Abstract
Respiration in mammals relies on the rhythmic firing of neurons within the Phrenic Motor Column (PMC), a motor neuron group that provides the sole source of diaphragm innervation. Despite their essential role in breathing, the specific determinants of PMC identity and patterns of connectivity are largely unknown. We show that two Hox genes, Hoxa5 and Hoxc5, control diverse aspects of PMC development including their clustering, intramuscular branching, and survival. In mice lacking Hox5 genes in motor neurons, axons extend to the diaphragm but fail to arborize, leading to respiratory failure. Genetic rescue of cell death fails to restore columnar organization and branching patterns, indicating these defects are independent of neuronal loss. Unexpectedly, late Hox5 removal preserves columnar organization but depletes PMC number and branches, demonstrating a continuous requirement for Hox function in motor neurons. These findings indicate that Hox5 genes orchestrate PMC development through deployment of temporally distinct wiring programs.
Introduction
Breathing is a basic motor behavior essential to all terrestrial vertebrates. The frequency and amplitude of respiratory contractions are driven by neural networks residing in the brainstem that coordinate the activation of dedicated sets of spinal motor neurons. Respiratory rhythm generation occurs primarily in the Pre-Bötzinger complex and can be modified by other brain stem nuclei in response to stimuli such as pH changes 1. This rhythm is transmitted via descending pathways to motor nuclei that directly drive the activity of inspiratory and expiratory muscles. Despite the complexity of the networks that regulate respiratory rhythms, contraction of the diaphragm is controlled by a single input supplied by motor neurons within the PMC. Phrenic nerve lesions or spinal cord injuries at or above the fourth cervical segment (C4) result in diaphragm paralysis and respiratory failure, underscoring the vital role of PMC neurons within the respiratory system.
Motor neurons within the PMC are generated in the cervical spinal cord where they form a single clustered population spanning ~3 segments 2. Most PMC axons exit the spinal cord at the C4 level, initially projecting along a medioventral path before converging with other cervical axons at the brachial plexus. Following their separation from limb-innervating axons, PMC axons extend ventrally through the thoracic cavity towards the primordial diaphragm. Upon reaching their target, phrenic axons defasciculate from the main nerve and split into multiple finer branches, prior to forming synapses across the muscle length 3. Although PMC neurons have a central role in respiration, and their columnar organization has been recognized for over 100 years 4, 5, surprisingly little is known about their developmental origins.
All motor neuron subtype identities emerge from the intersection of transcription factor-based programs acting along the dorsoventral and rostrocaudal axes of the spinal cord 6. Motor neurons as a class are produced as an outcome of signaling pathways acting along the dorsoventral axis that specify features common to all subtypes, such as exit of axons from the spinal cord and neurotransmitter phenotype 7. These signaling pathways generate motor neurons that initially express a common set of transcription factors (Hb9, Isl1/2, and Lhx3) which distinguish them from other neuronal classes 8–10. While mutation of transcription factors required for core motor neuron programs results in phrenic nerve loss, largely due to conversion to interneuron fates 10, no selective determinants of PMC identity have been described.
Given their discrete position within the spinal cord, the specification of PMC neurons could involve the same programs contributing to motor neuron diversity along the rostrocaudal axis. Members of the Hox gene family are critical in generating segmentally-restricted motor neuron subtypes at limb and thoracic levels 11. At limb levels, the diversification of lateral motor column (LMC) neurons employs a network of ~20 Hox genes 12, while thoracic level motor neuron fates are determined by the single Hoxc9 gene 13. All Hox gene activities in spinal motor neurons are thought to require the transcription factor FoxP1, as limb-level and thoracic Hox-dependent subtypes are lost in Foxp1 mutants 14, 15. PMC neurons are however not depleted in Foxp1 mutants, but instead appear to increase in number 15. These observations raise the question of whether PMC neurons are specified through mechanisms independent of Hox activities, or whether certain Hox proteins contribute to motor neuron specification independent of Foxp1.
We show here that Hoxa5 and Hoxc5 have critical roles in phrenic motor neuron development. PMC neurons are defined through a broader network of Hox factors that constrain their position and number. Selective deletion of Hox5 genes from motor neurons leads to an extinction of PMC molecular determinants, cell body disorganization, and the progressive loss of PMC numbers. Hox5 genes are also essential for a diaphragm-specific pattern of intramuscular branching, independent of their roles in cell survival. Temporal analysis of Hox5 function in motor neurons indicates that survival and intramuscular branching programs are distinct from those controlling columnar organization. These results define a specific transcriptional program for PMC neurons and indicate Hox activities are required throughout motor neuron ontogeny.
Results
Transcription factor profiles of phrenic motor neurons
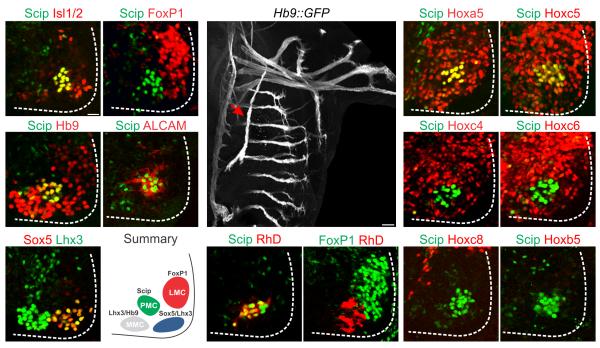
Anatomical studies have identified a column of neurons in segments CIII–CV that projects along the phrenic nerve and innervates the diaphragm 16. To define the molecular identity of this motor neuron group we analyzed transcription factor profiles at cervical levels at embryonic (e) day 11.5 in mice (Fig. 1a–f). Motor neurons within this region expressed combinations of Isl1/2, Hb9, or Lhx3, a core set of transcription factors expressed by all spinal motor neurons (Fig. 1a,c,e). Limb-innervating LMC neurons are distinguished from other subtypes by expression of the transcription factor FoxP1 (Fig. 1b) 14, 15, while medial motor column (MMC) neurons targeting axial muscles coexpress Hb9 and Lhx3 (Fig. 1c,e) 17. We also characterized two additional motor neuron groups at this level; a lateral group that coexpressed Lhx3 and Sox5 (Fig. 1e) 18, and a medial group that expressed Scip (also known as Pou3f1), a POU–class transcription factor (Fig. 1a) 19. Scip+ motor neurons expressed high levels of Isl1/2 and Hb9 (Fig. 1a,c and Supplementary Fig. 1a) and excluded FoxP1 (Fig. 1b). In addition, Scip+ motor neurons expressed ALCAM (Fig. 1d), a cell adhesion molecule expressed by motor neurons at rostral cervical levels 20.
Figure 1.
Transcription factor expression in cervical motor neurons. (a–b) A ventromedial column of Isl1/2+ neurons in the cervical spinal cord expresses Scip and excludes FoxP1. (c–d) Scip+ neurons express Hb9 and ALCAM. (e) A ventrolateral motor neuron population can be defined by the expression of Sox5 and Lhx3. (f) Summary of distribution of motor columns in the cervical spinal cord. (g–i) Retrograde labeling of motor neurons after rhodamine dextran (RhD) injection into the phrenic nerve (arrow in g) of e12.5 embryos, the age when phrenic axons approach the diaphragm. RhD+ motor neurons express Scip (h) but exclude FoxP1 (i). (j–o) Expression of Hox proteins in cervical/brachial spinal cord at e12.5. (j–k) Scip+ PMC neurons express Hoxa5 and Hoxc5. (l–o) Scip+ PMC neurons exclude Hoxc4, Hoxc6, Hoxc8 and Hoxb5. Scale bar=25μm in (a), 100μm in (g).
