Abstract
The development of the vertebrate jaw relies on a network of transcription factors that patterns the dorsal-ventral axis of the pharyngeal arches. Recent findings in both mouse and zebrafish illustrate that the basic-helix-loop-helix transcription factor, Hand2, is crucial in this patterning process. While Hand2 has functionally similar roles in these two species, little is known about the regulatory sequences controlling hand2 expression in zebrafish. Using bioinformatics and Tol2-mediated transgenesis, we have generated zebrafish transgenic reporter lines in which either the mouse or zebrafish arch-specific hand2 enhancer direct expression of a fluorescent reporter. We find that both the mouse and zebrafish enhancers drive early reporter expression in a hand2-specific pattern in the ventral pharyngeal arches of zebrafish embryos. These lines provide useful tools to follow ventral arch cells during vertebrate jaw development while also allowing dissection of hand2 transcriptional regulation during this process.
Keywords: Zebrafish, Hand2, bHLH, Transgenic, Enhancer, Mouse, Craniofacial
Introduction
Development of the mandibular portion of the first pharyngeal arch in jawed vertebrates (gnathostomes) is an intriguing process due to the diverse structures that arise from this arch, including Meckel’s cartilage and the dentary bone and in mammals, middle ear structures and the connective tissue of the tongue (Bronner-Fraser, 1995; Couly et al., 1996; Kimmel et al., 2001; Kontges and Lumsden, 1996; Le Douarin, 1982). A critical step in this process is the patterning of the neural crest cell (NCC)-derived mesenchyme that establishes a dorsal-ventral (D–V) (proximal-distal in mouse) polarity within the arch. This patterning is accomplished primarily through action of two ectodermal signals, bone morphogenetic proteins (Bmps) and endothelin-1 (Edn1) (Clouthier et al., 2010:Alexander et al., 2011). In zebrafish, Bmp signaling induces expression of Edn1 (Alexander et al., 2011), which in turn binds to the endothelin-A receptor (Ednra) on NCCs, initiating a hierarchical signaling cascade (Clouthier et al., 2000; Ruest et al., 2004). Loss of Ednra signaling in mice leads to the homeotic transformation of lower jaw elements into more maxillary-like derivatives (Clouthier et al., 1998; Kimmel et al., 2003; Kurihara et al., 1994; Miller et al., 2000; Ozeki et al., 2004; Ruest et al., 2004). While zebrafish have two Ednras (Ednra1 and Ednra2), one of which is found in the arch ectoderm, loss of both result in similar changes in jaw development as observed in mouse Ednra mutants (Nair et al., 2007).
During arch patterning, Ednra and Bmp signaling work in parallel with Mef2C to induce Dlx5/6 expression in the mandibular arch (Miller et al., 2007; Verzi et al., 2007), which in turn induces expression of the gene encoding the basic helix-loop-helix (bHLH) transcription factor Hand2 in the ventral arch (Charité et al., 2001). Recent work in both the mouse and zebrafish has established a role for Hand2 in proper D–V patterning, in part by acting in a negative feedback loop to downregulate Dlx5/6 in this region (Alexander et al., 2011; Barron et al., 2011;Talbot et al., 2010; Zuniga et al., 2011). NCC-specific loss of Hand2 in mice results in lower jaw defects that include bone and cartilage hypoplasia, the presence of duplicated maxillary structures in the mandible and aglossia (Barron et al., 2011). Hand2 loss in zebrafish results in an almost complete absence of ventral arch derived-cartilage (Miller et al., 2003; Talbot et al., 2010). Hand2 is therefore an excellent cellular marker of the distal/ventral domain of the pharyngeal arches.
In the mouse, a 210 bp pharyngeal arch-specific enhancer for Hand2 located 6.6 kb upstream from the start site of transcription is sufficient to drive transgene expression within the distal pharyngeal arches in a Hand2-specific fashion (Charité et al., 2001). A 7.6 kb upstream fragment of the Hand2 gene encompassing this enhancer was subsequently used to create a Hand2-Cre transgenic mouse strain; fate analyses with this strain illustrated that Hand2 daughter cells give rise to most of the mandible, middle ear and lower jaw connective tissue (Ruest et al., 2003). While fate mapping of cells within the pharyngeal arches of mice has been aided by Cre/loxP technology (Chai et al., 2000; Ruest et al., 2003), advances in zebrafish transgenesis and live imaging have greatly expanded the ability to study and follow the fate of arch cells within these domains in this model. Poss and colleagues have recently generated transgenic zebrafish in which enhanced green fluorescent protein (EGFP) was inserted in place of the first exon of hand2 in a BAC containing the hand2 genomic region (Kikuchi et al., 2011). In these Tg(hand2:EGFP)pd24 zebrafish, EGFP is expressed in the lateral plate mesoderm (Yin et al., 2010) and adult cardiac cells (Kikuchi et al., 2011). While GFP fluorescence also appears to be present in the pharyngeal arches (Yin et al., 2010), this has not been described.
We therefore set out to find the zebrafish pharyngeal arch enhancer regulating hand2 expression. Using bioinformatics analysis, we identified an enhancer element in the hand2 upstream region that shows strong conservation with the known mouse enhancer. This enhancer and the mouse enhancer are both capable of driving transgene expression in the ventral pharyngeal arches of zebrafish embryos in a hand2-specific pattern. In addition, the zebrafish enhancer is capable of driving transgene expression in the distal mandibular pharyngeal arch of mouse embryos. These findings illustrate that the genetic regulation of Hand2 expression is highly conserved among species. These new zebrafish lines will be useful both as markers of the ventral pharyngeal arch expression domain and as tools to dissect the mechanisms controlling the domain-specific regulation of hand2 expression.
Materials and methods
Fish breeding and maintenance
Fish were raised and maintained under standard conditions (Westerfield, 1993). Transgenic lines were created using a TAB wild-type line, which was generated by crossing AB and TL wild-type parents. Zebrafish embryos were collected from natural matings, cultured and staged as described (Kimmel et al., 1995).
Transgenic construct injection
Transposase mRNA was synthesized from the pCS2FA-transposase plasmid (Tol2 Kit plasmid 396) using the SP6 mMESSAGE mMACHINE In Vitro Transcription Kit (Ambion). Transposase mRNA and the appropriate hand2 reporter construct were mixed with 0.1% phenol red for a final concentration of 25 ng/μl of DNA and 25 ng/μl mRNA. 1 nl of solution was injected into wild-type TAB zebrafish embryos at the 1-cell stage using an Olympus SZX16 stereomicroscope and a Harvard Apparatus Pico Injector PLI-100.
Generation of a zhand2: Tag-RFP transgenic line
The zebrafish enhancer was amplified from genomic DNA with primers 5′-GCGCGGCCGGCCTTTAGAGCATTTTCCATGG-3′ and 5′-GCGCGGCGCGCCTGTTTGTGTGTGTGAAAG-3′ using High Fidelity Platinum Taq Polymerase (Invitrogen). These primers contain an FseI site added to the forward primer and an AscI site added to the reverse primer. The 600 bp PCR product was cloned into the pCRII plasmid using the TOPO TA Dual Promoter Cloning Kit (Invitrogen). The resulting plasmid (pCRII-hand2) was then cut with FseI and AscI and the enhancer fragment isolated using the MinElute PCR Purification Kit (Qiagen). The fragment was then cloned into the p5E-FAbas plasmid (Tol2 kit plasmid 549, a gift from Chi-Bin Chien), which contains the adenovirus E1b TATA box and the β-actin transcriptional start site. The resulting plasmid (p5E-zhand2-FAbas) was used with the Multisite Gateway Recombination Kit (Invitrogen) to recombine p5E-hand2-FAbas, pME-tag-RFP (a gift from Li-En Jao), p3E-polyA (Tol2 kit plasmid 302), and pDestTol2CG2 (Tol2 kit plasmid 395). The resulting plasmid, zhand2:tag-RFP;cmlc2:GFP, thus produces tag-RFP under control of the hand2 enhancer and GFP under the control of cmlc2 expressed in cardiomyocytes. Four stable Tg(hand2:tag-RFP,cmlc2:GFP) transgenic lines (co3, co4, co5 and co6) were isolated and all showed similar expression patterns. We chose Tg(hand2:tag-RFP,cmlc2:GFP)co3 for further analysis.
