Abstract
By precisely tracking the waves of elongating RNA polymerase II (Pol II) during gene activation, Danko et al. (2013) discovered a surprising diversity of elongation rates among and along human genes.
Since the 1950’s, fearless big wave riders at Hawaii’s Waimea Bay have contributed to the introduction of lighter boards and tow-in surfing that enabled 2011’s Guinness World Record, when Garrett McNamara caught a giant 24-meter wave off Portugal’s Nazaré Canyon. Similarly, Danko and colleagues (2013) have pushed the envelope in their dissection of transcription mechanisms using an elegant strategy to catch the waves of transcription elongation generated in the minutes following gene activation. As described in this issue of Molecular Cell, following these waves on many endogenous genes and under different activating conditions provides novel insights into gene regulation.
Pol II elongation rates have been measured previously at small sets of endogenous genes using qPCR or tiling arrays, and at integrated transgenes using imaging techniques (Ardehali and Lis, 2009; Darzacq et al., 2007; Singh and Padgett, 2009). These studies have reported a wide range of transcription elongation rates, from 1–6 kilobases per minute (kb/min), raising the possibility that the rate of elongation is varied to impact gene output. However, it has remained unclear whether the differing elongation rates reported reflect biological or technical variability. To address this issue, Danko et al. (2013) have systematically studied the initial waves of Pol II elongation in a human breast cancer cell line (MCF-7) exposed to an estrogenic steroid (17β-estradiol/E2) and a cardiomyocyte line (AC16) treated with a rapidly-acting cytokine (TNFα).
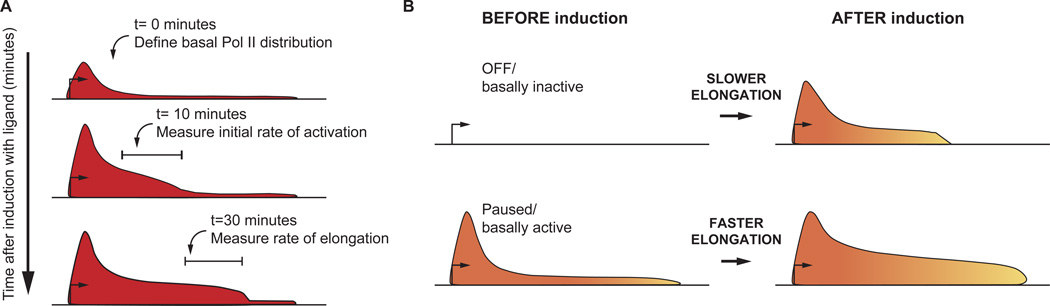
Danko et al. (2013) tracked the actively transcribing Pol II molecules using Global Run-on Sequencing (GRO-seq) coupled with a clever bioinformatics pipeline. This strategy allowed them to simultaneously measure the elongation rate at a significant number of genes during activation (~100 in MCF7 cells, 26 in AC16 cells), permitting direct comparisons of polymerase dynamics across the gene sets. By performing GRO-seq at a number of time points during gene activation, the authors could measure the elongation rate as Pol II moved across the gene body (Figure 1A). Interestingly, they found that the polymerase moves slowly initially and speeds up within the first 10–15 kb of the transcription unit. This result suggests that the factors that facilitate the polymerase’s journey through long genes take time to assemble, with Pol II reaching a maximally productive state well within the gene. A principal implication of this finding is that the availability of such factors, which might include chromatin modifying enzymes, elongation factors and RNA processing machineries, could impact elongation rate.
Figure 1.
Defining the waves of transcription elongation on endogenous genes under different activating conditions. (A) Using GRO-seq to map Pol II distribution at different time points following gene activation. (B) The observed elongation rate is linked to the basal activity level of a gene, with higher expression levels in uninduced cells correlating with faster elongation rates during gene activation.
Further supporting the idea that transcription elongation is a highly dynamic and regulated process, Danko et al. (2013) find a remarkable variability in the elongation rates from gene-to-gene following treatment with ligand. For the 81 genes measured following stimulation of Estrogen Receptor alpha (ERα) with E2, they find median rates of 0.37 to 3.57 kb/min, with a median rate of 2.1 kb/min. By comparison, the 26 genes activated by NF-κB after treatment with TNFα exhibit a median rate of 2.8 kb/min. Thus, elongation rates differ among genes induced by a common stimulus, and median rates are different between the two types of transcription activation. Importantly, the authors show that genes with the highest measured elongation rates also generate more mRNA, confirming a connection between the speed of elongation and gene output.