Scip has been suggested to be expressed by PMC neurons 15, and Scip mutant mice are not viable due to respiratory defects 19, although whether cervical Scip+ neurons correspond to the PMC is not known. To assess whether cervical Scip+ motor neurons target the diaphragm we used retrograde labeling assays. We injected rhodamine dextran (RhD) into the phrenic nerve and examined the transcriptional status of motor neurons that accumulated tracer by retrograde transport, using Hb9::GFP mice to aid in the identification of the phrenic nerve (Fig. 1g)8. After tracer injection, retrogradely labeled motor neurons expressed Scip (Fig. 1h and Supplementary Fig. 1c–d) and excluded FoxP1 (Fig. 1i), indicating that Scip expression at this level marks PMC neurons (Fig. 1f)13.
Hox protein activities are critical in motor neuron subtype differentiation along the rostrocaudal axis, and are thus potential determinants of PMC fate. At brachial levels (CIII– CVIII) several genes in the Hox4–Hox8 paralog groups are expressed by motor neurons targeting forelimb muscles12. To determine if a subset of these Hox genes selectively mark the PMC we examined Hox protein expression with respect to Scip+ neurons, and other cervical motor neuron subgroups (Fig. 1j–o). The majority of motor neurons generated at CII–CV expressed Hoxa5 and Hoxc5, including Scip+ PMC, rostral FoxP1+ LMC, and Sox5+ motor neurons (Fig. 1j–k and Supplementary Fig. 1b and data not shown). Unlike LMC neurons, however, Scip+ PMC neurons excluded Hoxc4, Hoxc6 and Hoxc8, although Hoxc6 and Hoxc4 were expressed by dorsal interneurons and other motor neuron subtypes at this level (Fig. 1l–n). PMC neurons can therefore be defined by the selective expression of Scip, ALCAM, Hoxa5 and Hoxc5, and the exclusion of FoxP1, Lhx3, Hoxc6, and Hoxc4.
Hox and FoxP1 activities restrict PMC position
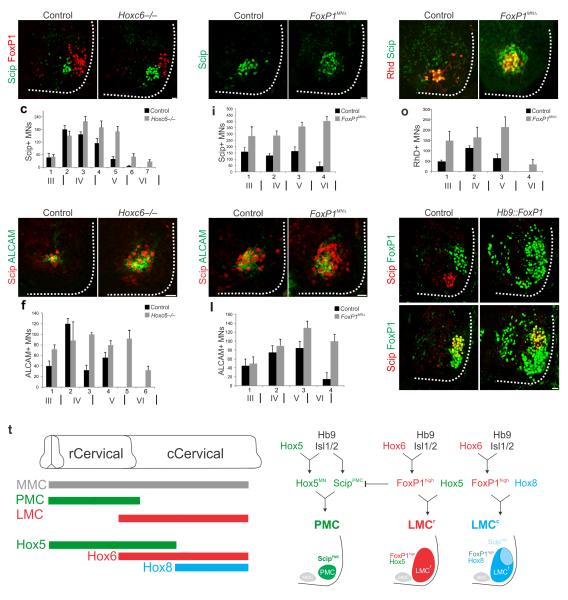
The restriction of FoxP1 and specific Hox proteins from PMC neurons raises the question of whether their actions contribute to PMC position and distribution. The absence of Hoxc4 and Hoxc6 from PMC neurons suggests a possible repressive influence of these proteins. We therefore examined the number of PMC neurons in mice lacking either Hoxc4 or Hoxc6. While the PMC was grossly normal in Hoxc4 mutants (data not shown), in Hoxc6 mutants we observed a significant increase in the number of motor neurons expressing Scip and ALCAM at e12.5. Specifically we found a ~30% caudal extension of Scip+/ALCAM+ motor neurons and an overall 65% increase in total PMC number (516±38 neurons in wt vs 852 ±44 neurons in Hoxc6 mutants, n=8, P<10−4, Fig. 2a–f). Expression of Hox5 proteins was unchanged in Hoxc6 mutants (data not shown), indicating the expansion is not due to alteration in other Hox genes.
Figure 2.
Hox and FoxP1 activities determine PMC neuron distribution. (a–c) In Hoxc6−/− mice there is an instrasegmental and rostrocaudal expansion of Scip+ motor neurons (MNs) at e12.5. (d–f) ALCAM expression expands in Hoxc6−/− mice. In panels c, f, i, l, and o total PMC numbers are extrapolated from serial sections, and roman numerals below section number indicate approximate position of cervical segments. (g–h) In Foxp1MNΔ mice there is an increase of Scip+ neurons in the rostral cervical spinal cord at e12.5 (505±51 motor neurons in wt vs 1330±45 motor neurons in Foxp1MNΔ mice, n=3, P<0.01; see also Supplementary Fig. 2c–f). (i) Quantification of Scip+ neurons in the Hox5 domain of Foxp1MNΔ mice. (j–k) ALCAM expression only expands within the Hox5+ domain in Foxp1MNΔ mice. Not all ectopically generated Scip+ neurons express ALCAM. (l) Quantification of ALCAM expression in Foxp1MNΔ mice. (m–n) Increase in the number of diaphragm projecting motor neurons in Foxp1MNΔ mice, as determined by RhD+/Scip+ cells after tracer injection into the phrenic nerve. (o) Distribution of RhD+ motor neurons in Foxp1MNΔ mice. Diaphragm projecting neurons are confined to the Hox5 domain. We also observed a 53% increase in the thickness of the phrenic nerve in Foxp1MNΔ mice at e12.5, in agreement with previous observations 15. By e18.5 the innervation pattern of the diaphragm in Foxp1MNΔ mice was grossly normal, although some branches overlapped (Supplementary Fig. 2g–j). (p–s) Ectopic FoxP1 expression eliminates Scip from PMC neurons in the rostral cervical spinal cord, but not from LMC neurons in the caudal cervical spinal cord. (t) Model for Hox and FoxP1 interactions controlling PMC specification. Scale bars=25μm. Error bars represent s.e.m.
A primary function of Hoxc6 in motor neurons is to promote FoxP1 expression 14, and in Foxp1 mutants Scip expression expands throughout the spinal cord 15. Increased PMC numbers in Hoxc6 mutants might therefore be a consequence of FoxP1 loss. Consistent with this idea, we find that the number of FoxP1+ motor neurons is reduced in Hoxc6 mutants (Supplementary Fig. 2a–b). This observation raises the possibility that the absence of FoxP1 would promote PMC fate, independent of Hox genes. We therefore analyzed how the loss of FoxP1 influences PMC development. To circumvent the early lethality of the global Foxp1 mutation, we analyzed mice in which Foxp1 is selectively deleted from all motor neurons (Foxp1MNΔ) 21. Similar to Foxp1 global mutants 15, Scip expression expanded throughout motor neurons (Fig. 2g–i). Expression of ALCAM, however, was extended only within the Hox5+ domain (Fig. 2j–l), suggesting that FoxP1 exclusion and Scip expression alone are insufficient to specify PMC fate. Furthermore, tracer injection into the phrenic nerve of Foxp1MNΔ mice labeled Scip+ motor neurons that were confined to the Hox5+ domain with no labeling of caudal Hoxc8+ neurons (Fig. 2m–o and data not shown).
To further test the hypothesis that PMC specification relies on the regulation of Scip expression selectively within Hox5+ populations, we analyzed mice that express FoxP1 in all motor neurons using Hb9 regulatory sequences 14. While FoxP1 misexpression had no effect on motor neuron generation or Hox patterns, Scip was extinguished from PMC neurons (Fig. 2p–q). In addition to the PMC, Scip is expressed by pools of LMC neurons (Hoxc8+ Hox5− FoxP1+) targeting the median and ulnar nerves 12. In Hb9::Foxp1 embryos Scip expression is maintained in these pools (Fig. 2r–s). Thus the restriction of Scip to PMC neurons by FoxP1 is constrained to Hox5+ levels. Collectively, these results indicate that Hox5+, Scip+, FoxP1− motor neurons have the capacity to acquire the molecular features and projection characteristics of PMC neurons (Fig. 2t).
