Abstract
The COP9 signalosome (CSN) is an eight subunit protein complex conserved in all higher eukaryotes. In Arabidopsis thaliana, the CSN regulates auxin response by removing the ubiquitin-like protein NEDD8/RUB1 from the CUL1 subunit of the SCFTIR1/AFB ubiquitin-ligase (deneddylation). Previously described null mutations in any CSN subunit result in the pleiotropic cop/det/fus phenotype and cause seedling lethality, hampering the study of CSN functions in plant development. In a genetic screen to identify enhancers of the auxin response defects conferred by the tir1-1 mutation, we identified a viable csn mutant of subunit 3 (CSN3), designated eta7/csn3-3. In addition to enhancing tir1-1 mutant phenotypes, the csn3-3 mutation alone confers several phenotypes indicative of impaired auxin signaling including auxin resistant root growth and diminished auxin responsive gene expression. Unexpectedly however, csn3-3 plants are not defective in either the CSN-mediated deneddylation of CUL1 or in SCFTIR1-mediated degradation of Aux/IAA proteins. These findings suggest that csn3-3 is an atypical csn mutant that defines a novel CSN or CSN3-specific function. Consistent with this possibility, we observe dramatic differences in double mutant interactions between csn3-3 and other auxin signaling mutants compared to another weak csn mutant, csn1-10. Lastly, unlike other csn mutants, assembly of the CSN holocomplex is unaffected in csn3-3 plants. However, we detected a small CSN3-containing protein complex that is altered in csn3-3 plants. We hypothesize that in addition to its role in the CSN as a cullin deneddylase, CSN3 functions in a distinct protein complex that is required for proper auxin signaling.
Introduction
The phytohormone auxin (indole-3-acetic acid or IAA) regulates numerous plant developmental processes, by modulating the expression of auxin responsive genes to control cell division, expansion and differentiation [1]. Two large families of transcriptional regulators play essential roles in auxin mediated gene expression: short-lived Aux/IAA proteins and Auxin Response Factor (ARF) transcription factors. In the absence of an auxin signal, Aux/IAAs heterodimerize with ARFs to repress the transcriptional activation of auxin responsive genes [2], [3]. To activate auxin responsive gene expression, Aux/IAA repression must be removed, which is accomplished by the SCFTIR1/AFB E3 ubiquitin ligase and the 26S proteasome mediated proteolysis of Aux/IAAs in an auxin-dependent manner [4]–[6].
Ubiquitin is a small conserved protein, which can be covalently conjugated to specific substrate proteins through the activity of a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin-ligase (E3) [7]. Proteins with a polyubiquitin chain can then be recognized and degraded by the 26S proteasome. A major class of E3 ubiquitin ligases are the cullin-RING ligases (CRLs), among which the SCF (for Skp1-Cullin-F-box) sub-type has been well-studied [8]. SCF ligases consist of four subunits: the cullin protein, CUL1, binds the RING box protein RBX1 with its C-terminus, while the SKP1 adaptor protein (ASK1 in Arabidopsis) recruits any of many F-box proteins (FBPs) to the N-terminus of CUL1 [9], [10]. The TIR1, AFB1, AFB2, and AFB3 F-box proteins of the SCFTIR1/AFB complex are auxin receptors, targeting Aux/IAAs for ubiquitination and degradation upon binding auxin [11]–[13]. Mutations in SCFTIR1/AFB subunits, such as axr6/cul1, ask1 and tir1, cause the stabilization of Aux/IAAs and a variety of auxin response defects including auxin resistant root growth and reduced lateral root development [4], [14], [15]. The proper function of SCF ligases, as well as other CRLs, also requires a process called neddylation, in which the ubiquitin-like protein RUB/NEDD8 is covalently conjugated to the cullin subunit [16]–[18]. The AXR1, ECR1, and RCE1 enzymes mediate cullin neddylation, and mutations in these factors result in a reduction in the level of neddylated cullin and pleiotropic growth defects, including diminished auxin signaling [16], [19]. The NEDD8 modification of CUL1 is a highly dynamic process [20], [21]. Cleavage of NEDD8 from cullins (deneddylation) is promoted by a protein complex called the COP9 signalosome (CSN), as the CSN complex purified from porcine spleen deneddylates Cul1 in vitro [21], [22]. Consistently, studies in various organisms have shown that neddylated cullin proteins accumulate in csn mutants [22]–[25].
The COP9 signalosome (CSN) is an evolutionarily conserved protein complex of eight subunits [26]. It was originally identified in Arabidopsis through genetic screens for mutants exhibiting a constitutive photomorphogenic/de-etiolated (cop/det) phenotype, and was subsequently biochemically purified from both plant and animal protein extracts [26], [27]. The CSN is structurally related to the 19S lid of the 26S proteasome and eukaryotic translation initiation factor 3 (eIF3), and is composed of six PCI (for Proteasome, COP9, eIF3) domain-containing subunits (CSN1-4, 7, and 8) and two MPN (for Mpr1p, Pad1p N-terminal) domain-containing subunits (CSN5 and CSN6) [27]. It has been established that the CSN regulates SCF activity by deneddylating the CUL1 subunit of the SCF [22], [28]. Besides CUL1, the CSN also deneddylates other cullin proteins and broadly regulates many CRLs, including those containing CUL2, CUL3 and CUL4 [21], [23], [29], [30]. In plants, the CSN has been implicated in a variety of processes, including auxin, jasmonate (JA), and gibberellin (GA) signaling, flower development, and light signaling via its interaction with many CRLs [29], [31], [32]. CSN-mediated cullin deneddylation is catalyzed by the JAMM metallozyme motif within the CSN5 subunit [22], [33]. However, CSN5 can only provide this isopeptidase activity after incorporation into the CSN holocomplex, as deneddylation activity is defective in all previously characterized CSN subunit mutants [23], [34], [35].
