Abstract
The structural origins of differences in susceptibilities of various DNA lesions to nucleotide excision repair (NER) are poorly understood. Here we compared, in the same sequence context, the relative NER dual incision efficiencies elicited by two stereochemically distinct pairs of guanine (N2-dG) and adenine (N6-dA) DNA lesions, derived from enantiomeric genotoxic diol epoxides of the highly tumorigenic fjord region polycyclic aromatic hydrocarbon dibenzo[a,l]pyrene (DB[a,l]P). Remarkably, in cell-free HeLa cell extracts, the guanine adduct with R absolute chemistry at the N2-dG linkage site is ~ 35 times more susceptible to NER dual incisions than the stereochemically identical N6-dA adduct. For the guanine and adenine adducts with S stereochemistry, a similar, but somewhat smaller effect (factor of ~15) is observed. The striking resistance of the bulky N6-dA in contrast to the modest to good susceptibilities of the N2-dG adducts to NER are interpreted in terms of the balance between lesion-induced DNA-distorting and DNA-stabilizing van der Waals interactions in their structures, that are partly reflected in the overall thermal stabilities of the modified duplexes. Our results are consistent with the hypothesis that the high genotoxic activity of DB[a,l]P is related to the formation of NER-resistant and persistent DB[a,l]P-derived adenine adducts in cellular DNA.
Keywords: Nucleotide excision repair; dibenzo[a,l]pyrene-derived DNA adducts
INTRODUCTION
Nucleotide excision repair (NER) is a versatile DNA repair mechanism that recognizes and removes bulky lesions from DNA by excising olgonucleotide sequences 24–32 nucleotides in length containing the lesion, in both transcription coupled repair 1 and global genomic repair (GGR).2–5 The versatility of GGR is achieved indirectly by the recognition of the DNA distortions/destabilizations caused by the lesions rather than the lesions themselves. 6, 7 Base sequence context can also have a pronounced impact on NER efficiencies,8–14 and can even result in the suppression of NER when the partner base opposite the lesion is deleted.15–17 The efficiency of NER is known to vary by two orders of magnitude or more, depending on the structural features of the lesions.2, 7 Bulky adducts derived from the binding of metabolically generated reactive intermediates of polycyclic aromatic hydrocarbons (PAH) to DNA 18 are particularly suitable for studying NER function-structure relationships 12 because of the availability of topologically and stereochemically different PAH-derived DNA adducts. Such studies are beginning to yield valuable insights into the features of DNA lesions that elicit efficient or inefficient NER excision efficiencies.17, 19–22
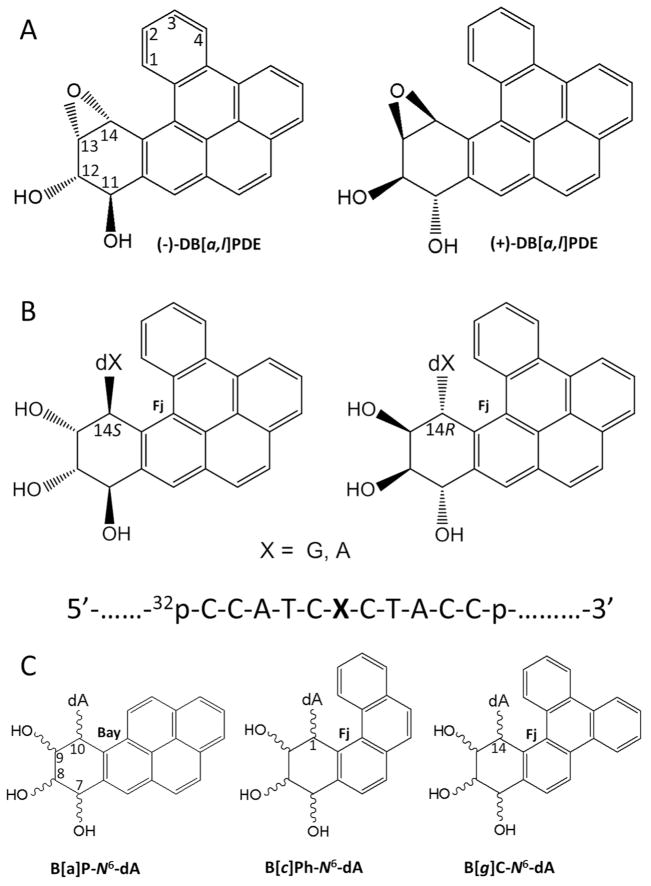
Benzo[a]pyrene (B[a]P) is the best known representative PAH carcinogen present in our environment. 23, 24 However, dibenzo[a,l]pyrene (DB[a,l]P), another PAH that is present in ~ 15-fold lower concentration in environmental complex mixtures,25 in vehicle exhaust and in cigarette smoke,26 is ~ 100 times more tumorigenic than B[a]P in rodent model systems.23, 27–33 The tumorigenic 33, 34 and mutagenic 35 activities of DB[a,l]P have been linked, in part, to their metabolic activation to the (−)-11R,12S,13S,14R diol epoxide (DB[a,l]PDE) shown in Figure 1A;36–38 it should be noted that the aldo-keto reductase mechanism is another metabolic pathway that activates fjord region PAHs to o-quinone derivatives that damage DNA by a redox cycling mechanism.39 The reactive diol epoxide intermediates react with purines in DNA by trans-addition of either N6-adenine or N2-guanine to their C14 positions. The stable DNA adducts thus formed are depicted in Figure 1B.40–43 Similarly, the (+)-7R,8S,9S,10R diol epoxide B[a]PDE also generates analogous DNA adducts with the exocyclic amino group of guanine, with minor proportions of N6-adenine adducts also being formed.44 However, the reactions of the (−)-DB[a,l]PDE enantiomer and other stereochemically identical fjord PAH diol epoxides with DNA generally yield greater proportions of adenine than guanine adducts.27, 45 Furthermore, mutations targeted predominantly to adenine derived from the reaction of DB[a,l]PDE with DNA have been observed in mouse oral tumor tissues.46 The significantly greater genotoxic activity of DB[a,l]P relative to B[a]P has been attributed to the differences in topology called bay and fjord 38, 47 defined in Figure 1C; the fjord aromatic 1,2,3,4 ring in DB[a,l]P (and analogous fjord PAH) is non-planar due to a steric clash between hydrogen atoms at the C14 and the C1 positions if it were planar. 48 By contrast, there are no steric hindrance effects in the case of B[a]P and other bay region PAH diol epoxide-guanine or adenine adducts, and the aromatic ring systems are planar and rigid. The twisted distal aromatic ring in fjord PAH dihydrodiol epoxides has been associated not only with the generally enhanced binding to N6-adenine in DNA, 45 but also to their higher tumorigenic activities. 28–31, 37, 38, 49–53
Figure 1.
(A) Structures and absolute configurations of the (−)- and (+)-anti-DB[a,l]PDE enantiomers that react with adenine or guanine in DNA. The “+” and “−” signs indicate the optical rotations of the two anti-DB[a,l]PDE enantiomer precursors. (B) Structures and configurations of the stereoisomeric S and R DB[a,l]P-dA or –dG adducts (X = A, or G); the sequence context of the 11-mer oligonucleotide sequence embedded in the double-stranded 135-mer oligonucleotide substrates is also shown. (C) Definitions of the bay regions of the stereoisomeric B[a]P-N6-dA adducts derived from (+)- or (−)B[a]PDE, and the fjord (Fj) regions of the stereoisomeric R and S adducts derived from the fjord region diol epoxides of benzo[c]phenanthrene (B[c]Ph) and benzo[g]chrysene (B[g]C) with absolute configurations analogous to those depicted in (A).
In this work we compared the NER dual incision efficiencies of the pairs of stereoisomeric R and S DB[a,l]P-N2-dG and DB[a,l]P-N6-dA adducts (Figure 1B) embedded in duplexes of otherwise identical sequence contexts incubated in cell-free human cell extracts. Remarkably, the dual efficiencies are dramatically lower by factors of ~ 35 (R adducts) and ~ 15 (S adducts) when either of the stereochemically identical DB[a,l]P residues are linked to adenine rather than to guanine in DNA. These differences are discussed in terms of the known conformational features of these DNA lesions, and the balance between DNA destabilizing and DNA stabilizing interactions between the bulky polycyclic aromatic residues and nearby DNA moieties.
EXPERIMENTAL MATERIALS AND METHODS
Racemic mixtures of the diol epoxides RSSR (−)-anti-DB[a,l]PDE and SRRS (+)-anti-DB[a,l]PDE were supplied by the National Cancer Institute (NCI) Chemical Repository for Chemical Carcinogenesis Research. This racemic mixture was incubated with the oligonucleotides 5′-CCATCXCTACC (X = G or A), and the trans-addition products were separated by repeated cycles of reversed phase HPLC described in detail elsewhere 54 and in Supporting Information, Figures S1, S2 and S3. The absolute configurations of the adenine adducts were obtained by digesting the modified oligonucleotides to the nucleoside levels and by comparing their CD spectra 54 to those of stereoisomeric R- or S-DB[a,l]P-N6-dA adducts prepared by total synthesis methods.42 A similar approach was used to match the experimental CD spectra of trans-anti-DB[a,l]P-N2-dG adducts 55 to the CD spectra of analogous adducts derived by phosphoramidite total synthesis methods. 43
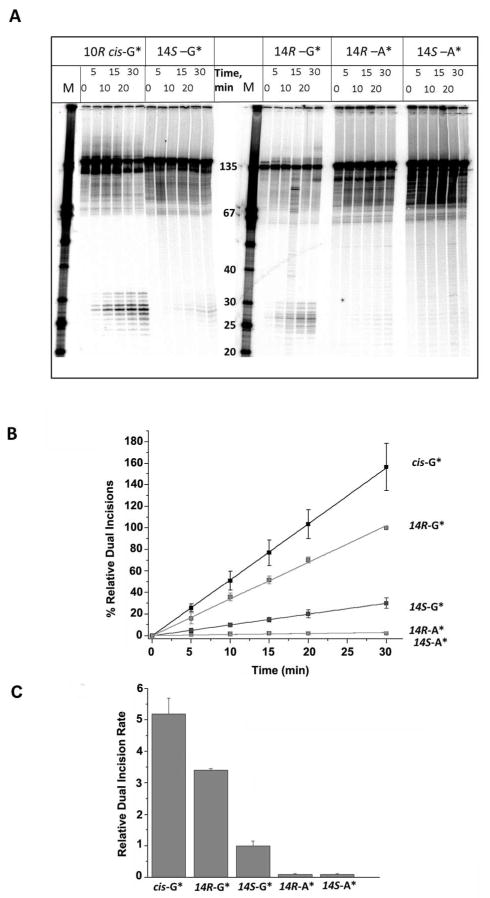
The modified or unmodified 32P-radioactively labeled 11-mer 5′-32P-CCATCXCTACC sequences with X = R- or S-DB[a,l]P-N2-dG, or R- or S-DB[a,l]P-N6-dA lesions, or G or A (as controls) positioned at the 67th position from the 5′-end of the modified strand, were generated by ligation methods as described in Supporting Information. The 135-mer DNA duplexes were incubated in human HeLa cell extracts that were prepared by methods adapted from those developed by Wood et al. 56, 57 as described earlier.13, 21 The dual incisions produced by the human NER apparatus give rise to a series of characteristic, radioactively labeled oligonucleotide fragments containing the lesion that are 24 – 32 nucleotides in length, which were visualized using 5% denaturing polyacrylamide gel electrophoresis methods. The appearance of this ladder of radioactive oligonucleotide excision fragments is characteristic of successful NER 2, 3 (Figure 2A). The fraction of doubly incised 135-mer oligonucleotide duplexes was obtained by scanning the gels using a Molecular Dynamics Storm 840 phosphorimager (GE Instruments).