Generation of a szhand2: mCherry transgenic line
The small zebrafish enhancer was cloned in a similar manner, first amplified from zebrafish genomic DNA using primers 5′-GCGCGGCCGGCCTCACGTTTTCATAAATTCTGAT-3′ and 5′-GCGCGGCGCGCCGTGTTGTGTGTGGGGTTCAG-3′. This PCR fragment was ligated into the pCRII plasmid and excised using the FseI/AscI sites that were added to the primers. This fragment was then ligated into the p5E-FAbas vector and the resulting vector recombined with pME-mCherry, p3E-polyA, and pDestTol2CG2 using the Gateway Multisite Recombination Kit. This gave a final plasmid, szhand2:m-Cherry;cmlc2:GFP, with the 280 bp hand2 enhancer driving expression of mCherry within the arches and cmlc2 regulatory regions driving GFP expression in the developing heart. Three stable Tg(shand2:mCherry,cmlc2:GFP) transgenic lines (co10, co11, co12) were isolated and all showed similar expression patterns. We chose Tg(shand2:mCherry,cmlc2:GFP)co10 for further analysis.
Generation of a mHand2: EGFP transgenic line
The mouse Hand2 arch-specific enhancer was excised from p0.6Hand2 using XhoI and BamHI. After purification using the MinElute PCR Purification Kit, the enhancer was blunted using Klenow enzyme (Invitrogen). The blunted fragment was then cloned into the p5E-FAbas plasmid previously cut with FseI and AsaII and blunted using Klenow enzyme to produce p5E-mHand2-FAbas. Gateway recombination was performed as described above with pME-EGFP (Tol2 kit plasmid 383), p3E-polyA and pDestTol2 (Tol2 kit plasmid 394). The resulting plasmid, mHand2:EGFP, yields an EGFP reporter under control of the mouse 757 bp arch specific enhancer. Three stable Tg(Mmu.Hand2:EGFP) transgenic lines (co7, co8, co9) were isolated and all showed similar expression patterns. We chose Tg(Mmu.Hand2;EGFP)co8 for further analysis.
Imaging and analysis of transgenic embryos
Injected embryos were visualized from 12 hpf to 7 dpf using an Olympus SZX16 stereomicroscope and a SPOT-RT slider digital camera fitted with a fluorescence unit containing GFP and Rhodamine cubes or an Olympus BX51 compound microscope and a DP-71 digital camera fitted with a fluorescence unit containing GFP and Texas Red cubes. Embryos displaying transgene expression were allowed to grow to maturity and then crossed with TAB fish to determine germ-line transmission. For analysis, live transgenic embryos were examined and photographed on our Olympus BX51 microscope fitted with a DP71 digital camera. For analysis of double fluorescent transgenic embryos, embryos were mounted in 1% low melting temperature agarose. Embryos were imaged live using a Leica SP5X line scanning confocal microscope fitted with an inverted 20x objective. The presented image represent a 20 μm compressed z-stack adjusted with Volocity software (Perkin-Elmer) and rotated and cropped using Photoshop CS5 (Adobe).
Morpholino oligonucleotide injection
2 nl of morpholino (diluted to 2.5 μg/μl) was injected into embryos at the 1–4 cell stage. edn1 translation blocking morpholino was synthesized by Gene Tools using previously reported sequence (Miller and Kimmel, 2001). This morpholino has previously been shown to illicit joint fusions and loss of cartilage consistent with loss of endothelin, as well as associated decreases in hand2 expression. To ensure that we analyzed only Tg(shand2:mCherry,cmlc2:GFP)co10 embryos, only embryos exhibiting GFP fluorescence in the heart were screened for mCherry fluorescence in the arches.
Whole mount in situ hybridization (ISH)
For ISH analysis in zebrafish, embryos were collected from wild-type matings and fixed in 4% paraformaldehyde. Embryos to be used for later stages were placed in phenylthiourea (PTU) at 24 hpf to prevent pigment formation. Antisense RNA probe was synthesized from hand2 cDNA and labeled with digoxigenin. Whole mount ISH was adapted from previously described protocols (Thisse, 1998). Embryos were mounted in 100% glycerol, coverslipped, and photographed under brightfield using an Olympus BX51 compound microscope fitted with a DP71 digital camera. Whole mount ISH analysis of Hand2 expression in mouse embryos was performed as previously described (Clouthier et al., 1998). Following staining, embryos were photographed as described above and then dehydrated through a graded series of ethanols and Cedarwood oil before being embedded in paraffin in a transverse orientation. 14 μm sections were cut, collected onto Plus-coated slides (Fisher, USA) rehydrated and mounted using Cytoseal 60. Sections were examined and photographed under Nomarski optics on our Olympus BX51 microscope.
Generation of transgenic mice
For production of transient transgenic mouse embryos, we used a pHsp68-lacZ vector (Charité et al., 2001; McFadden et al., 2000). pHsp68-lacZ was cut with HindIII (a unique site upstream of the Hsp68 promoter), purified, blunted with Klenow enzyme, again purified and then dephosphorylated using shrimp alkaline phosphatase (SAP; Invitrogen). The blunted zebrafish hand2 arch-specific enhancer used previously for Gateway cloning was then ligated into the blunted HindIII site, with the resulting plasmid verified by both restriction analysis and sequencing. The resulting plasmid, pzhand2-Hsp68-lacZ, was digested with SalI to excise the transgene from vector sequences. After agarose gel electrophoresis, the fragment was recovered by electroelution, precipitated and resuspended in injection buffer. The purified fragment was injected into the pronuclei of fertilized one-cell (C57Bl/6 × SJL) F2 hybrid mouse eggs as previously described (Clouthier et al., 1997). Pseudopregnant ICR recipients were sacrificed at either embryonic day (E) 10.5 or E13.5 and embryos collected and processed for β-galactosidase staining.
β-galactosidase (β-gal) staining
Embryos were processed for whole mount (E10.5) β-gal staining as previously described (Ruest et al., 2003). Whole mount-stained embryos were rinsed, fixed 4 h in 10% neutral buffered formalin and processed for paraffin embedding. Sectioning and counterstaining of β-gal stained slides was performed as previously described (Ruest et al., 2003).
Cell culture
C3H/10T1/2 cells (ATCC, Manassas, VA, USA) were maintained in Eagle’s Basal medium with 2 mM l-glutamine, 1.5 g/L sodium bicarbonate and Earle’s BSS (Invitrogen) supplemented with 10% fetal bovine serum (Sigma), penicillin/streptomycin (Invitrogen), and Fungizone (Invitrogen) at 37 °C in a humidified chamber with 5% CO2.
