Abstract
Efficient intracellular transport of the capsid of alphaherpesviruses, such as herpes simplex virus 1 (HSV-1), is known to be dependent upon the microtubule (MT) network. Typically, the MT network radiates from an MT-organizing center (MTOC), which is, in most cases, the centrosome. During herpesvirus egress, it has been assumed that capsids travel first from the nucleus to the centrosome and then from the centrosome to the site of envelopment. Here we report that the centrosome is no longer a primary MTOC in HSV-1-infected cells, but it retains this function in cells infected by another alphaherpesvirus, pseudorabies virus (PrV). As a result, MTs formed at late times after infection with PrV grow from a major, centralized MTOC, while those formed after HSV-1 infection arise from dispersed locations in the cytoplasm, indicating the presence of alternative and minor MTOCs. Thus, loss of the principal MT nucleating center in cells following HSV-1 infection raises questions about the mechanism of HSV-1 capsid egress. It is possible that, rather than passing via the centrosome, capsids may travel directly to the site of envelopment after exiting the nucleus. We suggest that, in HSV-1-infected cells, the disruption of centrosomal functions triggers reorganization of the MT network to favor noncentrosomal MTs and promote efficient viral spread.
INTRODUCTION
The microtubule (MT) network is commonly used for intracellular transport of virus particles (1), including the capsids of alphaherpesviruses such as herpes simplex virus 1 (HSV-1), which infects humans, and pseudorabies virus (PrV), a porcine herpesvirus (2, 3). This is important during entry, when capsids travel from the plasma membrane toward the nucleus, and during egress, when newly formed capsids exit the nucleus and travel toward the sites of envelopment at the trans-Golgi network (TGN) (4, 5). The MT network typically nucleates from a microtubule-organizing center (MTOC), which is most commonly the centrosome (6). The minus ends of MTs are embedded in the MTOC, while the highly dynamic plus ends extend throughout the cell. Because of this arrangement, it has been assumed that during entry, herpesvirus capsids travel from the plasma membrane to the centrosome, from which they are redirected to the nucleus, and that during egress, capsids are transported from the nucleus to the centrosome and then from the centrosome to the sites of envelopment (7). However, many viruses, such as adenovirus (8), rotavirus (9, 10), vaccinia virus (11, 12), and African swine fever virus (13), are known to reorganize the MT network and/or affect the centrosome during infection. Likewise, reorganization of the MT network and loss of the MTOC following HSV-1 infection have been reported (14, 15), leaving open the issue of what role the centrosome plays in HSV-1 capsid transport during egress. Although the interaction of capsids with the transport machinery is the subject of intensive research, the role of the centrosome in the process of capsid transport has not been fully evaluated.
In this paper, we investigate the effect of HSV-1 and PrV infections on the centrosome and on MT growth. We show that centrosome function is disrupted in HSV-1-infected cells but not in PrV-infected cells. Repolymerization of MTs following nocodazole treatment begins at the centrosome in mock- and PrV-infected cells, whereas it is absent in HSV-1-infected cells, thereby demonstrating that the centrosome no longer functions as an MTOC in these cells. Finally, EB3 labeling of MT plus ends revealed that whereas MTs no longer arise from a unique MTOC in HSV-1 cells, although they continue to do so in PrV-infected cells, they do still undergo growth, albeit slowly and from multiple locations in the cell.
MATERIALS AND METHODS
Cells and viruses.
African green monkey kidney (Vero), porcine kidney cells (PK15), and Human Fetal Foreskin Fibroblast (HFFF2) cells were grown at 37°C in Dulbecco's modified Eagle medium (DMEM; PAA Laboratories) supplemented with 8% fetal calf serum.
The 17syn+ strain of HSV-1 and the Kaplan strain of PrV were used as wild-type viruses. The ICP0-null virus dl1403 (16), the UL49-null virus Δ22 (which does not encode VP22 [17]), and the UL19-null virus K5ΔZ (18) were kindly provided by R. Everett (CVR, Glasgow, United Kingdom), G. Elliott (Imperial College, London, United Kingdom), and P. Desai (Johns Hopkins University, Baltimore, MD), respectively. The UL36- and UL37-null viruses ARΔUL36 and FRΔUL37 were described previously (19). To identify HSV-1-infected cells in live-cell microscopy experiments (see below), cells were infected with the vUL35RFP1D1 virus. This virus has a wild-type background (17syn+) except that it encodes a VP26 capsid protein fused to the monomeric Red Fluorescent Protein (mRFP) (20).
Antibodies and reagents.
The following antibodies were used: mouse monoclonal antibodies DM1A against alpha-tubulin (Sigma), GTU-88 against gamma-tubulin (Sigma), 11060 against HSV-1 ICP0 (SantaCruz Biotechnology), and DM165 against HSV-1 VP5 (21); rabbit polyclonal antibodies HPA016820 against pericentrin (Sigma), 14C10 against glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Cell Signaling), and PTNC, specific for HSV-1 capsid proteins VP23, VP26, and pUL36 (described in reference 22). Rabbit antibody 1702 directed against PrV capsids and mouse polyclonal antibody a5 against PrV pUL25 were gifts from K. Kaelin (VMS, Gif-sur-Yvette, France) (23). Secondary antibodies for immunofluorescence were goat anti-mouse or anti-rabbit Alexa 488 or Alexa 568 conjugated (GAM488, GAM568, GAR488, or GAR568) obtained from Molecular Probes. For Western blot analysis, secondary antibodies were goat anti-mouse DyLight680 and anti-rabbit DyLight800 (Cell Signaling). The plasmid pGFP-hEB3 encoding a green fluorescent protein (GFP) fusion of the human EB3 protein was a kind gift of John Victor Small (Institute of Molecular Biotechnology, Vienna, Austria). Nocodazole was obtained from Sigma, resuspended in dimethyl sulfoxide (DMSO) as a stock solution at a concentration of 10 mM, and used at a final concentration of 10 μM.
Nocodazole recovery experiments.
Cells were infected with 1 PFU/cell of the appropriate virus for 12 h before being incubated with 10 μM nocodazole for 1 h at 4°C. Cells were washed 2 times with cold DMEM followed by one wash with warm DMEM and then incubated for the indicated time in warm DMEM at 37°C. Cells were then fixed immediately as described below.
Fluorescence microscopy.
All immunofluorescence with alpha-tubulin staining was done as follows. Cells were fixed in a mix of 3.7% formaldehyde and 0.1% Triton X-100 in PEM buffer [100 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), 5 mM EGTA, 2 mM MgCl2, pH 6.8] for 5 min at room temperature as described in reference 24. Microtubules were stained using the alpha-tubulin antibody and GAM488.
To visualize centrosomes, cells were fixed with −20°C methanol and incubated at −20°C for 5 min. Methanol was discarded, and the cells were left to dry before being rehydrated with phosphate-buffered saline (PBS). Centrosomes were stained using the gamma-tubulin antibody and GAM488 or with the anti-pericentrin antibody and GAR488. Infected cells were identified using the anti-HSV-1 capsid antibody PTNC or with the anti-PrV capsid antibody 1702 and visualized with GAR568. Alternatively, infected cells were labeled with antibodies against HSV-1 ICP0 or PrV pUL25 and visualized with GAM568. When rabbit antibodies were used, 8% human serum (Sigma) was used to block the virus-encoded Fc-binding sites (25).