Figure 2.
(A) Autoradiogram of denaturing gels showing the appearance of NER dual incision products (~ 24 – 32 nucleotides in length) after incubating 135-mer duplexes with single DNA adducts in HeLa cell extracts for the time intervals indicated. The lanes containing unmodified single-stranded oligonucleotide size markers with 3′- and 5′-OH groups at the ends and the indicated number of nucleotides in lengths are shown in the lanes marked M. The results shown were all obtained with aliquots from the same incubated sample but the gel electrophoresis was conducted in two different gels (two samples on the left, and three samples on the right, respectively). The absolute % excision in this example was 8.7% in the case of the 14R-DB[a,l]P-N2-dG adduct (30 min time point), but varied from 6 – 17% in the other four cell extracts. Other details are discussed in Supporting Information. The densitometry tracings of some of the lanes are shown in Supporting Information, Figure S4. (B) Time course of NER dual incision product formation of 24 – 32 oligonucleotide fragments bearing either the R-cis-B[a]P-N2-dG (cis-G*), R-DB[a,l]P-N2-dG (14R-G*), S DB[a,l]P-N2-dG (14S-G*), R-DB[a,l]P-N6-dA (14R-A*), or the S-DB[a,l]P-N6-dA (14S-A*) adducts. The experimental data points are averages of five independent experiments in different cell extracts, and the error bars represent the standard deviations. (C) Comparisons of initial rates of dual incisions derived from the linear plots in panel B.
Molecular modeling and molecular dynamics (MD) simulations with full analyses have been previously published in detail for the DB[a,l]P-N6-dA adducts in Cai et al. 58 For the R-DB[a,l]P-N2-dG adduct, an NMR solution structure has recently been published. 59 In the case of the S-DB[a,l]P-N2-dG adduct, the conformational features have been investigated by NMR and molecular modeling methods.60, 61
RESULTS
DB[a,l]P-N2-dG adducts are excised while the DB[a,l]P-N6-dA adducts are strongly resistant to NER in human cell extracts
In order to assess the efficiencies of NER dual incisions, the 135-mer duplexes were incubated in human HeLa cell extracts, and the fractions of duplexes incised were determined by denaturing gel electrophoresis. The focus of this work was on establishing the relative efficiencies of dual incisions of the pairs of stereoisomeric adenine and guanine adducts depicted in Figure 1B. Since the NER activities of HeLa cell extracts prepared at different times are variable, we arbitrarily designated the relative NER efficiency exhibited by 135-mer duplexes containing the 14R-DB[a,l]P-N2-dG adduct in each experiment as 100 at the 30 min incubation time point. The dual efficiencies at all other time points and with all of the other three adducts were expressed relative to this value. In this manner, we were able to achieve significant improvements in reproducibility.13 In each set of experiments, 135-mer oligonucleotide duplexes embedded in the identical sequence context containing the 10R (+)-cis-B[a]P-N2-dG adduct were included as a comparison. This lesion is an excellent substrate of human NER in the DNA sequence context used here 16, 21 and is removed by the human NER system as efficiently as the 6–4 UV light-induced photodimers,13 and the bulky AAF-C8-dG adducts.14, 62 Similar high repair efficiencies are observed for these lesions in prokaryotic systems. 63–67 The central 11-mer sequence 5′-…CCATCG*CTACC with X = G* or A* was selected in this study because the NER efficiencies of (+)-cis-, and (+)- and (−)-trans-B[a]P-N2-dG embedded in 135-mer duplexes have been previously investigated,16, 21 thus allowing for comparisons with these previous studies.
A typical set of results showing the time course of dual incisions is shown in Figure 2B. In these experiments all duplexes were identical except for the lesions that included either a single R-cis-B[a]P-N2-dG adduct, or the stereoisomeric 14S- and 14R-DB[a,l]P-N2-dG and 14S- and 14R-DB[a,l]P-N6-dA adducts (in the following, the number 14 is omitted for simplicity). The ladder of bands in the 24 – 32-mer marker region correspond to successful NER excision reactions; these short oligonucleotide fragments contain the 32P-label at the 6th phosphate group on the 5′-side of the lesion counted from the modified base. Typical densitometry tracings of some of the individual lanes are shown in Figure S4, Supporting Information. The percent oligonucleotides incised successfully can be estimated from the areas under these traces in the 24 – 32 oligonucleotide range, divided by a similar integration of the area under the entire lane that includes the non-incised strands. The time dependence of the relative quantities of dual incision products, formed as a function of incubation time thus calculated, are plotted in Figure 2B. The initial kinetics of dual excision product formation are approximately linear as a function of time and are compared in Figure 2B. The experimental data points are averages of 5 independent experiments with different cell extracts, and the error bars represent the standard deviations. The relative dual incisions for the R and S DB[a,l]P-N6-dA adducts are at the level of the background as shown in Supporting Information.
The slopes of the straight-line plots in Figure 2B are of primary interest because they represent the relative initial rates of excisions and are represented in the bar graph shown in Figure 2C. The NER efficiency of the R-DB[a,l]P-N2-dG adduct is ~ 1.5 times smaller than that of the cis-B[a]P-N2-dG adduct, but ~ 3.5 times higher than the efficiency of the stereoisomeric S-DB[a,l]P-N2-dG adduct. The NER efficiencies are strikingly different for the pairs of stereoisomeric DB[a,l]P-N2-dG and DB[a,l]P-N6-dA adducts; the latter are strongly resistant to NER (Figure 2). These results are summarized in Table 1.
Table 1.
Summary of NER susceptibilities and Tm melting points of 11-mer duplexes.
| Adduct | Relative NER efficiencya (135-mers) | Conformationb | ΔTm (°C) (11-mers)c | Tm (°C)d |
|---|---|---|---|---|
| 10R cis-B[a]P-N2-dG | 5.2 ± 0.5 | Base-displaced intercalation; W-C pairing ruptured |
−12e | 39.4 ± 0.8e |
| 14R DB[a,l]P-N2-dG | 3.4 ± 0.1 | 3′-intercalation from the minor groove; W-C pairing at lesion site disrupted; but the bases are not displaced |
−10 | 40.7 ± 0.3 |
| 14S DB[a,l]P-N2-dG | 1.0 ± 0.1 | Conformational heterogeneity; 5′-minor groove aligned with two W-C pairs ruptured; |
−2 | 49.2 ± 0.1 |
| 14R DB[a,l]P-N6-dA | ≥ 0.10 | 5′-intercalation from the major groove | +10 | 56.2 ± 0.2 |
| 14S DB[a,l]P-N6-dA | ≥ 0.07 | 3′-intercalation from the major groove | −3 | 42.7 ± 0.3 |
% min−1 (estimated from the slopes of Figure 2B, and assigning a value of 100% to the magnitude of the dual incisions at the 30 min time point for the 135-mer duplex with the 14R DB[a,l]P-N2-dG adduct). The averages are results of 5 independent experiments in different cell extract preparations.
Structural properties were obtained via high resolution NMR for the R-cis-B[a]P-N2-dG 73, and R-DB[a,l]P-N2-dG 59 adducts. Structural properties were obtained from MD simulations based on NMR studies for the the S-DB[a,l]P-N2-dG 60, 61. Structural properties for the R- and S- DB[a,l]P-N6-dA adducts 58 were obtained from MD simulations based on NMR studies for the stereochemically identical fjord region R- and S- B[c]Ph-N6-dA adducts. 18
ΔTm (°C) = Tm (Lesion-containing duplex) − Tm (Unmodified).
For the unmodified duplex of 5′-d(CCATC XCTACC)•5′-d(GGTAGYGATGG), Tm is 51 ± 0.5 °C when X = G, and Y= C; Tm is 45.7 ± 0.1 °C when X = A, and Y= T.
Data from 17
Differences in thermal destabilizations of DNA duplexes by different adducts
The balance between destabilizing and stabilizing interactions between the bulky adducts and DNA residues is reflected partly in the change in thermal stabilities of the modified DNA duplexes. In order to qualitatively assess the effect of the different DNA lesions on the thermal stabilities of the modified DNA duplexes, the melting points Tm of the 11-mer duplexes 5′-d(CCATC[G*]CTACC)•5′-d(GGTAGCGATGG) or d(CCATC[A*]CTACC)•5′-d(GGTAGTGATGG) were determined (designated as duplexes I and II, respectively). The modified and unmodified duplexes are denoted by G*, A* or G and A, respectively. The Tm values of the unmodified duplexes I and II were 51 °C and 46 °C, respectively (Table 1).