Expression constructs and luciferase assay
The dlx5a cDNA was excised from pME18S-FL3 (Open Biosystems) using XhoI, gel extracted, blunted using Klenow Fragment (Invitrogen) and cloned into blunted pCS2 to generate the expression vector. C3H/10T1/2 cells seeded on 12-well plates were transfected with 0.8 μg pRL-TK (Promega), 0.8 μg pGL3-zhand2-luc [generated by placing the zhand2 enhancer 5′ of the firefly luciferase cDNA in the pGL3 (Promega) vector], and 0.8 μg of either pCS2-dlx5a or empty pCS2 (mock) using Fugene 6 (Roche). Experiments were performed in triplicate and collected 36 h after transfection. Lysate was prepared and firefly (experimental reporter) and renilla (normalizing reporter) luciferases were measured simultaneously using the Dual-Luciferase Reporter Assay System (Promega) and a Veritas Microplate Luminometer (Promega). Activity of the experimental reporter was calculated by dividing the firefly luciferase value by the renilla luciferase value. Fold change was then calculated by dividing the expression vector activity by the empty expression vector (mock) activity. Statistics were performed using unpaired two-tailed t-test with Prism software (Graphpad).
Results
Identification of a hand2 pharyngeal arch-specific enhancer in zebrafish
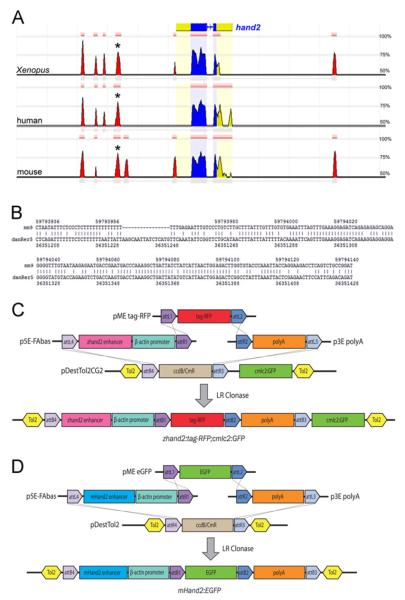
To determine the location of the zebrafish hand2 pharyngeal arch-specific enhancer, we examined the conserved genomic regions within the hand2 locus using the Evolutionary Conserved Region (ECR) Browser (http://ecrbrowser.dcode.org/). 12.4 kb of the zebrafish genome flanking the hand2 coding region was aligned with Xenopus tropicalis, mouse and human genomes, with alignment parameters set to identify regions greater than 75 base pairs (bp) long that exhibited at least 50% similarity to the zebrafish genome. Using this approach, four upstream ECRs were identified across these three species as well as one downstream ECR (Fig. 1A). Of these ECRs, a 226 bp peak located 2.1 kb upstream of hand2 showed 66.4% identity to mouse genomic sequence (Fig. 1B). Interestingly, this region of the mouse genome contained the previously identified mouse Hand2 pharyngeal arch specific enhancer (Charité et al., 2001). We therefore focused our search for the zebrafish arch specific enhancer on this region.
Fig. 1.
Identification and cloning of potentialhand2 regulatory regions. A. 12 kb of genomic sequence flanking the zebrafish hand2-coding region compared to Xenopus tropicalis, human and mouse genomes using dcode ECR browser. The zebrafish hand2 coding sequence is shown at the top, with blue indicating exons and yellow indicating 5′ and 3′ untranslated regions (UTRs). The red peaks identify conserved non-coding regions that show sequence similarity of at least 50%. The distance between vertical gray lines is 1 kb. Asterisks indicate the region chosen for further analysis. B. Nucleotide alignment of the mouse (mm9) and zebrafish (danRer5) ECRs identified in (A) illustrates 66.4% identity. C. The 600 bp zebrafish hand2 enhancer was cloned into p5E-FAbas upstream of a β-actin minimal promoter using FseI and AscI. The middle entry clone contains a tag-RFP reporter and the 3′ entry clone contains the SV40 poly-adenylation (pA) signal. These three plasmids were combined in a Multisite Gateway reaction with the destination vector pDestTol2CG2 containing cmlc2:GFP as a transgenesis marker, resulting in Hand2-directed tag-RFP expression. D. The previously described mouse Hand2 arch-specific enhancer was similarly cloned into p5E-FAbas and recombined with a middle entry clone containing EGFP and a 3′ SV40 pA entry clone in the Tol2 destination vector, pDestTol2, resulting in Hand2-directed EGFP expression. Plasmids are not drawn to scale.
The mouse Hand2 arch-specific enhancer is 210 bp (Charité et al., 2001), though mouse studies have used a larger genomic region (757 bp) that includes this minimal enhancer (Charité et al., 2001; Ruest et al., 2003). To maintain consistency with the mouse studies, we initially included additional flanking sequence in our enhancer analysis, bringing the overall sequence size of the region to 600 bp. Multisite Gateway Recombination was used to create a transgene in which the hand2 enhancer drives expression of tag-red fluorescence protein (RFP) (Fig. 1C). The destination vector, pDestTol2CG2, contains Tol2 integration sites as well as the cmlc2:GFP transgene, driving green fluorescent protein (GFP) expression in cardiomyocytes as a positive control for transgenesis (Fig. 1C). Three independent Tg(hand2:tag-RFP,cmlc2:GFP) founder lines demonstrating fluorescence in the ventral pharyngeal arches were established. We examined fluorescence in hemizygous F1 embryos from all three lines and confirmed hand2 enhancer-regulated tag-RFP fluorescence in the ventral arches, focusing on Tg(hand2:tag-RFP,cmlc2:GFP)co3 for our subsequent analysis (referred to hereafter as Tg(hand2:tag-RFP)).
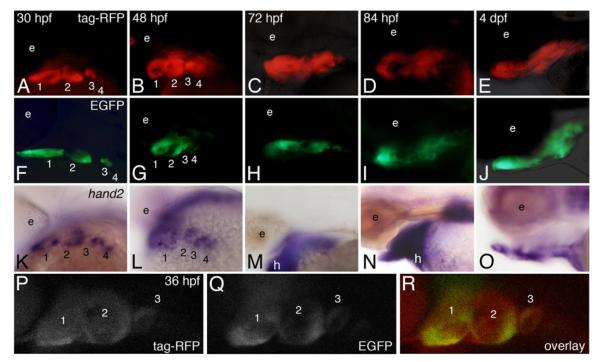
hand2-directed RFP fluorescence was first detected at 30 hpf in the pharyngeal arches (Fig. 2A), with strong fluorescence detected in arches 1–3, consistent with endogenous hand2 expression in those arches (Fig. 2K). In contrast, RFP fluorescence in arch 4 was far weaker (Fig. 2A) relative to the endogenous expression pattern (Fig. 2K), indicating a temporal lag in either expression or accumulation of RFP in this arch. While hand2 expression at this stage was also seen in the fin buds and heart tube (data not shown), RFP fluorescence was absent in these structures, suggesting that this enhancer drives hand2 expression specifically in the pharyngeal arches. By 48 hpf (Fig. 2B), strong fluorescence was observed in the ventral aspects of arches 1–4 in a pattern matching the endogenous expression pattern (Fig. 2L). Ventral RFP fluorescence remained at 72 hpf (Fig. 2C), though by 84 h, RFP fluorescence appeared to expand dorsally (Fig. 2D). While endogenous hand2 expression was present at both 72 (Fig. 2M) and 84 (Fig. 2N) hpf, the relative position in the developing jaw was difficult to ascertain. By 4 dpf, RFP fluorescence could be detected throughout the viscerocranium, including both ventral and dorsal arch derivatives (Fig. 2E and data not shown). Although the role of Hand2 at these later stages has not been examined, hand2 expression at 4 dpf was observed surrounding the ventral cartilages (Fig. 2O) in a pattern similar to that observed for Hand2 expression in the mouse (Ruest et al., 2003). These findings demonstrate that this enhancer is sufficient to drive tag-RFP expression in the ventral pharyngeal arches in a hand2-specific manner during early D–V patterning of the arches and that later enhancer regulation either requires additional regulatory sequences or occurs in a more complex manner than previously detected or appreciated.