Neddylation plays a positive role in regulating SCF activity, by promoting a conformational change in CUL1 that shortens the distance between FBP-bound substrates and the E2 ubiquitin-conjugating enzyme [36]–[38]. Although in vitro biochemical studies indicated that the CSN negatively regulates E3 ubiquitin ligase activity [21], [33], genetic evidence suggests otherwise [28], [39], [40]. Studies in yeast and Drosophila suggested that although the CSN returns CRL activities to basal levels, it can also facilitate CRL activities by either maintaining the stability of labile substrate adapters or by recycling unstable, neddylated cullins into the more stable deneddylated isoform [41], [42]. In Arabidopsis, the CSN is also required for the proper functions of SCFTIR1/AFB, as well as SCFSLY1, in the degradation of Aux/IAA proteins or the DELLA proteins, respectively [28], [31], [43]. Together, these findings suggest that cullin neddylation/deneddylation is a highly dynamic process essential for maintaining proper CRL function [44], [45]. In addition to regulating SCF activity by deneddylating cullins, recent biochemical findings indicate that the CSN also inhibits SCF complexes by a noncatalytic mechanism [46], [47]. Following deneddylation, the CSN remains stably associated with the SCF, sterically hindering both the re-neddylation of CUL1 and SCF interactions with the E2 enzyme.
Arabidopsis null mutants of any CSN subunit are phenotypically indistinguishable and exhibit the pleiotropic cop/det/fus phenotype, which is characterized by short hypocotyl and open cotyledons of dark-grown seedlings, accumulation of anthocyanin and seedling lethality [23], [27], [48], [49]. Severe deneddylation defects have been found in previously described null mutants of each of the CSN subunits [23], [48], but the associated seedling lethality hampers the further analyses of CSN functions. Recently however, a few weak, viable Arabidopsis csn mutants have been described. These include mutants lacking one of the two copies of CSN5 and CSN6 encoded in the Arabidopsis genome [23], [50], as well as hypomorphic missense alleles of CSN1 and CSN2 [40], [43]. Importantly however, these viable csn mutants still exhibit diminished deneddylation activity, resulting in the accumulation of neddylated CUL1 and reduced SCF activity [23], [28], [34], [35], [40], [43].
In a previously described genetic screen for enhancers of tir1-1 [15], [51]–[54], we identified two weak csn subunit mutants, designated as eta6/csn1-10 [43] and eta7/csn3-3. Our phenotypic characterization of these csn mutants, together with expression studies with auxin regulated reporters, demonstrate that csn1-10 and csn3-3 confer very similar reductions in auxin response. However, unlike csn1-10, which is a typical csn mutant with defects in CSN-mediated deneddylation and Aux/IAA protein degradation [43], neither of these defects were observed in the csn3-3 mutant. Furthermore, genetic interactions between these csn mutants and additional auxin signaling mutants also distinguish csn3-3 from other csn mutants. Our studies suggest that csn3-3 is a unique csn mutant that defines a novel functional activity for the CSN3 subunit of the COP9 signalosome in the regulation of auxin signaling.
Results
eta7 is a weak allele of COP9 signalosome subunit 3 (CSN3)
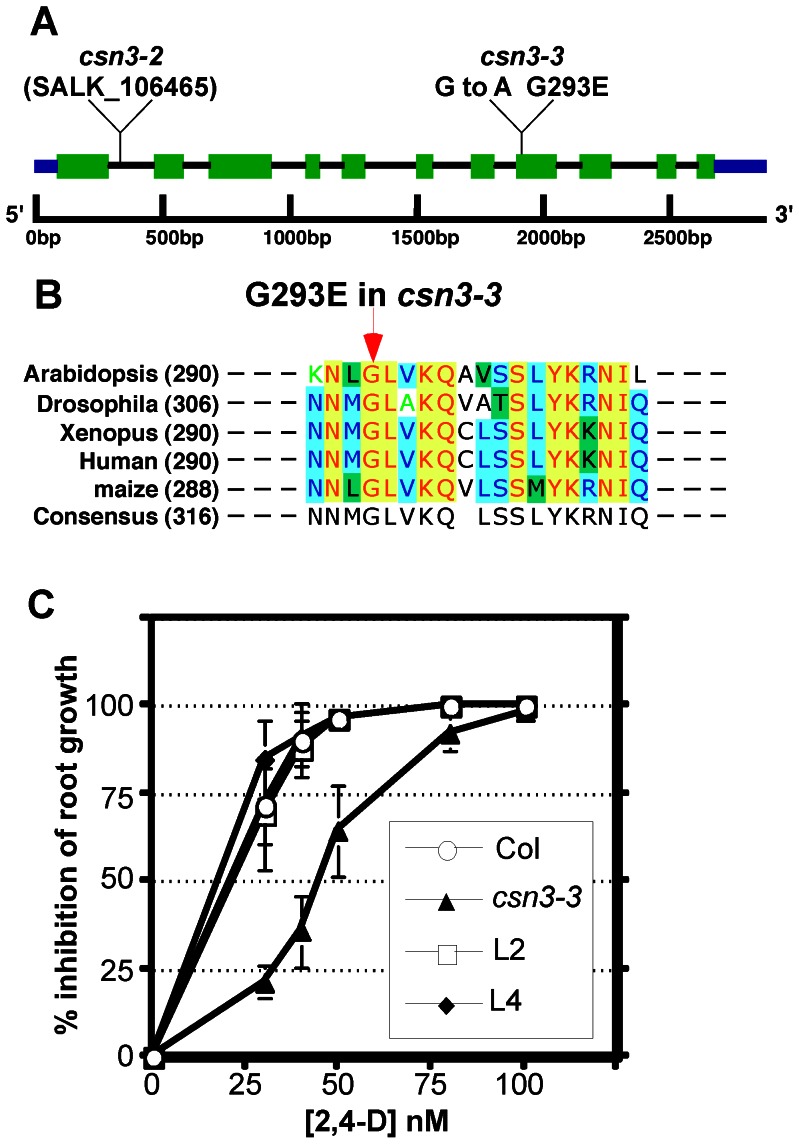
We have previously reported the identification of several auxin response mutants isolated from a genetic screen for enhancers of the tir1-1 auxin response defect (designated as eta mutants), including eta1/axr6-3 [15], eta2-1/cand1 [52], eta3/sgt1b [53], eta4/pdr9-1 [54], eta5/iar4 [51] and eta6/csn1-10 [43]. The eta7 mutant was also identified in this screen. Map based cloning positioned the eta7 mutation within a 330 kb interval on chromosome five. This interval contained 102 predicted genes, including CSN3/FUS11, which encodes subunit 3 of the COP9 signalosome (CSN) [55]. Given that csn subunit mutations are known to confer diminished auxin response phenotypes, we sequenced the CSN3 locus from eta7 plants and identified a single mutation ( Figure 1A ). This mutation results in a G293E missense mutation within the highly conserved PCI domain of CSN3. This domain is important for subunit interaction and CSN complex assembly [56]. Primary sequence alignment among the PCI domains of several CSN3 orthologs revealed that Gly293 is extremely highly conserved throughout eukaryotes ( Figure 1B ). To confirm that this mutation was responsible for the eta7 auxin response defect, we conducted complementation tests by transforming a genomic CSN3 construct into eta7 mutant plants. The CSN3 transgene fully restored auxin sensitivity to eta7 seedlings when tested in root growth assays ( Figure 1C ). We therefore renamed eta7 as csn3-3.