Some typical melting curves are depicted in Figure 3A. Duplex II with the S-DB[a,l]P-N6-dA adduct melts at a somewhat lower temperature than the unmodified duplex II; on the other hand, the stereoisomeric R-DB[a,l]P-N6-dA adduct strongly stabilizes duplex II. The difference in melting points, ΔTm = Tm (modified) - Tm (unmodified), is a manifestation of the effective balance between destabilizing effects associated with the lesions and the stabilizing interactions between the lesions and DNA residues. Negative values of ΔTm signify that the destabilizing effects of the lesion outweigh the stabilizing interactions, while positive values of ΔTm indicate that stabilizing interactions outweigh the destabilizing ones. The values of ΔTm for all modified duplexes studied are compared in Figure 3B, and the numerical values are shown in Table 1. The two DB[a,l]P-N2-dG adducts exhibit striking differences in ΔTm values, showing that duplex I with the R-guanine adduct is the most destabilized one (ΔTm = − 10 °C), while the S-guanine adduct is destabilized by only 2 °C; Similarly, ΔTm = − 3 °C in the case of the S-DB[a,l]P-N6-dA adduct with the same stereochemistry. However, in contrast to the strongly destabilizing R-DB[a,l]P-N2-dG adduct, the ΔTm value of the R-DB[a,l]P-N6-dA adduct with the identical stereochemistry is + 10 °C, in excellent agreement with the ΔTm of + 8 °C observed in a different sequence context. 54 In comparison, all known B[a]P-N6-dA 8, 22, 68 and B[a]P-N2-dG adducts 17, 18 in full duplexes, including the R-cis-B[a]P-N2-dG adduct (Figure 3B), are thermally destabilizing.
Figure 3.
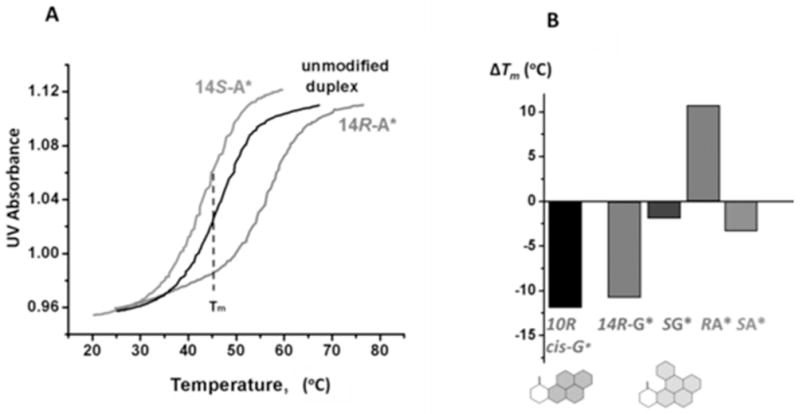
(A) Comparisons of normalized oligonucleotide melting curves of the unmodified and modified 11-mer duplexes with R DB[a,l]P-N6-dA and S DB[a,l]P-N6-dA adducts (X) in the sequence context 5′-d(CCATCXCTACC)•5′-d(GGTAGYGATGG) (Y = T). All measurements were conducted in 100 mM NaCl, 20mM Na2HPO4 solutions, DNA duplex concentration 4.9 μM, rate of temperature increase: 0.33 °C min−1 (B) Summary of ΔTm = Tm(modified) − Tm(unmodified) values of DB[a,l]P-derived stereoisomeric N6-dA (Y = T) and N2-dG (Y = C) adducts positioned at nucleotide X in the 11-mer duplexes. The ΔTm value of the 11-mer duplex with the R cis-B[a]P-N2-dG adduct is shown in black.
DISCUSSION
Replacing the modified base from guanine to adenine while maintaining the identical stereochemical properties of the fjord DB[a,l]P substituents, leads to a dramatic reduction in the NER efficiencies. The stereoisomeric R and S DB[a,l]P-N6-dA lesions exhibit efficiencies that are ~ 35 and ~15 times smaller than those of the R-DB[a,l]P-N2-dG and the stereoisomeric S-DB[a,l]P-N2-dG adducts, respectively. The NER signals for the two DB[a,l]P-N6-dA are close to the background levels (Supporting Information) and therefore cannot be accurately determined. Other fjord PAH-derived N6-dA adducts that differ in the number of aromatic rings, e.g., derived from benzo[c]phenanthrene (B[c]Ph, and benzo[g]chrysene (B[g]C (Figure 1C), have also been reported to be resistant to NER. 69 In order to gain insights into the underpinnings of these differences in NER activities, it is of interest to examine the known structural properties of these lesions and the extent of stabilization/destabilization of the DNA duplexes that they induce.
The NER-resistant R and S DB[a,l]P-N6-dA adducts
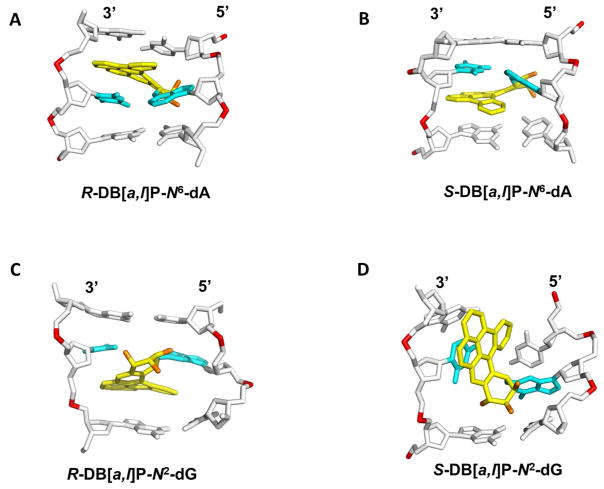
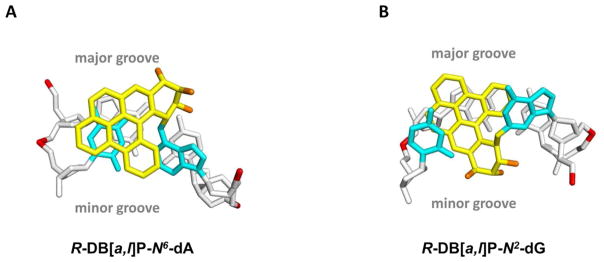
The NER resistance of the fjord PAH diol epoxide-N6-dA adducts can be interpreted in terms of conformations of the fjord B[c]Ph-N6-dA 70 and B[g]C-N6-dA adducts 71 that have been studied by solution NMR methods. The structures of the DB[a,l]P-N6-dA lesions were characterized by molecular modeling/dynamics methods based on the NMR studies of the B[c]Ph-N6-dA adducts.58 Figure 4A shows the R-DB[a,l]P-N6-dA structure, and Figure 4B shows the S-DB[a,l]P-N6-dA structure. All of these fjord PAH-N6-dA (A*) adducts adopt classical intercalation-type conformations without disruption of any of the Watson-Crick base pairs including the A*:T pair. Figure 5A presents a view looking down the helix axis for the R-DB[a,l]P-N6-dA showing the Watson-Crick alignment of the lesion-containing pair, and Figure S7-A, Supporting Information, shows a stereo view. In all cases, the adenine adducts with R absolute configurations at the linkage site are intercalated on the 5′-side of the modified adenine residue, while the S stereoisomers are intercalated on the 3′-side. In both cases intercalation occurs from the spacious major groove and the aromatic rings of the lesion are stacked in the intercalation pocket that is produced by local stretching and unwinding of the DNA duplex.58 The balance between destabilizing and stabilizing effects of these R- and S-lesions on double-stranded DNA was examined by molecular dynamics (MD) simulation methods.19 The local steric hindrance effects in the case of the S-adducts is partially relieved by local untwisting of adjacent base pairs. This effect results in lowered van der Waals stacking interactions between the DB[a,l]P residue and DNA bases, compared to the better-stacked R case. These characteristics are consistent with the observed differences in the ΔTm values of − 3 and + 10 °C for the S- and R- DB[a,l]P-N6-dA adducts, respectively, and previous observations that other 3′-intercalated PAH-N6-adenine adducts with S absolute configuration at the linkage site are generally more destabilizing than the stereoisomeric R adducts.22, 54, 68, 72 In spite of the differences in the thermal stabilities, both the R- and the S-DB[a,l]P-N6-dA adducts are similarly NER-resistant (discussed below).
Figure 4.
Best representative structures from MD simulations for the (A) R and (B) S-DB[a,l]P-N6-dA 58 adducts and NMR solution structures for the (C) R-DB[a,l]P-N2-dG 59 and (D) S-DB[a,l]P-N2-dG 60 adducts. The central trimers of the duplex 11-mers are shown. The full duplex 11-mers are shown in Supporting Information Figure S6. The view is looking into the minor groove. For the DB[a,l]P moiety, the carbon atoms are colored yellow and the oxygen atoms orange. The damaged base pairs are colored cyan and the DNA duplexes are colored white, except for the phosphorus atoms, which are colored red. Hydrogen atoms are not displayed for clarity. Coordinates for (D) are provided in Supporting Information Table S1.
Figure 5.
(A) View looking down the helix axis of the (A6*:T17)•(C7:G16) segment in the R-DB[a,l]P-N6-dA adduct. (B) View looking down the helix axis of the (G6*:C17)•(C7:G16) segment in the R-DB[a,l]P-N2-dG adduct. The view is in the 5′ to 3′ direction along the lesion-containing strand. Stereo views of (A) and (B) are provided in Supporting Information Figure S7.