Samples were mounted in ImmuMount (Thermo) containing 1 μg/ml 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) (Sigma) for DNA staining. All samples were visualized using a Zeiss Axio Observer Z1 microscope and a 63× Plan-apochromat oil-immersion objective (Zeiss) (numerical aperture = 1.4).
Live-cell microscopy.
Cells were seeded on glass-bottomed petri dishes (MatTek) and transfected with 0.5 μg of pGFP-hEB3 using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. At 6 h later, cells were infected with 1 PFU/cell of vUL35RFP1D1 or PrV for 12 h. Live-cell recording was done on the same microscope as described above in an incubation chamber with controlled temperature (37°C) and atmosphere (5% CO2, humidified). Movies of 60 frames were recorded at a rate of 1 frame every 2 s. Infected cells were identified through VP26RFP fluorescence (HSV-1) or subsequent immunostaining (PrV).
EB3 comet tracking.
Movies were imported into ImageJ (version 1.43u). EB3 comets were tracked using the Particles Detector&Tracker plug-in (version 1.5 [26]). Depending on the quality of each individual movie, detection parameters were set as Radius = 5, Cutoff = 0.0, and Percentile = 0.1 to 1.8% and linking parameters were Link Range = 2 and Displacement = 10 to 20. Comet motion was analyzed using the coordinates provided by the software to calculate the distance traveled and velocity.
RESULTS
Centrosomal localization of gamma-tubulin is lost in HSV-1- but not in PrV-infected cells.
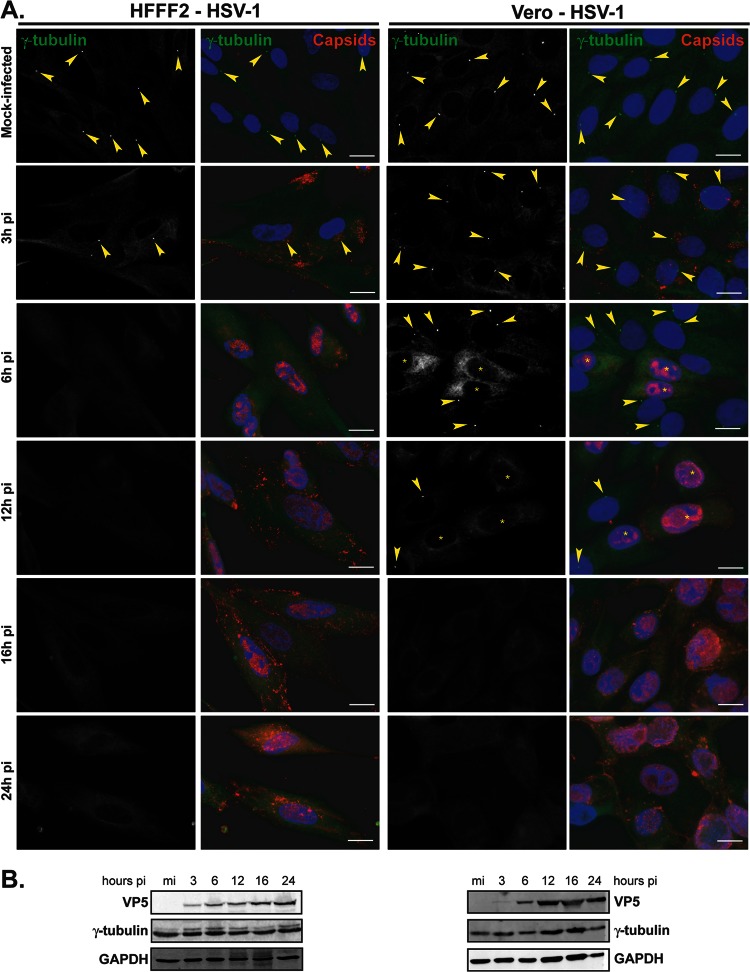
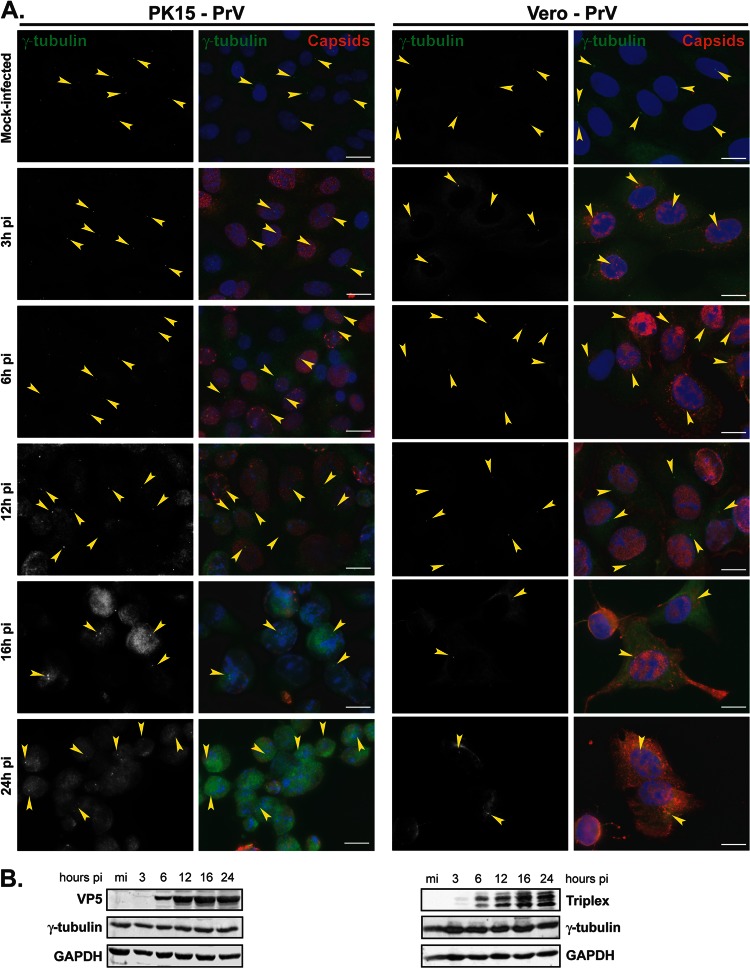
In the course of investigating the fate of the centrosome during HSV-1 infection, HFFF2, a human cell line, or Vero cells, a monkey cell line routinely used for alphaherpesviruses, were mock infected or infected with 1 PFU/cell of HSV-1 for 3 h, 6 h, 12 h, 16 h, and 24 h. The cells were analyzed using an antibody specific for gamma-tubulin, an integral component of the centrosome (27, 28), and infection was monitored using a capsid-directed rabbit antibody (PTNC). Gamma-tubulin staining of mock-infected HFFF2 or Vero cells showed typical centrosomal localization, with fluorescence localized as one or two spots close to the nucleus (arrowheads in Fig. 1A). This localization was lost by 6 h postinfection (pi), after which time no specific labeling of gamma-tubulin was evident. Western blotting showed that overall gamma-tubulin levels were unaffected by HSV-1 infection, thereby demonstrating that the loss of centrosomal fluorescence reflected changes in the distribution of the protein (Fig. 1B). To determine whether the dispersal of gamma-tubulin was a general property of alphaherpesviruses, we repeated the experiment with another virus from this subfamily, namely, PrV, a porcine herpesvirus. PK15 cells, a porcine cell line, or Vero cells were mock infected or infected with 1 PFU/cell of PrV, and gamma-tubulin was monitored as described for HSV-1-infected cells. PrV-infected cells were visualized using a PrV-capsid-directed rabbit antibody (1702). In contrast to HSV-1, gamma-tubulin localization was unaffected, at the level of immunocytochemistry, in PrV-infected cells (Fig. 2A), although strong cytopathic effects evident at 16 and 24 h pi made it increasingly difficult to assess localization of gamma-tubulin due to changes in the cell shape and high background fluorescence. As with HSV-1, levels of gamma-tubulin were unchanged over the course of the infection (Fig. 2B).