Figure 1. csn3-3 is a weak missense allele of CSN3.
(A) Schematic representation of the Arabidopsis CSN3 locus. Positions of the csn3-3 missense mutation and previously described csn3-2 T-DNA insertion are indicated. (B) Sequence alignment around the csn3-3 mutation among the CSN3 homologs from Arabidopsis thaliana, Drosophila melanogaster, Xenopus laevis, Homo sapiens (Human) and Zea mays (maize). The arrow indicates the position of the csn3-3 G293E missense mutation. (C) Complementation of the csn3-3 auxin resistance phenotype by introduction of a genomic CSN3 transgene. L2 and L4 are two independent csn3-3[gCSN3] transformants.
Characterization of csn3-3 phenotypes
The CSN plays a well-established role in auxin signaling, acting as a deneddylase to regulate SCFTIR1/AFB activity. Mutants in any CSN subunit exhibit auxin related phenotypes, such as auxin resistant root growth and reduced lateral root formation [28], [34], [43]. In a dose-response assay measuring the auxin inhibition of root elongation, we found csn3-3 was mildly resistant to exogenous auxins. After transfer to media supplemented with 0.05 µM 2,4-D, root growth of wild-type (Col) seedlings was nearly completely inhibited ( Figure 2A ). However, csn3-3 seedlings were resistant to this inhibition, displaying only 47% inhibition of root growth. A similar degree of auxin resistant root growth was observed with the csn1-10 and tir1-1 mutants. Additionally, both csn1-10 and csn3-3 enhanced the weak auxin resistance phenotype of tir1-1, with csn1-10 tir1-1 and csn3-3 tir1-1 double mutants exhibiting comparable dose-response profiles in root growth inhibition assays ( Figure 2A ). Similar assays using IAA-supplemented media also found that csn3-3 exhibited auxin resistance and enhanced the tir1-1 phenotype, demonstrating that the auxin response defect of csn3-3 is not 2,4-D specific ( Figure 2B ).
Figure 2. csn3-3 confers auxin response defects.
(A) Inhibition of root elongation by increasing concentrations of the synthetic auxin 2,4-D. 5-day-old (d.o.) seedlings (n≥15) grown on ATS medium were transferred to medium containing 2,4-D and grown for another four days. Data are presented as percent inhibition of root growth compared to growth on unsupplemented ATS. Error bars = SD. (B) IAA dose-response curve of inhibition of root growth. (C) Lateral root (LR) initiation was measured in 11-d.o. seedlings grown on unsupplemented ATS medium. Data were presented as number of the lateral root initiations per cm root length. Error bars = SD (n≥15). (D) Transgenic Col, csn1-10 and csn3-3 carrying either the BA3:GUS or DR5:GUS reporters. 6-d.o. seedlings were treated with 0.5 µM 2,4-D for 12 h (BA3:GUS) or 4 h (DR5:GUS) before histochemical staining for β-glucuronidase activity. (E) Complementation of the reduced BA3:GUS expression phenotype of csn3-3 seedlings by a genomic CSN3 transgene. L2 and L4 are two independent csn3-3[gCSN3] transformants.
Previous reports have shown that plants expressing a CSN5 antisense construct develop fewer lateral roots and reduced root hair elongation compared to wild-type controls [28]. Consistent with this finding, the csn3-3 and the csn1-10 mutations enhanced the weak lateral root defect of tir1-1 seedlings, with both double mutants developing fewer than 50% of the number of lateral roots observed in tir1-1 seedlings ( Figure 2C ). Furthermore, while lateral root numbers in csn3-3 and csn1-10 single mutants were comparable to wild-type controls, csn3-3 csn1-10 double mutants developed significantly fewer lateral roots, suggesting that auxin response is more severely impaired in the double mutant background.
To further compare the auxin response defects of csn3-3 and csn1-10 mutants, we introduced the BA3:GUS and DR5:GUS auxin responsive reporters [3], [57] to examine auxin mediated gene expression. Treatment of Col [BA3:GUS] control seedlings with 2,4-D or IAA triggered a strong GUS signal in the root elongation zone. In contrast, only a slight induction of BA3:GUS activity was observed in csn3-3 and csn1-10 mutants ( Figures 2D and S 1). Like the auxin-resistant root growth phenotype of csn3-3, the diminished BA3:GUS expression was complemented by a genomic CSN3 transgene ( Figure 2E ). Similar findings were obtained with csn3-3 and csn1-10 seedlings carrying the DR5:GUS reporter ( Figure 2D ). Together, our analysis of root growth inhibition, lateral root development, and auxin mediated gene expression suggest that the csn3-3 and csn1-10 mutations confer a very similar reduction in auxin sensitivity.