Fig. 2.
Both zebrafish and mouse enhancers drive transgene expression in a hand2-specific pattern within the pharyngeal arches. Dorsolateral views are shown at 30 and 48 hpf, and lateral views at 72 and 84 hpf and 4 dpf. A–E. Tg(hand2:tag-RFP) embryos were examined for RFP fluorescence between 30 h post fertilization (hpf) and 4 day post-fertilization (dpf). RFP was first detected at 30 hpf in the ventral aspect of pharyngeal arches 1–3 (A). At 48 hpf, RFP was present in the ventral aspects of arches 1–4 (B). At 72 hpf, RFP also appeared ventrally located (C). By 84 hpf (D) and 4 dpf (E), RFP was present in the developing cartilages while also appearing to expand dorsally. F-J. Tg(mHand2:EGFP) embryos were examined at the same stages as the Tg(hand2:tag-RFP) embryos. EGFP activity was observed in a hand2 specific expression pattern in the ventral aspects of arches 1–3 beginning around 30 hpf (F) and persisting in arches 1–4 through 48 hpf (G). EGFP activity continued as chondrogenesis began around 72–84 hpf (H, I) and was still present at 4 dpf (J), though EGFP activity expanded dorsally at both 84 hpf and 4 dpf. K–O. Whole mount in situ hybridization analysis of hand2 expression. Endogenous hand2 expression was present in arches 1–4 at 30 hpf (K) and 48 hpf (L). While endogenous hand2 expression remained at 72 and 84 hpf (M, N), expression began to be restricted by 4 dpf (O). P–R. Compressed confocal z-stacks of Tg(hand2:tag-RFP), Tg(mHand2:EGFP) embryos at 36 hpf in a lateral view, illustrating localization of tag-RFP (P), EGFP (Q) and co-localization of both reporters (R) within the ventral pharyngeal arches.
The mouse and zebrafish hand2 enhancers both drive transgene expression in the pharyngeal arches
To further investigate the functional equivalence of the mouse and zebrafish hand2 enhancers, we tested their ability to drive arch-specific expression across species. Again using Multisite Gateway Recombination, a transgene was created in which the 757 bp mouse arch-specific Hand2 enhancer drives expression of enhanced GFP (EGFP) (Fig. 1D). We identified three founders with similar expression patterns and chose Tg(Mmu.Hand2:EGFP)co8 for subsequent analysis (referred to hereafter as Tg(mHand2:EGFP)). Like the zebrafish hand2 enhancer, the mouse enhancer drove expression of EGFP specifically in the zebrafish hand2 domain of pharyngeal arches 1–3 at 30 hpf (Fig. 2F). Also like RFP fluorescence, EGFP was only faintly detected in arch 4. By 48 hpf, EGFP fluorescence was present in ventral aspects of arches 1–4 (Fig. 2G). EGFP fluorescence persisted in the ventral arches at 72 hpf (Fig. 2H), though by 84 hpf, fluorescence appeared to expand dorsally (Fig. 2I), similar to the pattern observed in Tg(hand2:tag-RFP) embryos. By 4 dpf, GFP was observed throughout the viscerocranium (Fig. 2J and data shown). These results suggest that the mouse and zebrafish hand2 arch-specific enhancers function in a similar manner.
To illustrate the similarities between the mouse and zebrafish transgenes, Tg(hand2:tag-RFP), Tg(mHand2:EGFP) double transgenic embryos were generated through pair-wise matings of single transgenic adults. Visualized by confocal microscopy at 36 hpf, RFP (Fig. 2O) and EGFP (Fig. 2Q) fluorescence showed extensive colocalization within cells in the ventral pharyngeal arches (Fig. 2R), with the overlapping pattern persisting from 30 hpf to 7 dpf (data not shown). While RFP fluorescence was in general weaker that GFP fluorescence, it appears that these enhancers drive transgene expression in the same cell population of the pharyngeal arches.
The zebrafish hand2 arch enhancer drives transgene expression in the mouse Hand2 expression domain
The above results indicate that sufficient evolutionary conservation exists to allow the mouse Hand2 arch-specific enhancer to function in an equivalent manner in zebrafish embryos. However, if this is true equivalence, the zebrafish hand2 enhancer should also be capable of driving transgene expression in mouse pharyngeal arches in a Hand2-specific pattern. To examine this question, the zebrafish hand2 enhancer was inserted into the basal lacZ expression vector Hsp68-lacZ, in which an enhancer is required to achieve expression of lacZ and hence β-galactosidase (β-gal) activity (Fig. 3A). Following injection of this zhand2-Hsp68-lacZ construct into fertilized 1-cell mouse ova, F0 embryos were collected at embryonic day (E) 10.5 and stained for β-gal activity. This staining was then compared to endogenous Hand2 expression in E10.5 control embryos. In E10.5 zhand2-Hsp68-lacZ embryos, β-gal staining was observed in the distal mesenchyme of mandibular arch 1 in a Hand2-specific pattern (Fig. 3B, D), though the staining appeared more faint at the distal-most region of the arch. Staining was also observed in arch 2 (Fig. 3B), though like in arch 1, the staining did not extend as far distally as did the endogenous expression (Fig. 3D). In sections through stained embryos, β-gal staining in zhand2-Hsp68-lacZ embryos was observed in the distal mandibular arch mesenchyme (Fig. 3C), generally resembling the endogenous Hand2 expression pattern (Fig. 3E) and that observed in E10.5 Hand2-lacZ embryos (Ruest et al., 2003), though a small disto-rostral region of the arch remained unstained. It should be noted that detailed analysis of Hand2-directed transgene expression in the mouse was never performed using a construct driven only by the arch-specific enhancer (Charité et al., 2001), so it is not clear if the more limited distal expression observed here is due to species-specific differences or lack of additional regulatory elements. In addition to the pharyngeal arch staining, β-gal activity was detected in a small, ventral population of neural progenitors along the entire length of the neural tube (data not shown), though the basis of this staining was not clear.
Fig. 3.
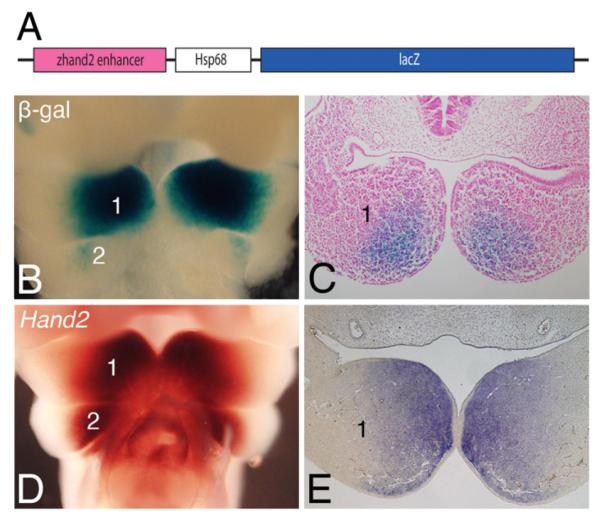
The zebrafish hand2 pharyngeal arch-specific enhancer drives lacZ expression in the mouse pharyngeal arches. A. Cloning of the zhand2 arch-specific enhancer into a minimal Hsp68 promoter-lacZ expression vector (Hsp68-lacZ). Plasmid is not drawn to scale. B. Ventral view of a transient transgenic embryo at embryonic day (E) 10.5 showed expression of the zhand2-Hsp68-lacZ transgene in the distal aspects of the mandibular arch (1). β-gal activity was also present in arch 2, but slightly more proximal to that observed in the mandibular arch. C. Transverse sectional analysis of the embryo shown in B illustrated the β-gal activity in the distal mandibular arch mesenchyme. D. Ventral view of endogenous Hand2 expression in control embryos at E10.5, illustrating the distal mesenchyme of the mandibular and second arches. E. Transverse sections through the mandibular arch of the embryos shown in D further illustrated the distal expression.
