Abstract
Preferential loss of heterozygosity at the rs1042522 locus of the tumor protein 53 gene (TP53) (Arg72Pro) is observed in several tumors. Genetic association studies in oncology often use tumor tissue rather than unaffected tissue for genotyping; in such cases, loss of heterozygosity at the TP53 locus could lead to differential misclassification and could bias estimates of association. We searched multiple databases (through March 8, 2011) for studies investigating the association of Arg72Pro with breast, lung, colorectal, ovarian, or endometrial cancer. Meta-analysis was performed with multilevel Bayesian models. Informative priors for the bias effect were derived from a meta-analysis of the same polymorphism in cervical cancer. Of 160 studies (68 breast, 42 lung, 26 colorectal, 16 ovarian, and 8 endometrial cancer), 22 used tumor tissue as the source of genotyping material for cases. Use of tumor tissue versus other sources of genotyping material was associated with an apparent protective effect of the proline allele (relative odds ratio = 0.78, 95% credible interval: 0.70, 0.88). The probability that use of tumor tissue induced bias was estimated to be higher than 99%. Use of tumor tissue as the source of genotyping material for cases is associated with significant bias in the estimate of the genetic effect in cancer genetic association studies.
Keywords: Arg72Pro, cancer, meta-analysis, rs1042522, TP53
Editor's note: This article also appears on the website of the Human Genome Epidemiology Network (http://www.cdc.gov/genomics/hugenet/default.htm).
Lung, breast, and colorectal cancers jointly account for the majority of new cancer cases and represent the top 3 causes of cancer-related death in Western countries (1, 2). Breast, ovarian, and endometrial cancers are major causes of cancer-related morbidity and mortality among women (2). Multiple lines of evidence, including studies of familial clustering and studies of heritability in twins, suggest that these common epithelial cancers have a substantial hereditary component (3). One of the most promising cancer-causing genes is the tumor protein 53 gene (TP53). TP53 encodes a 53-kDa transcription factor, tumor protein 53, which is involved in regulating apoptosis and cell-cycle control (4). Heritable mutations in TP53 are associated with the Li-Fraumeni syndrome, a mendelian disorder characterized by increased incidence of multiple types of cancer (5). In addition, the majority of epithelial cancers have been shown to carry somatic TP53 aberrations, mainly within the DNA-binding domain of the p53 protein (6). In cases in which TP53 mutations are not present, p53 function often is abrogated either through loss of heterozygosity (LOH) (by deletion or methylation of the 17p locus) or through inactivation of p53 downstream effectors. Furthermore, there is evidence that TP53 plays a role in modulating the frequency and mechanisms of mutagenesis during carcinogenesis (7). The high frequency of p53 inactivation in human cancers highlights the importance of its tumor suppressor function; for this reason, it has been called the “guardian of the genome” (8).
These observations have provided a strong biological rationale for the hypothesis that high-frequency functional TP53 polymorphisms can contribute to the population risk of developing common cancers (4). Most studies have focused on a nonsynonymous TP53 polymorphism in exon 4, where a guanine (G)-for-cytosine (C) substitution results in the substitution of arginine (Arg) for proline (Pro) at codon 72 of the p53 protein (Arg72Pro; rs1042522) (9). The 2 alleles at this locus encode protein isomorphs that differ in their capacities to induce target gene transcription, their ability to interact with p73 (another tumor suppressor protein), their targeting of the proteasome, and their susceptibility to degradation by human papillomavirus E6 protein (10–12). These observations have provided the rationale for a large number of genetic association studies investigating rs1042522 as a risk factor for various human malignancies (13). Nevertheless, most studies published to date have had small sample sizes, rendering them underpowered to detect small genetic effect sizes, and often have produced contradictory results.
With regard to cervical cancer, although initial evidence suggested a strong protective effect of the proline-encoding allele (14), a recent meta-analysis of individual patient data failed to identify any association with cancer risk (15). Intriguingly, a subgroup analysis suggested that a protective effect for the proline-encoding allele was observed in studies that used tumor tissue for genotyping cancer cases. Several lines of evidence indicate that epithelial cancers in heterozygotic individuals preferentially retain the arginine-encoding allele. This phenomenon, which represents nonrandom LOH, could cause directional genotype misclassification affecting only studies that use tumor tissue as the source of genotyping material for individuals with cancer (cases), resulting in a spurious protective effect of the proline-encoding allele (16–20).
We performed a systematic review of studies investigating rs1042522 and the risk of 5 common epithelial cancers: breast, lung, colorectal, ovarian, and endometrial. We explored the genetic effect of this polymorphism and evaluated whether a systematic bias due to differential genotype misclassification had affected study results across cancer subtypes.
MATERIALS AND METHODS
Search strategy and eligibility criteria
We searched PubMed to identify genetic association studies of the rs1042522 polymorphism and lung, breast, colorectal, ovarian, or endometrial cancer. We used combinations of the following keywords and their synonyms: “TP53,” “cancer,” “neoplasm,” “Arg72Pro,” and “rs1042522.” The full search strategy is given in the Web Appendix, available at http://aje.oxfordjournals.org/. These searches were complemented by searches of the Genetic Association Database (21) and the Human Genome Epidemiology Network's Literature Finder (22). We also used 2 TP53-specific databases that provide information on TP53 polymorphisms: the International Agency for Research on Cancer TP53 database (23, 24) (http://www-p53.iarc.fr/) and the p53 website (25, 26) (http://p53.free.fr/). Finally, we hand-searched the reference lists of all identified eligible articles and reviews of genetic association studies investigating TP53 polymorphisms. All searches were performed on March 8, 2011.