In Arabidopsis, loss of any of the eight CSN subunits results in an identical suite of phenotypes, including constitutive photomorphogenesis, anthocyanin accumulation, and seedling lethality [23], [35], [58]. We observed similar phenotypes upon examining the csn3-2 null mutant ( Figure 3A ). Previously described viable csn mutants include single mutants of the two MPN domain subunits, which are each encoded by two highly homologous genes (CSN5A/CSN5B [59] and CSN6A/CSN6B [50]), and the weak csn1-10 [43] and csn2-5 [40] missense mutants. Unlike previously described csn3 alleles, [23], [35], [55], csn3-3 is viable throughout development and does not exhibit a constitutive photomorphogenic (cop) phenotype ( Figure 3A-C ). Likewise, the csn1-10 and csn2-5 mutants also do not exhibit an obvious cop phenotype, although both mutants are mildly dwarfed compared to wild-type and csn3-3 adult plants [40], [43].
Figure 3. csn3-3 does not exhibit characteristic csn mutant phenoytpes.
(A) Seedling phenotypes of csn3-2 and csn3-3 mutants. (B) Phenotypes of Col, csn1-10 and csn3-3 adult (30-d.o.) plants. (C) 5-d.o. Col and csn3-3 etiolated seedlings. Size bars = 1 mm.
CUL1 deneddylation and Aux/IAA stability are not affected in csn3-3 seedlings
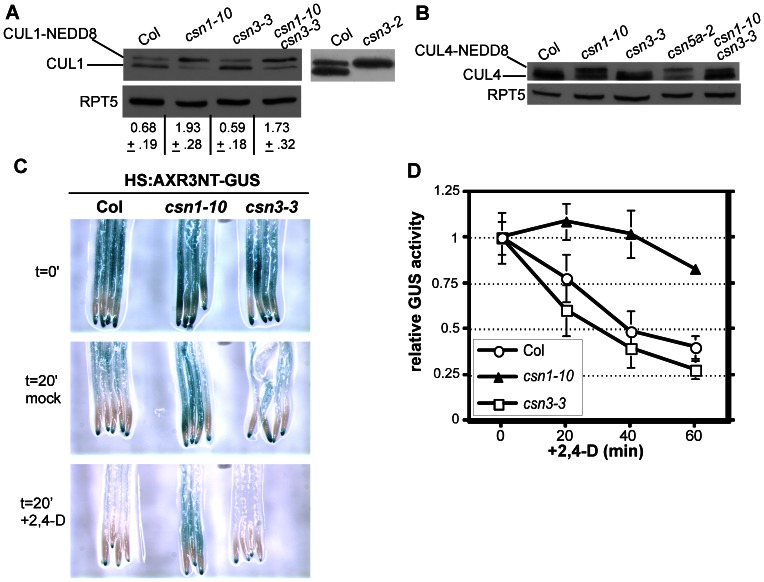
The CSN cleaves the RUB/NEDD8 peptide from the cullin subunit of CRL ubiquitin-ligases. All previously described csn mutants, including the weak csn1-10 and csn2-5 alleles, result in an increase in the CUL1-NEDD8 to unmodified CUL1 ratio [40], [43]. We therefore examined if the csn3-3 mutation affected CUL1 modification. We included csn1-10 for comparison with csn3-3, since both mutants are viable csn mutants and display similar auxin response defects ( Figure 2 ). While a clear increase in CUL1-NEDD8:CUL1 was seen in extracts prepared from csn1-10 seedlings, no change from wild-type was detected in α-CUL1 western blots of csn3-3 extracts ( Figure 4A ). In contrast, CUL1 was exclusively in its neddylated form (CUL1-NEDD8) in csn3-2 extracts ( Figure 4A ), as reported in previous studies [35].
Figure 4. csn3-3 affects auxin response independent of SCFTIR1/AFB.
(A) CUL1 western blot analysis of protein extracts prepared from Col and csn mutant seedlings. The upper band indicates the modified (neddylated) CUL1. RPT5 is shown as a loading control. Numbers below the blot indicate the ratio of CUL1-NEDD8:CUL1 ± SD from three experiments. (B) Western blot analysis of CUL4 neddylation status in Col and csn mutant seedling extracts. (C) Col, csn1-10 and csn3-3 carrying the HS:AXR3NT-GUS transgene were heat shocked for 2 h and stained immediately or following incubation with 1 µM 2,4-D for 20 min. (D) Quantitative measurement of the β-glucuronidase activity of the HS:AXR3NT-GUS reporter in Col, csn1-10, and csn3-3 seedlings. Seedlings were heat-shocked for 2 h, and then returned to room temperature and treated with 1 µM 2,4-D for 20, 40, or 60 min. β-glucuronidase activity is presented as the fraction remaining compared to the 0 min time point. Values shown depict the mean ± SD of 6 technical replicates. Similar results were obtained in two additional biological replicates.
Since the csn3-3 mutation unexpectedly did not affect the CUL1-NEDD8:CUL1 ratio, we examined double mutants with csn1-10 to test the possibility that csn3-3 might enhance the weak deneddylation phenotype of csn1-10. Once again however, the csn3-3 mutation did not increase the CUL1-NEDD8:CUL1 ratio ( Figure 4A ). Since the CSN also regulates other cullin-based E3 ubiquitin-ligases by deneddylating their cullin subunit, we applied a similar immunoblotting assay to examine CUL4 modification in csn3-3 seedlings. Similar to our findings with CUL1, we observed no accumulation of the CUL4-NEDD8 isoform in csn3-3 extracts, whereas neddylated CUL4 was readily detectable in csn1-10 and csn5a-2 extracts ( Figure 4B ). Together, our analyses of CUL1 and CUL4 suggest that the csn3-3 mutation does not affect the deneddylation activity of the CSN.
The reduced deneddylation activity of several previously characterized csn mutants has been found to result in diminished SCFTIR1/AFB activity, resulting in the stabilization of Aux/IAA proteins [28], [40]. Since our finding that csn3-3 plants exhibited no change in cullin deneddylation was quite surprising, we examined SCFTIR1/AFB activity by monitoring Aux/IAA stability using the previously described HS:AXR3NT-GUS reporter protein [4]. csn1-10 seedlings were again included for comparison. 6-d.o. Col, csn1-10 and csn3-3 seedlings, all of which carried the HS:AXR3NT-GUS construct, were heat-shocked at 37°C for 2 hours to induce expression, followed by return to ambient temperature and treatment with auxin. AXR3NT-GUS activity was examined both qualitatively and quantitatively at 20 min intervals during the treatment to monitor the remaining levels of AXR3NT-GUS fusion protein ( Figure 4C, D ). Following the 2h heat-shock induction, similar AXR3NT-GUS levels were observed in wild-type, csn3-3 and csn1-10 seedlings ( Figure 4C ). During the auxin treatment, AXR3NT-GUS activity in csn3-3 and wild-type seedlings diminished with comparable kinetics ( Figure 4D ), suggesting that SCFTIR1/AFB activity is unaffected in csn3-3 plants. In contrast, csn1-10 seedlings exhibited substantially slower AXR3NT-GUS degradation ( Figures 4C, D ).