PMC loss and respiratory failure in Hox5MNΔ mice
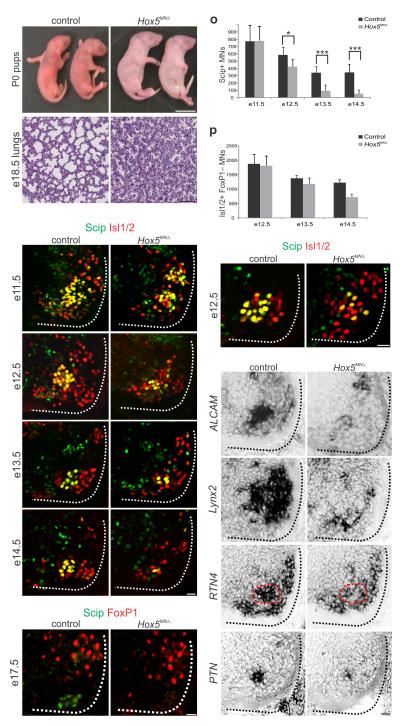
To determine whether Hox5 genes are required for PMC development we generated and analyzed mice lacking Hoxa5 and Hoxc5. Because Hoxa5−/– animals display non-neuronal defects in the respiratory system, including abnormal lung development 22, we generated mice in which Hoxa5 was selectively deleted from motor neurons by crossing a conditional Hoxa5 allele23 to Olig2::Cre mice (Hoxa5MNΔ) 24. We verified efficient Hoxa5 removal, finding that by e11.5 Hoxa5 is not expressed in motor neurons but is retained in neighboring cell types and cells of the lung (Supplementary Fig. 3a–f). Hoxa5 MNΔ mice were viable, and did not display discernable respiratory defects. Hoxc5−/− mice were also grossly normal and we therefore introduced the Hoxa5MNΔ mutant allele into a Hoxc5 mutant background. For simplicity we refer to Hoxa5 flox/flox; Hoxc5−/−; Olig2::Cre animals here as Hox5MNΔ mice, as Hoxb5 is normally excluded from motor neurons at this level of the neuraxis (Fig. 1o) and is not upregulated in PMC neurons of Hox5MNΔ mice (Supplementary Fig. 3g–j). At embryonic stages Hox5MNΔ mutants were recovered at the expected Mendelian ratios and showed no external anatomical differences when compared to controls (data not shown). However, all Hox5MNΔ neonates (from n>50 litters) failed to initiate breathing, were cyanotic, and died shortly after birth (Fig. 3a–b). Histological analysis revealed that the lungs of Hox5MNΔ mice were collapsed (Fig. 3c–d), and failed to surface when submerged in water (Supplementary Fig. 3k), indicating that they never inflated with air. Thus Hox5 genes are required in motor neurons for normal respiratory activity.
Figure 3.
Respiratory failure and PMC loss in Hox5 MNΔ mice. (a–b) All Hox5MNΔ mice are born cyanotic and perish at birth. (c–d) Histological analysis reveals that the lungs of e18.5 Hox5MNΔ embryos collapse before birth. (e–n) Progressive loss of Scip+ PMC neurons in Hox5MNΔ mice. At e11.5 Scip+ neuron numbers are similar between control and Hox5MNΔ mice but progressively decrease in the mutants. By e17.5 there are no detectable Scip+ motor neurons in Hox5MN mice. (o) Quantification of Scip+ motor neurons in control and Hox5MNΔ mice. At least 5 pairs of embryos were analyzed for each time point. *P<0.05, ***P<0.001 (p) Quantification of non-LMC Isl1/2+ motor neurons. There is selective loss of FoxP1− neurons in Hox5MNΔ mice. (q–r) PMC disorganization at e12.5 in Hox5MN mice. Isl1/2+ Scip– neurons are intercalated with PMC neurons. (s–t) ALCAM expression is reduced in Hox5MN mice, as early as e11.5 prior to loss of Scip. (u–z) Lynx2, RTN4 and PTN are downregulated in Hox5MN mice at e12.5. PMC position is outlined by dashed red line. Expression of Lynx2 is also lost from LMC neurons. Scale bars=25μm, except in (b)=1cm and (d)=100μm. Error bars represent s.e.m.
We next examined how Hox5 mutation affects the emergence of molecular features of PMC neurons. We first assessed the number and the distribution of Scip+ PMC neurons at multiple stages of development. Because Hoxc8 expression does not expand in Hox5MNΔ embryos (Supplementary Fig. 3l–m) 12, we used Hoxc8 as a landmark to define the caudal boundary of the former Hox5 domain and analyzed motor neurons within this region. At e11.5, when motor axons first exit the spinal cord, the number of Scip+ PMC neurons was similar between Hox5MNΔ and control animals (Fig. 3e–f). This number, however, began to decline at e12.5 in Hox5MNΔ mutants, and by e14.5 the number of Scip+ PMC neurons was reduced by 84% (Fig. 3g–l,o). By e17.5 Scip+ motor neurons were undetectable at cervical levels of the spinal cord (Fig. 3m–n). In contrast, the number of limb innervating LMC motor neurons was unchanged in Hox5MNΔ embryos, as assessed by FoxP1 expression at e13.5 (Supplementary Fig. 3n–o). To determine whether the attenuation of Scip expression reflects a selective depletion of PMC numbers, we also assessed the total number of non-LMC motor neurons in rostral cervical segments, by counting Isl1/2+, FoxP1− cells between e12.5–e14.5. This analysis revealed that the total number of non-LMC motor neurons also progressively declined, indicating that in Hox5MNΔ embryos PMC neurons selectively perish (Fig. 3p).
In addition to a reduction in Scip+ motor neurons in Hox5MNΔ mice, the remaining Scip+ neurons were disorganized at e12.5, and intercalated by other subtypes (Fig. 3q–r). However, they retained expression of Isl1/2 and Hb9 (Fig. 3q–r and Supplementary Fig. 3p–q). Expression of ALCAM was reduced at all developmental stages examined (Fig. 3s–t and Supplementary Fig. 3r–s), indicating PMC molecular features are lost prior to motor neuron death. This observation prompted us to examine whether there is an array of genes that are downregulated in PMC neurons in Hox5MNΔ mutants. We performed a microarray analysis of RNA purified from cervical motor neurons in control and Hox5MNΔ embryos, followed by in situ hybridization to verify candidates. Among the genes that showed reduced expression were lynx2, a modulator of nicotinic acetylcholine receptors (Fig. 3u–v) 25 and RTN4/NogoA, primarily known as an inhibitor of neuronal regeneration (Fig. 3w–x and Supplementary Table 1) 26.
Using a candidate approach, we also identified pleiotrophin (PTN), a known target of Hoxa5 that has been shown to have trophic effects in motor neurons 27, 28 as a PMC-restricted marker that is downregulated in Hox5MNΔ mutants (Fig. 3y–z). These observations indicate that Hox5 genes are required for multiple features of PMC fate.
