Abstract
Dentin sialophosphoprotein (DSPP) in the extracellular matrix of dentin is cleaved into dentin sialoprotein (DSP) and dentin phosphoprotein (DPP), which originate from the NH2-terminal and COOH-terminal regions of DSPP, respectively. In the proteolytic processing of mouse DSPP, the peptide bond at Gly451-Asp452 has been shown to be cleaved by bone morphogenetic protein 1 (BMP1)/Tolloid-like metalloproteinases. In the present study, we generated transgenic mice expressing a mutant DSPP in which Asp452 was substituted by Ala452. Protein chemistry analyses of extracts from the long bone of these transgenic mice showed that the D452A substitution partially blocked DSPP processing in vivo. When the full-length form of mutant DSPP (designated “D452A-DSPP”) isolated from the transgenic mice was treated with BMP1 in vitro, a portion of the D452A-DSPP was cleaved, suggesting the presence of secondary peptide bond(s) that can be broken by BMP1. To identify the potential secondary DSPP cleavage site(s), site-directed mutagenesis was performed to generate nine DNA constructs expressing DSPP bearing substitutions at potential scission sites. These different types of mutant DSPP made in eukaryotic cell lines were treated with BMP1 and the digestion products were assessed by Western immunoblotting. All of the mutant DSPP molecular species were partially cleaved by BMP1, giving rise to a protein band on SDS-PAGE similar to that of normal DSP. Taken together, we concluded that in addition to the peptide bond Gly451-Asp452, there must be a cryptic cleavage site or sites close to Asp452 in the mouse DSPP that can be cleaved by BMP1.
Keywords: DSPP, BMP1, Proteolytic Processing, Posttranslational Modification, Biomineralization
Introduction
In addition to type I collagen, dentin contains a set of non-collagenous proteins (NCPs), which include osteonectin, osteocalcin, osteopontin (OPN), bone sialoprotein (BSP), dentin matrix protein 1 (DMP1), dentin sialophosphoprotein (DSPP) and matrix extracellular phosphoglycoprotein (MEPE). OPN, BSP, DMP1, DSPP, MEPE belong to the SIBLING (Small Integrin-Binding Ligand, N-linked Glycoprotein) family [1], with similar properties in their genomic organization, posttranslational modification and tissue localization. These SIBLING proteins secreted into the extracellular matrix (ECM) during the formation and mineralization of dentin and boneare believed to play key biological roles in the formation of these tissues[2].
MacDougall et al. [3] and Feng et al. [4] discovered that the nucleotide sequences for mouse dentin sialoprotein (DSP) and dentin phosphoprotein (DPP) reside on the same gene coding for a single mRNA named DSPP. The full-length cDNA of mouse DSPP revealed a 934-amino acid open reading frame, including a 17-amino acid signal peptide. This transcript would result in a translational protein product termed dentin sialophosphoprotein (DSPP) that would be specifically cleaved into two proteins, DSP and DPP. Human genetic studies have shown the association of mutations in the DSPP gene with dentinogenesis imperfecta (DGI), an autosomal dominant disorder causing dentin hypomineralization and significant tooth decay [5], [6]. Dspp ablation studies showed that Dspp-deficient mice have defects in the mineralization of dentin [7] and bone [8]. The predentin in the Dspp null mice is hypomineralized (widening of predentin), giving rise to phenotypic changes similar to the manifestations of DGI in humans. This information indicates that DSPP is critical for dentin mineralization.
Although DSP and DPP are derived from the same protein precursor, their biochemical properties are very different. DSP, a sialic acid-rich glycoprotein with few or no phosphate groups [9], [10], accounts for 5–8% of the dentin non-collagenous proteins. This protein shares overall characteristics with other sialoproteins (OPN and BSP) present in dentin and bone. Recently we discovered that the majority of the NH2-terminal fragment of rat or mouse DSPP is present as the proteoglycan (PG) form containing two glycosaminoglycan (GAG) chains [11], [12]. DPP is the most abundant non-collagenous protein in the ECM of dentin, accounting for as much as 50% of the NCPs [13]. DPP contains a large number of aspartic acid (Asp) and serine (Ser) residues, with the majority of Ser being phosphorylated [14], [15]. The Asp and phosphoserine (Pse) residues are mostly present in the repeating sequences of DSS and DS. The rat DPP occurs in three forms with different levels of phosphorylation; the highly phosphorylated DPP has approximately 200 phosphates attached to the molecule [14]. The repeating sequences of (Asp-Pse-Pse)n and (Asp-Pse)n fit well with the purported function of DPP in the nucleation and modulation of hydroxyapatite crystal formation [2].