Fig 1.
Gamma-tubulin loses centrosomal localization in HFFF2 and Vero cells infected with HSV-1. (A) HFFF2 (left panels) or Vero (right panels) cells were infected with 1 PFU/cell of HSV-1 for the indicated times before fixation. Gamma-tubulin was visualized using mouse monoclonal antibody GTU-88 (green), and infected cells were visualized with the PTNC rabbit antibody (red). The nuclei were counterstained with DAPI. Arrowheads show centrosomal localization of gamma-tubulin. Asterisks indicate infected cells in panels also containing uninfected cells. Bars, 20 μm. (B) Western blot analysis of lysates obtained from cells infected for the indicated times. The progress of infection was visualized using the DM165 antibody directed against the major capsid protein VP5, gamma-tubulin levels were assessed using the GTU-88 antibody, and GAPDH levels were monitored as loading controls.
Fig 2.
Gamma-tubulin retains centrosomal localization in PK15 and Vero cells infected with PrV. (A) PK15 (left panels) or Vero (right panels) cells were infected with 1 PFU/cell of PrV for the indicated times before fixation. Gamma-tubulin was visualized using mouse monoclonal antibody GTU-88 (green), and infected cells were visualized with the 1702 antibody (red). The nuclei were counterstained with DAPI. Arrowheads show examples of centrosomal localization of gamma-tubulin. Bars, 20 μm. (B) Western blot analysis of lysates obtained from cells infected for the indicated times. The progress of infection was visualized using the anti-capsid 1702 antibody to show either VP5 or the two triplex proteins, gamma-tubulin levels were assessed using the GTU-88 antibody, and GAPDH levels were monitored as loading controls.
These results show that gamma-tubulin loses its centrosomal localization in HSV-1-infected cells but not in PrV-infected cells.
Pericentrin centrosomal localization is lost in HSV-1- but not in PrV-infected cells.
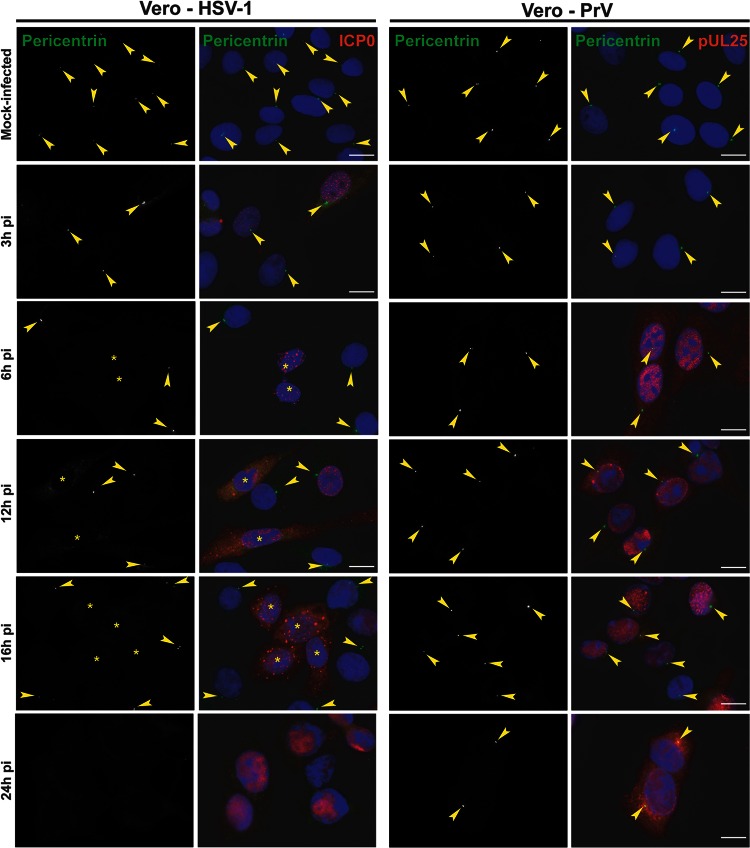
To determine whether the consequences of HSV-1 infection for gamma-tubulin were indicative of a more widespread effect, we monitored the localization of another centrosomal protein, pericentrin (29). Pericentrin was detected using the rabbit antibody HPA016820, which necessitated the use of mouse antibodies a5, directed against PrV pUL25 (a minor capsid protein [23]), and 11060, directed against HSV-1 ICP0, to monitor the infections. In mock-infected Vero cells, pericentrin localized to the centrosome, as expected, but in HSV-1-infected cells, the pericentrin behavior mimicked that of gamma-tubulin and this localization had disappeared by 6 h postinfection (Fig. 3). Once again, dispersion of the centrosome was not seen in PrV-infected cells, and pericentrin remained localized even at late times after infection.
Fig 3.
Differential loss of centrosomal localization of pericentrin in Vero cells infected with HSV-1 or PrV. Vero cells were infected with 1 PFU/cell of HSV-1 or PrV for the indicated times before fixation. Pericentrin was visualized using rabbit polyclonal antibody HPA016820 (green), and infected cells were visualized with the mouse antibodies 11060 against HSV-1 ICP0 and a5 against PrV pUL25 (red). The nuclei were counterstained with DAPI. Arrowheads show centrosomal localization of pericentrin. Asterisks indicate infected cells in panels also containing uninfected cells. Bars, 20 μm.
Thus, these results support the data obtained with gamma-tubulin staining and confirm that the centrosome is altered in HSV-1-infected cells but not in PrV-infected cells.
We quantified the loss of centrosomal markers in HFFF2 or Vero cells infected with HSV-1, and in PK15 or Vero cells infected with PrV, by determining the number of cells lacking detectable gamma-tubulin foci at different times after infection (Fig. 4). This showed that by 6 h pi, more than 50% of HFFF2 and more than 30% of Vero cells infected with HSV-1 had no detectable centrosomes. This number increased with time of infection to reach 89% in HFFF2 cells after 24 h. In contrast, PrV-infected Vero cells showed no significant centrosomal loss throughout the time of infection, while in PK15 cells, only ∼20% of cells had lost visible centrosomes by 16 h pi, and this number increased to ∼40% at 24 h pi. However, the extensive cytopathic effect evident in PK15 cells from 16 h pi (as shown in Fig. 2A) made it difficult to image these cells and could account for this apparent centrosomal loss. It is clear from these results that the effect of infection on the behavior of these centrosomal markers is much greater in HSV-1-infected cells than in cells infected by PrV, particularly at 6 h and 12 h pi.
Fig 4.