Studies were considered eligible if they used genotyping methods to determine rs1042522 genotype in patients with any of the cancers of interest and in controls with no neoplastic disease. We considered only studies with an analytical epidemiologic design (case-control, nested case-control, or cohort) that separately genotyped samples corresponding to each participant; studies that used DNA-pooling methods were excluded (27, 28). When overlapping patient groups were reported in multiple studies, we included information from the study with the largest number of cancer cases in our analyses. We identified overlap by comparing authors, research centers, recruitment periods, and patient demographic characteristics among otherwise eligible studies. We excluded studies in which all subjects had hereditary cancer syndromes. For example, we excluded studies enrolling exclusively BRCA1 (breast cancer 1, early-onset gene) or BRCA2 (breast cancer 2, early-onset gene) mutation carriers, studies of patients with familial adenomatous polyposis coli, and studies of patients with hereditary nonpolyposis colorectal cancer. We also did not consider family-based studies because of different design and analysis considerations. We limited inclusion to English-language studies. Finally, we did not consider editorials, narrative reviews, letters to the editor, or other manuscripts not reporting primary research results.
Study selection and data extraction
One reviewer screened all abstracts to identify potentially eligible studies, and a second reviewer independently screened abstracts excluded by the first reviewer; studies considered potentially eligible by at least 1 of the reviewers were retrieved and reviewed in full text. A single reviewer extracted the following information from each eligible study: author, year and journal of publication, numbers of cases and controls, participant ethnicity, study design, whether cases and controls were matched (for case-control studies), whether controls were sampled from specific disease groups, the genotyping method used, whether any genotyping quality control process was used, and whether genotyping was performed blinded to the disease status of participants. For our primary comparison of interest, we collected information on the source of genetic material that was used for genotyping cases (cancer tissue versus other DNA source, including buccal swabs, peripheral blood, and saliva). Finally, we extracted the rs1042522 genotype distributions in cases and controls. When studies did not report all the required information but instead cited relevant publications, we retrieved and extracted data from them. A second reviewer verified all extracted information, and discrepancies were resolved by consensus, involving a third reviewer.
Evidence synthesis across cancers
We performed meta-regressions based on generalized linear mixed-effects models to assess the potential biasing effect of using cancer tissue as the source of material for case genotyping across cancers. In all analyses, the genetic effect of rs1042522 was allowed to differ between cancers. Models were fitted by using a Bayesian approach (29), which enabled us to incorporate prior information on the bias arising from use of cancer tissue (30, 31). The prior was based on the results of a recent individual-patient data meta-analysis of the association between rs1042522 and cervical cancer (15) (details on how this prior distribution was derived are presented in the Web Appendix). We used an allele frequency comparison (proline-encoding vs. arginine-encoding allele odds ratios) to ensure consistency and the inclusion of the maximum possible number of studies. Details about the modeling approach are presented in the Web Appendix.
Sensitivity analyses and assessment of bias
We performed sensitivity analyses using alternative prior distributions for Bayesian analyses, including alternative distributions for the heterogeneity parameter. We also evaluated the effect of using a noninformative prior for the bias effect on our results (note that for all other model parameters, prior distributions were noninformative in all analyses). The model was also fitted by using a maximum-likelihood approach, which does not require the specification of prior distributions for the model parameters (32).
In our main analyses, studies that used nontumor tissue samples obtained at surgery as the source of genotyping material (for example, lymph nodes determined to be tumor-negative by pathological examination) were included with studies that used appropriate sources of genotyping material. In sensitivity analysis, we excluded these studies from the data set; this reflects an extreme scenario in which pathological examination is not considered informative. Furthermore, in our main analysis, when studies reported genotyping results from both tumor tissue and other nontumor sources, we used the genotype counts from the latter. In sensitivity analyses, we used the genotype counts from tumor tissue samples for these studies (thus increasing the number of studies using tumor tissue in the data set to 27).
Analyses stratified by cancer
In analyses performed separately for each cancer of interest, summary odds ratios were calculated with random-effects models (DerSimonian-Laird) under an allele frequency comparison (33, 34). Between-study heterogeneity was assessed with Cochran's Q statistic and the I2 index (35, 36).
For each of the cancers investigated, subgroup analyses were performed by stratifying studies on the following characteristics: ethnicity of participants, control selection, use of genotyping quality control, blind genotyping, and use of cancer tissue as the source of genetic material for case genotyping. We estimated the effect of these study-level covariates on the genetic effect by using random-effects meta-regression (37, 38). In view of the evidence that use of tumor tissue as the source of genotyping material for cases could introduce bias, subgroup and meta-regression analyses were performed only among studies that did not use tumor tissue as the source of genotyping material.
We assessed whether larger studies produced results different from those of smaller studies by using the Harbord modification of the Egger test for small study effects (39–41). To explore whether a single study affected estimates of the genetic effect in cancer-specific meta-analyses, we repeated each meta-analysis by sequentially dropping 1 study from the analysis and repeating the calculations. We also repeated the cancer-specific meta-analysis calculations using a fixed-effects model (Mantel-Haenszel) (42).