The NH2-terminal sequence of rat DPP determined by Edman degradation was Asp-Asp-Pro-Asn for the highly phosphorylated form of rat DPP [14], and to be Asp-Asp-Pro in human [16]. The beginning of the DPP portion of mouse DSPP was Asp-Asp-Pro [3]. The NH2-terminal sequences for rat [14], [17], [18] and mouse DPP [3], [4] establish that the cleavage sites of DSPP are NH2-terminal to residue 448 in rat or residue 452 in mouse DSPP. Earlier protein chemistry work in our laboratory indicated that proteolytic processing of rat DSPP to DSP and DPP involves the cleavage of the peptide bonds Gly447-Asp448, Tyr438-Asp439 and His423-Ser424[14], [19]. Recent studies indicated that the proteolytic processing of DSPP was catalyzed by bone morphogenetic protein 1 (BMP1)/Tolloid-like metalloproteinases [20], [21] and the astacin proteases [22]. However, the cleavage sites involved in the proteolytic processing of DSPP by BMP1 are not totally clear and the cleavage of DSPP by BMP1 has not been verified by in vivostudies.
The aims of this study were: 1) to examine if substitution of residue Asp452 by Ala452 blocks the in vivo processing of mouse DSPP, 2) to test if the D452A substitution blocks the cleavage of DSPP by BMP1, and 3) to determine if DSPP has peptide bonds, other than the one at the NH2-terminus of Asp452 in the mouse DSPP, which can be cleaved by BMP1.
Materials and Methods
Generation of Transgenic Mice Expressing Normal DSPP or D452A-DSPP
Site-directed mutagenesis was performed on pcDNA3-DSPP construct to generate the cDNA encoding a mutant DSPP in which Asp452 was substituted by Ala452 (designated “D452A-DSPP”) [20]. A pBCKS construct containing a 3.6-kb rat Col 1a1 promoter upstream of the mouse DMP1 cDNA [23] was used to generate the targeting transgene. The normal DSPP cDNA and D452A-DSPP cDNA were inserted downstream of the 3.6-kb rat Col 1a1 promoter in pBCKS construct, respectively. The restriction enzymes SacII and SalI (New England Biolabs) were used to release an 8.6-kb fragment containing the 3.6-kb rat Col 1a1 promoter, 1.6-kb intron l of the Col 1a1 gene, 3-kb mouse DSPP cDNA and a SV40 later poly A tail from the construct. This 8.6-kb fragment, referred to as the “normal DSPP transgene” or “ D452A-DSPP transgene”, was used for pronuclear injection at the University of Texas Southwestern Medical Center at Dallas to generate transgenic founders in the C57BL/6J background. We obtained four independent founders expressing the normal DSPP transgene (Tg) and three founders expressing DSPP-D452A Tg. DSPP mRNA in the long bone of the transgenic mice was detected using reverse transcription polymerase chain reaction (RT-PCR) analyses. The protocol of animal work was approved by the Animal Welfare Committee of Baylor College of Dentistry of the Texas A&M University Health Science Center.
Quantitative Real-time Reverse Transcription PCR (qRT-PCR) and Reverse Transcription Polymerase Chain Reaction (RT-PCR)
The qRT-PCR analyses were performed to measure the expression levels of DSPP in the long bone of the transgenic mice. The primers used for qRT-PCR were: forward, 5′AACTCTGTGGCTGTGCCTCT-3′ (in exon 3) and reverse, 5′TATTGACTCGGAGCCATTCC-3′ (in exon 4). The qRT-PCR reactions were performed using Brilliant SYBR Green QPCR Master Mix (Applied Biosystems; Foster City, California) and the Stratagene MX4000 Real-Time PCR Detection System. GAPDH was used as the control. The qRT-PCR conditions were: initial denaturation at 95°C for 10 min, followed by 45 cycles of 95°C for 30 s, 60°C for 30 s and 72°C for 30 s. The mean values from triplicate analyses were then calculated and compared. RT-PCR was performed to examine the expression of the exogenous DSPP transgene in bone and tooth. The primers used for RT-PCR were: forward, 5′-AGCTTGGTACCGAGCTCGGATCCCC (in the linker region between rat Col 1a1 promoter and DSPP cDNA) and reverse, 3′-TCATTGTGTTCTTCAGCAGTGTTCC (in exon 4). The reaction conditions were: 95°Cfor 30 s, 56 °Cfor 30 s and 72 °Cfor 30 s, 30 cycles.
Extraction of NCPs and Detection of DSPP-Related Proteins
NCPs were extracted from the long bones of 3-week-old normal DSPP Tg mice or D452A-DSPP Tg mice, as previously described [24]. The NCPs in the extracts were separated by a 10 ml Q-Sepharose (Amersham Biosciences) column connected to a fast protein liquid chromatography (FPLC) system (GE Healthcare). The gradient employed for the Q-Sepharose anion-exchange chromatography was 0.1–0.8 M NaCl in 6 M urea solution at pH 7.2. NCPs including DSPP-related proteins eluted into 120 0.5 ml fractions. The full-length form of DSPP and its processed fragments were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western immunoblotting. For Western immunoblotting, we used the polyclonal anti-DSP antibody. Alkaline phosphatase-conjugated anti-rabbit IgG (Sigma-Aldrich) was employed as the secondary antibody for the Western immunoblotting analysis. The blots were incubated in the chemiluminescent substrate CDP-star (Ambion) for 5 min and exposed to X-ray films (Kodak).