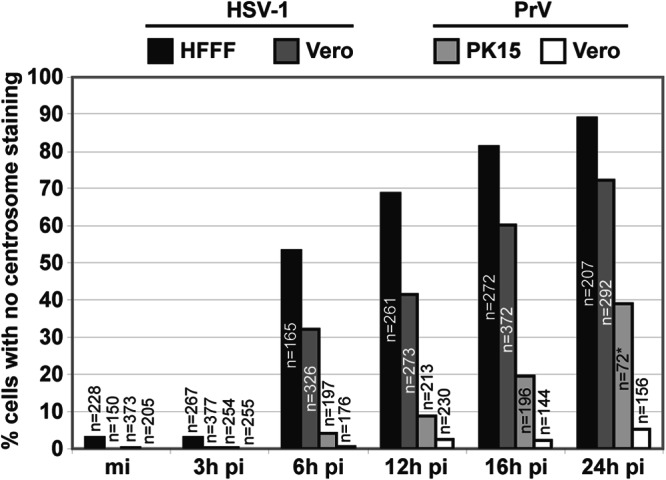
Quantification of centrosomal disruption in different cell lines infected by HSV-1 or PrV. A total number of 5661 cells, mock infected or infected with either HSV-1 or PrV for different times, were analyzed for the presence of the centrosome. Results are expressed as the percentage of cells with no detectable centrosomal gamma-tubulin staining. The asterisk indicates that due to extensive cell loss, fewer cells could be analyzed; most of the cells exhibited strong cytopathic effects.
ICP0, VP22, capsid formation, and the inner tegument are not necessary for centrosomal disruption.
To investigate candidate proteins that might influence centrosome function, we infected HFFF2 cells for 16 h with a range of HSV-1 mutants lacking specific functions. The HSV-1 immediate early protein ICP0 has been reported to dismantle the MTOC-based organization of the MT network (30). Therefore, we checked whether ICP0 was responsible for the loss of gamma-tubulin centrosomal localization using the ICP0-null virus dl1403 (16). As shown in Fig. 5, centrosomes could not be detected in dl1403 virus-infected cells, suggesting that ICP0 is not required. The tegument protein VP22 (pUL49) was reported to localize to MTs in transfected cells (31) and thus might be expected to influence the centrosome. However, gamma-tubulin dispersion was observed following infection with the VP22-null Δ22 virus (17), thus precluding a role for the protein in the process (Fig. 5). Since a major function of MTs at late times after infection is to transport capsids across the cytoplasm from the nucleus to the sites of envelopment, we examined whether this process might be necessary for centrosome disruption. The inner tegument proteins pUL36 and pUL37 have been shown to be important for capsid transport (32, 33). Therefore, we infected cells with the mutant viruses ARΔUL36 and FRΔUL37 (deleted for UL36 and UL37, respectively [19]). We used the UL19-null mutant, K5ΔZ, which is unable to produce capsids (18), as an additional control. Gamma-tubulin dispersion was observed in cells infected with each of these mutant viruses, which indicates that capsid transport and the inner tegument proteins pUL36 and pUL37 are not involved in changes to the centrosome.
Fig 5.
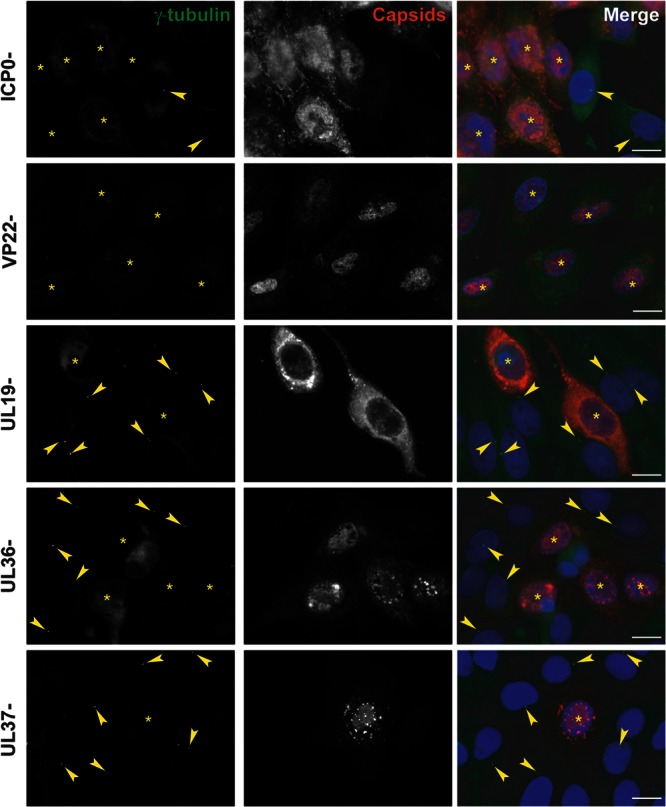
Centrosomal disruption in HFFF2 cells infected with ICP0-, VP22-, UL19-, UL36-, or UL37-null HSV-1 virus. HFFF2 cells were infected with 1 PFU/cell of dl1403 (ICP0-), Δ22 (VP22-), K5ΔZ (UL19-), ARΔUL36 (UL36-), or FRΔUL37 (UL37-) virus for 16 h before fixation. Centrosomes were visualized by staining for gamma-tubulin using mouse monoclonal antibody GTU-88 (green), and infected cells were visualized with the PTNC antibody (red). The nuclei were counterstained with DAPI. Arrowheads show centrosomes. Asterisks indicate infected cells. Bars, 20 μm.
MT recovery after nocodazole washout is impaired in HSV-1- but not in PrV-infected cells.