Analyses were carried out in Stata, version 11.1/SE (StataCorp LP, College Station, Texas); R, version 2.11.0 (R Foundation for Statistical Computing, Vienna, Austria); and WinBUGS, version 1.4.3 (Medical Research Council Biostatistics Unit, Cambridge, United Kingdom). For Bayesian analyses, we report the median values and 2.5th and 97.5th percentiles of the posterior distributions as 95% central credible intervals. For non-Bayesian analyses, we report 95% confidence intervals and P values. Statistical significance was defined as a 2-sided P value less than 0.05 with no adjustment for multiple comparisons (43).
RESULTS
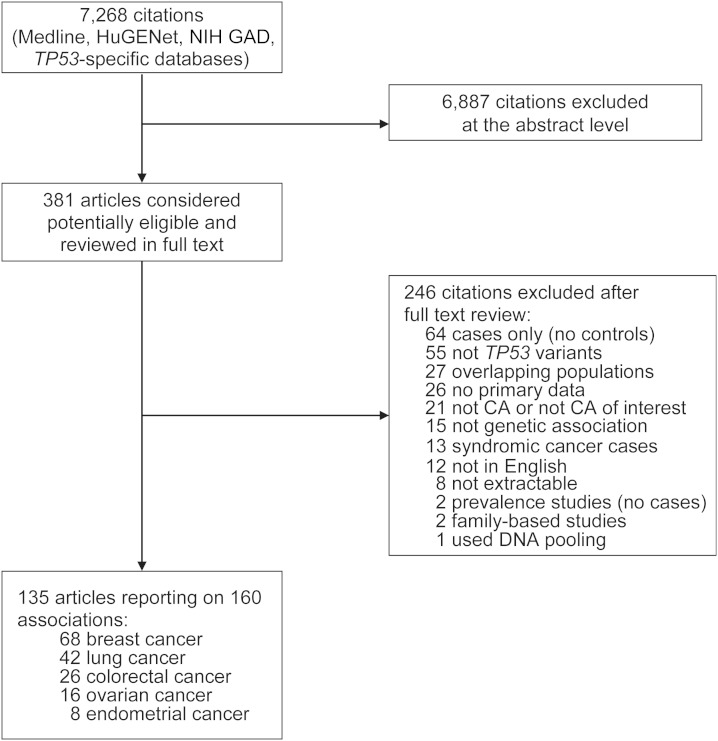
Our searches retrieved a total of 7,268 unique citations. Of these, 6,887 were excluded after screening of titles and abstracts, and 381 were retrieved and reviewed in full text. We further excluded 246 papers after full-text review. The most common reasons for exclusion were case-only designs, assessment of irrelevant genes or polymorphisms, and assessment of noncancer conditions or cancers other than breast, lung, colorectal, ovarian, or endometrial. Overall, 135 articles were considered eligible for this review. Figure 1 presents the details of the search flow. A list of included studies is provided in the Web Appendix.
Figure 1.
Search strategy and study eligibility flow in a systematic review of TP53 rs1042522 and 5 common epithelial cancers. CA, cancer; HuGENet, Human Genome Epidemiology Network; NIH GAD, National Institutes of Health Genetic Association Database.
In total, the eligible articles reported on 160 case-control substudies (some articles presented data on cases and controls sampled from different populations or reported on multiple cancers). We treated these 160 substudies as separate strata (“studies”) in our analyses because they pertained to different study bases (typically sampled from different geographical locations or belonging to different ethnicities). Overall, 68 studies investigated the association of rs1042522 with breast cancer, 42 with lung cancer, 26 with colorectal cancer, 16 with ovarian cancer, and 8 with endometrial cancer. The majority of studies reported on predominantly white (n = 96 (60%)) or East Asian (n = 35 (22%)) populations. Table 1 presents the characteristics of eligible studies stratified by the cancer type investigated, including details of the genotyping methods.
Table 1.