Substitutions of Amino Acid Residues at Potential DSPP Cleavage Sites
To identify the peptide bonds that were potential DSPP cleavage sites, we performed site-directed mutagenesis to substitute candidate residues in the mouse DSPP cDNA [20]. Totally, we generated nine mutant-DSPP constructs in which Asp residues at candidate sites were substituted with Ala. These substitution constructs (each representing one mutant DSPP) include: 1) D452A-DSPP [20], 2) H227A-D452A-DSPP, 3) E443A-D452A-DSPP, 4) D445A-D452A-DSPP, 5) D446A-D452A-DSPP, 6) D445A-D446A-D452A-DSPP, 7) D452A-D453A-DSPP, 8) D452A-D487A-DSPP, and 9) D452A-D491A-DSPP. The constructions of D452A-DSPP, H227A-D452A-DSPP and E443A-D452A-DSPP were based on our previous protein chemistry work [19], and the designs for the other six mutant constructs were based on the understanding of the sequences known to be cleaved by BMP1. These nine pcDNA-DSPP constructs were used to transfect human embryonic kidney 293-EBNA (HEK293-EBNA) cells using lipofectamine 2000 (Invitrogen). The cells were cultured in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum (Invitrogen). The medium was replaced with serum-free medium 16–18 hrs after transfection, and then the transfected cells were cultured for an additional 72 hrs. The medium was collected and analyzed for DSPP and/or its processed fragments.
Digestion of DSPP by BMP1
The cell culture medium or the protein sample from the chromatographic fractions containing the full-length form of DSPP was dialyzed for 72 hours at 4°C and then lyophilized. The lyophilized powder was reconstituted in BMP1 digestion buffer [25]. The prepared protein samples were incubated with recombinant human BMP1 (R&D Systems) for 21 hrs at 37°C. The digestion products were analyzed by Western immunoblotting using the polyclonal anti-DSP antibody [20].
Results
Expression of Normal DSPP and D452A-DSPP Transgenein the Transgenic Mice The expression of the normal DSPP and D452A-DSPP transgene in the transgenic mice was confirmed by qRT-PCR and RT-PCR, using RNAs extracted from the long bone of three lines of normal DSPP transgenic mice and four lines of the D452A-DSPP transgenic mice. Although different expression levels for the transgene were observed among the transgenic mice, they all contained higher levels of DSPP mRNA compared to the wild type mice (Fig. 1a), indicating that the transgene was active. It should be noted that the expression of endogenous DSPP in the long bone of wild type animals is very low [26]. The primers for RT-PCR were designed specifically to detect exogenous transgene as shown in Figure 1b, whereas the endogenous DSPP transcripts could not be detected using these primers (Fig. 1b).
Fig. 1.
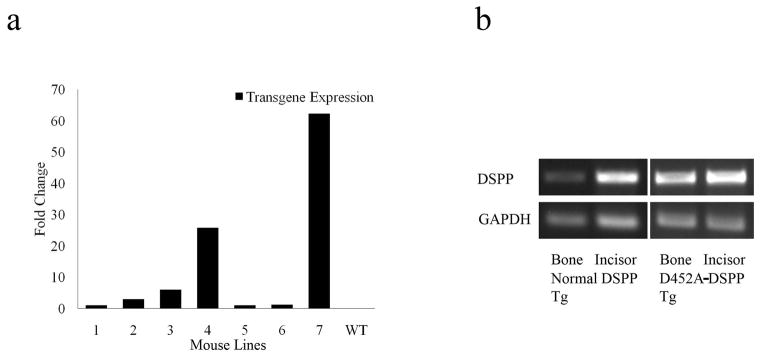
Expression of normal-DSPP and D452A-DSPP in the bone of transgenic mice. a: Real time PCR showing the different expression levels in the long bone from four lines of D452A-DSPP transgenic mice (1–4) and three lines of normal DSPP transgenic mice (5–7). The expression level of line 5 was set as 1. b: RT-PCR demonstrating the expression of normal DSPP transgene as well as D452A-DSPP transgene in the long bone and incisor of the transgenic mice. Note that only the exogenous DSPP mRNAs could be detected using the primers specifically designed for the transgenic transcripts
Full-length DSPP and its Fragments in the ECM Extractsof Mouse Long Bone
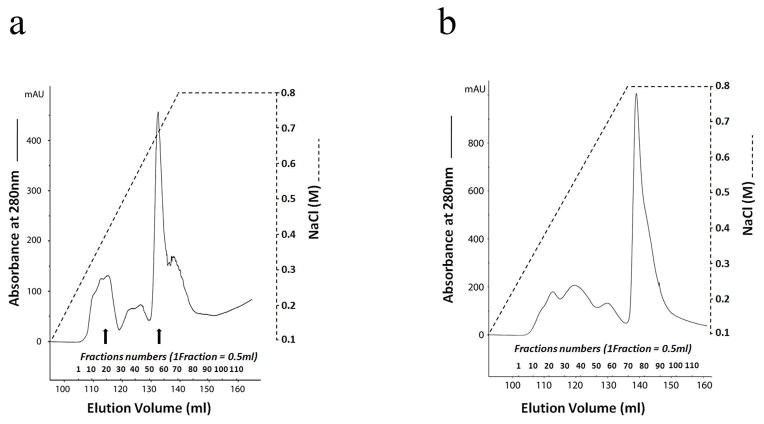