The highly NER-susceptible fjord R-DB[a,l]P-N2-dG and the bay R-CIS-B[a]P-N2-dG adducts
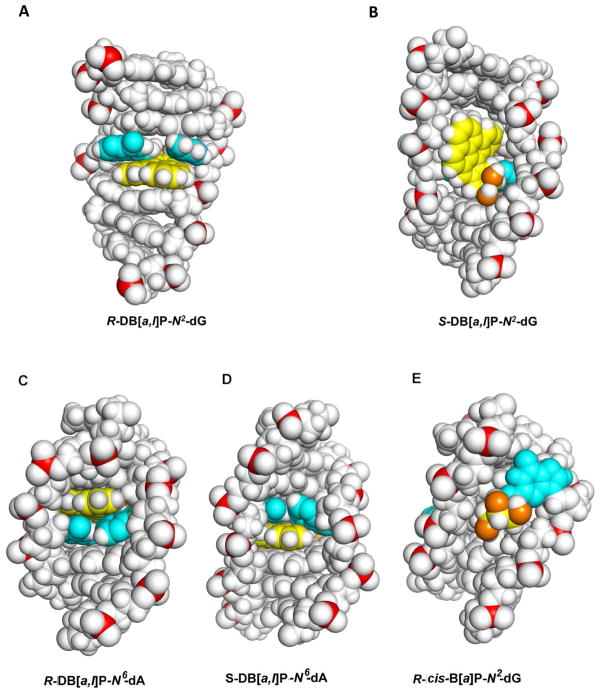
In contrast to the repair-resistant N6-adenine adducts, the guanine adducts considered in this study are modest to excellent substrates of NER. The best NER substrate is the R-cis-B[a]P-N2-dG adduct that assumes a base-displaced/intercalated adduct conformation, with the modified guanine displaced into the minor groove and the partner cytosine into the major groove, and the B[a]P residue intercalated within the space normally occupied by the displaced G:C base pair 73 (Figure S5-A, Supporting Information). The R-DB[a,l]P-N2-dG adduct is the next best substrate, and is characterized by the highest NER efficiency among the four DB[a,l]P adducts studied here. Recent solution NMR structural studies 59 revealed that the R-DB[a,l]P-N2-dG adduct adopts an intercalative conformation, but without base displacement as in the case of the stereochemically identical R-cis-B[a]P-N2-dG adduct. Figure 4C shows the structure of the R-DB[a,l]P-N2-dG adduct. The conformation of the R-DB[a,l]P-N2-dG adduct differs in key features from that of the stereochemically analogous R-DB[a,l]P-N6-dA and related fjord PAH-N6-dA adducts. Specifically, the intercalation in the former is from the narrow minor groove on the 3′-side of the damaged guanine. In order to accommodate this bulky lesion with five aromatic rings in the intercalation pocket, the DB[a,l]PDE-substituted guanine and its partner cytosine base are separated from one another so that Watson-Crick hydrogen bonding is lost at this base pair; this stretching is accompanied by severe opening of both the major groove and the minor groove.59 Figure 5B shows a view for this adduct looking down the helix axis which reveals this stretch in the lesion-containing base pair and Figure S7-B, Supporting Information, shows a stereo view. A CPK rendering is shown in Figure 6A and a stereo view in Figure S8-A, Supporting Information. It is notable that the modified guanine residue and its partner cytosine are not displaced as they are in the R-cis-B[a]P-N2-dG adduct 74(Figure S5-A, Supporting Information), and these bases retain modest stacking interactions with the DB[a,l]P aromatic ring system. By contrast, in the case of the R-DB[a,l]P-N6-dA adduct described above, the minor and major grooves are less distorted, Watson-Crick base pairing is maintained at the site of the lesion and at adjacent base pairs, and stacking between the DB[a,l]P aromatic rings and adjacent bases is strong.
Figure 6.
(A) CPK view into the major groove of the R-DB[a,l]P-N2-dG adduct structure. (B) CPK view into the minor groove of the S-DB[a,l]P-N2-dG adduct structure. (C) CPK view into the minor groove of the R-DB[a,l]P-N6-dA adduct structure (D) CPK view into the minor groove of the S-DB[a,l]P-N6-dA adduct structure (E) CPK view into the minor groove of the R-cis-B[a]P-N2-dG adduct structure. Central 7-mers are shown. Stereo views are provided in Supporting Information Figure S8. Color code is the same as in Figure 4.
Both the R-DB[a,l]P-N2-dG and the R-(+)-cis-B[a]P-N2-dG adducts strongly destabilize double-stranded DNA, which is indicated by the relatively large ΔTm values of −10 °C and −12 °C, respectively (Figure 3B). The shared features of ruptured Watson-Crick base pairing, intercalation from the minor groove, and similar degrees of thermal destabilization are consistent with their high NER susceptibilities (Figure 2C). By contrast, the NER efficiency of the stereoisomeric R-(−)-trans-B[a]P-N2-dG adduct is considered to be modest in comparison, being ~ 5 times smaller than that of the R-(+)-cis-B[a]P-N2-dG adduct, 16, 21 and it has a very different conformation: its aromatic ring system is not intercalated but is positioned in the minor groove pointing towards the 3′-end of the modified strand without rupturing any of the Watson-Crick base pairs (Figure S5-B, Supporting Information). 75
The S-DB[a,l]P-N2-dG adduct is characterized by a modest susceptibility to NER
NMR structural studies show that the bulky aromatic ring system of the S-DB[a,l]P-N2-dG adduct resides mostly in a widened minor groove, and that a minor conformation with poorly resolved NMR resonances is also present.61 MD simulations based on this NMR data 60 show that the aromatic rings of the DB[a,l]P are positioned in the minor groove, directed 5′ along the modified strand, where they interact strongly by van der Waals mechanisms with DNA residues on both strands; the bulky aromatic five-ring system forces a widening of the minor groove and causes rupture of the Watson-Crick hydrogen bonds of the two C:G base pairs, which is evident from the 2D-NMR data.61 The minor groove structure of the S-DB[a,l]P-N2-dG adduct is shown in Figures 4D and 6B; the latter shows a CPK rendering which highlights the interactions of the aromatic five-ring system with the minor groove wall and the distortion of the minor groove. Figure S8-B, Supporting Information shows a stereo view. Coordinates are provided in Supporting Information Table S1. Thus, while the S-DB[a,l]P-N2-dG adduct is highly distorting, the stabilizing contacts with the minor groove tend to compensate for the loss of hydrogen bonding and other distorting interactions. This conclusion is supported by the small decrease in the melting point associated with the 11-mer duplex (ΔTm ~ −2 °C) and the relatively modest repair susceptibility (Figure 2). Overall, we hypothesize that such minor groove conformations are associated with similar modest NER efficiencies; the analogous minor groove 76 S-(+)-trans-B[a]P-N2-dG adduct elicits a similar modest NER response.16, 21 The NMR solution structure of the S-(+)-trans-B[a]P-N2-dG is shown in Figure S5-C, Supporting Information. The NER efficiency of this S-(+)-trans-B[a]P-N2-dG adduct is ~ 5 times smaller than that of the R cis-B[a]P-N2-dG adduct 21, and is similar to the efficiency of the S-DB[a,l]P-N2-dG adduct, as already mentioned; the efficiency of the latter is ~ 3.5 times smaller than that of the R-adduct (Figure 2). An important insight that can be drawn from the similar NER efficiencies of the two stereochemically analogous minor groove adducts, the S-(+)-trans-B[a]P-N2-dG (Figure S5-C, Supporting Information) and S-DB[a,l]P-N2-dG adducts (Figure 4D), is that the loss of two hydrogen bonds in the S-DB[a,l]P-N2-dG adduct seems to be compensated by enhanced van der Waals interactions with adjacent DNA residues. The two ruptured base pairs may provide a local thermodynamic signal for the modest repair since flipping of two bases is a component of the recognition mechanism by Rad4-Rad23;77 however, stacking interactions between the lesion and the groove wall likely present some hindrance for β-hairpin intrusion between the two strands at the lesion site, discussed below.
The balance between DNA lesion-induced destabilizing and stabilizing interactions and NER
The most NER-resistant R-DB[a,l]P-N6-dA adduct exhibits a ΔTm of +10 °C, while the best repaired R-DB[a,l]P-N2-dG adduct has a ΔTm of −10 °C. Clearly, helix destabilizing factors are dominant in the case of the G adduct, while stabilizing effects are dominant in the case of the A adduct. This difference is well correlated with the high NER efficiency of the G-adduct, and the NER-resistance of the A-adduct. On the other hand, both the R- and the S-N6-dA adducts exhibit similar NER susceptibilities, but there is a large difference in the ΔTm, values (−3 °C and +10 °C, respectively), for reasons discussed above. The melting of modified DNA duplexes involves a global cooperative dissociation of the entire duplex and base sequence-dependent disruption of hydrogen bonding and base stacking interactions. 78, 79 However, based on the crystal structure of Rad4-Rad23, the yeast homolog 77 of the human NER recognition factor XPC, it is plausible that the recognition of the DNA lesions is a local phenomenon that presumably involves the insertion of a β-hairpin of the NER recognition factor between the two strands at the lesion site; this is accompanied by the flipping out of the helix of two bases opposite the lesion in the complementary strand and their interactions with amino acids in the protein.77 The present results support the hypothesis that these critical steps can occur once the local balance between destabilizing and stabilizing interactions exceeds a certain threshold that can trigger these events and thus lead to stabilization of the XPC-DNA complex, the first 80 and rate-limiting step 81 in the multistep NER process. However, it is possible that the affinity of binding of XPC-RAD23B or the characteristics of the DNA complexes may be different in the case of the DB[a,l]P-N6-dA and -dG adducts, and that downstream events such as unwinding or cleavage also may play important roles in conferring NER resistance. 4, 5, 82, 83. Another consideration in repair resistance may be the possible role of repair shielding proteins such as HMGB1 that recognize and bind DNA lesions and shield them from the NER machinery. 84 While a quantitative correlation between NER efficiencies and ΔTm values cannot always be expected, parallel trends between thermal destabilization and NER susceptibility have been observed in the case of some lesions that belong to the same conformational families.19, 22
There is an increasing realization that DNA lesions that cause a minimal decrease, or even an enhancement in the stabilities of double-stranded DNA, may also be resistant to DNA repair. The B[a]P diol epoxide-derived N2-dG adducts are susceptible to NER in normal duplexes, but in sequence contexts that involve the deletion of one base opposite the lesion in the complementary strand, the same lesions are fully resistant to NER 16, 69 and the ΔTm value is large and positive. 17 The R-fjord PAH diol epoxide-N6-adenine adduct described here is yet another example of this behavior, but the modified duplex is fully complementary and also exhibits a large positive ΔTm value. Other DNA lesions that exhibit similar behavior include those derived from aristolochic acid,85, 86 AAF-N2-dG, 87, 88 3-benzanthrone, 86, 89 and potentially some of the thermodynamically stable Fapy-dG adducts derived from aflatoxin B1. 90 More accurate evaluations of the local thermodynamic effects that are relevant to NER but are not quantitatively correlated with global melting stabilities of DNA duplexes derived from Δ Tm measurements are amenable to temperature-dependent studies by NMR methods. 86, 91
Biological implications: NER resistance and tumorigenicity
The relative in vitro NER efficiencies reported here provide novel insights, at a molecular level, into the startling poor susceptibility to NER of the DB[a,l]P-N6-dA adducts alluded to earlier in the case of a different sequence context, 69 and the modest to strong NER susceptibilities of the DB[a,l]P-N2-dG lesions investigated in the present work. It has been reported that the levels of stable covalent DNA adduct formation are up to ten times greater in the case of the major (−)-anti-DB[a,l]P diol epoxide than in the case of the stereochemically identical bay region diol epoxide (+)-anti-BP[a]DE. 92 Indeed, Dreij et al.92 and Lagerqvist et al. 93 found that fjord DB[a,l]P-DNA adducts are overall more persistent in A549 lung epithelial and hamster cell lines, respectively, than B[a]P-DNA adducts. It has been shown that the genotoxic activities of similar fjord PAH diol epoxides is well correlated with DNA adduct levels in V79 cells35, 94 and that mutations are observed in higher proportions at adenines than in the case of B[a]P-derived adducts. Analogous correlations between DNA adduct levels and tumorigenicity have been reported in various mammalian systems. 30, 95, 96 Zhang et al. found that DB[a,l]P gives rise to significant and persistent S-DB[a,l]P-N6-dA adducts in the oral tissues of mice that are correlated with the formation of mutations 97 and tumorigenesis in mice. 33, 53 However guanine adducts may also contribute to the tumorigenicity of DB[a,l]P in the light of their established mutagenicities, 35 and the persistence of guanine adducts of unknown stereochemistry in cells.98 Future studies are needed to establish whether the remarkable differences in NER susceptibilities are correlated with the interactions of these lesions with XPC-HR23B and the properties of the DNA-protein complexes formed.