Characteristics of Eligible Studies in a Systematic Review of TP53 rs1042522 and 5 Common Epithelial Cancers (n = 160)
| Study Characteristic | Breast Cancer (68 Studies) |
Lung Cancer (42 Studies) |
Colorectal Cancer (26 Studies) |
Ovarian Cancer (16 Studies) |
Endometrial Cancer (8 Studies) |
All Studies (160 Studies) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | % of Studiesa | No. | % of Studies | No. | % of Studies | No. | % of Studies | No. | % of Studies | No. | % of Studies | |
| No. of cases | 30,586 | 16,743 | 7,377 | 1,982 | 726 | 57,414 | ||||||
| No. of controls | 36,213 | 16,504 | 10,011 | 5,226 | 1,292 | 69,246 | ||||||
| Median no. of cases (IQR) | 166 (94–436) | 147 (91–307) | 121 (76–345) | 109 (48–193) | 94 (43–118) | 137 (78–293) | ||||||
| Median no. of controls (IQR) | 215 (109–486) | 176 (133–379) | 220 (140–347) | 281 (74–446) | 78 (31–310) | 209 (109–423) | ||||||
| Ethnicity | ||||||||||||
| White | 45 | 66 | 16 | 38 | 16 | 62 | 13 | 81 | 6 | 75 | 96 | 60 |
| East Asian | 10 | 15 | 13 | 31 | 8 | 31 | 2 | 13 | 2 | 25 | 35 | 22 |
| Black | 1 | 1 | 2 | 5 | 0 | 0 | 1 | 6 | 0 | 0 | 4 | 3 |
| Latino | 1 | 1 | 3 | 7 | 1 | 4 | 0 | 0 | 0 | 0 | 5 | 3 |
| Other/mixed/NR | 11 | 16 | 8 | 19 | 1 | 4 | 0 | 0 | 0 | 0 | 20 | 13 |
| Control selection | ||||||||||||
| Healthy | 61 | 90 | 24 | 57 | 19 | 73 | 15 | 94 | 6 | 75 | 125 | 78 |
| Diseased | 7 | 10 | 18 | 43 | 7 | 27 | 1 | 6 | 2 | 25 | 35 | 22 |
| Matched controls | ||||||||||||
| Yes | 29 | 43 | 22 | 52 | 10 | 38 | 6 | 37 | 2 | 25 | 69 | 43 |
| No/not applicable | 39 | 57 | 20 | 48 | 16 | 62 | 10 | 63 | 6 | 75 | 91 | 57 |
| Blinding to case-control status | ||||||||||||
| Yes | 7 | 10 | 9 | 21 | 3 | 12 | 2 | 12 | 0 | 0 | 21 | 13 |
| No/NR | 61 | 90 | 33 | 79 | 23 | 88 | 14 | 88 | 8 | 100 | 139 | 87 |
| Use of genotyping quality control | ||||||||||||
| Yes | 26 | 38 | 17 | 40 | 11 | 42 | 9 | 56 | 2 | 25 | 65 | 41 |
| No/NR | 42 | 62 | 25 | 60 | 15 | 57 | 7 | 44 | 6 | 75 | 95 | 59 |
| Genotyping methods | ||||||||||||
| RFLP | 32 | 47 | 24 | 57 | 13 | 50 | 2 | 12 | 3 | 38 | 74 | 46 |
| Other methods | 36 | 53 | 18 | 43 | 13 | 50 | 14 | 88 | 5 | 63 | 86 | 54 |
| Hardy-Weinberg equilibrium | ||||||||||||
| Compliant | 53 | 78 | 36 | 86 | 24 | 92 | 13 | 81 | 7 | 88 | 133 | 83 |
| In violation | 15 | 22 | 6 | 14 | 2 | 8 | 3 | 19 | 1 | 12 | 27 | 17 |
| Use of only tumor tissue for genotypingb | ||||||||||||
| Yes | 9 | 13 | 3 | 7 | 6 | 23 | 2 | 12 | 2 | 25 | 22 | 14 |
| No | 59 | 87 | 39 | 93 | 20 | 77 | 14 | 88 | 6 | 75 | 138 | 86 |
Abbreviations: IQR, interquartile range; NR, not reported; RFLP, restriction fragment length polymorphism.
a Percentages have been rounded to the nearest integer. Percentages might not sum to 100 because of rounding.
b In all cases, studies provided adequate data to evaluate the source of DNA for case genotyping.
All studies included in our analyses had a case-control design. Sixty-nine (43%) of the studies reported matching participants for at least 1 characteristic; age and sex (in lung and colorectal cancer studies) were the most commonly matched variables. The median number of cases was 137 (interquartile range, 78–293), and the median number of controls was 209 (interquartile range, 109–423). Studies of breast cancer were generally larger than studies of other cancers; across cancers, the number of participants increased over time (P < 0.001 for the numbers of both cases and controls).
Few studies reported that genotyping was blinded to the disease status of subjects (n = 21 (13%)). Genotyping quality control procedures were used in 65 (41%) of the studies. Twenty-two of the studies used tumor tissue as the only source of genotyping material for cases, 133 studies used nontumor sources, and 5 studies used both. Specifically, 9 studies on breast cancer, 3 on lung cancer, 6 on colorectal cancer, 2 on ovarian cancer, and 2 on endometrial cancer explicitly stated that tumor tissue was used as the source of genotyping material and did not report the use of any technique or method that suggested inclusion of an adequate amount of normal tissue (e.g., pathological examination or microdissection).
rs1042522 and cancer risk
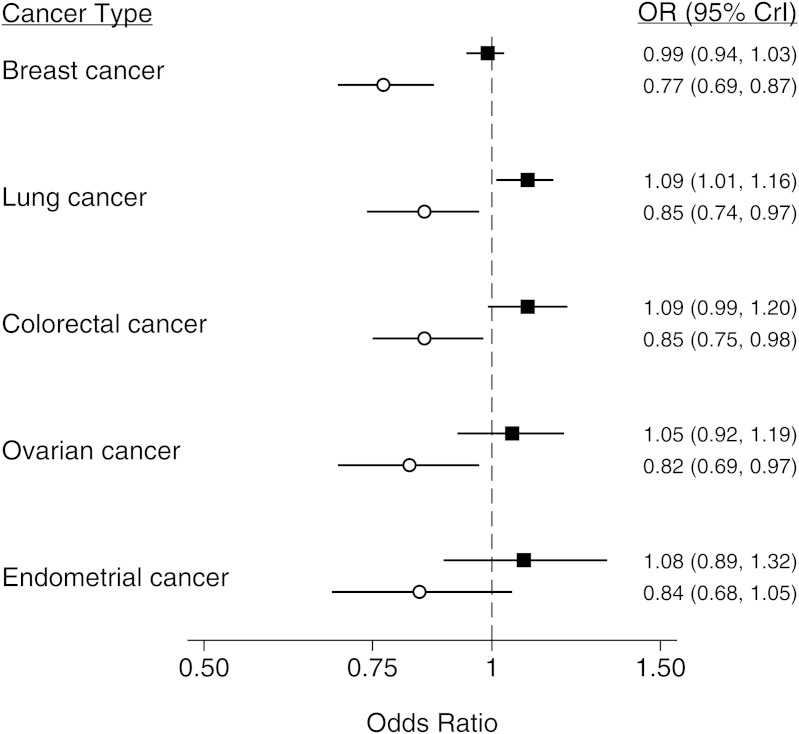
Results of the Bayesian meta-analysis in which an informative prior (derived from the cervical cancer meta-analysis) was used suggested that, in analyses in which nontumor tissue was the source of genotyping material, the Pro allele could be associated with an increased risk of lung cancer (odds ratio (OR) = 1.09, 95% credible interval (CrI): 1.01, 1.16) but not breast (OR = 0.99, 95% CrI: 0.94, 1.03), colorectal (OR = 1.09, 95% CrI: 0.99, 1.20), ovarian (OR = 1.05, 95% CrI: 0.91, 1.19), or endometrial (OR = 1.08, 95% CrI: 0.88, 1.32) cancers (Figure 2 and Web Table 1). These results were confirmed in Bayesian meta-analyses with a noninformative prior (Web Table 2).