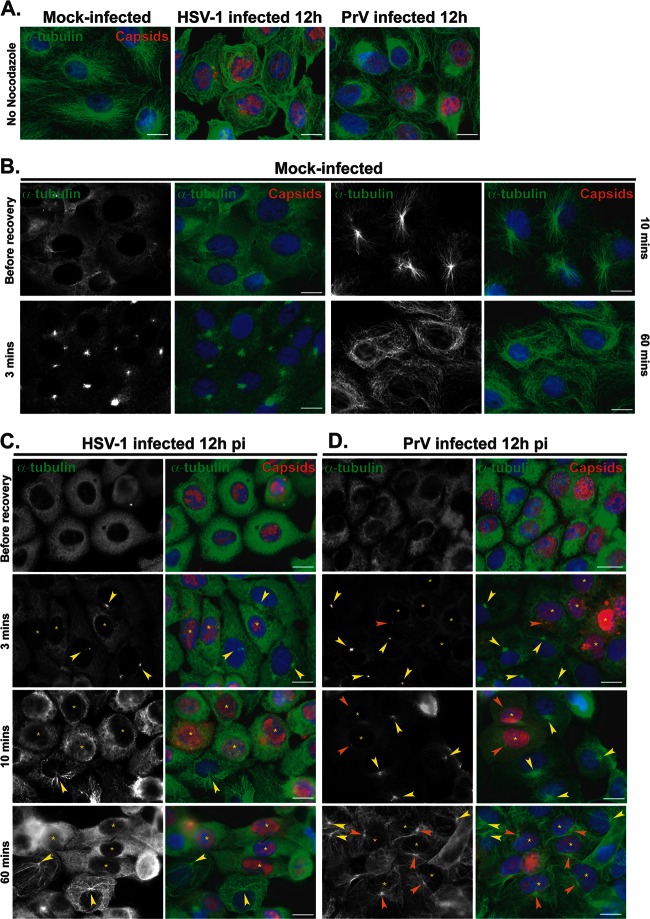
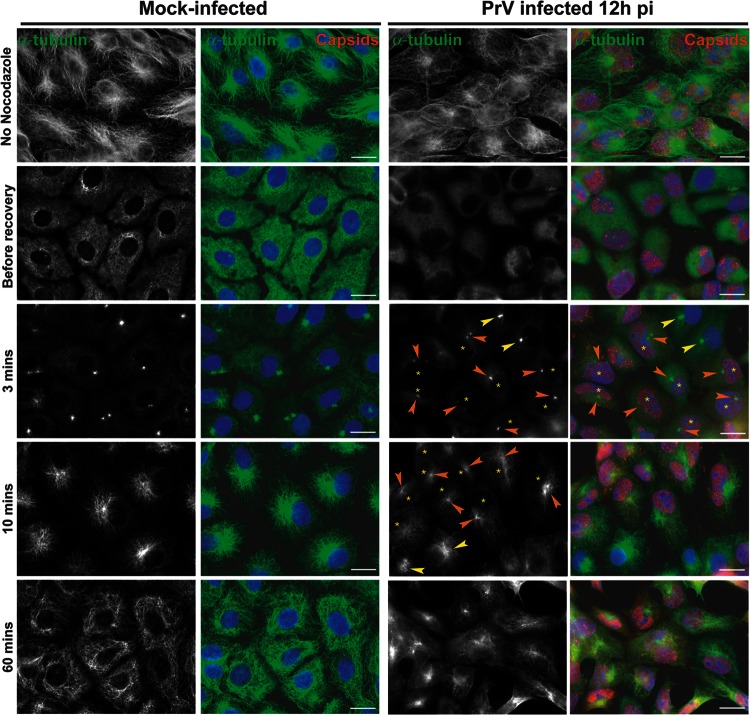
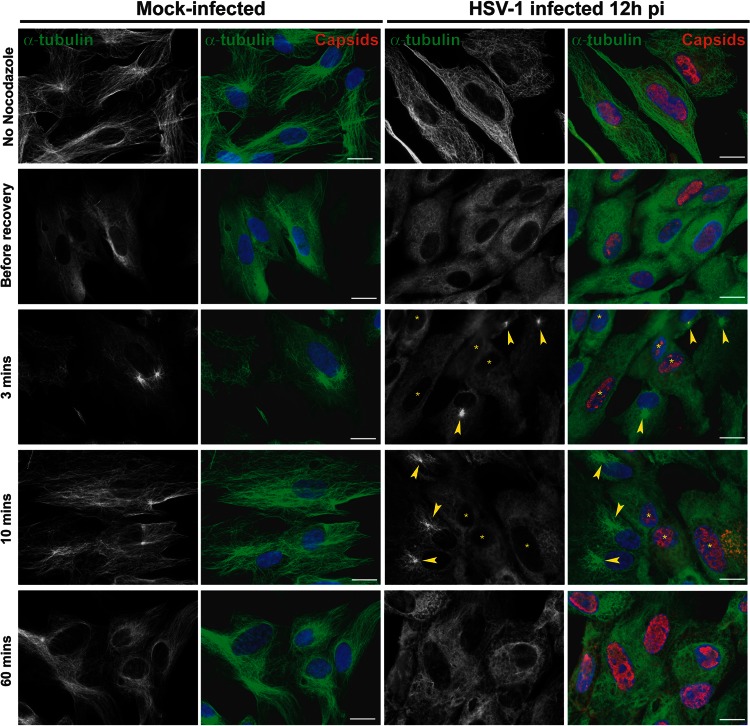
Loss of the centrosome during HSV-1 infection would imply that it no longer functioned as the major MTOC, meaning that MT growth from the centrosome would not occur. To monitor this, nocodazole recovery assays were carried out. Nocodazole is a drug which triggers MT depolymerization. This effect is reversible, and upon nocodazole washout, MTs polymerize de novo from MTOCs, which in most cells are provided by the centrosome. Vero cells were mock infected or infected with 1 PFU/cell of HSV-1 or PrV. In order to observe a significant number of cells with centrosomal disruption while avoiding severe cytopathic effects, infection was carried out for 12 h. Cells were then either fixed directly or incubated with 10 μM nocodazole for 1 h. Then, cells were fixed immediately (“before recovery”) or 3, 10, or 60 min after the drug was washed out. As shown in Fig. 6A, the MT network underwent reorganization in cells infected for 12 h in the absence of nocodazole. In both HSV-1- and PrV-infected cells, the MT network was less dense than in mock-infected cells. In PrV-infected cells, MT filaments were usually observed concentrated in a perinuclear area suggestive of the presence of a centrosome whereas most HSV-1-infected cells showed no such organization (Fig. 6A).
Fig 6.
Recovery of nocodazole-disrupted MTs in Vero cells infected with HSV-1 or PrV. (A) Comparison of MT networks in untreated Vero cells that were mock infected or infected with HSV-1 or PrV for 12 h. (B to D) Nocodazole recovery experiments. Vero cells were mock infected (B) or infected with HSV-1 (C) or PrV (D) for 12 h before being treated with 10 μM nocodazole. One hour later, the drug was washed out and cells were fixed after the indicated times. MTs were visualized using a mouse antibody directed against alpha-tubulin (green). HSV-1-infected cells were visualized using the PTNC rabbit antibody and PrV-infected cells using the 1702 rabbit antibody (red). Arrowheads in panels C and D indicate centers of MT growth in infected cells (orange) or uninfected cells (yellow). Asterisks indicate infected cells in panels also containing uninfected cells. Note that MT growth is absent (C) or limited (D) in infected cells compared to uninfected cells. Bars, 20 μm.
Treatment of mock-infected and infected cells with 10 μM nocodazole for 1 h was sufficient to depolymerize the majority of Vero cell MTs (Fig. 6B to D). Three minutes after nocodazole washout, MT polymerization had started in mock-infected cells and was clearly centralized at an MTOC, presumably the centrosome (Fig. 6B). By 10 min, MT recovery was almost complete, with the MTOC organization very evident, and after 1 h the MT network was fully recovered. This contrasted sharply with the behavior in cells infected with HSV-1 for 12 h, where no de novo polymerized MT filaments were detected even at 1 h after nocodazole washout (Fig. 6C).
In PrV-infected Vero cells, initiation of MT growth was delayed compared to mock-infected cells, with little visible 3 min after nocodazole washout. By 10 min after washout, MT growth from visible MTOCs had started (Fig. 6D), although it was much more limited than in mock-infected cells. One hour after nocodazole washout, MTOC-based networks of MTs radiated throughout the infected cells, although the growth was less dense than that seen in mock-infected cells (Fig. 6A).
These effects were not limited to Vero cells. In PrV-infected PK15 cells, the same behavior was observed, although the MT repolymerization was more efficient than in infected Vero cells (Fig. 7). Nevertheless, as with the Vero cells, the MT network did not fully recover after 1 h, which suggests that PrV infection has an inhibitory effect on MT polymerization and/or MT stability but not on centrosomal-based nucleation of MTs. Similarly, the influence of HSV-1 infection on MT polymerization was not cell specific, since MTOC-based MT repolymerization following nocodazole treatment and washout was also lacking in HFFF2 cells (Fig. 8).
Fig 7.
Recovery of nocodazole-disrupted MTs in PK15 cells infected with PrV. PK15 cells were mock infected or infected with PrV for 12 h before being treated with 10 μM nocodazole. One hour later, the drug was washed out and cells were fixed after the indicated time. Immunolabeling was carried out as described for Fig. 6. Arrowheads indicate centers of MT growth in infected cells (orange) or uninfected cells (yellow). Asterisks indicate infected cells in panels also containing uninfected cells. Note that MTs are centrally nucleated in infected cells. Bars, 20 μm.