The hand2 enhancer is partially regulated by endothelin signaling
To narrow and refine our understanding of hand2 transcriptional regulation, we focused on the 280 bp zebrafish sequence (Fig. 1B) showing the highest conservation to the known mouse enhancer that is located within the larger 600 bp fragment described above. This smaller enhancer was cloned into a mCherry reporter vector using Multisite Gateway Recombination (Fig. 4A). We established three independent Tg(shand2:mCherry, cmlc2:GFP) founder lines showing stable germline transmission of the transgene and comparable reporter expression patterns, with Tg(shand2:mCherry,cmlc2:GFP)co10 subsequently analyzed in detail (referred to hereafter as Tg(shand2:mCherry)). While mCherry fluorescence was the weakest of the fluorophores we examined, mCherry was first detected in the ventral domain of arches 1 and 2 around 30 hpf (Fig. 4B), similar to what was found with the previous enhancers. mCherry continued to be present in a hand2-like expression pattern at 48 hpf, when fluorescence was also detected in arch 3 (Fig. 4C). Weak fluorescence could also be observed in arch 4. As seen in the other enhancer lines, Tg(shand2:mCherry) embryos continued to display reporter expression throughout the ventral and dorsal arch at 4 dpf (Fig. 4D) and 7 dpf (Fig. 4E). The shand2 enhancer was also able to drive reporter expression in the pectoral fin buds (data not shown). Other regions of endogenous hand2 expression, such as the gut and heart, did not display reporter activity with any of the enhancers examined. Thus, this 280 bp region is sufficient to drive early reporter expression within the ventral arch in Tg(shand2:m-Cherry) embryos, with fluorescence later observed in dorsal and ventral cartilage derivatives.
Fig. 4.
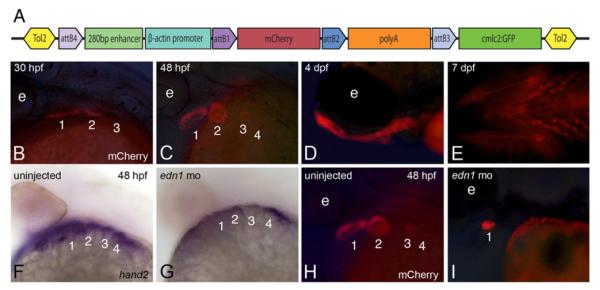
The minimal hand2 arch-specific enhancer is regulated by endothelin signaling. A. A 280 bp region of sequence contained within the larger zhand2 enhancer shown in Fig. 1C was cloned into p5E-FAbas and recombined with a middle entry clone containing a mCherry fluorescent reporter into the Tol2 destination vector pDestTol2CG2, resulting in hand2-directed mCherrry expression. Plasmid is not drawn to scale. B–E. In Tg(shand2:mCherry) embryos, mCherry fluorescence was observed in the Hand2 domain beginning around 30 hpf (B) and continuing through arch development (C, D). mCherry fluorescence can still be detected in the ventral cartilages as late as 7 dpf (E). F, G. Injecting 5 ng of an edn1 morpholino led to decreased hand2 expression at 48 hpf (G) compared to wild-type expression (F). H, I. mCherry activity was similarly decreased in Tg(shand2:mCherry) embryos following injection of the edn1 morpholino (I) compared to uninjected Tg(shand2:mCherry) embryos (H).
Loss or downregulation of any Edn1/Ednra signaling component leads to early loss of Hand2 in both mouse and zebrafish mutant embryos (Clouthier et al., 1998; Miller et al., 2007; Miller et al., 2003; Ozeki et al., 2004; Ruest et al., 2004; Walker et al., 2007; Walker et al., 2006; Yanagisawa et al., 1998). This effect is due in part to loss of Dlx5 and Dlx6 expression following disruption of endothelin signaling (Charité et al., 2001), as Dlx5 and Dlx6 contribute to induce Hand2 expression in the arches (Charité et al., 2001). Similarly, β-gal staining was not observed in most of the lower jaw of Ednra−/−;R26R;Hand2-Cre embryos, indicating that the enhancer was not induced in most of the mandibular arch (Ruest et al., 2004). To prove that our zebrafish hand2 arch-specific enhancer was indeed the enhancer downstream of Ednra1/2 signaling, we injected a morpholino against edn1 into Tg(shand2:mCherry) zebrafish embryos. Injection of this morpholino was previously shown to result in arch defects that phenocopy the defects observed in edn1/sucker mutants (Miller and Kimmel, 2001). When Tg(shand2:mCherry) morphant embryos were examined for hand2 message at 48 hpf, only the most ventral hand2 expression domain remained (Fig. 4G). This is consistent with previous studies in both mice and zebrafish showing that this region of hand2 expression occurs in an Ednra-independent manner (Alexander et al., 2011; Ozeki et al., 2004; Ruest et al., 2004; Talbot et al., 2010). Likewise, mCherry fluorescence in these morphant embryos was observed only in the ventral most region of the arch (Fig. 4H, I). This indicates that this enhancer is indeed regulated by Ednra-mediated signaling.
Activity of the hand2 enhancer is upregulated in vitro by Dlx5a
The mouse Hand2 enhancer is activated by Dlx5 and Dlx6, downstream of Ednra signaling (Barron et al., 2011; Charité et al., 2001). We therefore wished to determine whether this regulation by Dlx members is conserved in the zebrafish arch enhancer. We screened the zebrafish enhancer using MatInspector (Genomatix), a bioinformatics program that identifies predicted transcription factor binding sites based on known consensus recognition sequences. This analysis yielded multiple predicted sites for Dlx binding in addition to other predicted binding sites for several transcription factors not previously associated with Hand2 regulation (Fig. 5A). To verify that this enhancer can be activated by Dlx family members, the 600 bp zebrafish enhancer was cloned into a luciferase reporter vector (zhand2-luc). This vector was transfected into 10T1/2 mouse embryonic fibroblast cells along with either a mock expression vector or a dlx5a expression vector, with cell lysates subsequently analyzed for relative luciferase expression. Co-transfection of zhand2-luc with dlx5a led to a 2.6-fold increase in luciferase activity, suggesting that the zhand2 enhancer is regulated in a similar manner to that of the mouse Hand2 arch-specific enhancer (Fig. 5B).
Fig. 5.
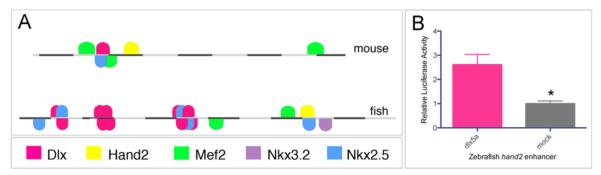
Transcriptional regulation of the hand2 arch-specific enhancer. A. The zebrafish enhancer (bottom) is shown, with the location of several potential regulators as predicted by MatInspector marked by colored boxes. The identities of the colored boxes are shown below the enhancer; a similar map of the mouse Hand2 enhancer is shown for comparison (top). Each gray or black bar represents 50 bp. B. A hand2-luciferase vector was transfected into 10T1/2 mouse embryonic fibroblasts along with a dlx5a expression vector. Transfection efficiency was controlled by co-transfection with a renilla reporter construct. Activation of zhand2 by Dlx5a was normalized to the activation obtained when using an empty expression vector (mock). The error is presented as S.E.M * p=0.0026.
