Abstract
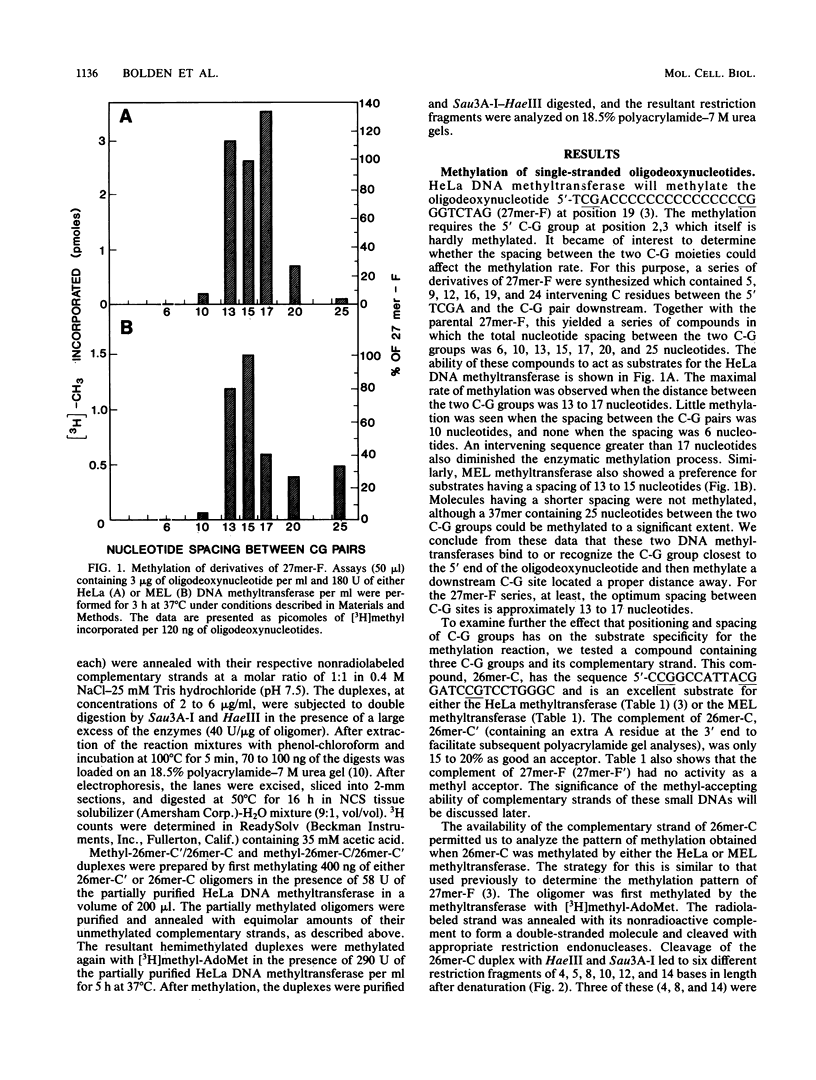
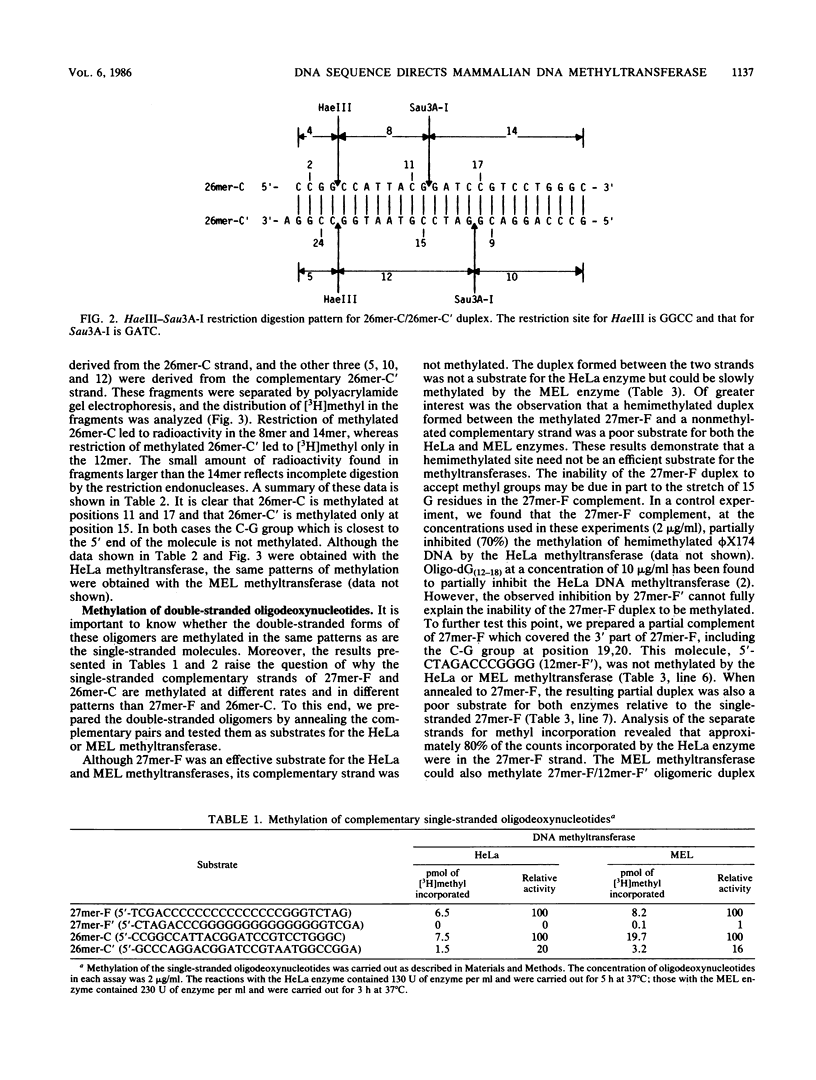
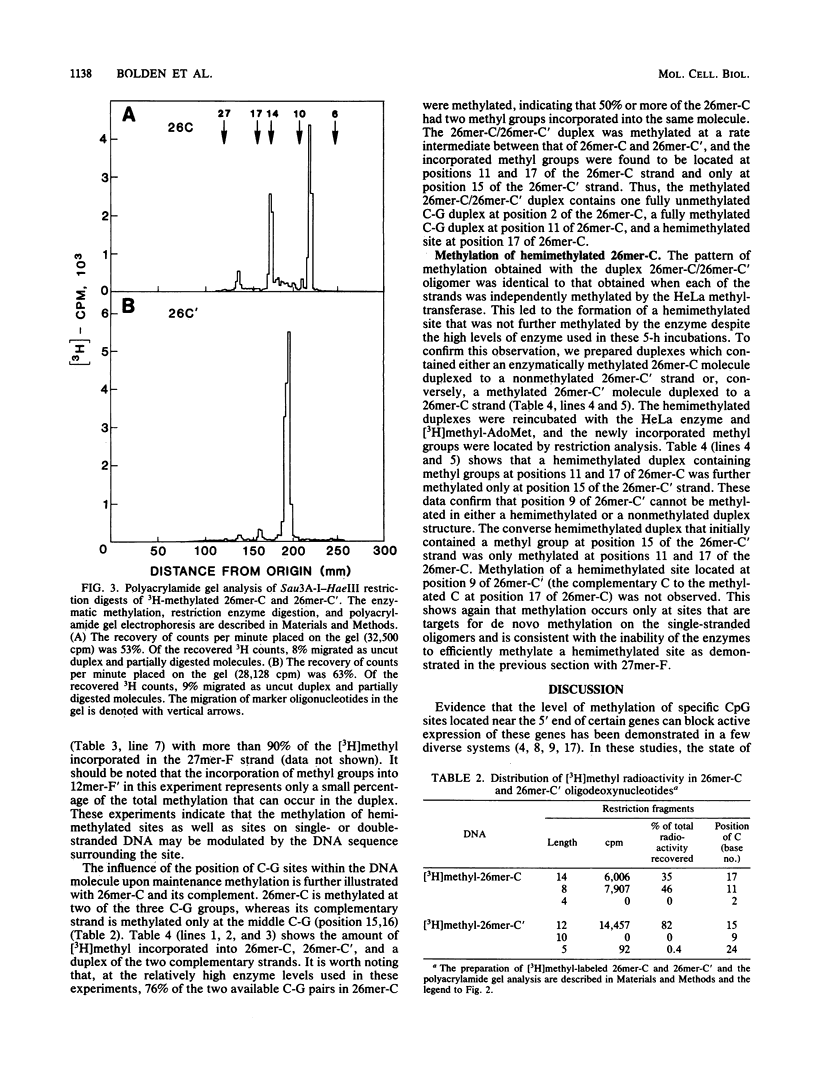
Analysis of the enzymatic methylation of oligodeoxynucleotides containing multiple C-G groups showed that hemimethylated sites in duplex oligomers are not significantly methylated by human or murine DNA methyltransferase unless those sites are capable of being methylated de novo in the single- or double-stranded oligomers. Thus, the primary sequence of the target strand, rather than the methylation pattern of the complementary strand, determines maintenance methylation. This suggests that de novo and maintenance methylation are the same process catalyzed by the same enzyme. In addition, the study revealed that complementary strands of oligodeoxynucleotides are methylated at different rates and in different patterns. Both primary DNA sequence and the spacing between C-G groups seem important since in one case studied, maximal methylation required a specific spacing of 13 to 17 nucleotides between C-G pairs.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolden A. H., Nalin C. M., Ward C. A., Poonian M. S., McComas W. W., Weissbach A. DNA methylation: sequences flanking C-G pairs modulate the specificity of the human DNA methylase. Nucleic Acids Res. 1985 May 24;13(10):3479–3494. doi: 10.1093/nar/13.10.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolden A., Ward C., Siedlecki J. A., Weissbach A. DNA methylation. Inhibition of de novo and maintenance methylation in vitro by RNA and synthetic polynucleotides. J Biol Chem. 1984 Oct 25;259(20):12437–12443. [PubMed] [Google Scholar]