Diaphragm innervation defects in Hox5MNΔ mice
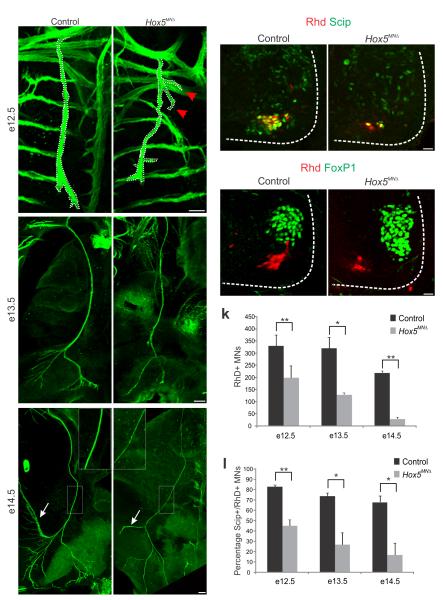
To further assess the impact of Hox5 deletion on PMC development, we monitored the projection of motor axons along the phrenic nerve. We introduced the Hox5MNΔ allele into a Hb9::GFP background and compared projections of control and mutant mice at multiple stages. At e12.5 the phrenic nerve trajectory was similar between Hox5MNΔ and control mice (Fig. 4a–b). However, while all axons were directed to the diaphragm in control mice, in Hox5MNΔ mutants we observed branches straying from the main trunk that appeared to be directed towards the forelimb (Fig. 4b). These branches were transient, as they were not seen at later stages. At e13.5 and later stages we observed a thinning of the phrenic nerve, and by e14.5 the nerve diameter was reduced to 67% of wt (16.47 ±1.9 μm in wt vs 11.04 ±0.3 μm in mutants, P<0.05, Fig. 4c–f), reflecting the progressive loss of PMC neurons.
Figure 4.
Fidelity of PMC axon projections in Hox5MNΔ mutants. (a–f) The phrenic nerve progressively thins in Hox5MNΔ mutants. At e12.5 phrenic nerve diameter is similar between control and Hox5MNΔ mice (a–b). Some axons stray from the phrenic nerve in Hox5MNΔmice (arrows in b). By e14.5 the phrenic nerves become thinner in Hox5MNΔmice (see inserts e–f) and lack arbors seen in control nerves (arrows in e–f). (g–j) Retrogradely labeled motor neurons after RhD injection in the phrenic nerve are reduced in Hox5MNΔ mice, but retain some aspects of their molecular identity, such as Scip expression (g–h) and FoxP1 exclusion (i–j). (k–l) Quantification of RhD retrograde transport after RhD injection in the phrenic nerve. There is a decrease in the number of RhD+ neurons in Hox5MNΔ mice (k), as well as a decrease in the percentage of RhD+ neurons that express Scip (l). Scale bars=100μm (a–f), 25 μm (g–j). Error bars represent s.e.m., *P<0.05,**P<0.01.
To assess the molecular identity of motor neurons projecting along the phrenic nerve we performed retrograde tracing experiments in Hox5MNΔ mice. Consistent with a reduction in Scip+ neurons, we observed a 40% decrease in the number of motor neurons that were retrogradely labeled with RhD at e12.5 in Hox5MNΔ mice (Fig. 4g–h,k) and a further reduction at later developmental stages (Supplementary Fig. 4a–d). At this stage labeled cells retained Scip expression and excluded FoxP1, although they were not tightly clustered (Fig. 4g–j). However, by e14.5, when Scip is further downregulated in Hox5MNΔ mice, 85% of retrogradely labeled neurons in Hox5MNΔ mice were Scip–, compared to 35% in control mice (Fig. 4l). These observations indicate a progressive loss of PMC identity in Hox5MNΔ mice, reflecting both a decline of PMC numbers and an altered molecular identity in the remaining motor neurons that have selected a phrenic trajectory.
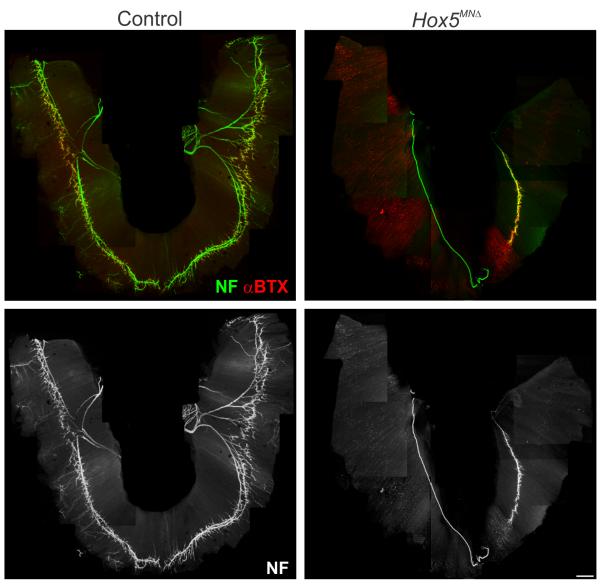
Because axons extend to the diaphragm in Hox5MNΔ mice we next examined the behavior of PMC neurons at their target muscle. As axons from the phrenic nerve arrive at the diaphragm they split into three main branches, the two largest of which enter the muscle and project in opposing directions along its length. Subsequently, these main branches split further into finer arbors prior to establishing synapses. At e14.5 primary branches appeared to form normally in Hox5MNΔ mice, although there was noticeable absence of secondary and tertiary branches (Fig. 4e–f and Supplementary Fig. 4e–f). By e18.5, intramuscular branches were dramatically reduced in Hox5MNΔ mice, and in the most severe cases only a single branch was observed on one side of the muscle (Fig. 5a–d). Some synapses are formed between the phrenic nerves and the diaphragm as assessed by coincidence of synaptophysin and α-bungarotoxin staining (Fig. 5a–b), but they are localized to a small portion of the muscle. Therefore, the lack of sufficient synapses with the diaphragm is likely the cause of perinatal lethality in Hox5MNΔ mice.
Figure 5.
Loss of synaptic contacts between PMC neurons and diaphragms in Hox5MNΔ mice. (a– d) Analysis of diaphragm innervation patterns at e18.5. (a–b) Diaphragms of Hox5MNΔ mutants display a marked reduction in terminal branches and neuromuscular synapses, as revealed by neurofilament and bungarotoxin staining (n=10). (c–d) While the phrenic nerve establishes contacts with the muscle and forms a primary branch, secondary and tertiary branches fail to form. As a consequence the number of synapses formed at the diaphragm muscle is dramatically reduced in Hox5MNΔ mutants. Scale bar=500 μm. Images shown are tiled composites of individual panels. The pattern of forelimb innervation was not appreciably affected in Hox5MNΔ embryos, and the biceps, a muscle supplied by Hox5+ LMC neurons, was innervated normally (Supplementary Fig. 5a–d).
Preventing PMC loss fails to rescue innervation defects
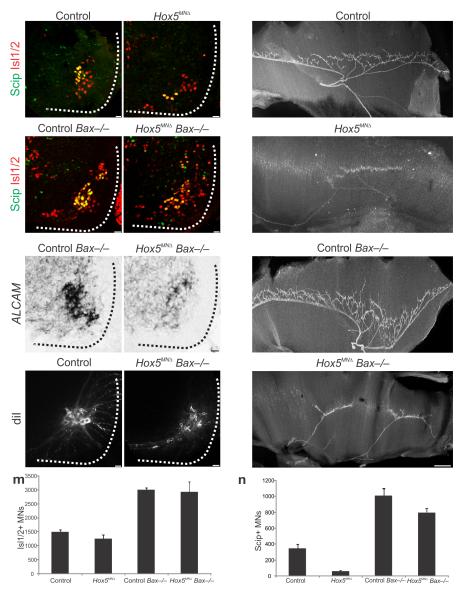
The loss of diaphragm innervation in Hox5MNΔ mice could be a consequence of the absence of a specific molecular program initiated by Hox5 proteins, or simply a manifestation of the loss of PMC neurons. To address this question we introduced the Hox5MNΔ allele into a Bax mutant background to prevent programmed cell death. As reported previously, Bax−/− mice displayed a global increase in motor neuron number, marked by Isl1/2 expression (Fig. 6a–d,m)29. As a result, phrenic neuron number also increased, as evident by Scip and ALCAM expression at e15.5 (Fig. 6a–f,n). Deletion of Bax in a Hox5MNΔ background also dramatically increased Scip+ neurons (Fig. 6d,n). However these motor neurons were scattered (Fig. 6d) and ALCAM expression was not significantly recovered (Fig. 6f). We verified that motor neurons project towards the diaphragm, by DiI retrograde labeling in Hox5MNΔ Bax−/− mice at e18.5 (Fig. 6g–h), indicating rescued cells are not directed to other muscle targets.