In summary, the persistence of mutagenic 27, 99, 100 stable fjord region adducts in various cells and tissues 34, 101, 102 and the correlation with their high tumorigenicities, 33, 53 suggests that the higher genotoxic activity of DB[a,l]P, relative to B[a]P, may be partly related to the formation of NER-resistant, and thus persistent DB[a,l]P-derived adenine adducts.
Supplementary Material
Acknowledgments
Funding Sources
This research was supported by the National Cancer Institute (NCI) of the National Institutes of Health under award numbers RO1-CA099194 (N.E.G.) and RO1-CA28038 (S.B.), and in part by the NCI Chemical Carcinogen Storage Facility at Pennsylvania State University (NCI Contract NO2-CB-81013-74 to S.A.). The computational infrastructure and systems management was partially supported by NIH Grant RO1-CA75449 to S.B.. Components of this work were conducted in the Shared Instrumentation Facility at NYU that was constructed with support from the Research Facilities Improvement Grant C06RR-16572 from the National Center for Research Resources, NIH.
The DB[a,l]P diol epoxides were obtained from the National Cancer Institute (NCI) Chemical Repository for Chemical Carcinogenesis Research. This work used computational resources of the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant MCB060037. We thank NYU-ITS for its support and use of computational resources. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
ABBREVIATIONS
- NER
nucleotide excision repair
- PAHs
polycyclic aromatic hydrocarbons
- DB[a,l]P
dibenzo[a,l]pyrene
- B[a]P
benzo[a]pyrene
- B[c]Ph
benzo[c]phenanthrene
- B[g]C
benzo[g]chrysene
- DE
diol epoxide
- (+)-B[a]PDE
(+)-(7R,8S,9S,10R)-7,8-dihydroxy-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene
- (−)-DB[a,l]
PDE, (−)-(11R,12S)-dihydroxy-(13S,14R)-epoxy-11,12,13,14-tetrahydrodibenzo[a,l]pyrene
- (+)-DB[a,l]
PDE, (+)-(11S,12R)-dihydroxy-(13R,14S)-epoxy-11,12,13,14-tetrahydrodibenzo[a,l]pyrene
- R-DB[a,l]
P-N2-dG, 14R (+)-trans-anti-DB[a,l]P-N2-dG
- S-DB[a,l]
P-N2-dG, 14S (−)-trans-anti-DB[a,l]P-N2-dG
- R-DB[al]
P-N6-dA, 14R (+)-trans-anti-DB[a,l]P-N6-dA
- S-DB[a,l]
P-N6-dA, 14S (−)-trans-anti-DB[a,l]P-N6-dA
- R-cis-B[a]P-N2-dG
10R (+)-cis-anti-B[a]P-N2-dG
- R-B[a]P-N2-dG
10R (−)-trans-anti-B[a]P-N2-dG
- S-B[a]P-N2-dG
10S (+)-trans-anti-B[a]P-N2-dG
- R-B[a]P-N6-dA
10R (−)-trans-anti-B[a]P-N6-dA
- S-B[a]P-N6-dA
10S (+)-trans-anti-B[a]P-N6-dA
- MD
molecular dynamics
Footnotes
Supporting Information available: Supporting Methods and Supporting Figures S1–S8. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 2.Gillet LC, Scharer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem Rev. 2006;106:253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 3.Reardon JT, Sancar A. Nucleotide excision repair. Prog Nucleic Acid Res Mol Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 4.Peng Y, Wang H, Santana dos Santos L, Kisker C, Van Houten B. Nucleotide excision repair from bacteria to humans: Structure–function studies. In: Penning TM, editor. Chemical Carcinogenesis, Current Cancer Research. Chapter 13. Springer Science+Business Media, LLC; 2011. pp. 267–296. [Google Scholar]
- 5.Scharer OD. Mechanisms of base excision repair and nucleotide excision repair In. In: Geacintov NE, Broyde S, editors. The Chemical Biology of DNA Damage. Chapter 11. Wiley-VCH; Weinheim, Germany: 2010. pp. 239–260. [Google Scholar]
- 6.Gunz D, Hess MT, Naegeli H. Recognition of DNA adducts by human nucleotide excision repair. Evidence for a thermodynamic probing mechanism. J Biol Chem. 1996;271:25089–25098. doi: 10.1074/jbc.271.41.25089. [DOI] [PubMed] [Google Scholar]
- 7.Wood RD. DNA damage recognition during nucleotide excision repair in mammalian cells. Biochimie. 1999;81:39–44. doi: 10.1016/s0300-9084(99)80036-4. [DOI] [PubMed] [Google Scholar]
- 8.Yan S, Wu M, Buterin T, Naegeli H, Geacintov NE, Broyde S. Role of base sequence context in conformational equilibria and nucleotide excision repair of benzo[a]pyrene diol epoxide-adenine adducts. Biochemistry. 2003;42:2339–2354. doi: 10.1021/bi0270081. [DOI] [PubMed] [Google Scholar]
- 9.Cai Y, Kropachev K, Xu R, Tang Y, Kolbanovskii M, Kolbanovskii A, Amin S, Patel DJ, Broyde S, Geacintov NE. Distant neighbor base sequence context effects in human nucleotide excision repair of a benzo[a]pyrene-derived DNA lesion. J Mol Biol. 2010;399:397–409. doi: 10.1016/j.jmb.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai Y, Patel DJ, Broyde S, Geacintov NE. Base sequence context effects on nucleotide excision repair. J Nucleic Acids. 2010;2010 doi: 10.4061/2010/174252. Article ID 174252, 9 pages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai Y, Patel DJ, Geacintov NE, Broyde S. Dynamics of a benzo[a]pyrene-derived guanine DNA lesion in TGT and CGC sequence contexts: enhanced mobility in TGT explains conformational heterogeneity, flexible bending, and greater susceptibility to nucleotide excision repair. J Mol Biol. 2007;374:292–305. doi: 10.1016/j.jmb.2007.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai Y, Kropachev K, Kolbanovskiy M, Kolbanovskiy A, Broyde S, Patel DJ, Geacintov NE. Recognition and removal of bulky DNA lesions by the nucleotide excision repair system In The Chemical Biology of DNA. In: Geacintov NEaBS., editor. Damage. Chapter 12. Wiley-VCH; Weinheim, Germany: 2010. pp. 261–298. [Google Scholar]
- 13.Kropachev K, Kolbanovskii M, Cai Y, Rodriguez F, Kolbanovskii A, Liu Y, Zhang L, Amin S, Patel D, Broyde S, Geacintov NE. The sequence dependence of human nucleotide excision repair efficiencies of benzo[a]pyrene-derived DNA lesions: insights into the structural factors that favor dual incisions. J Mol Biol. 2009;386:1193–1203. doi: 10.1016/j.jmb.2008.12.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mu H, Kropachev K, Wang L, Zhang L, Kolbanovskiy A, Kolbanovskiy M, NEG, Broyde S. Nucleotide excision repair of 2-acetylaminofluorene- and 2-aminofluorene-(C8)-guanine adducts: molecular dynamics simulations elucidate how lesion structure and base sequence context impact repair efficiencies. Nucleic Acids Res. 2012;19:9675–9690. doi: 10.1093/nar/gks788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buterin T, Meyer C, Giese B, Naegeli H. DNA quality control by conformational readout on the undamaged strand of the double helix. Chem Biol. 2005;12:913–922. doi: 10.1016/j.chembiol.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 16.Hess MT, Gunz D, Luneva N, Geacintov NE, Naegeli H. Base pair conformation-dependent excision of benzo[a]pyrene diol epoxide-guanine adducts by human nucleotide excision repair enzymes. Mol Cell Biochem. 1997;17:7069–7076. doi: 10.1128/mcb.17.12.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reeves DA, Mu H, Kropachev K, Cai Y, Ding S, Kolbanovskiy A, Kolbanovskiy M, Chen Y, Krzeminski J, Amin S, Patel DJ, Broyde S, Geacintov NE. Resistance of bulky DNA lesions to nucleotide excision repair can result from extensive aromatic lesion-base stacking interactions. Nucleic Acids Res. 2011;39:8752–8764. doi: 10.1093/nar/gkr537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geacintov NE, Cosman M, Hingerty BE, Amin S, Broyde S, Patel DJ. NMR solution structures of stereoisometric covalent polycyclic aromatic carcinogen-DNA adduct: principles, patterns, and diversity. Chem Res Toxicol. 1997;10:111–146. doi: 10.1021/tx9601418. [DOI] [PubMed] [Google Scholar]
- 19.Cai Y, Geacintov NE, Broyde S. Nucleotide excision repair efficiencies of bulky carcinogen-DNA adducts are governed by a balance between stabilizing and destabilizing interactions. Biochemistry. 2012;51:1486–1499. doi: 10.1021/bi201794x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai Y, Patel DJ, Geacintov NE, Broyde S. Differential nucleotide excision repair susceptibility of bulky DNA adducts in different sequence contexts: hierarchies of recognition signals. J Mol Biol. 2009;385:30–44. doi: 10.1016/j.jmb.2008.09.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mocquet V, Kropachev K, Kolbanovskiy M, Kolbanovskiy A, Tapias A, Cai Y, Broyde S, Geacintov NE, Egly JM. The human DNA repair factor XPC-HR23B distinguishes stereoisomeric benzo[a]pyrenyl-DNA lesions. The EMBO J. 2007;26:2923–2932. doi: 10.1038/sj.emboj.7601730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geacintov NE, Broyde S, Buterin T, Naegeli H, Wu M, Yan S, Patel DJ. Thermodynamic and structural factors in the removal of bulky DNA adducts by the nucleotide excision repair machinery. Biopolymers. 2002;65:202–210. doi: 10.1002/bip.10239. [DOI] [PubMed] [Google Scholar]