Discussion
Using comparative genomics and Tol2-mediated transgenesis, we have identified a zebrafish pharyngeal arch-specific enhancer for hand2 that drives transgene expression in the early ventral pharyngeal arches, with fluorescence later observed in the cartilaginous skeleton of the viscerocranium. This enhancer appears homologous to the arch-specific enhancer for Hand2 that has been described in mouse (Charité et al., 2001). Using this enhancer, it will be possible to better understand the early transcriptional regulation of hand2 during D–V patterning of the arches in zebrafish. In addition, the enhancer will drive targeted gene expression in the early ventral zebrafish arch, allowing a more detailed investigation of the regulatory mechanisms that pattern this domain during facial morphogenesis. In addition, these studies may uncover functions of Hand2 not previously recognized.
While we have focused on the ECR that shows homology to the known mouse Hand2 pharyngeal arch enhancer, several other ECRs exist within the hand2 locus that likely contribute to the regulation of hand2 expression during the development of other organs and tissues (Fig. 1A). One of these may be the other pharyngeal arch enhancer that appears to act redundantly with the identified arch enhancer during mouse development (Ruest et al., 2004; Yanagisawa et al., 2003); discussed below). In addition, one or more of the ECRs may regulate hand2 expression in the sympathetic and enteric nervous systems, as the location of these enhancers in either the mouse or zebrafish is not known. Based on sequence similarity alone, none of the ECRs appear to be the hand2 heart-specific enhancer, which in the mouse lies 2 kb downstream of the arch-specific enhancer (Charité et al., 2001;McFadden et al., 2000). However, we are currently analyzing the functions of the other ECRs in zebrafish embryos to understand their potential regulatory roles and whether they drive hand2 expression in these other tissues.
Enhancers as a mechanism to understand the transcriptional regulation of arch development
Proper temporospatial hand2 expression has been established as a vital component of D-V patterning in the pharyngeal arches and several upstream signals act in conjunction to regulate this expression. Ednra signaling is a crucial regulator of Hand2 expression, as loss of Edn1/Ednra signaling in both mice and zebrafish leads to downregulation of Hand2/hand2 expression, due primarily to loss of Dlx5/Dlx6 expression (Clouthier et al., 2000; Miller et al., 2003; Ozeki et al., 2004; Ruest et al., 2004). In addition, Schilling and colleagues have recently shown that Bmp signaling works both in parallel and in series with Edn1 to regulate hand2 expression (Alexander et al., 2011). It is not known if the Ednra-independent expression domain of Hand2 observed in the most distal aspect of the mandibular arch of mouse (Ozeki et al., 2004; Ruest et al., 2004) and zebrafish (Miller et al., 2003) can be explained by this action of Bmps. However, the conserved enhancer we have isolated in both mouse (Charité et al., 2001) and zebrafish (this study) contains all regulatory sequences required to achieve this complex regulation. This is demonstrated both by the finding that Dlx5 and Dlx5a can activate the mouse (data not shown) and zebrafish Hand2/hand2 enhancer (this study), respectively, and by the fact that hand2 expression remains in the most ventral region of the zebrafish arch following morpholino knockdown of edn1. We have also identified a number of other potential transcription factor binding sequences that could contribute to the positive and negative regulation of hand2 in both species (Fig. 5A). Since testing the function of putative sites in vivo using zebrafish is relatively fast and efficient, these new transgenic fish lines could rapidly increase our understanding of hand2 regulation in both wild type and mutant embryos. Additionally, by utilizing both the Tg(hand2:tag-RFP) and Tg(mHand2:GFP) lines, it is now possible to use the zebrafish model to understand the similarities and differences in mouse and zebrafish hand2 transcriptional regulation.
Regulation of Hand2 expression appears complex in both species. One nuance described above involves an additional Hand2 enhancer in the mouse that appears to act redundantly in inducing or maintaining Hand2 expression in the distal mandibular arch, though little is known about it (Yanagisawa et al., 2003). Here we have shown that both our fish and mouse arch enhancers initially drive transgene expression in the ventral hand2 domain during early D-V patterning, though later fluorescence is also observed in more dorsal derivatives, suggesting several possibilities. First, other regulatory elements may be required to restrict hand2 expression to the ventral arch and its derivatives during later arch morphogenesis. Indeed, the mouse Hand2 upstream region used to create both Hand2-lacZ (Charité et al., 2001) and Hand2-Cre (Ruest et al., 2003) embryos contains a strong repressor region that lies several kilobases downstream of the arch-specific enhancer (Marthe Howard, personal communication), This may explain why neither proximal arch nor CNS expression of Hand2-driven transgenes has been observed in transgenic mouse embryos, as we used only a small region of the Hand2/hand2 upstream region that does not contain this repressor.
Two other possibilities are that hand2 expression normally spreads dorsally as part of later differentiation (i.e., chondrogenesis) or that some of the cells that express hand2 during early arch patterning are later located more dorsally. While our in situ hybridization analysis did not reveal obvious dorsal hand2 expression, we cannot rule out the possibility that such dorsal expression occurs in a transient manner. Likewise, the exact boundaries of dorsal, intermediate and ventral domains during later arch jaw development are difficult to define and have not been finely mapped. However, fate mapping studies in zebrafish of cells in the mandibular and second arches have shown that cells located near the ventral-intermediate domain boundary can contribute to dorsal arch derivatives (Crump et al., 2004; Kimmel and Eberhart, 2008). If such cell movement occurred, fluorescence perdurance could explain the fluorescence observed in dorsal elements in all transgenics we tested. It will be interesting to examine the dorsal and ventral fluorescence in a hand2 BAC transgenic fish line recently described (Kikuchi et al., 2011).
Further analysis of the enhancer and surrounding regulatory sequences will be required to better understand how normal regulation of hand2 expression in the developing pharyngeal arches is achieved. Until this point is better understood, the fact that hand2-driven fluorescence expands dorsally may initially limit the utility of the hand2 arch-specific enhancer later in facial development. However, our results also illustrate that the enhancer will be quite useful as a tool to mark the ventral arch domain during early D-V patterning or to drive transgene expression to this domain.
Evolutionary conservation of Hand2 regulation and function
Loss of Hand2 leads to severe ventral craniofacial defects in both mouse and zebrafish (Barron et al., 2011; Miller et al., 2003), illustrating that the requirement for Hand2 in proper facial morphogenesis is conserved amongst gnathostomes. Our current results suggest that the regulation of hand2 expression is highly conserved as well. This point is evident in the finding that each enhancer can function in the other species, driving transgene expression within the early hand2 expression domain in an almost identical pattern to the endogenous pattern. The identity of factors besides Dlx proteins that might bind to the hand2 enhancer are not known, though the similarity of predicted transcription factor binding sites across the mouse and fish enhancers (Fig. 5A) is further evidence for conservation of transcriptional regulation. Dlx proteins are predicted to bind to multiple sites in the zebrafish hand2 enhancer, something that occurs in the mouse Hand2 enhancer (Charité et al., 2001). Perhaps more interesting are the putative binding sites for Hand2 and Mef2. While it is not known whether Hand2 can positively regulate its own expression during early arch patterning, such a positive feedback loop could explain in part how arch expression of Hand2 is maintained when Ednra signaling is lost subsequent to Hand2 induction (Ruest and Clouthier, 2009). Mef2c (mef2ca in zebrafish) is also expressed in the arches and appears to work in concert with Ednra signaling to induce Dlx5/Dlx6 expression during arch patterning and thus indirectly affects Hand2 expression (Miller et al., 2007; Verzi et al., 2007). Hand2/hand2 expression is downregulated in both mouse and zebrafish embryos following loss of Mef2c or mef2ca function, respectively, even though Dlx5/Dlx6 (dlx5a/dlx6a) expression is only partially lost (Miller et al., 2007; Verzi et al., 2007). Direct regulation of Hand2/hand2 expression by Mef2c/mef2ca is a possible explanation for this Dlx5/Dlx6 independent change in expression, as Hand2 and Mef2c have been shown to act in conjunction in other developmental processes. Hand2 and Mef2c act as cofactors for transcriptional activation of Nppa (the gene encoding atrial natriuretic peptide) in vitro and as an essential component of cardiac development in vivo (Zang et al., 2004). Similar interactions have been described between Mef2 family members and other bHLH factors, namely Mef2a and MyoD, in skeletal muscle development (Kaushal et al., 1994). As the Hand2 and Mef2c predicted binding sites are closely associated in both the mouse and zebrafish enhancers (Fig. 5A), it is possible that these two proteins may be acting together to regulate Hand2 expression during pharyngeal arch development. Further biochemical analysis will be required to understand this relationship.