Regulated pausing of Pol II elongation has surfaced as a prevalent and rate-limiting step for many metazoan genes (Adelman and Lis, 2012; Moore and Proudfoot, 2009). The most prominent pausing observed across metazoan genomes occurs promoter-proximally, when an early elongation complex comes under control of pause-inducing factors such as the Negative Elongation Factor (NELF) complex. Release of this paused Pol II into the gene body is accomplished through recruitment of the kinase Positive Transcription Elongation factor-b (P-TEFb), which stimulates productive elongation in part through dissociation of NELF (Adelman and Lis, 2012). However, even during productive synthesis, the polymerase is known to pause temporarily, generating significant variations in elongation rates (Darzacq et al., 2007; Landick, 2006).
By performing GRO-seq in both cell types prior to stimulation with ligand, Danko et al. (2013) defined the baseline Pol II distribution for each activated gene. They determined the level of Pol II recruitment and promoter-proximal pausing that occurs at each gene, as well as the level of transcription experienced prior to stimulation. Interestingly, Danko et al. (2013) found that the elongation rate observed at an activated gene is linked to its basal activity level (Figure 1B). For example, genes activated by TNFα/NF-κB tend to exhibit a higher level of expression prior to stimulation and are transcribed more rapidly during activation than E2/ERα-induced genes. Further, the authors noted that genes activated by TNFα/NF-κB are more likely to harbor paused Pol II in uninduced cells than are E2/ERα targets.
These results suggest a potential explanation for the connection between paused Pol II and rapid, robust activation observed at some genes. Genes with paused Pol II display inherently ‘leaky’ expression, with nearly all highly paused genes undergoing detectable transcription (Adelman and Lis, 2012). Perhaps this continued Pol II release into genes ‘primes the pump’, such that additional Pol II molecules can elongate more efficiently? One potential mechanism for this might be the establishment of a particular chromatin structure, with histone modifications that facilitate elongation (e.g. H3-K36me3). Another possibility might be that a high density of Pol II molecules across a gene enables cooperation between elongation complexes. In situations where a leading polymerase encounters obstacles that cause it to pause or backtrack, a helpful collision or push from a trailing polymerase has been shown to restart productive synthesis (Epshtein et al., 2003). The results of Danko et al. (2013) suggest that this kind of cooperation could be more prevalent on the paused genes activated by TNFα/NF-κB, than on the less Pol II-dense genes induced by E2/ERα, leading to differences in rates of mRNA production.
Finally, the data in Danko et al. (2013) strongly support a model wherein different transcription factors can target distinct steps in the transcription cycle, with some factors promoting pre-initiation complex formation and others facilitating release of promoter-proximally paused Pol II (Adelman and Lis, 2012). The authors found that activation of ERα primarily increased the levels of initiating Pol II, but did not prevent pausing or the recruitment of NELF. As a result, recruited Pol II experienced an extended residence time near promoters and a delay in its release into productive synthesis. In contrast, NF-κB-induced genes did not exhibit a delay between Pol II initiation and release into the gene, consistent with previous data indicating that NF-κB recruits P-TEFb (Barboric et al., 2001) and that increases in Pol II levels at NF-κB-activated genes are not accompanied by increases in NELF occupancy (Adelman et al., 2009). Thus, in addition to stimulating the formation of new initiation complexes, activation by NF-κB increases the rate of pause release, allowing for a more rapid entry of recruited Pol II into elongation and faster synthesis of mRNAs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adelman K, Kennedy MA, Nechaev S, Gilchrist DA, Muse GW, Chinenov Y, Rogatsky I. Proc. Natl. Acad. Sci. USA. 2009;106:18207–18212. doi: 10.1073/pnas.0910177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman K, Lis JT. Nat. Rev. Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardehali MB, Lis JT. Nat. Struct. Mol. Biol. 2009;16:1123–1124. doi: 10.1038/nsmb1109-1123. [DOI] [PubMed] [Google Scholar]
- Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. Mol. Cell. 2001;8:327–337. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- Danko, et al. (not sure what the full reference will be) [Google Scholar]
- Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy S, Phair R, Singer R. Nat. Struct. Mol. Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epshtein V, Toulme F, Rahmouni AR, Borukhov S, Nudler E. EMBO J. 2003;22:4719–4727. doi: 10.1093/emboj/cdg452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landick R. Biochem. Soc. Transact. 2006;34:1062–1066. doi: 10.1042/BST0341062. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Singh J, Padgett RA. Nat. Struct. Mol. Biol. 2009;16:1128–1133. doi: 10.1038/nsmb.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]