- 23.Bostrom CE, Gerde P, Hanberg A, Jernstrom B, Johansson C, Kyrklund T, Rannug A, Tornqvist M, Victorin K, Westerholm R. Cancer risk assessment, indicators, and guidelines for polycyclic aromatic hydrocarbons in the ambient air. Environ Health Perspect. 2002;110(Suppl 3):451–488. doi: 10.1289/ehp.110-1241197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luch A. Nature and nurture – Lessons from chemical carcinogenesis. Nat Rev Cancer. 2005;5:113–125. doi: 10.1038/nrc1546. [DOI] [PubMed] [Google Scholar]
- 25.Hutzler C, Luch A, Filser JG. Analysis of carcinogenic polycyclic aromatic hydrocarbons in complex environmental mixtures by LC-APPI-MS/MS. Anal Chim Acta. 2011;702:218–224. doi: 10.1016/j.aca.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 26.Seidel A, Frank H, Behnke A, Schneider D, Jacob J. Determination of dibenzo[a,l]pyrene and other fjord region PAH isomers with MW 302 in environmental samples. Polycyclic Aaromat Compd. 2004;24:759–772. [Google Scholar]
- 27.Luch A. On the impact of the molecule structure in chemical carcinogenesis. EXS. 2009;99:151–179. doi: 10.1007/978-3-7643-8336-7_6. [DOI] [PubMed] [Google Scholar]
- 28.Amin S, Desai D, Dai W, Harvey RG, Hecht SS. Tumorigenicity in newborn mice of fjord region and other sterically hindered diol epoxides of benzo[g]chrysene, dibenzo[a,l]pyrene (dibenzo[def,p]chrysene), 4H-cyclopenta[def]chrysene and fluoranthene. Carcinogenesis. 1995;16:2813–2817. doi: 10.1093/carcin/16.11.2813. [DOI] [PubMed] [Google Scholar]
- 29.Amin S, Krzeminski J, Rivenson A, Kurtzke C, Hecht SS, El-Bayoumy K. Mammary carcinogenicity in female CD rats of fjord region diol epoxides of benzo[c]phenanthrene, benzo[g]chrysene and dibenzo[a,l]pyrene. Carcinogenesis. 1995;16:1971–1974. doi: 10.1093/carcin/16.8.1971. [DOI] [PubMed] [Google Scholar]
- 30.Prahalad AK, Ross JA, Nelson GB, Roop BC, King LC, Nesnow S, Mass MJ. Dibenzo[a,l]pyrene-induced DNA adduction, tumorigenicity, and Ki-ras oncogene mutations in strain A/J mouse lung. Carcinogenesis. 1997;18:1955–1963. doi: 10.1093/carcin/18.10.1955. [DOI] [PubMed] [Google Scholar]
- 31.Cavalieri EL, Higginbotham S, RamaKrishna NV, Devanesan PD, Todorovic R, Rogan EG, Salmasi S. Comparative dose-response tumorigenicity studies of dibenzo[alpha,l]pyrene versus 7,12-dimethylbenz[alpha]anthracene, benzo[alpha]pyrene and two dibenzo[alpha,l]pyrene dihydrodiols in mouse skin and rat mammary gland. Carcinogenesis. 1991;12:1939–1944. doi: 10.1093/carcin/12.10.1939. [DOI] [PubMed] [Google Scholar]
- 32.Cavalieri EL, Higginbotham S, Rogan EG. Dibenzo[a,l]pyrene: the most potent carcinogenic aromatic hydrocarbon. Polycyclic Aromat Compd. 1994;6:177–183. [Google Scholar]
- 33.Zhang SM, Chen KM, Aliaga C, Sun YW, Lin JM, Sharma AK, Amin S, El-Bayoumy K. Identification and quantification of DNA adducts in the oral tissues of mice treated with the environmental carcinogen dibenzo[a,l]pyrene by HPLC-MS/MS. Chem Res Toxicol. 2011;24:1297–1303. doi: 10.1021/tx200188j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melendez-Colon VJ, Luch A, Seidel A, Baird WM. Cancer initiation by polycyclic aromatic hydrocarbons results from formation of stable DNA adducts rather than apurinic sites. Carcinogenesis. 1999;20:1885–1891. doi: 10.1093/carcin/20.10.1885. [DOI] [PubMed] [Google Scholar]
- 35.Leavitt SA, George MH, Moore T, Ross JA. Mutations induced by benzo[a]pyrene and dibenzo[a,l]pyrene in lacI transgenic B6C3F1 mouse lung result from stable DNA adducts. Mutagenesis. 2008;23:445–450. doi: 10.1093/mutage/gen033. [DOI] [PubMed] [Google Scholar]
- 36.Conney AH. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons: G. H. A. Clowes Memorial Lecture. Cancer Res. 1982;42:4875–4917. [PubMed] [Google Scholar]
- 37.Gill HS, Kole PL, Wiley JC, Li KM, Higginbotham S, Rogan EG, Cavalieri EL. Synthesis and tumor-initiating activity in mouse skin of dibenzo[a,l]pyrene syn- and anti-fjord-region diolepoxides. Carcinogenesis. 1994;15:2455–2460. doi: 10.1093/carcin/15.11.2455. [DOI] [PubMed] [Google Scholar]
- 38.Ralston SL, Lau HH, Seidel A, Luch A, Platt KL, Baird WM. The potent carcinogen dibenzo[a,l]pyrene is metabolically activated to fjord-region 11,12-diol 13,14-epoxides in human mammary carcinoma MCF-7 cell cultures. Cancer Res. 1994;54:887–890. [PubMed] [Google Scholar]
- 39.Zhang L, Jin Y, Huang M, Penning TM. The Role of Human Aldo-Keto Reductases in the Metabolic Activation and Detoxication of Polycyclic Aromatic Hydrocarbons: Interconversion of PAH Catechols and PAH o-Quinones. Front in Pharmacol. 2012;3:193. doi: 10.3389/fphar.2012.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li KM, George M, Gross ML, Lin CH, Jankowiak R, Small GJ, Seidel A, Kroth H, Rogan EG, Cavalieri EL. Structure elucidation of the adducts formed by fjord region Dibenzo[a,l]pyrene-11,12-dihydrodiol 13,14-epoxides with deoxyguanosine. Chem Res Toxicol. 1999;12:778–788. doi: 10.1021/tx980234k. [DOI] [PubMed] [Google Scholar]
- 41.Li KM, George M, Gross ML, Seidel A, Luch A, Rogan EG, Cavalieri EL. Structure elucidation of the adducts formed by fjord-region Dibenzo[a,l]pyrene 11,12-dihydrodiol 13,14-epoxides and deoxyadenosine. Chem Res Toxicol. 1999;12:758–767. doi: 10.1021/tx980197x. [DOI] [PubMed] [Google Scholar]
- 42.Yagi H, Frank H, Seidel A, Jerina DM. Revised assignment of absolute configuration of the cis- and trans-N6-deoxyadenosine adducts at C14 of (+/−)-11beta,12alpha-dihydroxy-13alpha,14alpha-epoxy-11,12,13,14-tetrahydr odibenzo[a,l]pyrene by stereoselective synthesis. Chem Res Toxicol. 2008;21:2379–2392. doi: 10.1021/tx800268f. [DOI] [PubMed] [Google Scholar]
- 43.Wang B, Sayer JM, Yagi H, Frank H, Seidel A, Jerina DM. Facile interstrand migration of the hydrocarbon moiety of a dibenzo[a,l]pyrene 11,12-diol 13,14-epoxide adduct at N2 of deoxyguanosine in a duplex oligonucleotide. J AM Chem Soc. 2006;128:10079–10084. doi: 10.1021/ja0608038. [DOI] [PubMed] [Google Scholar]
- 44.Cheng SC, Hilton BD, Roman JM, Dipple A. DNA adducts from carcinogenic and noncarcinogenic enantiomers of benzo[a]pyrene dihydrodiol epoxide. Chem Res Toxicol. 1989;2:334–340. doi: 10.1021/tx00011a011. [DOI] [PubMed] [Google Scholar]
- 45.Ralston SL, Seidel A, Luch A, Platt KL, Baird WM. Stereoselective activation of dibenzo[a,l]pyrene to (−)-anti (11R,12S,13S,14R)- and (+)-syn(11S,12R,13S,14R)-11,12-diol-13,14- epoxides which bind extensively to deoxyadenosine residues of DNA in the human mammary carcinoma cell line MCF-7. Carcinogenesis. 1995;16:2899–2907. doi: 10.1093/carcin/16.12.2899. [DOI] [PubMed] [Google Scholar]
- 46.Chen KM, Guttenplan JB, Zhang SM, Aliaga C, Cooper TK, Sun YW, Deltondo J, Kosinska W, Sharma AK, Jiang K, Bruggeman R, Ahn K, Amin S, El-Bayoumy K. Mechanisms of oral carcinogenesis induced by dibenzo[a,l]pyrene: An environmental pollutant and a tobacco smoke constituent. Int J Cancer. 2013 doi: 10.1002/ijc.28152. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szeliga J, Dipple A. DNA adduct formation by polycyclic aromatic hydrocarbon dihydrodiol epoxides. Chem Res Toxicol. 1998;11:1–11. doi: 10.1021/tx970142f. [DOI] [PubMed] [Google Scholar]
- 48.Katz AK, Carrell HL, Glusker JP. Dibenzo[a,l]pyrene (dibenzo[def,p]chrysene): fjord-region distortions. Carcinogenesis. 1998;19:1641–1648. doi: 10.1093/carcin/19.9.1641. [DOI] [PubMed] [Google Scholar]
- 49.Luch A, Friesel H, Seidel A, Platt KL. Tumor-initiating activity of the (+)-(S,S)- and (−)-(R,R)- enantiomers of trans-11,12-dihydroxy-11,12-dihydrodibenzo[a,l]pyrene in mouse skin. Cancer Lett. 1999;136:119–128. doi: 10.1016/s0304-3835(98)00265-1. [DOI] [PubMed] [Google Scholar]