Figure 2.
Summary results from a meta-analysis of the association between TP53 rs1042522 and breast, lung, colorectal, ovarian, and endometrial cancer, stratified by the source of genotyping material for cases. Estimates were derived from a Bayesian 2-level logistic regression model using an informative prior. The regression model incorporated evidence from all cancers to estimate the bias effect. Point estimates from studies using appropriate DNA sources are shown as black squares, and point estimates from studies using tumor tissue as the source of genotyping material are shown as white circles. Bars represent the 95% credible interval (CrI) for each estimate. OR, odds ratio.
Assessment of the bias effect across cancers
The use of multilevel models allowed us to borrow strength by combining information across cancers to better quantify the bias arising from use of cancer tissue as the sole source of genotyping material. With external information from the cervical cancer meta-analysis incorporated, the bias effect was estimated to be 0.78 (95% CrI: 0.70, 0.88), and the probability that use of cancer tissue as a source of genotyping material led to bias was higher than 99%. When a noninformative prior was used, the bias was estimated to have a relative odds ratio of 0.79 (95% CrI: 0.69, 0.89). Again, the probability that use of cancer tissue as a source of genotyping material led to underestimation of the “true” genetic effect of the Pro allele was estimated to be higher than 99%.
Sensitivity analyses
Sensitivity analyses using alternative prior distributions for Bayesian analyses, including alternative distributions for the heterogeneity parameter, produced results similar to those of the main analyses, which highlighted the robustness of our results to model specification. Exclusion of studies that used nontumor tissue obtained by surgery for genotyping cases did not affect our results. Similarly, using the genotype counts obtained from analyses of tumor tissue from the 5 studies that used both tumor tissue and other sources of genetic material did not qualitatively affect our findings. Maximum-likelihood methods produced results very similar to those of the main analyses for both cancer risk and bias (Web Table 3).
Analyses stratified by cancer type
Details from meta-analyses stratified by cancer type are presented in Web Tables 4–8. Overall, these analyses were consistent with the results based on multilevel models, although they were less precise: We found no significant effect of the Pro allele for breast, colorectal, or endometrial cancer but did find some evidence of an increase in the risk of lung (OR = 1.09, 95% confidence interval: 1.03, 1.15) and ovarian (OR = 1.10, 95% confidence interval: 1.01, 1.19) cancers. The result for ovarian cancer was based on a small number of studies. Stratified and regression analyses of studies that used nontumor tissue as the source of genotyping material did not indicate any significant modification of the genetic effect by the majority of covariates assessed (Web Tables 4–9).
Assessment of systematic differences across studies
There was no evidence of a systematic difference in the effect sizes reported in smaller versus larger studies for any of the cancers of interest according to the Harbord test. There was also no evidence that the first published study assessing the association of rs1042522 with any of the cancers we evaluated produced more extreme results than all subsequent studies. Finally, among studies that used appropriate sources of genotyping material, leave-1-out meta-analyses and analyses using a fixed-effects model produced inferences similar to those from our main analyses (data not shown).
DISCUSSION
TP53 rs1042522 is one of the most commonly investigated variants in cancer genetic epidemiology (13). The present systematic review suggests that this polymorphism is unlikely to be a risk factor for breast, colorectal, ovarian, or endometrial cancer and that the Pro allele at this locus might cause a small increase in the risk of lung cancer. More importantly, our work provides evidence that some of the findings of genetic association studies of this variant could have been driven by differential genotype misclassification, such as in cases where tumor tissue was used as the source of genotyping material for cancer cases. We used different analytical approaches that allowed us to borrow strength across cancers and increase the precision of the estimate of this bias effect. According to the published data on all 5 of the cancers we evaluated, use of tumor tissue appears to lead to underestimation of the genetic effect by approximately 20%. The probability that use of tumor tissue actually biases estimates of the genetic effect downward was higher than 99% when we incorporated prior evidence from a recent meta-analysis of the same polymorphism in cervical cancer and was higher than 95% in all analyses. Non-Bayesian analyses produced results very similar to those of analyses that used a noninformative prior, further indicating that the prior distributions used were “overwhelmed” by the data. That studies using cancer tissue are susceptible to bias is biologically plausible and has empirical support from studies of other cancers.