Figure 6.
Preventing apoptosis fails to rescue diaphragm innervation in Hox5MNΔ mutants. (a–d) Deletion of Bax increases Scip+ motor neuron numbers both in control and in Hox5MNΔ mice at e14.5 (P<0.001). However, in Hox5MNΔ Bax−/− mice the PMC still appears disorganized, despite rescue of Scip+ neuron numbers (d). (e–f) ALCAM expression is increased in Bax−/− mice while it is only slightly recovered in Hox5MNΔ Bax−/− mice at e14.5 (f). (g–h) Retrograde labeling of phrenic neurons with DiI at e18.5 shows that motor neurons innervate the diaphragm in Hox5MNΔ Bax−/− mice. (i–l) Deletion of Bax in a Hox5MNΔ background does not rescue the diaphragm innervation defects seen in Hox5MNΔ mice. Both control and control Bax−/− mice exhibit a stereotypical diaphragm innervation pattern (i,k) while Hox5MNΔ and Hox5MNΔ Bax−/− mice show similar defects in diaphragm innervation at e15.5 (j,l), as seen by neurofilament staining. (m–n) Scip+ and total Isl1/2+ neuron numbers in the different strains. Scale bars=25μm (a–h), 200μm (i–l). Error bars represent s.e.m.
An examination of the diaphragm in Bax−/− mice in a wildtype background revealed that the increase in Scip+ PMC neurons does not lead to diaphragm hyperinnervation, consistent with the observation that the overall pattern is not sensitive to elevated motor neuron number 30. Importantly in Hox5MNΔ Bax−/− we did not observe a recovery of the innervation defects seen in Hox5MNΔ mutants (Fig. 6i–l). These results indicate that the defects in Hox5MNΔ mice are not merely a consequence of decreased PMC number, but reflect a specific action of Hox5 genes in determining a molecular program required for diaphragm innervation.
Temporal roles of Hox5 genes during PMC development
Deletion of Hox5 genes from motor neuron progenitors results in a progressive decrease in the size of the PMC, cell body disorganization, and most strikingly, a loss of diaphragm innervation. These phenotypes become apparent at distinct stages of embryonic development, with cell loss and disorganization occurring at the time of PMC clustering (e11.5–12.5) and innervation defects manifesting during the onset of terminal arborization (e14.5–15.5). Hoxa5 expression persists in PMC neurons until late embryonic stages (e17.5, Supplementary Fig. 6a–b), suggesting a possible late role in PMC development.
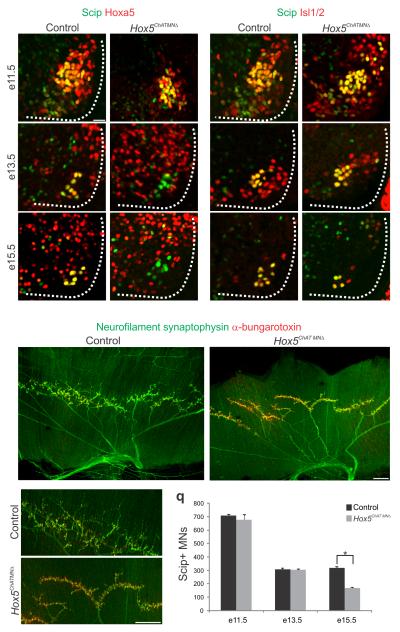
To investigate the temporal role of Hox5 genes in PMC neurons we removed Hoxa5 function in a Hoxc5 mutant background by using a Cre recombinase controlled by ChAT regulatory sequences, a gene which is activated shortly after cervical motor neurons differentiate beginning at ~e9.5–e10.5. This strategy effectively removes Hoxa5 protein from motor neurons by e13.5, but preserves expression over a short window between e9.5 and e11.5 (Fig. 7a–d), during the peak of motor neuron generation and as axons select their initial trajectories. Unlike Hox5MNΔ mice, Hox5ChATMNΔ mice are viable, suggesting a less severe defect in PMC development. While the PMC was clustered at e13.5 in Hox5ChATMNΔ mice, there was a progressive reduction in PMC size which was reduced by 48% at e15.5 (Fig. 7a–l,q). Interestingly, while expression of ALCAM was normal at e14.5, PTN expression was attenuated (Supplementary Fig. 6c–f). These results indicate that late removal of Hox5 genes preserves some but not all molecular features of PMC neurons, and that Hox5 genes are required at later stages for maintaining PMC number.
Figure 7.
Late removal of Hox5 genes leads to PMC loss and reduced terminal branching. (a–f) In Hox5ChATMNΔ mice Hoxa5 expression persists at e11.5 (b) but is effectively removed from motor neurons by e13.5 (d). (g–l) While Scip+ PMC neuron number is similar between control and Hox5ChATMNΔ mice at e11.5 (g–h) and e13.5 (i–j), there is a 48% reduction by e15.5 in the mutant (k–l). The remaining motor neurons appear to remain clustered at the correct PMC position. (m–p) Diaphragms of Hox5ChATMNΔmice exhibit grossly normal neuronal innervation, with synapses on the entire muscle. However, at high magnification, a reduction in terminal arborization is observed (p). (q) Quantification of Scip+ PMC neurons in Hox5ChATMNΔ mice. Scale bars=25μm (a–l), 200μm (m–p). Error bars represent s.e.m.,*P<0.05
Analysis of Hox5ChATMNΔ mice at e18.5 showed that the diaphragm was fully innervated, with the initial projection patterns similar to control mice. However there was a reduction in the number of terminal branches, resulting in a higher concentration of Ach receptors at individual axon terminals (Fig. 7m–p). While this branching phenotype does not affect viability, it nevertheless indicates that continuous Hox5 activity is required in PMC neurons for the stereotypic pattern of diaphragm innervation.
Discussion
The neural networks that control breathing rely on the coordinate activation of a select set of muscle groups, which in mammals are supplied by motor neurons within the phrenic and hypaxial motor columns. While the steps that contribute to the specification of hypaxial projecting motor neurons are well documented 9, 14, 31, the programs that distinguish PMC neurons from other motor neuron subtypes are largely unknown. In this study we have found that Hox5 genes are expressed by PMC neurons and are required for multiple aspects of their development, including their organization, survival, and patterns of peripheral connectivity. Unexpectedly, sustained Hox5 activity is necessary for key aspects of PMC maturation subsequent to their initial differentiation. We discuss these findings in the context of Hox networks in the spinal cord and the role of Hox5 genes in respiratory motor neuron specification.
Hox networks and the emergence of PMC identity
Innervation of the diaphragm is part of a motor circuit unique to mammals, and likely evolved to meet the increased metabolic demands of terrestrial life. How the PMC appeared during mammalian evolution is not known. Terrestrial tetrapods that lack a diaphragm muscle, and therefore a PMC, express Hox5 proteins in domains very similar to mammals 12. In non-mammalian vertebrates, such as chick, Hox5 proteins contribute to the diversity of limb-innervating motor neurons within the rostral half of the LMC 12, a program that requires the Hox cofactor FoxP1 14, 15. However, unlike other Hox-dependent motor neurons, we find that the development of the PMC does not require FoxP1, but rather that FoxP1 inhibits PMC differentiation. These findings indicate a novel FoxP1-independent strategy for motor neuron diversification.