- Busslinger M., Hurst J., Flavell R. A. DNA methylation and the regulation of globin gene expression. Cell. 1983 Aug;34(1):197–206. doi: 10.1016/0092-8674(83)90150-2. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y., Cedar H., Razin A. Substrate and sequence specificity of a eukaryotic DNA methylase. Nature. 1982 Feb 18;295(5850):620–622. doi: 10.1038/295620a0. [DOI] [PubMed] [Google Scholar]
- Holliday R., Pugh J. E. DNA modification mechanisms and gene activity during development. Science. 1975 Jan 24;187(4173):226–232. [PubMed] [Google Scholar]
- Jones P. A., Taylor S. M. Hemimethylated duplex DNAs prepared from 5-azacytidine-treated cells. Nucleic Acids Res. 1981 Jun 25;9(12):2933–2947. doi: 10.1093/nar/9.12.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruczek I., Doerfler W. The unmethylated state of the promoter/leader and 5'-regions of integrated adenovirus genes correlates with gene expression. EMBO J. 1982;1(4):409–414. doi: 10.1002/j.1460-2075.1982.tb01183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langner K. D., Vardimon L., Renz D., Doerfler W. DNA methylation of three 5' C-C-G-G 3' sites in the promoter and 5' region inactivate the E2a gene of adenovirus type 2. Proc Natl Acad Sci U S A. 1984 May;81(10):2950–2954. doi: 10.1073/pnas.81.10.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- Orlofsky A., Chasin L. A. A domain of methylation change at the albumin locus in rat hepatoma cell variants. Mol Cell Biol. 1985 Jan;5(1):214–225. doi: 10.1128/mcb.5.1.