We hypothesize that the misclassification arises because of preferential LOH of the Pro allele in heterozygous individuals (16, 17, 19, 20). A substantial body of evidence, in which matched samples of peripheral blood (or other sources of genotyping material, including buccal swabs and saliva) and tumor tissue from cancer patients were used, demonstrates that LOH at the TP53 locus is nonrandom and preferentially involves the Pro allele. This phenomenon has been documented in the cancers considered in the present review as well as in other cancer types, such as cervical, urothelial, and head and neck cancer (17, 18, 20, 44–47). The biological mechanisms underlying this phenomenon are largely unknown, but it appears that the occurrence of TP53 mutations is more common on the DNA strand carrying the arginine-encoding allele (16, 48). It has been suggested that preferential retention of the Arg allele might be the result of an antiapoptotic advantage conferred by TP53 mutations.
Our findings are consistent with a large individual-patient data meta-analysis of 49 studies investigating the association between rs1042522 and cervical cancer risk (15). In that meta-analysis, studies in which the genotype of cases was determined from white blood cells produced null results; in contrast, studies in which genotype was determined from tumor tissue suggested a protective effect of the Pro allele. Our Bayesian analyses incorporated these findings in the form of an informative prior distribution and demonstrated that the bias toward a protective effect of the Pro allele operates across several cancers.
Our work suggests that use of tumor-derived DNA in genetic association studies should be avoided because it can appreciably bias the results of genetic association studies. This could be true particularly for variants in regions where LOH is known to occur, and it might present an important concern for pharmacogenetic studies where the only available source of genotyping material is often tumor tissue. When the interest is in identifying germline (i.e., nonsomatic) variants potentially associated with treatment outcomes, it might be prudent to obtain paired normal–tumor samples from at least a random sample of participants, to establish the extent of LOH as well as to gauge the effect it could have on the overall study results.
Our analyses consistently demonstrated an association between the Pro allele and a small increase in lung cancer risk. This finding is in agreement with previously published meta-analyses on this association (49, 50). However, given the small effect size we observed (OR < 1.10) and the fact that in Bayesian analyses the 95% credible interval for the lung cancer odds ratio was very close to 1, further studies could be necessary to confirm our results.
Several limitations need to be considered for interpretation of our results. Our analysis was based on published data, which prevented any statistical adjustment for individual-level factors that might modify the genetic effect, such as sex (for lung and colorectal cancer), age, or smoking. However, we note that the potential for confounding bias is limited because of the random assortment of alleles at meiosis (51). Furthermore, we could not assess gene-environment interactions because data on potential exposures of interest were unavailable from most studies. Nonetheless, it is unlikely that such factors would also influence the estimate of the bias effect. Finally, our findings should be viewed primarily as hypothesis-generating; large studies reporting on paired samples of normal tissue and tumor tissue obtained from the same patient are needed to confirm our observations. Also, it is unclear whether the phenomenon described herein occurs in other cancers or other genetic loci, but some caution could be warranted in evaluation of variants in genomic regions where little is known about the prevalence of LOH.
In conclusion, our analyses demonstrate that TP53 rs1042522 is unlikely to be associated with breast, colorectal, or endometrial cancer but that a weak association with lung cancer could exist. Across cancer types, there is compelling evidence that use of genetic material obtained from tumor tissue to genotype cases can bias the estimate of the genetic effect, leading to a 20% underestimation of the Pro allele's effect; the probability that bias is toward a protective effect for the Pro allele was at least 95% in our analyses. This finding, along with laboratory evidence indicating that LOH at the TP53 locus in many epithelial cancers is nonrandom, suggests that studies that used tumor tissue as the source of genotyping material for cases have been affected by differential genotype misclassification. In future studies, the use of tumor tissue as the primary source of genetic material should be avoided, particularly when the genetic loci of interest are known to exhibit LOH.
Supplementary Material
ACKNOWLEDGMENTS
Author affiliations: Center for Clinical Evidence Synthesis, Institute for Clinical Research and Health Policy Studies, Tufts Medical Center, Boston, Massachusetts (Issa J. Dahabreh, Joseph Lau, Thomas A. Trikalinos); Biostatistics Research Center, Institute for Clinical Research and Health Policy Studies, Tufts Medical Center, Boston, Massachusetts (Christopher H. Schmid); Department of Occupational and Environmental Medicine and Epidemiology, Harvard School of Public Health, Boston, Massachusetts (Vasileia Varvarigou); Oncology Division, GeneKOR AE, Athens, Greece (Samuel Murray); Biomarker Solutions Ltd., London, United Kingdom (Samuel Murray); and Center for Evidence-Based Medicine, Program in Public Health, Brown University, Providence, Rhode Island (Thomas A. Trikalinos).
This study was supported in part by the National Institute of Research Resources (grant UL1 RR025752) and a research scholarship from the “Maria P. Lemos” Foundation to Dr. Dahabreh.
This article does not represent the opinions of the National Institute of Research Resources or the National Institutes of Health. The funders did not participate in the study design; the collection, analysis, or interpretation of the data; the writing of the report; or the decision to submit the manuscript for publication.
Conflict of interest: none declared.