To further confirm that csn3-3 had no effect on SCFTIR1/AFB-mediated proteolysis, we examined another Aux/IAA reporter, IAA28-myc [60]. The IAA28-myc construct was crossed into csn3-3 and csn1-10 backgrounds. Abundance of the IAA28-myc protein was then examined by treating seedlings with IAA and immunoblotting root protein extracts. As previously reported [60], IAA28-myc was rapidly degraded in wild-type seedlings and was nearly undetectable after a 10 minute auxin treatment (Figure S2). While IAA28-myc was clearly more stable in csn1-10 seedlings, it was rapidly degraded in the csn3-3 background (Figure S2). Combined with our HS:AXR3NT-GUS degradation data, this strongly suggests that SCFTIR1/AFB activity is unaltered by the csn3-3 mutation.
Genetic interactions distinguish csn3-3 and csn1-10
The CSN regulates auxin signaling by deneddylating CUL1 to modulate SCFTIR1/AFB activity [22], [28], [44]. Our findings with the csn1-10 mutant are consistent with this model. The csn3-3 mutation on the other hand, affects neither CUL1 deneddylation nor SCFTIR1/AFB-mediated Aux/IAA degradation, yet confers auxin response defects virtually identical in severity to csn1-10. These findings strongly suggest that the CSN, or at least the CSN3 subunit, plays a second role in the regulation of auxin signaling in addition to deneddylating CUL1. If so, we reasoned that csn3-3 and csn1-10 may exhibit distinct genetic interactions when combined with other mutations affecting auxin signaling.
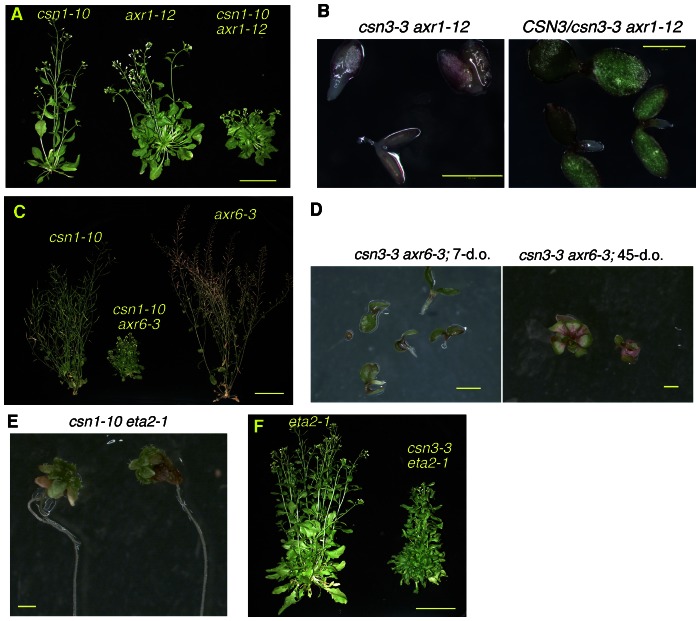
We therefore crossed axr1-12 plants with csn1-10 and csn3-3 to generate double mutants. The axr1-12 mutation affects a subunit of the NEDD8 activating enzyme. This mutation confers a reduction in CUL1 neddylation and strong auxin response defects [16], [17], [61]. The interaction between axr1-12 and csn1-10 appeared additive, as double mutants exhibited a moderately more severe dwarf phenotype than the single mutant parents ( Figure 5A ). In sharp contrast, csn3-3 axr1-12 doubles displayed a seedling-lethal phenotype ( Figure 5B ), indicating that axr1-12 and csn3-3 interact synergistically. Double mutant seedlings exhibited cotyledon morphogenic defects and lacked a basal pole similar to loss-of-function mutants of the monopteros (mp) auxin response factor and the dominant negative axr6-1 and axr6-2 alleles of CUL1 [14], [62]. Furthermore, a slightly less severe phenotype was observed in CSN3/csn3-3 axr1-12/axr1-12 seedlings ( Figure 5B ). A similar set of genetic interactions was observed when csn1-10 and csn3-3 were combined with axr6-3. axr6-3 is a temperature-sensitive allele of CUL1 that exhibits reduced CUL1 neddylation and severe auxin response defects [15]. While csn1-10 axr6-3 plants were viable, flowered, and produced a few seeds, csn3-3 axr6-3 plants exhibited arrested development, with virtually no root growth occurring even after 45 days of growth ( Figure 5C, D ).
Figure 5. csn3-3 and csn1-10 exhibit distinct double mutant interactions.
(A) Adult csn1-10 axr1-12 double mutant exhibit a slightly more severe dwarf phenotype than either of the single mutants. Size bar = 4 cm. (B) csn3-3 interacts with axr1-12 synergistically, resulting in the seedling lethality. Right panel shows the seedling-lethal phenotype of heterozygous csn3-3/CSN3 in the axr1-12 background. Size bar = 1 mm. (C–D) While csn1-10 axr6-3 plants are viable and complete the life cycle, csn3-3 interacts with axr6-3 synergistically, with double mutants exhibiting developmental arrest at the seedling stage. Size bars = 4 cm (C) and 1 mm (D), respectively. (E–F) csn1-10 exhibits a stronger interaction with eta2-1 than does csn3-3. As previously reported [43], csn1-10 eta2-1 seedlings fail to develop past the early seedling stage. Size bars = 1 mm (E) and 4 cm (F), respectively.