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M. O., Sperling L., Cassio D., Levilliers J., Sala-Trepat J., Weiss M. C. Undermethylation at the 5' end of the albumin gene is necessary but not sufficient for albumin production by rat hepatoma cells in culture. Cell. 1982 Oct;30(3):825–833. doi: 10.1016/0092-8674(82)90287-2. [DOI] [PubMed] [Google Scholar]
- Pfeifer G. P., Grünwald S., Boehm T. L., Drahovsky D. Isolation and characterization of DNA cytosine 5-methyltransferase from human placenta. Biochim Biophys Acta. 1983 Aug 2;740(3):323–330. doi: 10.1016/0167-4781(83)90141-0. [DOI] [PubMed] [Google Scholar]
- Riggs A. D. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14(1):9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- Roy P. H., Weissbach A. DNA methylase from HeLa cell nuclei. Nucleic Acids Res. 1975 Oct;2(10):1669–1684. doi: 10.1093/nar/2.10.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon D., Grunert F., von Acken U., Döring H. P., Kröger H. DNA-methylase from regenerating rat liver: purification and characterisation. Nucleic Acids Res. 1978 Jun;5(6):2153–2167. doi: 10.1093/nar/5.6.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein R., Razin A., Cedar H. In vitro methylation of the hamster adenine phosphoribosyltransferase gene inhibits its expression in mouse L cells. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3418–3422. doi: 10.1073/pnas.79.11.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]