REFERENCES
- 1.Jemal A, Siegel R, Xu J, et al. Cancer statistics, 2010. CA Cancer J Clin. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343(2):78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 4.Whibley C, Pharoah PD, Hollstein M. p53 polymorphisms: cancer implications. Nat Rev Cancer. 2009;9(2):95–107. doi: 10.1038/nrc2584. [DOI] [PubMed] [Google Scholar]
- 5.Li FP, Fraumeni JF., Jr Soft-tissue sarcomas, breast cancer, and other neoplasms. A familial syndrome? Ann Intern Med. 1969;71(4):747–752. doi: 10.7326/0003-4819-71-4-747. [DOI] [PubMed] [Google Scholar]
- 6.Hollstein M, Sidransky D, Vogelstein B, et al. p53 mutations in human cancers. Science. 1991;253(5015):49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 7.Morris SM. A role for p53 in the frequency and mechanism of mutation. Mutat Res. 2002;511(1):45–62. doi: 10.1016/s1383-5742(01)00075-8. [DOI] [PubMed] [Google Scholar]
- 8.Lane DP. p53, guardian of the genome. Nature. 1992;358(6381):15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 9.Matlashewski GJ, Tuck S, Pim D, et al. Primary structure polymorphism at amino acid residue 72 of human p53. Mol Cell Biol. 1987;7(2):961–963. doi: 10.1128/mcb.7.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas M, Kalita A, Labrecque S, et al. Two polymorphic variants of wild-type p53 differ biochemically and biologically. Mol Cell Biol. 1999;19(2):1092–1100. doi: 10.1128/mcb.19.2.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marin MC, Jost CA, Brooks LA, et al. A common polymorphism acts as an intragenic modifier of mutant p53 behaviour. Nat Genet. 2000;25(1):47–54. doi: 10.1038/75586. [DOI] [PubMed] [Google Scholar]
- 12.Dumont P, Leu JI, Della Pietra AC, 3rd, et al. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33(3):357–365. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 13.Vineis P, Manuguerra M, Kavvoura FK, et al. A field synopsis on low-penetrance variants in DNA repair genes and cancer susceptibility. J Natl Cancer Inst. 2009;101(1):24–36. doi: 10.1093/jnci/djn437. [DOI] [PubMed] [Google Scholar]
- 14.Storey A, Thomas M, Kalita A, et al. Role of a p53 polymorphism in the development of human papillomavirus-associated cancer. Nature. 1998;393(6682):229–234. doi: 10.1038/30400. [DOI] [PubMed] [Google Scholar]
- 15.Klug SJ, Ressing M, Koenig J, et al. TP53 codon 72 polymorphism and cervical cancer: a pooled analysis of individual data from 49 studies. Lancet Oncol. 2009;10(8):772–784. doi: 10.1016/S1470-2045(09)70187-1. [DOI] [PubMed] [Google Scholar]
- 16.Tada M, Furuuchi K, Kaneda M, et al. Inactivate the remaining p53 allele or the alternate p73? Preferential selection of the Arg72 polymorphism in cancers with recessive p53 mutants but not transdominant mutants. Carcinogenesis. 2001;22(3):515–517. doi: 10.1093/carcin/22.3.515. [DOI] [PubMed] [Google Scholar]
- 17.Schneider-Stock R, Boltze C, Peters B, et al. Selective loss of codon 72 proline p53 and frequent mutational inactivation of the retained arginine allele in colorectal cancer. Neoplasia. 2004;6(5):529–535. doi: 10.1593/neo.04178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brooks LA, Tidy JA, Gusterson B, et al. Preferential retention of codon 72 arginine p53 in squamous cell carcinomas of the vulva occurs in cancers positive and negative for human papillomavirus. Cancer Res. 2000;60(24):6875–6877. [PubMed] [Google Scholar]
- 19.Dahabreh IJ, Linardou H, Bouzika P, et al. TP53 Arg72Pro polymorphism and colorectal cancer risk: a systematic review and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2010;19(7):1840–1847. doi: 10.1158/1055-9965.EPI-10-0156. [DOI] [PubMed] [Google Scholar]
- 20.Schneider-Stock R, Mawrin C, Motsch C, et al. Retention of the arginine allele in codon 72 of the p53 gene correlates with poor apoptosis in head and neck cancer. Am J Pathol. 2004;164(4):1233–1241. doi: 10.1016/S0002-9440(10)63211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Becker KG, Barnes KC, Bright TJ, et al. The genetic association database. Nat Genet. 2004;36(5):431–432. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]
- 22.Yu W, Yesupriya A, Wulf A, et al. An automatic method to generate domain-specific investigator networks using PubMed abstracts. BMC Med Inform Decis Mak. 2007;7:17. doi: 10.1186/1472-6947-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hainaut P, Hernandez T, Robinson A, et al. IARC database of p53 gene mutations in human tumors and cell lines: updated compilation, revised formats and new visualisation tools. Nucleic Acids Res. 1998;26(1):205–213. doi: 10.1093/nar/26.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hainaut P, Soussi T, Shomer B, et al. Database of p53 gene somatic mutations in human tumors and cell lines: updated compilation and future prospects. Nucleic Acids Res. 1997;25(1):151–157. doi: 10.1093/nar/25.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamroun D, Kato S, Ishioka C, et al. The UMD TP53 database and website: update and revisions. Hum Mutat. 2006;27(1):14–20. doi: 10.1002/humu.20269. [DOI] [PubMed] [Google Scholar]
