan PL, Briese T, Street C, Jabado O, Conlan S, Ali Khan S, Verdugo D, Hossain MJ, Hutchison SK, Egholm M, Luby SP, Daszak P, Lipkin WI. 2010. Identification of GBV-D, a novel GB-like flavivirus from Old World frugivorous bats (Pteropus giganteus) in Bangladesh. PLoS Pathog. 6:e1000972 doi: 10.1371/journal.ppat.1000972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Simons JN, Pilot-Matias TJ, Leary TP, Dawson GJ, Desai SM, Schlauder GG, Muerhoff AS, Erker JC, Buijk SL, Chalmers ML, Van Sant CL, Mushahwar IK. 1995. Identification of two flavivirus-like genomes in the GB hepatitis agent. Proc. Natl. Acad. Sci. U. S. A. 92:3401–3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Waheed Y, Saeed U, Anjum S, Afzal MS, Ashraf M. 2012. Development of global consensus sequence and analysis of highly conserved domains of the HCV NS5B protein. Hepat. Mon. 12:e6142 doi: 10.5812/hepatmon.6142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113 doi: 10.1186/1471-2105-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Simmonds P. 2012. SSE: a nucleotide and amino acid sequence analysis platform. BMC Res. Notes 5:50 doi: 10.1186/1756-0500-5-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 15. Knudsen B, Hein J. 1999. RNA secondary structure prediction using stochastic context-free grammars and evolutionary history. Bioinformatics 15:446–454 [DOI] [PubMed] [Google Scholar]
- 16. Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. 2008. The Vienna RNA websuite. Nucleic Acids Res. 36:W70–W74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Simons JN, Desai SM, Schultz DE, Lemon SM, Mushahwar IK. 1996. Translation initiation in GB viruses A and C: evidence for internal ribosome entry and implications for genome organization. J. Virol. 70:6126–6135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chandriani S, Skewes-Cox P, Zhong W, Ganem DE, Divers TJ, Van Blaricum AJ, Tennant BC, Kistler AL. 2013. Identification of a previously undescribed divergent virus from the Flaviviridae family in an outbreak of equine serum hepatitis. Proc. Natl. Acad. Sci. U. S. A. 110:E1407–E1415 [DOI] [PMC free article] [PubMed] [Google Scholar]