Fig 8.
Recovery of nocodazole-disrupted MTs in HFFF2 cells infected with HSV-1. HFFF2 cells were mock infected or infected with HSV-1 for 12 h before being treated with 10 μM nocodazole. One hour later, the drug was washed out and cells fixed after the indicated time. Immunolabeling was carried out as described for Fig. 6. Arrowheads indicate centers of MT growth. Asterisks indicate infected cells in panels also containing uninfected cells. Note the absence of centralized MT growth in infected cells. Bars, 20 μm.
These experiments show that in cells infected by HSV-1, the growth, and possibly the stability, of MTs originating from the centrosome is impaired, thereby indicating that the centrosome is functionally disrupted. This does not appear to be the case for PrV.
Growth of MTs is less efficient and originates from noncentrosomal sites in HSV-1-infected cells.
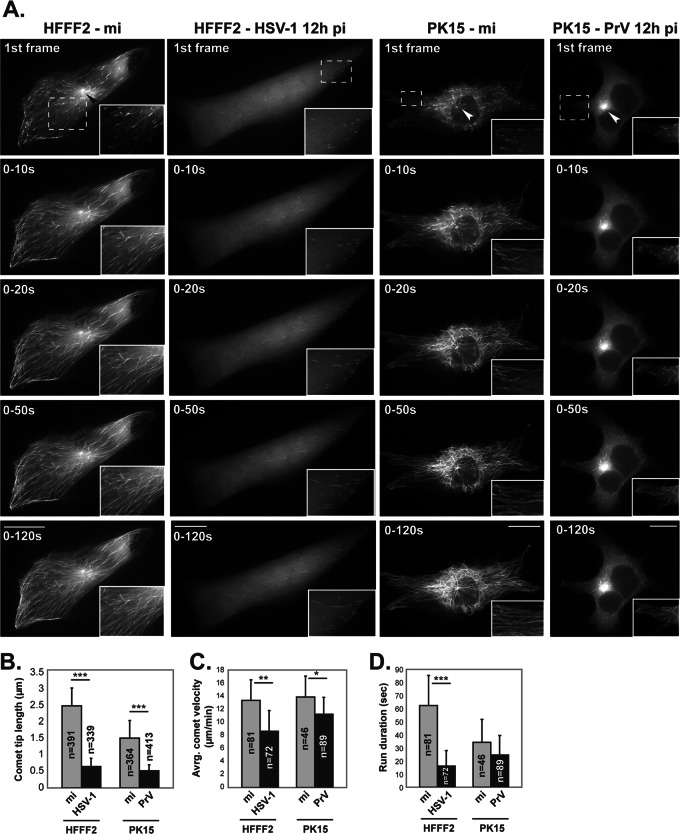
To investigate MT nucleation in HSV-1- and PrV-infected cells, MT dynamics were followed in living cells by tracking the distribution of End-Binding protein 3 (EB3), which predominantly binds to the plus-end tip of growing MTs (34). To illustrate the pattern of MT growth, time-lapse movies were collected and projections of the first 5, 10, 25, and 60 time-lapse frames, covering periods of 10, 20, 50, and 120 s, respectively, were produced (Fig. 9; see also Movies S1 to S4 in the supplemental material). Mock-infected HFFF2 cells expressing EB3 fused to GFP (GFP-EB3) showed strong fluorescence with a typical “comet” shape at the plus-end tip of each growing MT (see Movie S1 in the supplemental material). MT growth was active throughout cells, with most originating at an obvious nucleating center near the nucleus (arrowheads in Fig. 9; see also Movie S1 in the supplemental material). In cells infected with HSV-1 for 12 h, MT growth was very limited. It was mostly localized near the plasma membrane at cell extremities, and no obvious nucleation centers were visible (Fig. 9; see also Movie S2 in the supplemental material). The GFP-EB3 signal was weak and diffuse, and EB3 comets were reduced in number. Specifically, individual comets were almost five times shorter (Fig. 9B) and the rate of MT growth appeared to be ∼40% lower than in mock-infected cells (Fig. 9C; see also Movie S2 in the supplemental material). Moreover, the comets in infected cells appear to be less stable, since many of them were observed in only ∼7 to 8 consecutive frames before they disappeared, whereas those in mock-infected cells were frequently present throughout the period of observation (more than 30 consecutive frames; Fig. 9D). Similar observations were made at later times after infection, including 16 h and 24 h pi (data not shown).
Fig 9.
MT growth in living HFFF2 and PK15 cells that were mock infected or infected with HSV-1 or PrV. HFFF2 and PK15 cells were transfected with a plasmid encoding GFP-EB3, a protein that localizes at plus-end tips of MTs. At 6 h later, HFFF2 cells were mock infected (mi) or infected with vUL35RFP1D1 (an HSV-1 virus in which the capsid protein VP26 is fused to the mRFP) and PK15 cells were infected with wild-type PrV. After a further 12 h, GFP-EB3 localization was recorded by live-cell microscopy at a rate of one frame every 2 s. Infected HFFF2 cells were identified by mRFP fluorescence (not shown). Infection of PK15 cells by PrV was subsequently checked by immunostaining for capsids (not shown). (A) Individual panels show cumulative projections of frames obtained over the indicated periods after the start of recording. Arrowheads show MT nucleating centers in the first frame images. An area from each cell image (dashed boxes) is enlarged. To observe MT growth, see Movies S1 and S2 (mock-infected and HSV-1-infected HFFF2 cells, respectively) and Movies S3 and S4 (mock-infected and PrV-infected PK15 cells, respectively) in the supplemental material. Bars, 20 μm. Movies were taken at a rate of 1 frame every 2 s for 120 s and are shown at a rate of 10 frames/s. (B) The length of 730 EB3 comets was measured in single time frames from movies of nine mock-infected (mi) and 10 HSV-1-infected HFFF2 cells. The length of 777 EB3 comets was measured in single time frames from movies of four uninfected and eight PrV-infected PK15 cells. (C and D) A total of 153 (HFFF2) and 135 (PK15) comets were tracked, and their velocity was measured (C) as well as the number of consecutive frames where they could be tracked (run duration) (D). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The dispersed pattern of MT growth and lack of a single focal site support the idea that the centrosome is lost during HSV-1 infection and suggest the possibility that MT growth originates from multiple noncentrosomal nucleating centers throughout the infected cell.
Live-cell experiments were also carried out in PK15 cells. These showed smaller comet tips and shorter runs than HFFF2 cells but had similar comet velocities (Fig. 9B to D). As with HSV-1 in HFFF2 cells, infection of PK15 cells by PrV resulted in shorter comet tips than in mock-infected cells. The reduction was less than that seen in HSV-1-infected HFFF2 cells (∼3 times shorter compared to ∼5 times shorter). However, their length of 0.5 μm (± 0.17) is very similar to that in HSV-infected HFFF2 cells (0.64 μm ± 0.24). In contrast to HSV-1-infected cells, comet velocity was only slightly lower in PrV-infected cells than in mock-infected cells (Fig. 9C), and run duration was not significantly altered (Fig. 9D). As would be expected from the nocodazole washout experiments, MT growth in PrV-infected PK15 cells predominantly originated from a major MTOC, presumably the centrosome (Fig. 9A; see also Movie S4 in the supplemental material). In this respect, PrV-infected cells resemble mock-infected cells and differ from HSV-1-infected cells (see Movie S2 in the supplemental material).