We extracted NCPs from the long bone of line 4 (Fig. 1a) for D452A-DSPP analysis and that of line 7 (Fig. 1a) for analyzing the normal DSPP. Q-Sepharose chromatography separated the long bone extracts into 120 fractions (Fig. 2). Stains-All staining and Western immunoblotting were used to detect normal DSPP as well as D452A-DSPP and to determine if the mutant D452A-DSPP was processed into DSP and DPP fragments. All of the chromatographic fractions from the bone extracts that might potentially contain DSPP-related products were analyzed; to illustrate the results, fraction 34 was shown in this report. Both the fragment (DSP) and the full-length form of DSPP were observed in the long bone extracts of D452A-DSPP Tg as well as in the normal DSPP Tg mice. The ratio of full-length form of DSPP to DSP in the D452A-DSPP Tg mice was much greater than that in the normal DSPP-Tg mice. Significant amounts of the full-length DSPP were detected in the D452A-DSPP Tg mice, while very weak protein bands matching the NH2-terminal fragment of DSPP were observed, indicating that D452A-DSPP was present mainly as the full-length form. Only a small portion of the mutant D452A-DSPP was cleaved in vivo(Fig. 3).
Fig. 2.
FPLC profile of Q-Sepharose chromatography for acidic proteins extracted from the mouse long bone. Protein extracts was separated and collected into continuous 120 fractions for biochemistry analyses. a: FPLC chromatogram for bone extracts from D452A-DSPP transgenic mice. b: FPLC chromatogram for bone extracts from wild type mice. Western immunoblotting and Stains-All staining illustrated that the fractions between the two arrows (fractions 20 to 58) in(a) contained DSPP and its fragments, DSP and DPP.
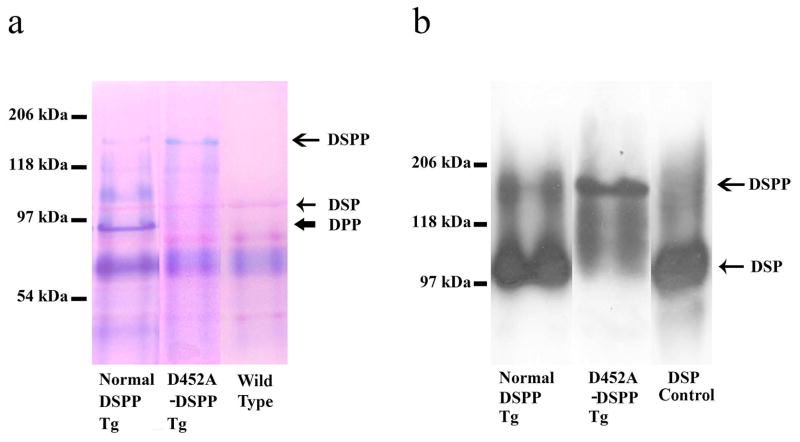
Fig. 3.
Fraction 34 from the Q-Sepharose chromatography separations was used to demonstrate the acidic proteins between the three types of mice. We loaded the normal DSPP lane and D452A DSPP lane with equal volume (60 μl) of the sample from the fraction 34 of the long bone extracts of normal DSPP or D452A-DSPP transgenic mice and for each sample, the non-collagenous proteins were extracted from the long bone of six mice. a: acidic proteins (including the SIBLING members) were visualized by Stains-All staining (blue and purple bands). An extra blue band (DPP) and weak staining of full-length form of DSPP were detected in the long bone extracts of normal-DSPP transgenic mice, whereas a weak staining of DPP and a stronger full-length form of DSPP were detected in the D452A-DSPP transgenic mice. The normal DSPP sample was from transgenic mice expressing the normal DSPP. The D452A-DSPP was from the transgenic mice expressing D452A-DSPP. The wild type sample was from the C57BL/6J mice. DSP (less acidic than DPP) is much less sensitive to Stains-All staining. The purple protein band indicated by the thin short arrow was likely to be DSP. In each lane, 60 μl of sample was loaded. b: DSPP-related proteins in the long bone of the transgenic mice (Western blotting). The ratio of DSP to DSPP in the normal-DSPP transgenic mice is much greater than that in the D452A-DSPP transgenic mice, indicating that the D452A substitution effectively blocked DSPP processing in vivo. Normal DSPP or D452A-DSPP lane: 60 μl of sample. DSP lane: 0.1 μg of rat DSP
BMP1 Completely Cleaves Normal DSPP, and Partially Cleaves D452A-DSPP
The in vitro enzyme assay experiments showed that the full-length form of DSPP extracted from the wild type C57BL/6 mice was completely processed by BMP1, whereas the full-length form of DSPP extracted from the long bone of D452A-DSPP Tg mice was partially cleaved (Fig. 4).