At thoracic levels FoxP1 misexpression inhibits the differentiation of ventrally projecting hypaxial motor column (HMC) neurons, but not dorsally projecting medial motor column (MMC) neurons 14. Thus it is intriguing that the two columns that are selectively inhibited by FoxP1, the HMC and PMC, both project ventrally and are involved in respiratory motor function, suggesting a common origin. At most levels of the spinal cord motor neurons revert to a ground state of the HMC subtype in Foxp1 mutants 14, 15. Our studies indicate that at rostral cervical levels Hox5+ motor neurons acquire molecular features and projection characteristics of PMC neurons in the absence of Foxp1. The PMC therefore likely emerged from an HMC-like precursor that excluded LMC Hox determinants and acquired sensitivity to Hox5 activity.
Hox5 genes control multiple aspects of PMC development
Deletion of Hox5 genes in motor neuron progenitors leads to a variety of PMC defects, consistent with the idea that Hox genes control diverse facets of motor neuron identity. While Scip+ neurons are generated in Hox5 mutant mice, these motor neurons fail to cluster and expression of ALCAM is dramatically reduced. PMC neurons are progressively lost, and the motor axons that do reach the diaphragm fail to arborize. This innervation phenotype appears to be unique to the diaphragm, as in Foxp1 mutants the majority of limb muscles receive innervation, despite the loss of LMC molecular identity 14, 21. While this suggests a unique program of muscle-specific innervation by PMC neurons, similar phenotypes have been observed in genetic manipulations that affect motor pool specification within the LMC. Mutation in the transcription factor Pea3, for example, leads to motor pool disorganization and loss of intramuscular branches at target muscles 32. Pea3 expression requires peripheral signals from its target provided by glial-derived neurotrophic factor (GDNF) 33. The phenotypes in Hox5 mutants likely reflect a similar defect in nerve-muscle communication, due to an inability of PMC neurons to respond to trophic signals, leading to cell death and nerve degeneration.
Several gene mutations are known to affect diaphragm innervation patterns. For example, mutations in the netrin receptor Unc5c and the neuregulin receptor ErbB2 cause a loss of synapses at the diaphragm 34, 35 and mice lacking ephrins-A2/A5 exhibit impaired topographic innervation 36. Conversely mutations in robo/slit genes increase the number of phrenic branches as a consequence of premature defasciculation 37. Many of these genes are expressed broadly by motor neurons, and whether they contribute to Hox-dependent PMC programs is unclear. Given the variety of defects in Hox5 mutants, it is plausible that Hox5 proteins regulate both PMC-restricted target effectors, such as ALCAM and PTN, as well as determinants shared by many motor neuron subtypes, including RTN4 and lynx2. ALCAM and RTN4 are required for the branching of peripheral neurons 38, 39, while PTN appears to contribute to motor neuron survival28. The concurrent loss of these genes likely underlies the multiple PMC defects observed in Hox5 mutants. Finally, while the precise role of Scip in controlling PMC gene programs is unclear, it likely acts coordinately with Hox5 proteins, as Scip mutants also perish at birth due to respiratory deficiencies.
Hox5 proteins act at distinct phases of PMC development
The actions of Hox factors in motor neuron differentiation are thought to be mediated through the induction of downstream transcription factors, which in turn control distinct aspects of their specification. An early target of Hox proteins is the Foxp1 gene, which is subsequently required for the expression of all LMC and PGC determinants 14, 15. Within LMC motor pools, Hox proteins control the transcription factors Nkx6.1 and Pea3, each contributing to different facets of pool identity. Mutation of Pea3 does not affect the trajectory of motor axons to their target but scrambles cell body positioning and intramuscular branching 32. In contrast, mutation of Nkx6.1 preserves cell body positioning but motor axons fail to project to their correct target 40. These findings raise the question of whether there are Hox-dependent actions in motor neurons that are independent of intermediate transcription factors, and whether Hox function is required subsequent to their induction.
Our findings indicate that Hox5 gene activity is required in PMC neurons subsequent to their differentiation. Motor neuron clustering and Scip expression are not affected under conditions of late depletion using ChAT::Cre, indicating early programs are preserved. However by e15.5 PMC number is decreased by half and terminal branches at the diaphragm are reduced (Supplementary Fig. 7a–b). While it is unclear whether PMC loss is a consequence of reduced branching, a branching defect could in principle limit the accessibility of terminals to trophic molecules leading to motor neuron death41. Alternatively, late Hox5-dependent genes like PTN could act in an autocrine fashion to promote PMC survival after differentiation. Thus, the observed defects could reflect the need for Hox5 genes to maintain expression of effectors required for terminal branching, and preserving communication between motor neurons and muscle.
Hox5 genes control phrenic intramuscular branching patterns
Our data indicate that Hox5 genes control the branching patterns of PMC neurons, independent of their role in maintaining cell viability. Rescue of the cell death phenotype in the Hox5 mutant fails to restore the normal pattern of diaphragm innervation. While cell number may not directly influence nerve branching, the concurrence of both phenotypes could reflect a common upstream mechanism. For example, peripheral nerve growth factor (NGF) is required both for sympathetic neuron survival and peripheral target organ innervation 42. GDNF, a potent survival factor for some classes of motor neurons, is also required for the correct innervation of the cutaneous maximus and lattisimus dorsi muscles 32, 33. In the absence of Hox5 genes, PMC neurons may lose responsiveness to a muscle-derived signal that regulates both survival and nerve branching.
Some synapses are present at the diaphragm in Hox5 mutants although they are apparently insufficient to drive normal respiratory motor function, as all mutants perish shortly after birth. A contributing factor to the perinatal lethality of Hox5 mutants may be the disorganization of the PMC, which could affect the strength of inputs from hindbrain respiratory networks. Recent studies have suggested that cell position is a critical determinant in shaping the synaptic inputs from premotor sources 21, 43. Loss of Hox5 activity may additionally deplete the inputs from supraspinal networks, thereby exacerbating the defects caused by the loss of diaphragm innervation.
The establishment of connections between phrenic motor neurons and the diaphragm is a key step in the assembly of the neural networks that control breathing. In recent years significant progress has been made in defining the transcriptional codes that contribute to the assembly of the circuits that provide rhythm and modulation to respiratory motor networks 44-46. However there still remain significant gaps in our understanding of basic features of their wiring. For example, the sources of the neuronal subtypes that mediate the connections between respiratory networks to phrenic motor neurons are not clearly defined. Phrenic motor neurons are also known to fire in synchrony with HMC neurons 47, 48, and whether specificity of these connections relies on shared molecular determinants between PMC and HMC neurons is unknown. The definition of a selective molecular code for PMC neurons should aid in attempts to define the mechanisms that shape the specificity of connections between respiratory rhythm networks and the spinal cord.
Methods
Mouse Genetics
The Hoxa5 floxed alleles 23, Hb9::GFP 8, ChAT::Cre 49, Olig2::Cre 24, Foxp1MNΔ 21, Hoxc5, Hoxc6 mutant strains 50, Hb9::Foxp1 14 and Bax −/− 51 lines were generated as described. Mouse colony maintenance and handling was performed in compliance with the protocols approved by the Institutional Animal Care and Use Committee of the New York University School of Medicine. Mice were housed in a 12hr light/dark cycle in cages containing no more than 5 animals at a time.
In Situ Hybridization and Immunohistochemistry
In situ hybridization and immunohistochemistry were performed as described 9. Whole-mount GFP staining was performed as described 40 and motor axons were visualized in projections of confocal Z-stacks (500–1000 μm). Whole mounts of diaphragm muscles from e18.5 mice were stained as described 52. Antibodies were generated as described 9, 12, 14. Other antibodies were used as follows: rabbit anti-GFP (1:1000; Invitrogen), rat anti-ALCAM (1:250; eBioscience), rabbits anti-Neurofilament (1:1000; Millipore), rabbit anti-synaptophysin (1:5; Invitrogen), anti-bungarotoxin Alexa594 conjugate (1:1000; Invitrogen). A Hoxb5 antibody was generated in rabbit using the peptide sequence: ESSRAFPASAQEPRFRQATSSC. For lung histology, lungs were dissected, fixed in 4% paraformaldehyde, paraffin-embedded, sectioned at 4μm and stained with hematoxylin and eosin. Images were obtained with a Zeiss (LSM 510 Meta) confocal microscope and analysed with LSM Image Browser.