- 26.Beroud C, Soussi T. The UMD-p53 database: new mutations and analysis tools. Hum Mutat. 2003;21(3):176–181. doi: 10.1002/humu.10187. [DOI] [PubMed] [Google Scholar]
- 27.Zou G, Zhao H. The impacts of errors in individual genotyping and DNA pooling on association studies. Genet Epidemiol. 2004;26(1):1–10. doi: 10.1002/gepi.10277. [DOI] [PubMed] [Google Scholar]
- 28.Jawaid A, Sham P. Impact and quantification of the sources of error in DNA pooling designs. Ann Hum Genet. 2009;73(1):118–124. doi: 10.1111/j.1469-1809.2008.00486.x. [DOI] [PubMed] [Google Scholar]
- 29.Smith TC, Spiegelhalter DJ, Thomas A. Bayesian approaches to random effects meta analysis: a comparative study. Stat Med. 1995;14(24):2685–2699. doi: 10.1002/sim.4780142408. [DOI] [PubMed] [Google Scholar]
- 30.Gelman A, Hill J. Data Analysis Using Regression and Multilevel/Hierarchical Models. Cambridge, United Kingdom: Cambridge University Press; 2007. Multilevel generalized linear models; pp. 325–342. [Google Scholar]
- 31.Spiegelhalter D, Abrams K, Myles J. Bayesian Approaches to Clinical Trials and Health-Care Evaluation. Chichester, United Kingdom: John Wiley & Sons Ltd; 2004. Evidence synthesis; pp. 267–303. [Google Scholar]
- 32.Turner RM, Omar RZ, Yang M, et al. A multilevel model framework for meta-analysis of clinical trials with binary outcomes. Stat Med. 2000;19(24):3417–3432. doi: 10.1002/1097-0258(20001230)19:24<3417::aid-sim614>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 33.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 34.Higgins J, Thompson SG, Spiegelhalter DJ. A re-evaluation of random-effects meta-analysis. J R Stat Soc Ser A Stat Soc. 2009;172(1):137–159. doi: 10.1111/j.1467-985X.2008.00552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cochran W. The combination of estimates from different experiments. Biometrics. 1954;10(1):101–129. [Google Scholar]
- 36.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 37.Thompson SG, Higgins JP. How should meta-regression analyses be undertaken and interpreted? Stat Med. 2002;21(11):1559–1573. doi: 10.1002/sim.1187. [DOI] [PubMed] [Google Scholar]
- 38.Knapp G, Hartung J. Improved tests for a random effects meta-regression with a single covariate. Stat Med. 2003;22(17):2693–2710. doi: 10.1002/sim.1482. [DOI] [PubMed] [Google Scholar]
- 39.Harbord RM, Egger M, Sterne JA. A modified test for small-study effects in meta-analyses of controlled trials with binary endpoints. Stat Med. 2006;25(20):3443–3457. doi: 10.1002/sim.2380. [DOI] [PubMed] [Google Scholar]
- 40.Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lau J, Ioannidis JP, Terrin N, et al. The case of the misleading funnel plot. BMJ. 2006;333(7568):597–600. doi: 10.1136/bmj.333.7568.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. [PubMed] [Google Scholar]
- 43.Rothman KJ. No adjustments are needed for multiple comparisons. Epidemiology. 1990;1(1):43–46. [PubMed] [Google Scholar]
- 44.Nelson HH, Wilkojmen M, Marsit CJ, et al. TP53 mutation, allelism and survival in non-small cell lung cancer. Carcinogenesis. 2005;26(10):1770–1773. doi: 10.1093/carcin/bgi125. [DOI] [PubMed] [Google Scholar]
- 45.Papadakis E, Soulitzis N, Spandidos D. Association of p53 codon 72 polymorphism with advanced lung cancer: the Arg allele is preferentially retained in tumours arising in Arg/Pro germline heterozygotes. Br J Cancer. 2002;87(9):1013–1018. doi: 10.1038/sj.bjc.6600595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Furihata M, Takeuchi T, Matsumoto M, et al. p53 mutation arising in Arg72 allele in the tumorigenesis and development of carcinoma of the urinary tract. Clin Cancer Res. 2002;8(5):1192–1195. [PubMed] [Google Scholar]
- 47.Bonafe M, Ceccarelli C, Farabegoli F, et al. Retention of the p53 codon 72 arginine allele is associated with a reduction of disease-free and overall survival in arginine/proline heterozygous breast cancer patients. Clin Cancer Res. 2003;9(13):4860–4864. [PubMed] [Google Scholar]
- 48.Kawaguchi H, Ohno S, Araki K, et al. p53 polymorphism in human papillomavirus-associated esophageal cancer. Cancer Res. 2000;60(11):2753–2755. [PubMed] [Google Scholar]
- 49.Dai S, Mao C, Jiang L, et al. P53 polymorphism and lung cancer susceptibility: a pooled analysis of 32 case-control studies. Hum Genet. 2009;125(5):633–638. doi: 10.1007/s00439-009-0664-3. [DOI] [PubMed] [Google Scholar]
- 50.Matakidou A, Eisen T, Houlston R. TP53 polymorphisms and lung cancer risk: a systematic review and meta analysis. Mutagenesis. 2003;18(4):377–385. doi: 10.1093/mutage/geg008. [DOI] [PubMed] [Google Scholar]
- 51.Lawlor DA, Harbord RM, Sterne JA, et al. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27(8):1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.