In contrast to the double mutants with axr1-12 and axr6-3, the phenotypic severity was reversed when csn1-10 and csn3-3 were combined with the eta2-1 mutation. eta2-1 is a missense mutation in CAND1, a cullin binding protein that mediates cycles of SCF complex assembly and disassembly [43], [52]. While the eta2-1 mutation abolishes CUL1 binding activity and thus disrupts SCF cycling, it has no detectable effect on CUL1 neddylation. Whereas csn1-10 eta2-1 double mutants failed to progress past the seedling stage [43], csn3-3 only slightly enhanced the developmental defects of eta2-1 ( Figure 5F ). Together, the highly differential genetic interactions conferred by the csn1-10 and csn3-3 mutations in combination with axr1-12, axr6-3, and eta2-1 strongly suggest that these two mutations in CSN subunits affect distinct aspects of auxin signaling. This notion is consistent with our finding that only csn1-10 exhibits defects in cullin deneddylation and SCFTIR1/AFB-mediated regulation of Aux/IAA protein stability.
A novel CSN3 complex, but not the CSN holocomplex, is altered in csn3-3 plants
The CSN deneddylase activity is catalyzed by the CSN5 subunit, in which the JAMM motif is the catalytic center required for the isopeptidase activity to remove NEDD8 from the cullin proteins [33], [44]. However, the proper assembly of the CSN holocomplex is required for deneddylase activity. Consequently, null mutations in any of the eight CSN subunits abolish both CSN assembly and deneddylase activity, and confer similar seedling-lethal phenotypes [23], [48]–[50], [55], [59].
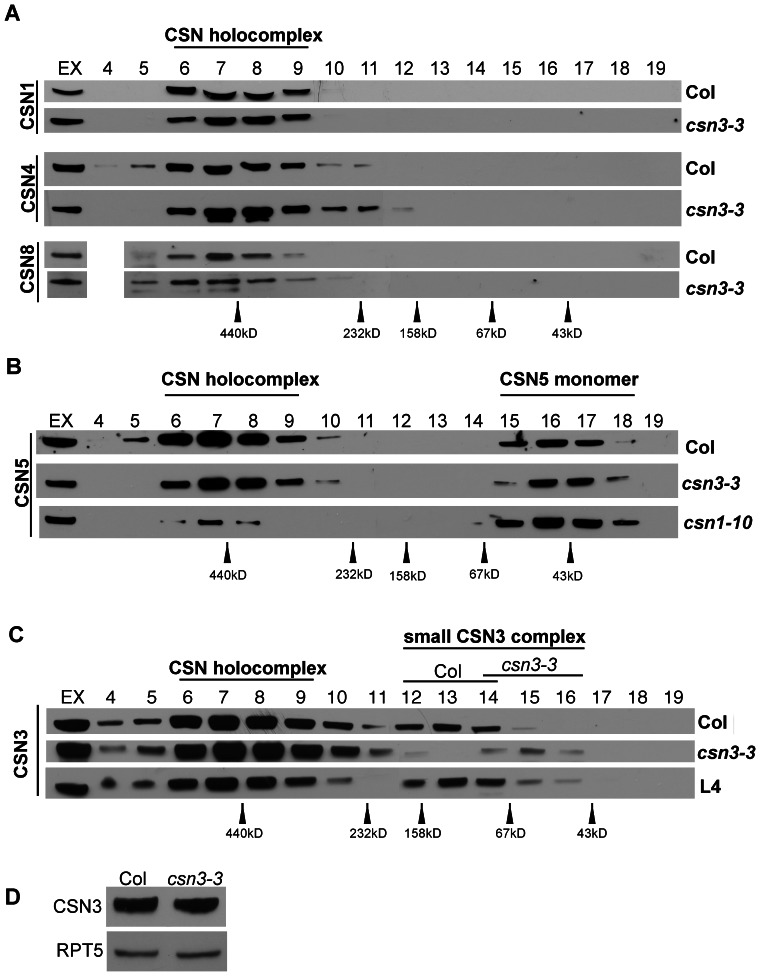
Since csn3-3 confers reduced auxin signaling but does not affect the deneddylase activity of the CSN, we examined whether the csn3-3 mutation had any effect on CSN complex assembly. Protein extracts from Col and csn3-3 seedlings were fractionated by gel filtration chromatography as previously described [43], and analyzed by immunoblotting with antibodies against several CSN subunits. With wild-type extracts, the CSN1, CSN4, and CSN8 subunits were detected primarily in high molecular mass fractions (#6-9) corresponding to 450-700 kD ( Figure 6A ), consistent with the previously observed molecular mass of the CSN holocomplex [23], [58]. No obvious differences from this pattern were detected with csn3-3 extracts. Unlike other CSN subunits, prior studies in both plants and animals have demonstrated that the CSN5 subunit is present in both the CSN and as a monomer [34], [59], [63]. Consistent with these findings, we detected CSN5 in both high and low molecular mass fractions corresponding the CSN holocomplex and CSN5 monomer, respectively ( Figure 6B ). Again however, no differences between Col and csn3-3 were observed. In contrast, the csn1-10 mutation resulted in a clear shift in CSN5 fractionation, with the majority of CSN5 fractionating in the monomeric form ( Figure 6B ). This finding suggests that the csn1-10 mutation impairs incorporation of CSN5 into the CSN holocomplex and is consistent with the reduction in cullin deneddylation we observe in this mutant.
Figure 6. A small CSN3-containing complex, but not the CSN, is affected by the csn3-3 mutation.
Protein extracts from 7-d.o. Col, csn1-10 and csn3-3 seedlings were fractionated on a Superdex-200 gel filtration column and fractions (4 to 19) were collected and blotted with α-CSN1, α-CSN4, and α-CSN8 (A), α-CSN5 (B) and α-CSN3 (C). Fraction #4 was lost in the experiment examining CSN8. In (C), the assembly of a small CSN3-containing protein complex was observed in fractions 12-14. This complex was absent in csn3-3 extracts, but was restored by introduction of a genomic CSN3 transgene (L4 complementation line). Molecular mass standards are shown at the bottom of each panel. EX indicates the protein crude extract before gel filtration. (D) CSN3 protein level is unaffected by the csn3-3 mutation. RPT5 is shown as a loading control.