Retrograde Labeling of Motor Neurons
Retrograde labeling of motor neurons was performed as described 14. Lysine-fixable dextran-tetramethylrhodamine (RhD, Molecular Probes) was injected into transected phrenic nerves of e12.5-e14.5 embryos. To aid in the identification of nerves, we used GFP fluorescence from Hb9::GFP transgenic mouse embryos, visualized using a MVX10 wide-field fluorescent microscope (Olympus). Nerves were severed using Vannas Spring Scissors (FST) and RhD was injected onto the cut terminal. Embryos were incubated for 5 to 6 hours in oxygenated F12/DMEM (50:50) solution at 32–34°C and subsequently fixed in 4% paraformaldehyde. For labeling of phrenic motor neurons at e18.5, crystals of carbocyanine dye, DiI were pressed onto the left phrenic nerve of eviscerated embryos and the embryos were incubated in 4% paraformaldehyde at 37°C in the dark for 2 weeks. Subsequently, spinal cords were dissected, embedded in 4% low melting point agarose (Invitrogen) and sectioned using a Leica VT1000S vibratome at 100μm.
Microarray screening
Cervical spinal cords were dissected from 3 control and 3 Hox5MNΔ mutant embryos in a Hb9::GFP background at e12.5 and motor neurons were sorted by Fluorescence–activated cell sorting (FACS). RNA was extracted using the PicoPure RNA isolation system (Arcturus) and amplified using the Illumina TotalPrep RNA Amplification kit (Ambion). The cRNA from each sample was then hybridized to a MouseWG–6 v2.0 Expression BeadChip (Illumina).
Data quantitation/Statistical analysis
For all experiments n always refers to the number of embryos used. Unless otherwise specified, a minimum of 3 embryos per genotype were used for all reported results. In experiments where motor neuron numbers are reported, cells were counted as motor neurons if they expressed Isl1/2 and as PMC motor neurons if they expressed both Scip and Isl1/2. Statistical analysis was performed by using Student’s t–test (two–tailed distribution, homoscedastic).
Supplementary Material
Acknowledgements
We thank Christopher Henderson, Kevin Kanning, and Ivo Lieberam for discussions and sharing unpublished observations, Myungin Baek, Heekyung Jung and Catarina Catela for comments on the manuscript, and Peter Hallock and Steve Burden for assistance with diaphragm preparations. L.J is supported by a grant from the Canadian Institutes of Health Research (MOP-15139). J.S.D is supported by grants from the McKnight Foundation, Alfred P. Sloan, Project ALS, NYSTEM, HHMI and NIH R01 NS062822.
Footnotes
Author Contributions P.P. performed all experiments described apart from the Hoxc6−/− and Hb9::Foxp1 analyses which were performed by J.S.D. C.W. provided technical assistance with in situ experiments and serial sectioning, J.A. performed lung histology, L.J. provided the Hoxa5 flox/flox mouse line, P.P. and J.S.D. designed the study, analyzed the data and wrote the manuscript.
Competing financial interests The authors declare no competing financial interests.
References
- 1.Smith JC, Abdala AP, Rybak IA, Paton JF. Structural and functional architecture of respiratory networks in the mammalian brainstem. Philos Trans R Soc Lond B Biol Sci. 2009;364:2577–2587. doi: 10.1098/rstb.2009.0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allan DW, Greer JJ. Development of phrenic motoneuron morphology in the fetal rat. J Comp Neurol. 1997;382:469–479. doi: 10.1002/(sici)1096-9861(19970616)382:4<469::aid-cne4>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 3.Allan DW, Greer JJ. Embryogenesis of the phrenic nerve and diaphragm in the fetal rat. J Comp Neurol. 1997;382:459–468. doi: 10.1002/(sici)1096-9861(19970616)382:4<459::aid-cne3>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 4.Bruce A. A Topographical atlas of the spinal cord. Williams & Norgate; London: 1901. [Google Scholar]
- 5.Romanes GJ. The development and significance of the cell columns in the ventral horn of the cervical and upper thoracic spinal cord of the rabbit. J Anat. 1941;76:112–130. 115. [PMC free article] [PubMed] [Google Scholar]
- 6.Shirasaki R, Pfaff SL. Transcriptional codes and the control of neuronal identity. Annu Rev Neurosci. 2002;25:251–281. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- 7.Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- 8.Arber S, et al. Requirement for the homeobox gene Hb9 in the consolidation of motor neuron identity. Neuron. 1999;23:659–674. doi: 10.1016/s0896-6273(01)80026-x. [DOI] [PubMed] [Google Scholar]
- 9.Tsuchida T, et al. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79:957–970. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- 10.Thaler J, et al. Active suppression of interneuron programs within developing motor neurons revealed by analysis of homeodomain factor HB9. Neuron. 1999;23:675–687. doi: 10.1016/s0896-6273(01)80027-1. [DOI] [PubMed] [Google Scholar]
- 11.Dasen JS, Liu JP, Jessell TM. Motor neuron columnar fate imposed by sequential phases of Hox-c activity. Nature. 2003;425:926–933. doi: 10.1038/nature02051. [DOI] [PubMed] [Google Scholar]
- 12.Dasen JS, Tice BC, Brenner-Morton S, Jessell TM. A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell. 2005;123:477–491. doi: 10.1016/j.cell.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 13.Jung H, et al. Global control of motor neuron topography mediated by the repressive actions of a single hox gene. Neuron. 2010;67:781–796. doi: 10.1016/j.neuron.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dasen JS, De Camilli A, Wang B, Tucker PW, Jessell TM. Hox repertoires for motor neuron diversity and connectivity gated by a single accessory factor, FoxP1. Cell. 2008;134:304–316. doi: 10.1016/j.cell.2008.06.019. [DOI] [PubMed] [Google Scholar]
- 15.Rousso DL, Gaber ZB, Wellik D, Morrisey EE, Novitch BG. Coordinated actions of the forkhead protein Foxp1 and Hox proteins in the columnar organization of spinal motor neurons. Neuron. 2008;59:226–240. doi: 10.1016/j.neuron.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goshgarian HG, Rafols JA. The phrenic nucleus of th albino rat: a correlative HRP and Golgi study. J Comp Neurol. 1981;201:441–456. doi: 10.1002/cne.902010309. [DOI] [PubMed] [Google Scholar]
- 17.Sharma K, Leonard AE, Lettieri K, Pfaff SL. Genetic and epigenetic mechanisms contribute to motor neuron pathfinding. Nature. 2000;406:515–519. doi: 10.1038/35020078. [DOI] [PubMed] [Google Scholar]
- 18.Stolt CC, et al. SoxD proteins influence multiple stages of oligodendrocyte development and modulate SoxE protein function. Dev Cell. 2006;11:697–709. doi: 10.1016/j.devcel.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 19.Bermingham JR, Jr., et al. Tst-1/Oct-6/SCIP regulates a unique step in peripheral myelination and is required for normal respiration. Genes Dev. 1996;10:1751–1762. doi: 10.1101/gad.10.14.1751. [DOI] [PubMed] [Google Scholar]
- 20.Dillon AK, et al. Molecular control of spinal accessory motor neuron/axon development in the mouse spinal cord. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2005;25:10119–10130. doi: 10.1523/JNEUROSCI.3455-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Surmeli G, Akay T, Ippolito GC, Tucker PW, Jessell TM. Patterns of spinal sensory-motor connectivity prescribed by a dorsoventral positional template. Cell. 2011;147:653–665. doi: 10.1016/j.cell.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aubin J, Lemieux M, Tremblay M, Berard J, Jeannotte L. Early postnatal lethality in Hoxa-5 mutant mice is attributable to respiratory tract defects. Dev Biol. 1997;192:432–445. doi: 10.1006/dbio.1997.8746. [DOI] [PubMed] [Google Scholar]