- 50.Nesnow S, Ross JA, Mass MJ, Stoner GD. Mechanistic relationships between DNA adducts, oncogene mutations, and lung tumorigenesis in strain A mice. Exp Lung Res. 1998;24:395–405. doi: 10.3109/01902149809087376. [DOI] [PubMed] [Google Scholar]
- 51.Nesnow S, Mass MJ, Ross JA, Galati AJ, Lambert GR, Gennings C, Carter WH, Jr, Stoner GD. Lung tumorigenic interactions in strain A/J mice of five environmental polycyclic aromatic hydrocarbons. Environ Health Perspect. 1998;106(Suppl 6):1337–1346. doi: 10.1289/ehp.98106s61337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Higginbotham S, RamaKrishna NV, Johansson SL, Rogan EG, Cavalieri EL. Tumor-initiating activity and carcinogenicity of dibenzo[a,l]pyrene versus 7,12-dimethylbenz[a]anthracene and benzo[a]pyrene at low doses in mouse skin. Carcinogenesis. 1993;14:875–878. doi: 10.1093/carcin/14.5.875. [DOI] [PubMed] [Google Scholar]
- 53.Chen KM, Zhang SM, Aliaga C, Sun YW, Cooper T, Gowdahalli K, Zhu J, Amin S, El-Bayoumy K. Induction of ovarian cancer and DNA adducts by Dibenzo[a,l]pyrene in the mouse. Chem Res Toxicol. 2012;25:374–380. doi: 10.1021/tx2004322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ruan Q, Kolbanovskiy A, Zhuang P, Chen J, Krzeminski J, Amin S, Geacintov NE. Synthesis and characterization of site-specific and stereoisomeric fjord dibenzo[a,l]pyrene diol epoxide-N(6)-adenine adducts: unusual thermal stabilization of modified DNA duplexes. Chem Res Toxicol. 2002;15:249–261. doi: 10.1021/tx010157k. [DOI] [PubMed] [Google Scholar]
- 55.Dreij K, Bajak E, Sundberg K, Cotgreave I, Jernstrom B. DNA adducts of benzo[a]pyrene- and dibenzo[a,l]pyrene-diol epoxides in human lung epithelial cells: Kinetics of adduct removal, effects on cell cycle checkpoints, and gene expression. Polyclic Arom Comp. 2004;24:549–566. [Google Scholar]
- 56.Shivji MK, Moggs JG, Kuraoka I, Wood RD. Dual-incision assays for nucleotide excision repair using DNA with a lesion at a specific site. Methods Mol Biol. 1999;113:373–392. doi: 10.1385/1-59259-675-4:373. [DOI] [PubMed] [Google Scholar]
- 57.Huang JC, Svoboda DL, Reardon JT, Sancar A. Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5′ and the 6th phosphodiester bond 3′ to the photodimer. Proc Natl Acad Sci U S A. 1992;89:3664–3668. doi: 10.1073/pnas.89.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cai Y, Ding S, Geacintov NE, Broyde S. Intercalative conformations of the 14R (+)- and 14S (−)-trans-anti-DB[a,l]P-N(6)-dA adducts: Molecular modeling and MD simulations. Chem Res Toxicol. 2011;24:522–531. doi: 10.1021/tx1004002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang Y, Liu Z, Ding S, Rodriguez FA, Sayer JM, Jerina DM, Cai Y, Amin S, Broyde S, Geacintov NE. NMR solution structure of an adduct derived from the potent tumorigen dibenzo[a,l]pyrene linked to N2-guanine: intercalation from the B-DNA minor groove with ruptured Watson-Crick pairing at the lesion site. Biochemistry. 2012;51:9751–9762. doi: 10.1021/bi3013577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cai Y. Chemistry Department. New York University; New York, N.Y: 2008. Elucidating structure-function relationships in DNA repair: Molecular dynamics and NMR studies of polycyclic aromatic hydrocarbon-derived DNA lesions. [Google Scholar]
- 61.Rodriguez F. Chemistry Department. New York University; New York, N.Y: 2007. Nuclear Magnetic Resonance Solution Structure of Covalent Polycyclic Aromatic Carcinogen–DNA adducts: Influence of Base Sequence Contexts and Carcinogen Topology. [Google Scholar]
- 62.Yeo JE, Khoo A, Fagbemi AF, Scharer OD. The efficiencies of damage recognition and excision correlate with duplex destabilization induced by acetylaminofluorene adducts in human nucleotide excision repair. Chem Res Toxicol. 2012;25:2462–2468. doi: 10.1021/tx3003033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chandrasekhar D, Van Houten B. In vivo formation and repair of cyclobutane pyrimidine dimers and 6–4 photoproducts measured at the gene and nucleotide level in Escherichia coli. Mutat Res. 2000;450:19–40. doi: 10.1016/s0027-5107(00)00014-2. [DOI] [PubMed] [Google Scholar]
- 64.Jain V, Hilton B, Lin B, Patnaik S, Liang F, Darian E, Zou Y, Mackerell AD, Jr, Cho BP. Unusual sequence effects on nucleotide excision repair of arylamine lesions: DNA bending/distortion as a primary recognition factor. Nucleic Acids Res. 2012;41:869–880. doi: 10.1093/nar/gks1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jiang GH, Skorvaga M, Croteau DL, Van Houten B, States JC. Robust incision of benoz[a]pyrene-7,8-dihyrodiol-9,10-epoxide-DNA adducts by a recombinant thermoresistant interspecies combination UvrABC endonuclease system. Biochemistry. 2006;45:7834–7843. doi: 10.1021/bi052515e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu Y, Reeves D, Kropachev K, Cai Y, Ding S, Kolbanovskiy M, Kolbanovskiy A, Bolton JL, Broyde S, Van Houten B, Geacintov NE. Probing for DNA damage with beta-hairpins: similarities in incision efficiencies of bulky DNA adducts by prokaryotic and human nucleotide excision repair systems in vitro. DNA Repair (Amst) 2011;10:684–696. doi: 10.1016/j.dnarep.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zou Y, Shell SM, Utzat CD, Luo C, Yang Z, Geacintov NE, Basu AK. Effects of DNA adduct structure and sequence context on strand opening of repair intermediates and incision by UvrABC nuclease. Biochemistry. 2003;42:12654–12661. doi: 10.1021/bi034446e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schurter EJ, Yeh HJ, Sayer JM, Lakshman MK, Yagi H, Jerina DM, Gorenstein DG. NMR solution structure of a nonanucleotide duplex with a dG mismatch opposite a 10R adduct derived from trans addition of a deoxyadenosine N6-amino group to (−)-(7S,8R,9R,10S)-7,8-dihydroxy-9,10-epoxy-7,8,9,10- tetrahydrobenzo[a]pyrene. Biochemistry. 1995;34:1364–1375. doi: 10.1021/bi00004a031. [DOI] [PubMed] [Google Scholar]
- 69.Buterin T, Hess MT, Luneva N, Geacintov NE, Amin S, Kroth H, Seidel A, Naegeli H. Unrepaired fjord region polycyclic aromatic hydrocarbon-DNA adducts in ras codon 61 mutational hot spots. Cancer Res. 2000;60:1849–1856. [PubMed] [Google Scholar]
- 70.Lin CH, Huang X, Kolbanovskii A, Hingerty BE, Amin S, Broyde S, Geacintov NE, Patel DJ. Molecular topology of polycyclic aromatic carcinogens determines DNA adduct conformation: a link to tumorigenic activity. J Mol Biol. 2001;306:1059–1080. doi: 10.1006/jmbi.2001.4425. [DOI] [PubMed] [Google Scholar]
- 71.Suri AK, Mao B, Amin S, Geacintov NE, Patel DJ. Solution conformation of the (+)-trans-anti-benzo[g]chrysene-dA adduct opposite dT in a DNA duplex. J Mol Biol. 1999;292:289–307. doi: 10.1006/jmbi.1999.2974. [DOI] [PubMed] [Google Scholar]
- 72.Yan SX, Shapiro R, Geacintov NE, Broyde S. Stereochemical, structural, and thermodynamic origins of stability differences between stereoisomeric benzo[a]pyrene diol epoxide deoxyadenosine adducts in a DNA mutational hot spot sequence. J Am Chem Soc. 2001;123:7054–7066. doi: 10.1021/ja0043035. [DOI] [PubMed] [Google Scholar]
- 73.Cosman M, de los Santos C, Fiala R, Hingerty BE, Ibanez V, Luna E, Harvey R, Geacintov NE, Broyde S, Patel DJ. Solution conformation of the (+)-cis-anti-[BP]dG adduct in a DNA duplex: intercalation of the covalently attached benzo[a]pyrenyl ring into the helix and displacement of the modified deoxyguanosine. Biochemistry. 1993;32:4145–4155. doi: 10.1021/bi00067a001. [DOI] [PubMed] [Google Scholar]
- 74.Cosman M, Fiala R, Hingerty BE, Amin S, Geacintov NE, Broyde S, Patel DJ. Solution conformation of the (+)-cis-anti-[BP]dG adduct opposite a deletion site in a DNA duplex: intercalation of the covalently attached benzo[a]pyrene into the helix with base displacement of the modified deoxyguanosine into the minor groove. Biochemistry. 1994;33:11518–11527. doi: 10.1021/bi00204a014. [DOI] [PubMed] [Google Scholar]