Fig. 4.
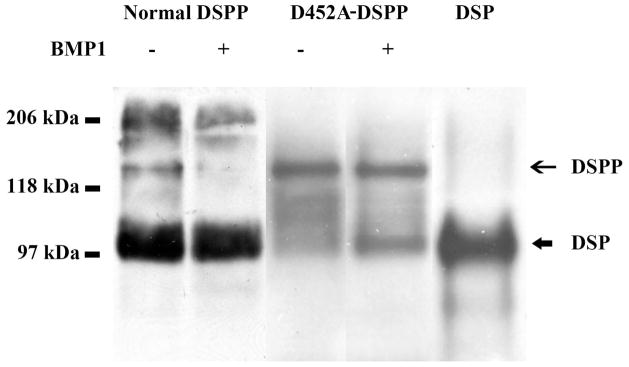
Full-length form of DSPP from the long bone of the D452A-DSPP transgenic mice as well as from wild type mouse dentin was treated with BMP1. DSPP-related proteins were detected by Western immunoblotting using anti-DSP polyclonal antibody. The full-length form of normal DSPP was totally processed, whereas the full-length form of D452A-DSPP were partially cleaved, indicating that D452A substitution could partially block DSPP processing by BMP1. The differences in the ratio of full-length DSPP to DSP indicated that more DSPP molecules were cleaved into DSP in the wild type mice than in the transgenic mice expressing the mutant D452A-DSPP. Normal DSPP or D452A-DSPP lane: 60 μl of sample. DSP lane: 0.1 μg of rat DSP.
Substitutions of Amino Acid Residues at Potential DSPP cleavage Sites Were Unable to Completely Block DSPP Processing
Western immunoblotting analyses showed that the culture medium of the HEK-293 EBNA cells transfected with the normal pcDNA3-DSPP or the mutant constructs contained significant amounts of full-length DSPP and/or its fragments. When treated with BMP1, the full-length form of normal DSPP disappeared, whereas the majority of the full length-form of mutant D452A-DSPP was still present, indicating that this substitution partially blocked the proteolytic processing of DSPP. Nevertheless, a portion of mutant DSPP were cleaved suggesting that there might be unidentified “secondary” or “cryptic” cleavage site(s), in addition to the peptide bond at the NH2-terminus of Asp452. To uncover the secondary cleavage sites, we generated and analyzed eight more DNA constructs expressing DSPP with substitutions at the potential secondary cleavage sites. None of the substitutions of could completely block DSPP processing (Fig. 5).
Fig. 5.
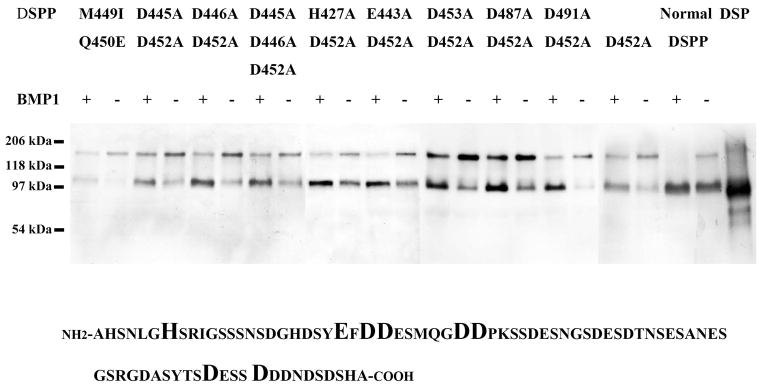
Mutant mouse DSPP from HEK-293 EBNA cells transfected with pcDNA3-mutant DSPP constructs was treated with BMP1. DSPP-related proteins were identified using the anti-DSP polyclonal antibody in Western immunoblotting analyses. All of these different types of DSPP proteins were partially cleaved by BMP1 digestion, implying that BMP1 cleaves DSPP at secondary, unidentified peptide bond(s), in addition to that at the NH2-terminus of Asp452. The sample for each lane was equal to 1 ml of culture medium (concentrated by dialysis and lyophilization). 0.1 μg of rat DSP was loaded in DSP lane. The substitution sites we designed in the mouse DSPP linker region were shown in bold and enlarged font
Discussion
DSPP belongs to the SIBLING family. Although details concerning their precise functions are unknown, the common features of SIBLING proteins indicated that they may share some common mechanisms in the biomineralization of dentin and bone. DMP1, DSPP, MEPE are all cleaved into NH2-terminal and COOH- terminal fragments, indicating that the proteolytic processing of these proteins may play important roles in the formation of dentin and bone. We have hypothesized that the proteolytic processing of DSPP and DMP1 is an activation step converting the protein precursors to functional fragments. This hypothesis has been proven in our previous study showing that failure to process DMP1 into fragments leads to its loss of function in osteogenesis [27]and dentinogenesis [28].