- 23.Tabaries S, Lemieux M, Aubin J, Jeannotte L. Comparative analysis of Hoxa5 allelic series. Genesis. 2007;45:218–228. doi: 10.1002/dvg.20292. [DOI] [PubMed] [Google Scholar]
- 24.Dessaud E, et al. Interpretation of the sonic hedgehog morphogen gradient by a temporal adaptation mechanism. Nature. 2007;450:717–720. doi: 10.1038/nature06347. [DOI] [PubMed] [Google Scholar]
- 25.Dessaud E, Salaun D, Gayet O, Chabbert M, deLapeyriere O. Identification of lynx2, a novel member of the ly-6/neurotoxin superfamily, expressed in neuronal subpopulations during mouse development. Molecular and cellular neurosciences. 2006;31:232–242. doi: 10.1016/j.mcn.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 26.GrandPre T, Nakamura F, Vartanian T, Strittmatter SM. Identification of the Nogo inhibitor of axon regeneration as a Reticulon protein. Nature. 2000;403:439–444. doi: 10.1038/35000226. [DOI] [PubMed] [Google Scholar]
- 27.Chen H, et al. Identification of transcriptional targets of HOXA5. The Journal of biological chemistry. 2005;280:19373–19380. doi: 10.1074/jbc.M413528200. [DOI] [PubMed] [Google Scholar]
- 28.Mi R, Chen W, Hoke A. Pleiotrophin is a neurotrophic factor for spinal motor neurons. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4664–4669. doi: 10.1073/pnas.0603243104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun W, Gould TW, Vinsant S, Prevette D, Oppenheim RW. Neuromuscular development after the prevention of naturally occurring neuronal death by Bax deletion. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2003;23:7298–7310. doi: 10.1523/JNEUROSCI.23-19-07298.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Misgeld T, et al. Roles of neurotransmitter in synapse formation: development of neuromuscular junctions lacking choline acetyltransferase. Neuron. 2002;36:635–648. doi: 10.1016/s0896-6273(02)01020-6. [DOI] [PubMed] [Google Scholar]
- 31.Agalliu D, Takada S, Agalliu I, McMahon AP, Jessell TM. Motor neurons with axial muscle projections specified by Wnt4/5 signaling. Neuron. 2009;61:708–720. doi: 10.1016/j.neuron.2008.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livet J, et al. ETS gene Pea3 controls the central position and terminal arborization of specific motor neuron pools. Neuron. 2002;35:877–892. doi: 10.1016/s0896-6273(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 33.Haase G, et al. GDNF acts through PEA3 to regulate cell body positioning and muscle innervation of specific motor neuron pools. Neuron. 2002;35:893–905. doi: 10.1016/s0896-6273(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 34.Burgess RW, Jucius TJ, Ackerman SL. Motor axon guidance of the mammalian trochlear and phrenic nerves: dependence on the netrin receptor Unc5c and modifier loci. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2006;26:5756–5766. doi: 10.1523/JNEUROSCI.0736-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woldeyesus MT, et al. Peripheral nervous system defects in erbB2 mutants following genetic rescue of heart development. Genes Dev. 1999;13:2538–2548. doi: 10.1101/gad.13.19.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng G, et al. Roles for ephrins in positionally selective synaptogenesis between motor neurons and muscle fibers. Neuron. 2000;25:295–306. doi: 10.1016/s0896-6273(00)80895-8. [DOI] [PubMed] [Google Scholar]
- 37.Jaworski A, Tessier-Lavigne M. Autocrine/juxtaparacrine regulation of axon fasciculation by Slit-Robo signaling. Nat Neurosci. 2012;15:367–369. doi: 10.1038/nn.3037. [DOI] [PubMed] [Google Scholar]
- 38.Weiner JA, et al. Axon fasciculation defects and retinal dysplasias in mice lacking the immunoglobulin superfamily adhesion molecule BEN/ALCAM/SC1. Molecular and cellular neurosciences. 2004;27:59–69. doi: 10.1016/j.mcn.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 39.Petrinovic MM, et al. Neuronal Nogo-A regulates neurite fasciculation, branching and extension in the developing nervous system. Development. 2010;137:2539–2550. doi: 10.1242/dev.048371. [DOI] [PubMed] [Google Scholar]
- 40.De Marco Garcia NV, Jessell TM. Early Motor Neuron Pool Identity and Muscle Nerve Trajectory Defined by Postmitotic Restrictions in Nkx6.1 Activity. Neuron. 2008;57:217–231. doi: 10.1016/j.neuron.2007.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oppenheim RW, Bursztajn S, Prevette D. Cell death of motoneurons in the chick embryo spinal cord. XI. Acetylcholine receptors and synaptogenesis in skeletal muscle following the reduction of motoneuron death by neuromuscular blockade. Development. 1989;107:331–341. doi: 10.1242/dev.107.2.331. [DOI] [PubMed] [Google Scholar]
- 42.Glebova NO, Ginty DD. Heterogeneous requirement of NGF for sympathetic target innervation in vivo. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2004;24:743–751. doi: 10.1523/JNEUROSCI.4523-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tripodi M, Stepien AE, Arber S. Motor antagonism exposed by spatial segregation and timing of neurogenesis. Nature. 2011;479:61–66. doi: 10.1038/nature10538. [DOI] [PubMed] [Google Scholar]
- 44.Bouvier J, et al. Hindbrain interneurons and axon guidance signaling critical for breathing. Nat Neurosci. 2010;13:1066–1074. doi: 10.1038/nn.2622. [DOI] [PubMed] [Google Scholar]
- 45.Caubit X, et al. Teashirt 3 regulates development of neurons involved in both respiratory rhythm and airflow control. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:9465–9476. doi: 10.1523/JNEUROSCI.1765-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rose MF, et al. Math1 is essential for the development of hindbrain neurons critical for perinatal breathing. Neuron. 2009;64:341–354. doi: 10.1016/j.neuron.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iizuka M. Rostrocaudal distribution of spinal respiratory motor activity in an in vitro neonatal rat preparation. Neurosci Res. 2004;50:263–269. doi: 10.1016/j.neures.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 48.Smith JC, Greer JJ, Liu GS, Feldman JL. Neural mechanisms generating respiratory pattern in mammalian brain stem–spinal cord in vitro. I. Spatiotemporal patterns of motor and medullary neuron activity. J Neurophysiol. 1990;64:1149–1169. doi: 10.1152/jn.1990.64.4.1149. [DOI] [PubMed] [Google Scholar]
- 49.Lowell BB, O. D, Yu J. Development and phenotype of ChAT-IRES-Cre mice MGI Direct Data Submission. Jackson laboratory. 2006 [Google Scholar]
- 50.McIntyre DC, et al. Hox patterning of the vertebrate rib cage. Development. 2007;134:2981–2989. doi: 10.1242/dev.007567. [DOI] [PubMed] [Google Scholar]
- 51.Knudson CM, Tung KS, Tourtellotte WG, Brown GA, Korsmeyer SJ. Bax– deficient mice with lymphoid hyperplasia and male germ cell death. Science. 1995;270:96–99. doi: 10.1126/science.270.5233.96. [DOI] [PubMed] [Google Scholar]
- 52.Hallock PT, et al. Dok-7 regulates neuromuscular synapse formation by recruiting Crk and Crk-L. Genes Dev. 2010;24:2451–2461. doi: 10.1101/gad.1977710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.