- 75.de los Santos C, Cosman M, Hingerty BE, Ibanez V, Margulis LA, Geacintov NE, Broyde S, Patel DJ. Influence of benzo[a]pyrene diol epoxide chirality on solution conformations of DNA covalent adducts: the (−)-trans-anti-[BP]G.C adduct structure and comparison with the (+)-trans-anti-[BP]G.C enantiomer. Biochemistry. 1992;31:5245–5252. doi: 10.1021/bi00138a002. [DOI] [PubMed] [Google Scholar]
- 76.Cosman M, de los Santos C, Fiala R, Hingerty BE, Singh SB, Ibanez V, Margulis LA, Live D, Geacintov NE, Broyde S, et al. Solution conformation of the major adduct between the carcinogen (+)-anti-benzo[a]pyrene diol epoxide and DNA. Proc Natl Acad Sci U S A. 1992;89:1914–1918. doi: 10.1073/pnas.89.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Min JH, Pavletich NP. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. 2007;449:570–575. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- 78.Breslauer KJ, Frank R, Blocker H, Marky LA. Predicting DNA duplex stability from the base sequence. Proc Natl Acad Sci U S A. 1986;83:3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.SantaLucia J, Jr, Allawi HT, Seneviratne PA. Improved nearest-neighbor parameters for predicting DNA duplex stability. Biochemistry. 1996;35:3555–3562. doi: 10.1021/bi951907q. [DOI] [PubMed] [Google Scholar]
- 80.Sugasawa K, Ng JM, Masutani C, Iwai S, van der Spek PJ, Eker AP, Hanaoka F, Bootsma D, Hoeijmakers JH. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 81.Luijsterburg MS, von Bornstaedt G, Gourdin AM, Politi AZ, Mone MJ, Warmerdam DO, Goedhart J, Vermeulen W, van Driel R, Hofer T. Stochastic and reversible assembly of a multiprotein DNA repair complex ensures accurate target site recognition and efficient repair. J Cell Biol. 2010;189:445–463. doi: 10.1083/jcb.200909175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kuper J, Kisker C. Damage recognition in nucleotide excision DNA repair. Curr Opin Struct Biol. 2012;22:88–93. doi: 10.1016/j.sbi.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 83.Naegeli H, Sugasawa K. The xeroderma pigmentosum pathway: decision tree analysis of DNA quality. DNA Repair. 2011;10:673–683. doi: 10.1016/j.dnarep.2011.04.019. [DOI] [PubMed] [Google Scholar]
- 84.Lange SS, Vasquez KM. HMGB1: the jack-of-all-trades protein is a master DNA repair mechanic. Mol Carcinog. 2009;48:571–580. doi: 10.1002/mc.20544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sidorenko VS, Yeo JE, Bonala RR, Johnson F, Scharer OD, Grollman AP. Lack of recognition by global-genome nucleotide excision repair accounts for the high mutagenicity and persistence of aristolactam-DNA adducts. Nucleic Acids Res. 2012;40:2494–2505. doi: 10.1093/nar/gkr1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lukin M, Zaliznyak T, Johnson F, de los Santos C. Structure and stability of DNA containing an aristolactam II-dA lesion: implications for the NER recognition of bulky adducts. Nucleic Acids Res. 2012;40:2759–2770. doi: 10.1093/nar/gkr1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zaliznyak T, Bonala R, Johnson F, de Los Santos C. Structure and stability of duplex DNA containing the 3-(deoxyguanosin-N2-yl)-2-acetylaminofluorene (dG(N2)-AAF) lesion: a bulky adduct that persists in cellular DNA. Chem Res Toxicol. 2006;19:745–752. doi: 10.1021/tx060002i. [DOI] [PubMed] [Google Scholar]
- 88.Cui XS, Eriksson LC, Moller L. Formation and persistence of DNA adducts during and after a long-term administration of 2-nitrofluorene. Mutat Res. 1999;442:9–18. doi: 10.1016/s1383-5718(99)00053-4. [DOI] [PubMed] [Google Scholar]
- 89.Bieler CA, Cornelius MG, Stiborova M, Arlt VM, Wiessler M, Phillips DH, Schmeiser HH. Formation and persistence of DNA adducts formed by the carcinogenic air pollutant 3-nitrobenzanthrone in target and non-target organs after intratracheal instillation in rats. Carcinogenesis. 2007;28:1117–1121. doi: 10.1093/carcin/bgl219. [DOI] [PubMed] [Google Scholar]
- 90.Mao H, Deng Z, Wang F, Harris TM, Stone MP. An intercalated and thermally stable FAPY adduct of aflatoxin B1 in a DNA duplex: structural refinement from 1H NMR. Biochemistry. 1998;37:4374–4387. doi: 10.1021/bi9718292. [DOI] [PubMed] [Google Scholar]
- 91.Rodriguez FA, Cai Y, Lin C, Tang Y, Kolbanovskiy A, Amin S, Patel DJ, Broyde S, Geacintov NE. Exocyclic amino groups of flanking guanines govern sequence-dependent adduct conformations and local structural distortions for minor groove-aligned benzo[a]pyrenyl-guanine lesions in a GG mutation hotspot context. Nucleic acids research. 2007;35:1555–1568. doi: 10.1093/nar/gkm022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dreij K, Seidel A, Jernstrom B. Differential removal of DNA adducts derived from anti-diol epoxides of dibenzo[a,l]pyrene and benzo[a]pyrene in human cells. Chem Res Toxicol. 2005;18:655–664. doi: 10.1021/tx0497090. [DOI] [PubMed] [Google Scholar]
- 93.Lagerqvist A, Hakansson D, Prochazka G, Lundin C, Dreij K, Segerback D, Jernstrom B, Tornqvist M, Seidel A, Erixon K, Jenssen D. Both replication bypass fidelity and repair efficiency influence the yield of mutations per target dose in intact mammalian cells induced by benzo[a]pyrene-diol-epoxide and dibenzo[a,l]pyrene-diol-epoxide. DNA Repair. 2008;7:1202–1212. doi: 10.1016/j.dnarep.2008.03.022. [DOI] [PubMed] [Google Scholar]
- 94.Phillips DH, Hewer A, Seidel A, Steinbrecher T, Schrode R, Oesch F, Glatt H. Relationship between mutagenicity and DNA adduct formation in mammalian cells for fjord- and bay-region diol-epoxides of polycyclic aromatic hydrocarbons. Chem Biol Interact. 1991;80:177–186. doi: 10.1016/0009-2797(91)90023-z. [DOI] [PubMed] [Google Scholar]
- 95.Arif JM, Gupta RC. Microsome-mediated bioactivation of dibenzo[a,l]pyrene and identification of DNA adducts by 32P-postlabeling. Carcinogenesis. 1997;18:1999–2007. doi: 10.1093/carcin/18.10.1999. [DOI] [PubMed] [Google Scholar]
- 96.Arif JM, Smith WA, Gupta RC. Tissue distribution of DNA adducts in rats treated by intramammillary injection with dibenzo[a,l]pyrene, 7,12-dimethylbenz[a]anthracene and benzo[a]pyrene. Mutat Res. 1997;378:31–39. doi: 10.1016/s0027-5107(97)00095-x. [DOI] [PubMed] [Google Scholar]
- 97.Guttenplan JB, Kosinska W, Zhao ZL, Chen KM, Aliaga C, DelTondo J, Cooper T, Sun YW, Zhang SM, Jiang K, Bruggeman R, Sharma AK, Amin S, Ahn K, El-Bayoumy K. Mutagenesis and carcinogenesis induced by dibenzo[a,l]pyrene in the mouse oral cavity: a potential new model for oral cancer. Int J Cancer. 2011;130:2783–2790. doi: 10.1002/ijc.26344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Spencer WA, Singh J, Orren DK. Formation and differential repair of covalent DNA adducts generated by treatment of human cells with (+/−)-anti-dibenzo[a,l]pyrene-11,12-diol-13,14-epoxide. Chem Res Toxicol. 2009;22:81–89. doi: 10.1021/tx8001675. [DOI] [PubMed] [Google Scholar]
- 99.Mahadevan B, Dashwood WM, Luch A, Pecaj A, Doehmer J, Seidel A, Pereira C, Baird WM. Mutations induced by (−)-anti-11R,12S-dihydrodiol 13S,14R-epoxide of dibenzo[a,l]pyrene in the coding region of the hypoxanthine phosphoribosyltransferase (Hprt) gene in Chinese hamster V79 cells. Environ Mol Mutagen. 2003;41:131–139. doi: 10.1002/em.10136. [DOI] [PubMed] [Google Scholar]
- 100.Yoon JH, Besaratinia A, Feng Z, Tang MS, Amin S, Luch A, Pfeifer GP. DNA damage, repair, and mutation induction by (+)-syn and (−)-anti-dibenzo[a,l]pyrene-11,12-diol-13,14-epoxides in mouse cells. Cancer Res. 2004;64:7321–7328. doi: 10.1158/0008-5472.CAN-04-1094. [DOI] [PubMed] [Google Scholar]
- 101.Melendez-Colon VJ, Smith CA, Seidel A, Luch A, Platt KL, Baird WM. Formation of stable adducts and absence of depurinating DNA adducts in cells and DNA treated with the potent carcinogen dibenzo[a,l]pyrene or its diol epoxides. Proc Natl Acad Sci USA. 1997;94:13542–13547. doi: 10.1073/pnas.94.25.13542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mahadevan B, Luch A, Bravo CF, Atkin J, Steppan LB, Pereira C, Kerkvliet NI, Baird WM. Dibenzo[a,l]pyrene induced DNA adduct formation in lung tissue in vivo. Cancer Lett. 2005;227:25–32. doi: 10.1016/j.canlet.2004.11.056. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.