These results show that infection by both HSV-1 and PrV alters MT growth, although to differing degrees, but only infection by HSV-1 disrupts the MTOC function of the centrosome.
DISCUSSION
This paper reports on two main findings. First, the centrosome's function as an MTOC is disrupted in the course of infection by HSV-1 but not by another alphaherpesvirus, PrV. Second, MT dynamics are modified in cells infected with either HSV-1 or PrV, although to different degrees. These observations raise questions regarding egress of alphaherpesviruses and their interactions with the MT network.
The capsids of alphaherpesviruses are known to rely on MTs for transport throughout the cell (3). In particular, MTs have been shown to be important for rapid capsid transport at late stages of infection (32, 35). Since the MT network is typically organized around MTOCs, the most prominent of which is the centrosome, it was reasonably assumed that capsids would have to pass through MTOCs during entry and egress (7). Traversing the centrosome represents an important phase in capsid transport since it would require a change of transport polarity. However, this step and the fate of the centrosome during herpesvirus infection have not been extensively analyzed. One study described loss of the MTOC in Vero cells at 6 h pi and its reformation at 25 h pi (15), while MT reorganization at 6 h pi has also been reported (14). Another study reported that the MT network is disrupted in HSV-1-infected Vero cells, that this requires ICP0, and that tubulin colocalizes into cytoplasmic globular bodies with ICP0 (30). In this paper, we demonstrate using centrosome-specific markers and functional assays that the centrosome ceases to function as the primary MTOC following HSV-1 infection and confirm that the disruption of centrosomal activity starts by 6 h pi. In contrast to Kotsakis and collaborators (15), we did not observe reformation of the centrosome at 24 h pi but were unable to check later times given the extent of the cytopathic effect. Unlike Liu et al. (30), we saw no accumulation of tubulin into globular bodies in HSV-1-infected Vero or HFFF2 cells and we show that deletion of ICP0 did not prevent loss of the centrosome.
As already stated, loss of the centrosomal MTOC function is apparent by about 6 h after HSV-1 infection, which is around the time when capsids start to exit the nucleus and have to be transported across the cytoplasm to the sites of envelopment. It is interesting, therefore, to speculate as to how these changes might affect viral egress. In most cell types, the majority of MT minus ends are embedded in the centrosome and uncapped MT minus ends typically disassemble quickly in the cytoplasm. In the centrosome, MTs nucleate from their minus ends through γ-tubulin ring complexes (or γ-TuRCs), in which γ-tubulin plays a critical role (36). Therefore, it is likely that if the centrosome or its nucleating material is lost as a result of HSV-1 infection (as is suggested by the absence of centrosomal γ-tubulin), the population of MTs with their minus ends anchored at the centrosome will also be lost. However, MTs with free and stable minus ends, independent of the centrosome, are found in some cell lines (37). In addition, there are specialized cell types, such as epithelial and neuronal cells, in which “noncentrosomal” MTOCs have been described as performing the MT-stabilizing role (6, 37, 38). It is not clear whether either of these mechanisms accounts for some or all of the noncentrosomal MTs observed in both Vero and HFFF2 cells infected with HSV-1. However, we did observe growth of noncentrosomal MTs in HFFF2 cells infected with HSV-1 (Fig. 9) and observations in live cells confirmed that MTs were growing from many different points, predominantly localized in the vicinity of the plasma membrane. One noncentrosomal MTOC that has previously been identified is the TGN (39). This suggests the possibility that disruption of the centrosome and destabilization of centrosomal MTs could favor noncentrosomal MTs emanating from alternative MTOCs such as the TGN. Since the TGN is an important site of envelopment for the virus, such noncentrosomal MTs could provide a direct route from the nucleus to the TGN for the many newly formed capsids. The retention of centrosomal organization would suggest that this is not the case for PrV.
In addition to the loss of centrosomal organization in HSV-1-infected cells, live-cell microscopy of MT plus ends in HSV-1-infected cells revealed that the rate and duration of MT growth were also reduced following infection (Fig. 9). Although MTs were observed growing from centrosomal sites in PrV-infected PK15 and Vero cells, the rate and extent of this growth also seemed to be limited compared to mock-infected cells (Fig. 6, 7, and 9). These observations suggest that MT growth is affected in cells infected with either HSV-1 or PrV and that the effects on MT growth may be at least partly independent of those on the centrosome.
HSV-1 is not the only virus reported to have a disruptive effect on the centrosome. Infections by vaccinia virus and African swine fever virus also trigger centrosome loss (12, 13). Moreover, Bystrevskaya and colleagues reported on centriole alteration in dividing human embryo lung fibroblasts (HEL) and Vero cells productively infected with human cytomegalovirus (40). It is therefore plausible that centrosomal disruption is part of a general viral strategy to optimize the MT network in a way that benefits viral egress. How this works is yet to be determined.
In summary, the data presented here suggest that alphaherpesviruses can adopt differing egress strategies and that investigating the role of the centrosome will provide interesting insights into their nature.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yves Gaudin for critical reading of the manuscript. We are grateful to John Victor Small (Institute of Molecular Biotechnology, Vienna, Austria) for providing the pGFP-hEB3 plasmid, to Karin Kaelin (VMS, Gif-sur-Yvette, France) for antibodies 1702 and a5, to Roger Everett for the dl1403 virus, to Gillian Elliott (Imperial College, London, United Kingdom) for the Δ22 virus, and to Prashant Desai (John Hopkins University, Baltimore, MD) for the K5ΔZ virus.
This work was supported by funds from the CNRS.
Footnotes
Published ahead of print 17 April 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00764-13.