Understanding the basis of hand2 transcriptional regulation may also shed light on the evolution of the jaw structure in gnathostomes. In cyclostomes (such as lampreys), there is some degree of D–V patterning of the pharyngeal arches, including ventral-restricted expression of HandA (Cerny et al., 2010; Kuraku et al., 2010). This suggests that the Hand enhancer regulating this ventral expression preceded jaw formation. While there may be some variation among lamprey species (Cerny et al., 2010), P. marinus lampreys exhibit a nested pattern of Dlx gene expression in the arches in addition to ventral HandA expression (Cerny et al., 2010). Thus, the Dlx/Hand2 pathway appears to be a very early event in the evolution of facial development. However, this D–V pattern of gene expression is not sufficient to induce jaw formation in cyclostomes, suggesting that other factors are required for the evolution of the jaw. One major difference in arch gene expression between lampreys and gnathostomes is the presence of nkx3.2 (formerly bapx1) expression in the gnathostome intermediate arch domain. This expression, induced by Ednra signaling in both zebrafish (Miller et al., 2003; Zuniga et al., 2011) and mouse (Tavares and Clouthier, unpublished) and by Bmp4 upstream of Edn1 in zebrafish (Alexander et al., 2011), establishes the joint between the upper and lower jaws in zebrafish (Miller et al., 2003) and chick (Wilson and Tucker, 2003). That Hand gene expression is ventrally restricted in lampreys indicates that Hand2 by itself is not directly responsible for evolution of the jaw apparatus. However, Hand2 confines nkx3.2 to the intermediate domain in the zebrafish (Miller et al., 2003; Talbot et al., 2010) and mouse (Tavares and Clouthier, unpublished), with disruption of hand2 expression in zebrafish leading to ventral expansion of nkx3.2 and disruption of the joint between Meckel’s cartilage and the palatoquadrate (Miller et al., 2003). Thus, while ventral restriction of Hand is not sufficient for jaw formation, the transcriptional regulation leading to its ventral restriction may have been critical for later defining the boundary of jaw joint formation.
Acknowledgements
The authors would like to thank Richard Cripps and Tom Schilling for helpful comments, Chi-Bin Chien and Li-En Jao for plasmids, Robert E. Hammer (University of Texas Southwestern Medical Center) for assistance with mouse embryo injections, Crystal Woods and Alicia Navetta for technical assistance and Alex Blasky for assistance with confocal microscopy. The work was supported by NIH grant DE018899 (D.E.C.). J.M.I. was supported by NIH T32 GM008497-S1 as a NIH Graduate Partnership Program student with NHLBI.
References
- Alexander C, Zuniga E, Blitz IL, Wada N, LePabic P, Javidan Y, Zhang T, Cho KW, Crump JG, Schilling TF. Combinatorial roles for Bmps and Endothelin 1 in patterning the dorsal-ventral axis of the craniofacial skeleton. Development. 2011;138:5135–5146. doi: 10.1242/dev.067801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron F, Woods C, Kuhn K, Bishop J, Howard MJ, Clouthier DE. Downregulation of Dlx5 and Dlx6 expression by Hand2 is essential for initiation of tongue morphogenesis. Development. 2011;138:2249–2259. doi: 10.1242/dev.056929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner-Fraser M. Origins and developmental potential of the neural crest. Exp. Cell. Res. 1995;218:405–417. doi: 10.1006/excr.1995.1173. [DOI] [PubMed] [Google Scholar]
- Cerny R, Cattell M, Sauka-Spengler T, Bronner-Fraser M, Yu F, Medeiros DM. Evidence for the prepattern/cooption model of vertebrate jaw evolution. Proc. Nat. Acad. Sci. USA. 2010;107:17262–17267. doi: 10.1073/pnas.1009304107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Jiang X, Ito Y, Bringas P, Jr., Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- Charité J, McFadden DG, Merlo GR, Levi G, Clouthier DE, Yanagisawa M, Richardson JA, Olson EN. Role of Dlx6 in regulation of an endothelin-1-dependent, dHAND branchial arch enhancer. Genes Dev. 2001;15:3039–3049. doi: 10.1101/gad.931701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouthier DE, Comerford SA, Hammer RE. Hepatic fibrosis, glomerulo-sclerosis, and a lipodystrophy-like syndrome in PEPCK-TGF-β1 transgenic mice. Journal of Clinical Investigation. 1997;100:2697–2713. doi: 10.1172/JCI119815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouthier DE, Garcia E, Schilling TF. Regulation of facial morphogenesis by endothelin signaling: insights from mouse and fish. Am. J. Med. Genet. A. 2010;152A:2962–2973. doi: 10.1002/ajmg.a.33568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouthier DE, Hosoda K, Richardson JA, Williams SC, Yanagisawa H, Kuwaki T, Kumada M, Hammer RE, Yanagisawa M. Cranial and cardiac neural crest defects in endothelin-A receptor-deficient mice. Development. 1998;125:813–824. doi: 10.1242/dev.125.5.813. [DOI] [PubMed] [Google Scholar]
- Clouthier DE, Williams SC, Yanagisawa H, Wieduwilt M, Richardson JA, Yanagisawa M. Signaling pathways crucial for craniofacial development revealed by endothelin-A receptor-deficient mice. Dev. Biol. 2000;217:10–24. doi: 10.1006/dbio.1999.9527. [DOI] [PubMed] [Google Scholar]
- Couly GF, Grapin-Botton A, Coltey P, Le Douarin NM. The regeneration of the cephalic neural crest, a problem revisited: the regenerating cells originate from the contralateral or from the anterior and posterior neural fold. Development. 1996;122:3393–3407. doi: 10.1242/dev.122.11.3393. [DOI] [PubMed] [Google Scholar]
- Crump JG, Swartz ME, Kimmel CB. An integrin-dependent role of pouch endoderm in hyoid cartilage development. PLoS Biol. 2004;2:1432–1445. doi: 10.1371/journal.pbio.0020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushal S, Schneider JW, Nadal-Ginard B, Mahdavi V. Activation of the myogenic lineage of MEF2A, a factor that induces and cooperates with MyoD. Science. 1994;266:1236–1240. doi: 10.1126/science.7973707. [DOI] [PubMed] [Google Scholar]
- Kikuchi K, Holdaway I, Major RJ, Blum N, Dahn RD, Begemann G, Poss KD. Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev. Cell. 2011;15:397–404. doi: 10.1016/j.devcel.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel CB, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Eberhart JK. The midline, oral ectoderm, and the arch-0 problem. Integr. Comp. Biol. 2008;48:668–680. doi: 10.1093/icb/icn048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Miller CT, Moens CB. Specification and morphogenesis of the zebrafish larval head skeleton. Dev. Biol. 2001;233:239–257. doi: 10.1006/dbio.2001.0201. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ullmann B, Walker M, Miller CT, Crump JG. Endothelin 1-mediated regulation of pharyngeal bone development in zebrafish. Development. 2003;130:1339–1351. doi: 10.1242/dev.00338. [DOI] [PubMed] [Google Scholar]