Several research groups have performed studies to analyze the cleavage sites during DSPP processing. Earlier work in our group revealed that three peptide bonds were cleaved during the processing of rat DSPP [14], [19]. An in vitro study showed that matrix metalloproteinases (MMP-2 and MMP-20) cleaved the NH2-terminal portion of porcine DSPP (namely, DSP and dentin glycoprotein) at multiple sites, yielding several low molecular-weight fragments [29]. Another in vitro study [30] showed that a recombinant rat DSP-PP (a shorter form of the rat DSPP variants) underwent self-processing; one of the self-cleavages appeared to be at Gly447-Asp448, which corresponds to Gly451-Asp452 in the mouse DSPP. In a previous investigation, we and another group have found that BMP1 is responsible for the proteolytic processing of DSPP into DSP and DPP [20], [21]. The astacin proteases were also reported to cleave DSPP to generate DPP [22]. We have found that the substitution of Asp452 to Ala452 could block the proteolytic processing of DSPP in HEK293-EBNA cells [20]. In this study, we observed that the D452A-DSPP from transfected HEK293-EBNA cells was partially processed when treated with recombinant human BMP1 in vitro, while normal DSPP was completely cleaved by BMP1 [20].
In this study, transgenic mice were generated to express normal DSPP or D452A-DSPP (mutant) driven by the type I collagen promoter. A great level of variation in the transgene expression was observed among different transgenic mouse lines (particularly with regard to the expression of the normal DSPP transgene). In the generation of these transgenic mice, the normal DSPP or D452A-DSPP transgene (cDNA) was randomly inserted into the mouse genome. The variation of in the expression level of the transgene was likely caused by the difference in the localization of the cDNA inserted in the mouse genome. Different insertion sites might have either enhancing or inhibiting effects on the transcription of the exogenous genes due to the local genomic environment. In the D452A-DSPP transgenic mice, the D452A substitution could not completely block the proteolytic processing of mouse DSPP and the full-length D452A-DSPP isolated from the mutant mice were also partially cleaved when treated with BMP1 in vitro. As we were performing the mutation analyses, another group reported that DSPP was processed into DSP and DPP by BMP1, and a combined substitution of M449I and Q450E could completely block DSPP processing in vitro [21]. In the present study, we also included M449I and Q450E substitutions to test if BMP1 could digest DSPP with the double mutations; we found that this combination of mutations (M449I+Q450E) was unable to completely block DSPP processing. These results showed strong evidence that there must be secondary, cryptic cleavage site(s), in addition to Gly451-Asp452.
According to the sequence characteristics of the known substrates of BMP1, the presence of aspartate residues at P1 positions and of residues with aromatic side chains or methionines at P2–P4 positions are the most common features of the cleavage sites in the substrates of the BMP1/Tolloid-like proteinases [25]. Based on these rules, we searched the sequence surrounding the Asp452a nd found several amino acid sequence clusters showing some degree of similarity to those known cleavage-sequences of BMP1. We did site-directed mutagenesis to substitute these potential secondary cleavage sites. The different types of mutant DSPP molecular species made by HEK293-EBNA cells were treated with BMP1 in vitro to see if the cleavage of these mutant DSPP molecules was totally blocked. However, we found that none of the combined mutations, containing potential secondary cleavage site mutations and the D452A substitution could completely block DSPP processing. Interestingly, in the Western immunoblotting analyses, after the dialysis and freeze-drying of the mutant DSPP samples, we detected a certain portion of cleaved fragment (DSP) even though these samples were not treated with BMP1 (Fig. 5), whereas no DSP fragments could be detected before dialysis and freeze-drying (data not shown). One interpretation is that before the dialysis and freeze-drying, the concentration of the endogenous BMP1 in the medium was too low to initiate the cleavage reaction. After the dialysis, freeze-drying and reconstitution, the concentration of the endogenous BMP1 from the culture medium was increased and the salt concentration was lowered, which helped to increase the BMP1 activity [21]. Another possibility is that besides BMP1, other enzymes in the astacin family may also cleave DSPP [22], or perhaps DSPP might have undergone self-processing [30] at sites other than the NH2-terminus of Asp452.
It should be noted that the molecular weight of the newly formed DSP fragments by BMP1 digestion was consistent with those of the endogenous DSP generated from the HEK-293-EBNA cells transfected with normal DSPP construct, indicating that the unidentified secondary cleavage site(s) must be very close to Asp452in mouse DSPP.