Together with our finding that CSN deneddylase activity is unaltered in csn3-3 mutants, the above gel filtration findings with CSN1, CSN4, CSN5, and CSN8 indicate that CSN holocomplex assembly is unaffected by csn3-3. Consistent with this conclusion, we found that the abundance of the csn3-3 mutant protein is similar to wild-type, and the mutant protein also incorporates into the CSN holocomplex ( Figures 6C, D ). However, during our gel filtration studies with CSN3, we noticed an additional CNS3 elution peak centered around 130 kD in wild-type extracts ( Figure 6C , fractions 12-14). Interestingly, this small CSN3-containing complex (sCSN3c) was absent in csn3-3 seedlings, indicating that the csn3-3 missense mutation specifically disrupts the formation of this complex. Furthermore, the sCSN3c was restored by expression of a PCSN3:CSN3 transgene in csn3-3 mutant plants (L4 in Figure 6C ).
Our results suggest that sCSN3c represents a novel CSN3-containing complex and not an intermediate in CSN holocomplex assembly. First, a prior CSN subunit interaction mapping study concluded that the CSN3 subunit primarily contacts CSN1, CSN8, and probably CSN4 [64]. However, none of these subunits co-eluted with sCNS3c in our gel filtration experiments ( Figure 6A ). Likewise, no overlap in the elution profile of CSN5 with sCSN3c was observed ( Figure 6A, B ). Secondly, we tested the possibility that sCSN3c may be due to partial disassembly of the CSN that occurred during our in vitro fractionations. Fractions containing the CSN holocomplex were isolated and subjected to a second round of gel filtration and subsequently immunoblotted with α-CSN3 antibody (Figure S3). However, no CSN3 protein was detected in the low molecular mass fractions corresponding to sCSN3c, demonstrating that the CSN is stable under our conditions. Together, these findings suggest that CSN3 is the only canonical CSN subunit present in sCSN3c, although the possibility that subunits for which we do not have antibodies (CSN2/6/7) are sCSN3c components cannot be eliminated.
Discussion
In plants, animals, and fungi, the CSN has a well-established role as a cullin deneddylase that regulates CRL ubiquitin-ligase activity [44]. Prior reports in Arabidopsis have demonstrated that reduced deneddylase activity in various csn mutants affects SCFTIR1/AFB ubiquitin-ligase activity and consequently results in impaired auxin signaling [28], [35], [40]. In this study, we identified the csn3-3 missense mutation as an enhancer of the auxin resistant root growth phenotype conferred by tir1-1. Consistent with diminished auxin signaling, csn3-3 also enhanced the tir1-1 lateral root development defect and conferred diminished auxin mediated expression of the DR5: and BA3:GUS reporters. While these findings were hardly surprising given the aforementioned studies of Arabidopsis CSN subunit mutants, we quite unexpectedly found that neither the deneddylation activity of the CSN nor the SCFTIR1/AFB-mediated regulation of Aux/IAA protein stability was affected by the csn3-3 mutation. These findings clearly distinguish csn3-3 from all previously characterized csn mutants.
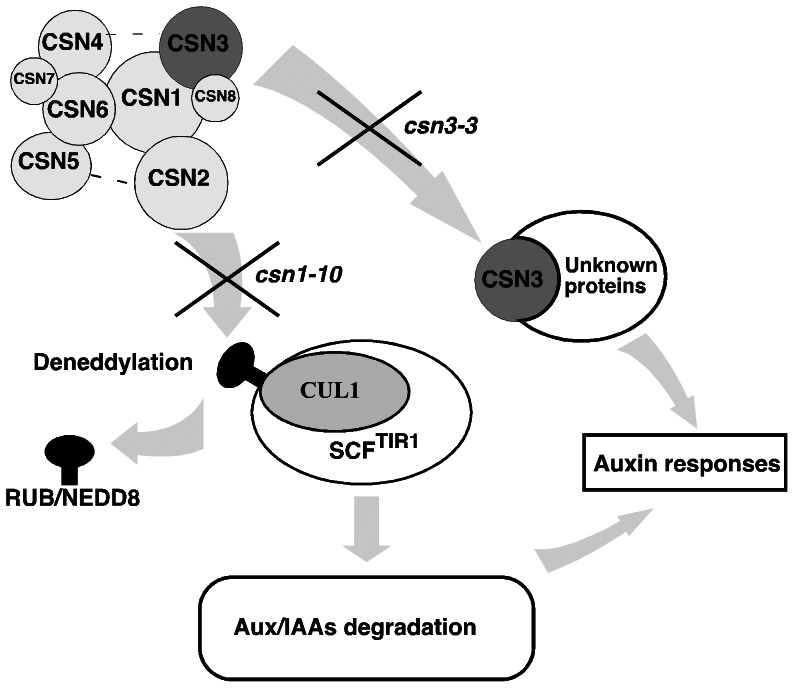
It could be argued that csn3-3 is simply a weak mutation that does not confer sufficient defects to be detected in assays examining neddylation status of cullins or the SCFTIR1/AFB activity. However, our comparison of csn3-3 phenotypes to those of a second weak CSN subunit mutant, csn1-10, strongly argues against this possibility. csn3-3 and csn1-10 confer virtually identical dose-response curves in assays examining auxin-mediated inhibition of root growth, with both mutants exhibiting 50% growth inhibition at ∼50 nM 2,4-D. Likewise, both mutants enhance the tir1-1 auxin resistant root growth and reduced lateral root development phenotypes to a similar extent. Furthermore, both csn3-3 and csn1-10 confer comparable reductions in auxin mediated expression of the BA3: and DR5:GUS reporters. Together, these findings indicate that csn3-3 and csn1-10 impair auxin signaling to a similar extent. However, whereas cullin deneddylation and Aux/IAA degradation were unaffected in csn3-3 seedlings, both of these molecular defects were clearly apparent in csn1-10 mutants. Prior studies have clearly demonstrated that the CSN3 subunit is required for CSN deneddylase activity [35], which we have confirmed with the csn3-2 null allele. The csn3-3 missense mutation however, confers a reduction in auxin response without affecting cullin deneddylation, suggesting that CSN3 plays a second role in auxin signaling in addition to its role in regulating the SCFTIR1/AFB ubiquitin ligase ( Figure 7 ).