REFERENCES
- 1. Greber UF, Way M. 2006. A superhighway to virus infection. Cell 124:741–754 [DOI] [PubMed] [Google Scholar]
- 2. Mabit H, Nakano MY, Prank U, Saam B, Dohner K, Sodeik B, Greber UF. 2002. Intact microtubules support adenovirus and herpes simplex virus infections. J. Virol. 76:9962–9971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sodeik B, Ebersold MW, Helenius A. 1997. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 136:1007–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Harley CA, Dasgupta A, Wilson DW. 2001. Characterization of herpes simplex virus-containing organelles by subcellular fractionation: role for organelle acidification in assembly of infectious particles. J. Virol. 75:1236–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Turcotte S, Letellier J, Lippe R. 2005. Herpes simplex virus type 1 capsids transit by the trans-Golgi network, where viral glycoproteins accumulate independently of capsid egress. J. Virol. 79:8847–8860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bartolini F, Gundersen GG. 2006. Generation of noncentrosomal microtubule arrays. J. Cell Sci. 119:4155–4163 [DOI] [PubMed] [Google Scholar]
- 7. Radtke K, Dohner K, Sodeik B. 2006. Viral interactions with the cytoskeleton: a hitchhiker's guide to the cell. Cell Microbiol. 8:387–400 [DOI] [PubMed] [Google Scholar]
- 8. Warren JC, Rutkowski A, Cassimeris L. 2006. Infection with replication-deficient adenovirus induces changes in the dynamic instability of host cell microtubules. Mol. Biol. Cell 17:3557–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brunet JP, Jourdan N, Cotte-Laffitte J, Linxe C, Geniteau-Legendre M, Servin A, Quero AM. 2000. Rotavirus infection induces cytoskeleton disorganization in human intestinal epithelial cells: implication of an increase in intracellular calcium concentration. J. Virol. 74:10801–10806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zambrano JL, Sorondo O, Alcala A, Vizzi E, Diaz Y, Ruiz MC, Michelangeli F, Liprandi F, Ludert JE. 2012. Rotavirus infection of cells in culture induces activation of RhoA and changes in the actin and tubulin cytoskeleton. PLoS One 7:e47612 doi: 10.1371/journal.pone.0047612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arakawa Y, Cordeiro JV, Way M. 2007. F11L-mediated inhibition of RhoA-mDia signaling stimulates microtubule dynamics during vaccinia virus infection. Cell Host Microbe 1:213–226 [DOI] [PubMed] [Google Scholar]
- 12. Ploubidou A, Moreau V, Ashman K, Reckmann I, Gonzalez C, Way M. 2000. Vaccinia virus infection disrupts microtubule organization and centrosome function. EMBO J. 19:3932–3944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jouvenet N, Wileman T. 2005. African swine fever virus infection disrupts centrosome assembly and function. J. Gen. Virol. 86:589–594 [DOI] [PubMed] [Google Scholar]
- 14. Avitabile E, Di Gaeta S, Torrisi MR, Ward PL, Roizman B, Campadelli-Fiume G. 1995. Redistribution of microtubules and Golgi apparatus in herpes simplex virus-infected cells and their role in viral exocytosis. J. Virol. 69:7472–7482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kotsakis A, Pomeranz LE, Blouin A, Blaho JA. 2001. Microtubule reorganization during herpes simplex virus type 1 infection facilitates the nuclear localization of VP22, a major virion tegument protein. J. Virol. 75:8697–8711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stow ND, Stow EC. 1986. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J. Gen. Virol. 67(Pt 12):2571–2585 [DOI] [PubMed] [Google Scholar]
- 17. Elliott G, Hafezi W, Whiteley A, Bernard E. 2005. Deletion of the herpes simplex virus VP22-encoding gene (UL49) alters the expression, localization, and virion incorporation of ICP0. J. Virol. 79:9735–9745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Desai P, DeLuca NA, Glorioso JC, Person S. 1993. Mutations in herpes simplex virus type 1 genes encoding VP5 and VP23 abrogate capsid formation and cleavage of replicated DNA. J. Virol. 67:1357–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Roberts AP, Abaitua F, O'Hare P, McNab D, Rixon FJ, Pasdeloup D. 2009. Differing roles of inner tegument proteins pUL36 and pUL37 during entry of herpes simplex virus type 1. J. Virol. 83:105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pasdeloup D, Beilstein F, Roberts AP, McElwee M, McNab D, Rixon FJ. 2010. Inner tegument protein pUL37 of herpes simplex virus type 1 is involved in directing capsids to the trans-Golgi network for envelopment. J. Gen. Virol. 91:2145–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McClelland DA, Aitken JD, Bhella D, McNab D, Mitchell J, Kelly SM, Price NC, Rixon FJ. 2002. pH reduction as a trigger for dissociation of herpes simplex virus type 1 scaffolds. J. Virol. 76:7407–7417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pasdeloup D, Blondel D, Isidro AL, Rixon FJ. 2009. Herpesvirus capsid association with the nuclear pore complex and viral DNA release involve the nucleoporin CAN/Nup214 and the capsid protein pUL25. J. Virol. 83:6610–6623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaelin K, Dezelee S, Masse MJ, Bras F, Flamand A. 2000. The UL25 protein of pseudorabies virus associates with capsids and localizes to the nucleus and to microtubules. J. Virol. 74:474–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vielkind U, Swierenga SH. 1989. A simple fixation procedure for immunofluorescent detection of different cytoskeletal components within the same cell. Histochemistry 91:81–88 [DOI] [PubMed] [Google Scholar]
- 25. Baucke RB, Spear PG. 1979. Membrane proteins specified by herpes simplex viruses. V. Identification of an Fc-binding glycoprotein. J. Virol. 32:779–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sbalzarini IF, Koumoutsakos P. 2005. Feature point tracking and trajectory analysis for video imaging in cell biology. J. Struct. Biol. 151:182–195 [DOI] [PubMed] [Google Scholar]
- 27. Stearns T, Evans L, Kirschner M. 1991. Gamma-tubulin is a highly conserved component of the centrosome. Cell 65:825–836 [DOI] [PubMed] [Google Scholar]
- 28. Zheng Y, Jung MK, Oakley BR. 1991. Gamma-tubulin is present in Drosophila melanogaster and Homo sapiens and is associated with the centrosome. Cell 65:817–823 [DOI] [PubMed] [Google Scholar]
- 29. Doxsey SJ, Stein P, Evans L, Calarco PD, Kirschner M. 1994. Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell 76:639–650 [DOI] [PubMed] [Google Scholar]
- 30. Liu M, Schmidt EE, Halford WP. 2010. ICP0 dismantles microtubule networks in herpes simplex virus-infected cells. PLoS One 5:e10975 doi: 10.1371/journal.pone.0010975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Elliott G, O'Hare P. 1998. Herpes simplex virus type 1 tegument protein VP22 induces the stabilization and hyperacetylation of microtubules. J. Virol. 72:6448–6455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Luxton GW, Lee JI, Haverlock-Moyns S, Schober JM, Smith GA. 2006. The pseudorabies virus VP1/2 tegument protein is required for intracellular capsid transport. J. Virol. 80:201–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wolfstein A, Nagel CH, Radtke K, Dohner K, Allan VJ, Sodeik B. 2006. The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro. Traffic 7:227–237 [DOI] [PubMed] [Google Scholar]
- 34. Nakagawa H, Koyama K, Murata Y, Morito M, Akiyama T, Nakamura Y. 2000. EB3, a novel member of the EB1 family preferentially expressed in the central nervous system, binds to a CNS-specific APC homologue. Oncogene 19:210–216 [DOI] [PubMed] [Google Scholar]
- 35. Pasdeloup D, McElwee M, Beilstein F, Labetoulle M, Rixon FJ. 2013. Herpesvirus tegument protein pUL37 interacts with dystonin/BPAG1 to promote capsid transport on microtubules during egress. J. Virol. 87:2857–2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kollman JM, Merdes A, Mourey L, Agard DA. 2011. Microtubule nucleation by gamma-tubulin complexes. Nat. Rev. Mol. Cell Biol. 12:709–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dammermann A, Desai A, Oegema K. 2003. The minus end in sight. Curr. Biol. 13:R614–R624 [DOI] [PubMed] [Google Scholar]
- 38. Döhner K, Nagel CH, Sodeik B. 2005. Viral stop-and-go along microtubules: taking a ride with dynein and kinesins. Trends Microbiol. 13:320–327 [DOI] [PubMed] [Google Scholar]
- 39. Efimov A, Kharitonov A, Efimova N, Loncarek J, Miller PM, Andreyeva N, Gleeson P, Galjart N, Maia AR, McLeod IX, Yates JR, III, Maiato H, Khodjakov A, Akhmanova A, Kaverina I. 2007. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev. Cell 12:917–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bystrevskaya VB, Lobova TV, Smirnov VN, Makarova NE, Kushch AA. 1997. Centrosome injury in cells infected with human cytomegalovirus. J. Struct. Biol. 120:52–60 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.