Acknowledgments
This work was supported by Grant DE005092 (to CQ) from the National Institutes of Health and Grant 30700943 from National Natural Science Foundation of China.
Footnotes
DECLARATION OF INTEREST
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- 1.Fisher LW, Torchia DA, Fohr B, Young MF, Fedarko NS. Flexible structures of SIBLING proteins, bone sialoprotein, and osteopontin. Biochem Biophys Res Commun. 2001;280:460–465. doi: 10.1006/bbrc.2000.4146. [DOI] [PubMed] [Google Scholar]
- 2.Butler WT. Dentin matrix proteins. Eur J Oral Sci. 1998;106 (Suppl 1):204210. doi: 10.1111/j.1600-0722.1998.tb02177.x. [DOI] [PubMed] [Google Scholar]
- 3.MacDougall M, Simmons D, Luan X, Nydegger J, Feng J, Gu TT. Dentin phosphoprotein and dentin sialoprotein are cleavage products expressed from a single transcript coded by a gene on human chromosome 4. Dentin phosphoprotein DNA sequence determination. J Biol Chem. 1997;272:835–842. doi: 10.1074/jbc.272.2.835. [DOI] [PubMed] [Google Scholar]
- 4.Feng JQ, Luan X, Wallace J, Jing D, Ohshima T, Kulkarni AB, D’Souza RN, Kozak CA, MacDougall M. Genomic organization, chromosomal mapping, and promoter analysis of the mouse dentin sialophosphoprotein (Dspp) gene, which codes for both dentin sialoprotein and dentin phosphoprotein. J Biol Chem. 1998;273:9457–9464. doi: 10.1074/jbc.273.16.9457. [DOI] [PubMed] [Google Scholar]
- 5.Xiao S, Yu C, Chou X, Yuan W, Wang Y, Bu L, Fu G, Qian M, Yang J, Shi Y, Hu L, Han B, Wang Z, Huang W, Liu J, Chen Z, Zhao G, Kong X. Dentinogenesis imperfecta 1 with or without progressive hearing loss is associated with distinct mutations in DSPP. Nat Genet. 2001;27:201–204. doi: 10.1038/84848. [DOI] [PubMed] [Google Scholar]
- 6.Zhang X, Zhao J, Li C, Gao S, Qiu C, Liu P, Wu G, Qiang B, Lo WH, Shen Y. DSPP mutation in dentinogenesis imperfecta Shields type II. Nat Genet. 2001;27:151–152. doi: 10.1038/84765. [DOI] [PubMed] [Google Scholar]
- 7.Sreenath T, Thyagarajan T, Hall B, Longenecker G, D’Souza R, Hong S, Wright JT, MacDougall M, Sauk J, Kulkarni AB. Dentin sialophosphoprotein knockout mouse teeth display widened predentin zone and develop defective dentin mineralization similar to human dentinogenesis imperfecta type III. J Biol Chem. 2003;278:24874–24880. doi: 10.1074/jbc.M303908200. [DOI] [PubMed] [Google Scholar]
- 8.Verdelis K, Ling Y, Sreenath T, Haruyama N, MacDougall M, van der Meulen MC, Lukashova L, Spevak L, Kulkarni AB, Boskey AL. DSPP effects on in vivo bone mineralization. Bone. 2008;43:983–990. doi: 10.1016/j.bone.2008.08.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butler WT. Dentin-specific proteins. Methods Enzymol. 1987;145:290–303. doi: 10.1016/0076-6879(87)45017-9. [DOI] [PubMed] [Google Scholar]
- 10.Butler WT, Bhown M, Brunn JC, D’Souza RN, Farach-Carson MC, Happonen RP, Schrohenloher RE, Seyer JM, Somerman MJ, Foster RA. Isolation, characterization and immunolocalization of a 53-kDal dentin sialoprotein (DSP) Matrix. 1992;12:343–351. doi: 10.1016/s0934-8832(11)80030-2. [DOI] [PubMed] [Google Scholar]
- 11.Qin C, Brunn JC, Baba O, Wygant JN, McIntyre BW, Butler WT. Dentin sialoprotein isoforms: detection and characterization of a high molecular weight dentin sialoprotein. Eur J Oral Sci. 2003;111:235–242. doi: 10.1034/j.1600-0722.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- 12.Zhu Q, Sun Y, Prasad M, Wang X, Yamoah AK, Li Y, Feng J, Qin C. Glycosaminoglycan chain of dentin sialoprotein proteoglycan. J Dent Res. 2010;89:808–812. doi: 10.1177/0022034510366902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacDougall M, Zeichner-David M, Slavkin HC. Production and characterization of antibodies against murine dentine phosphoprotein. Biochem J. 1985;232:493–500. doi: 10.1042/bj2320493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butler WT, Bhown M, DiMuzio MT, Cothran WC, Linde A. Multiple forms of rat dentin phosphoproteins. Arch Biochem Biophys. 1983;225:178–186. doi: 10.1016/0003-9861(83)90021-8. [DOI] [PubMed] [Google Scholar]