- Kontges G, Lumsden A. Rhombencephalic neural crest segmentation is preserved throughout craniofacial ontogeny. Development. 1996;122:3229–3242. doi: 10.1242/dev.122.10.3229. [DOI] [PubMed] [Google Scholar]
- Kuraku S, Takio Y, Sugahara F, Takechi M, Kuratani S. Evolution of oropharyngeal patterning mechanisms involving Dlx and endothelins in vertebrates. Dev. Biol. 2010;341:315–323. doi: 10.1016/j.ydbio.2010.02.013. [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Kurihara H, Suzuki H, Kodama T, Maemura K, Nagai R, Oda H, Kuwaki T, Cao W-H, Kamada N, Jishage K, Ouchi Y, Azuma S, Toyoda Y, Ishikawa T, Kumada M, Yazaki Y. Elevated blood pressure and craniofacial abnormalities in mice deficient in endothelin-1. Nature. 1994;368:703–710. doi: 10.1038/368703a0. [DOI] [PubMed] [Google Scholar]
- Le Douarin NM. The Neural Crest. Cambridge Univ. Press; Cambridge: 1982. [Google Scholar]
- McFadden DG, Charité J, Richardson JA, Srivastava D, Firulli AB, Olson EN. A GATA-dependent right ventricular enhancer controls dHAND transcription in the developing heart. Development. 2000;127:5331–5341. doi: 10.1242/dev.127.24.5331. [DOI] [PubMed] [Google Scholar]
- Miller CT, Kimmel CB. Morpholino phenocopies of endothelin 1 (sucker) and other anterior arch class mutations. Genesis. 2001;30:186–187. doi: 10.1002/gene.1061. [DOI] [PubMed] [Google Scholar]
- Miller CT, Schilling TF, Lee K-H, Parker J, Kimmel CB. sucker encodes a zebrafish Endothelin-1 required for ventral pharyngeal arch development. Development. 2000;127:3815–3838. doi: 10.1242/dev.127.17.3815. [DOI] [PubMed] [Google Scholar]
- Miller CT, Swartz ME, Khuu PA, Walker MB, Eberhart JK, Kimmel CB. mef2ca is required in cranial neural crest to effect Endothelin1 signaling in zebrafish. Dev. Biol. 2007;308:144–157. doi: 10.1016/j.ydbio.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CT, Yelon D, Stainier DY, Kimmel CB. Two endothelin 1 effectors, hand2 and bapx1, pattern ventral pharyngeal cartilage and the jaw joint. Development. 2003;130:1353–1365. doi: 10.1242/dev.00339. [DOI] [PubMed] [Google Scholar]
- Nair S, Li W, Cornell R, Schilling TF. Requirements for endothelin type-A receptors and endothelin-1 signaling in the facial ectoderm for the patterning of skeletogenic neural crest cells in zebrafish. Development. 2007;134:335–345. doi: 10.1242/dev.02704. [DOI] [PubMed] [Google Scholar]
- Ozeki H, Kurihara Y, Tonami K, Watatani S, Kurihara H. Endothelin-1 regulates the dorsoventral branchial arch patterning in mice. Mech. Dev. 2004;121:387–395. doi: 10.1016/j.mod.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Ruest L-B, Dager M, Yanagisawa H, Charité J, Hammer RE, Olson EN, Yanagisawa M, Clouthier DE. dHAND-Cre transgenic mice reveal specific potential functions of dHAND during craniofacial development. Dev. Biol. 2003;257:263–277. doi: 10.1016/s0012-1606(03)00068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruest LB, Clouthier DE. Elucidating timing and function of endothelin-A receptor signaling during craniofacial development using neural crest cell-specific gene deletion and receptor antagonism. Dev. Biol. 2009;328:94–108. doi: 10.1016/j.ydbio.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruest LB, Xiang X, Lim KC, Levi G, Clouthier DE. Endothelin-A receptor-dependent and -independent signaling pathways in establishing mandibular identity. Development. 2004;131:4413–4423. doi: 10.1242/dev.01291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot JC, Johnson SL, Kimmel CB. hand2 and Dlx genes specify dorsal, intermediate and ventral domains within zebrafish pharyngeal arches. Development. 2010;137:2507–2517. doi: 10.1242/dev.049700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C.a.T.B. High resolution whole-mount in situ hybridization. Zebrafish Science Monitor. 1998;15:8–9. [Google Scholar]
- Verzi MP, Agarwar P, Brown C, McCulley DJ, Schwarz JJ, Black BL. The transcription factor MEF2C is required for craniofacial development. Dev. Cell. 2007;12:645–652. doi: 10.1016/j.devcel.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker MB, Miller CT, Swartz ME, Eberhart JK, Kimmel CB. phospholipase C, beta 3 is required for Endothelin1 regulation of pharyngeal arch patterning in zebrafish. Dev. Biol. 2007;304:194–207. doi: 10.1016/j.ydbio.2006.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker MB, Miller CT, Talbot JC, Stock DW, Kimmel CB. Zebrafish furin mutants reveal intricacies in regulating Endothelin1 signaling in craniofacial patterning. Dev. Biol. 2006;295:194–205. doi: 10.1016/j.ydbio.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book: A Guide for the Laboratory use of Zebrafish (Danio rerio) University of Oregon Press; Eugene: 1993. [Google Scholar]
- Wilson J, Tucker AS. Fgf and Bmp signals repress the expression of Bapx1 in the mandibular mesenchyme and control the position of the developing jaw joint. Dev. Biol. 2003;266:138–150. doi: 10.1016/j.ydbio.2003.10.012. [DOI] [PubMed] [Google Scholar]
- Yanagisawa H, Clouthier DE, Richardson JA, Charité J, Olson EN. Targeted deletion of a branchial arch-specific enhancer reveals a role of dHAND in craniofacial development. Development. 2003;130:1069–1078. doi: 10.1242/dev.00337. [DOI] [PubMed] [Google Scholar]
- Yanagisawa H, Yanagisawa M, Kapur RP, Richardson JA, Williams SC, Clouthier DE, de Wit D, Emoto N, Hammer RE. Dual genetic pathways of endothelin-mediated intercellular signaling revealed by targeted disruption of endothelin converting enzyme-1 gene. Development. 1998;125:825–836. doi: 10.1242/dev.125.5.825. [DOI] [PubMed] [Google Scholar]
- Yin C, Kikuchi K, Hochgreb T, Poss KD, Stainier DY. Hand2 regulates extracellular matrix remodeling essential for gut-looping morphogenesis in zebrafish. Dev. Cell. 2010;18:973–984. doi: 10.1016/j.devcel.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang MX, Li Y, Xue LX, Jia HT, Jing H. Cooperative activation of atrial naturetic peptide promoter by dHAND and MEF2C. J. Cell Biochem. 2004;93:1255–66. doi: 10.1002/jcb.20225. [DOI] [PubMed] [Google Scholar]
- Zuniga E, Rippen M, Alexander C, Schilling TF, Crump JG. Gremlin2 regulates distinct roles of Bmp and Endothelin 1 signaling in dorsoventral patterning of the facial skeleton. Development. 2011;138:5147–5156. doi: 10.1242/dev.067785. [DOI] [PMC free article] [PubMed] [Google Scholar]