- 15.Lee SL, Veis A, Glonek T. Dentin phosphoprotein: an extracellular calcium-binding protein. Biochemistry. 1977;16:2971–2979. doi: 10.1021/bi00632a026. [DOI] [PubMed] [Google Scholar]
- 16.Chang SR, Chiego D, Jr, Clarkson BH. Characterization and identification of a human dentin phosphophoryn. Calcif Tissue Int. 1996;59:149–153. doi: 10.1007/s002239900101. [DOI] [PubMed] [Google Scholar]
- 17.Ritchie HH, Wang LH. Sequence determination of an extremely acidic rat dentin phosphoprotein. J Biol Chem. 1996;271:21695–21698. doi: 10.1074/jbc.271.36.21695. [DOI] [PubMed] [Google Scholar]
- 18.George A, Srinivasan R, Thotakura S, Veis A. The phosphophoryn gene family: identical domain structures at the carboxyl end. Eur J Oral Sci. 1998;106 (Suppl 1):221–226. doi: 10.1111/j.1600-0722.1998.tb02179.x. [DOI] [PubMed] [Google Scholar]
- 19.Qin C, Cook RG, Orkiszewski RS, Butler WT. Identification and characterization of the carboxyl-terminal region of rat dentin sialoprotein. J Biol Chem. 2001;276:904–909. doi: 10.1074/jbc.M006271200. [DOI] [PubMed] [Google Scholar]
- 20.Sun Y, Lu Y, Chen S, Prasad M, Wang X, Zhu Q, Zhang J, Ball H, Feng J, Butler WT, Qin C. Key proteolytic cleavage site and full-length form of DSPP. J Dent Res. 2010;89:498–503. doi: 10.1177/0022034510363109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.von Marschall Z, Fisher LW. Dentin sialophosphoprotein (DSPP) is cleaved into its two natural dentin matrix products by three isoforms of bone morphogenetic protein-1 (BMP1) Matrix Biol. 2010;29:295–303. doi: 10.1016/j.matbio.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsuchiya S, Simmer JP, Hu JC, Richardson AS, Yamakoshi F, Yamakoshi Y. Astacin proteases cleave dentin sialophosphoprotein (Dspp) to generate dentin phosphoprotein (Dpp) J Bone Miner Res. 2011;26:220–228. doi: 10.1002/jbmr.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu Y, Ye L, Yu S, Zhang S, Xie Y, McKee MD, Li YC, Kong J, Eick JD, Dallas SL, Feng JQ. Rescue of odontogenesis in Dmp1-deficient mice by targeted re-expression of DMP1 reveals roles for DMP1 in early odontogenesis and dentin apposition in vivo. Dev Biol. 2007;303:191–201. doi: 10.1016/j.ydbio.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang B, Maciejewska I, Sun Y, Peng T, Qin D, Lu Y, Bonewald L, Butler WT, Feng J, Qin C. Identification of full-length dentin matrix protein 1 in dentin and bone. Calcif Tissue Int. 2008;82:401–410. doi: 10.1007/s00223-008-9140-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steiglitz BM, Ayala M, Narayanan K, George A, Greenspan DS. Bone morphogenetic protein-1/Tolloid-like proteinases process dentin matrix protein-1. J Biol Chem. 2004;279:980–986. doi: 10.1074/jbc.M310179200. [DOI] [PubMed] [Google Scholar]
- 26.Qin C, Brunn JC, Cadena E, Ridall A, Tsujigiwa H, Nagatsuka H, Nagai N, Butler WT. The expression of dentin sialophosphoprotein gene in bone. J Dent Res. 2002;81:392–394. doi: 10.1177/154405910208100607. [DOI] [PubMed] [Google Scholar]
- 27.Sun Y, Prasad M, Gao T, Wang X, Zhu Q, D’Souza R, Feng JQ, Qin C. Failure to process dentin matrix protein 1 (DMP1) into fragments leads to its loss of function in osteogenesis. J Biol Chem. 2010;285:31713–31722. doi: 10.1074/jbc.M110.137059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun Y, Lu Y, Chen L, Gao T, D’Souza R, Feng JQ, Qin C. DMP1 Processing is Essential to Dentin and Jaw Formation. J Dent Res. 2011;90:619–624. doi: 10.1177/0022034510397839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamakoshi Y, Hu JC, Iwata T, Kobayashi K, Fukae M, Simmer JP. Dentin sialophosphoprotein is processed by MMP-2 and MMP-20 in vitro and in vivo. J Biol Chem. 2006;281:38235–38243. doi: 10.1074/jbc.M607767200. [DOI] [PubMed] [Google Scholar]
- 30.Godovikova V, Ritchie HH. Dynamic processing of recombinant dentin sialoprotein-phosphophoryn protein. J Biol Chem. 2007;282:31341–31348. doi: 10.1074/jbc.M702605200. [DOI] [PubMed] [Google Scholar]