Figure 7. The CSN3 subunit plays multiple roles in auxin signaling.
We hypothesize that in addition to its role in the CSN as a cullin deneddylase, the CSN3 subunit also regulates auxin signaling independently of the SCFTIR1/AFB ubiquitin-ligase. This second regulatory mechanism may involve the small CSN3-containing complex whose assembly is disrupted by the csn3-3 point mutation.
The dramatically different double mutant phenotypes exhibited when the csn1-10 and csn3-3 mutations were combined with axr6-3, axr1-12, or eta2-1 also indicate that csn1-10 and csn3-3 affect distinct aspects of auxin signaling. While csn3-3 confers seedling lethality when combined with axr6-3 or axr1-12, the csn1-10 mutation does not. In contrast, csn1-10, but not csn3-3, confers a seedling arrest phenotype in the eta2-1 background. Given that SCFTIR1/AFB activity appears unaffected in csn3-3 plants, it seems unlikely that the lethality of csn3-3 axr1-12 and csn3-3 axr6-3 seedlings is due to a further reduction in SCFTIR1/AFB activity. Instead, we speculate that csn3-3 specifically affects auxin signaling downstream or perhaps independently of SCFTIR1/AFB ( Figure 7 ), such that the combination of csn3-3 with axr6-3 or axr1-12 causes auxin sensitivity to fall below the threshold required for early seedling development. Although it is unclear what these differential genetic interactions might mean mechanistically, it is interesting to note that both axr6-3 and axr1-12 result in a reduction in neddylated CUL1 [15], [16], [19], [37]. On the other hand, the eta2-1 mutation has no effect on CUL1 neddylation status [52]. Rather, the eta2-1 mutation abolishes the CUL1 binding activity of CAND1, resulting in the disruption of CAND1-mediated cycling of SCF complexes [43].
Consistent with the hypothesis that an SCFTIR1/AFB-independent pathway may regulate auxin signaling to control gene expression, like csn3-3, the previously described ibr5 mutants of Arabidopsis also exhibit diminished auxin response without inhibiting SCFTIR1/AFB-mediated degradation of Aux/IAA proteins [60]. IBR5 encodes a putative dual-specificity protein phosphatase. However, although Aux/IAA proteins are highly unstable in both csn3-3 and ibr5 mutants, ibr5 seedlings exhibited reduced steady-state levels of the AXR3NT-GUS and IAA28-myc reporter proteins. In our analysis of csn3-3 mutants, however, both of these reporter proteins were present at levels comparable to wild-type controls. Furthermore, unlike csn3-3, ibr5 does not interact with axr1 in a synergistic manner [60]. Together, these findings suggest that it is unlikely that csn3-3 and ibr5 share a common auxin signaling defect.
The fact that the csn3-3 mutation did not diminish CSN deneddylase activity, yet conferred reduced auxin response phenotypes, suggests that this mutation defines a novel function for CSN3. Therefore, a major question is whether csn3-3 defines a new role in auxin signaling for the CSN holocomplex or a distinct CSN3-containing complex. While deneddylation is the only known biochemical activity of the CSN itself, additional activities including deubiquitinylating and protein kinase activities have been reported to be associated with the CSN [44], [65], [66]. Furthermore, in animal systems some CSN subunits have been found to be DNA associated and suggested to regulate transcription [67], [68]. Also, whether or not all CSN subunits function solely within the CSN holocomplex is unclear. On one hand, null mutations in any of the eight Arabidopsis subunits confer identical seedling-lethal phenotypes [23] and transcription profiles [35], suggesting that each subunit only functions within the CSN. However, it is possible that CSN subcomplexes or individual subunits have additional functions that are masked by the early seedling lethality of these null mutants. Consistent with this possibility, fission yeast csn1 and csn2 mutations confer DNA replication defects whereas other subunit mutations do not [25]. Similarly, while both Drosophila csn4null and csn5null mutants are embryo-lethal, these two mutants exhibit distinct developmental arrest phenotypes [63], [69] and differentially affect gene expression [70].
Given that csn3-3 confers no apparent defects in cullin deneddylation, SCFTIR1/AFB activity, or CSN holocomplex assembly, but does specifically abolish the ∼130 kD sCSN3c complex, we hypothesize that a defective sCSN3c may be the basis of the auxin signaling defects displayed by csn3-3 mutant plants ( Figure 7 ). Consistent with this possibility, expression of a PCSN3:CSN3 transgene in csn3-3 mutant plants restored both the auxin response defects ( Figures 1C , 2E ) and the sCSN3c complex ( Figures 6C ). Prior studies have reported CSN subunits in complexes smaller than the CSN holocomplex. While some support has been presented for CSN5 functioning autonomously of other CSN subunits [71], [72], these smaller CSN complexes have generally been proposed to be mini-CSN complexes containing several, but not all subunits [64]. Whether these represent intermediates in CSN holocomplex assembly or functionally distinct complexes is uncertain. Interestingly, one study examining these mini-CSN complexes from animal cells by non-denaturing polyacrylamide electrophosesis detected CSN3-containing complexes that appeared to lack other CSN subunits [71]. In Arabidopsis, Rubio et al. [73] also detected, but did not discuss or characterize, small CSN3-containing complexes.
Recent analyses of CSN subunit interactions using in vitro reconstituted human CSN subunits suggest that the CSN consists of two symmetrical modules; CSN1/2/3/8 and CSN4/5/6/7 [64]. With this subunit topology, CSN3 would directly interact with CSN1/8 and possibly CSN4, which is consistent with prior two-hybrid studies [74]. However, our gel filtration analysis indicates that neither CSN1/4/8 nor CSN5 are components of the sCSN3c complex. While we cannot eliminate the possibility that CSN2/6/7 are sCSN3c components, this seems unlikely given these prior findings. Thus, together with our finding that CSN holocomplex assembly is unaffected by csn3-3, we hypothesize that sCSN3c represents a novel complex rather than a mini-CSN complex. Identification of the other components within sCSN3c may provide crucial information into what role this complex might play in auxin signaling. Furthermore, since the residue affected by the csn3-3 missense mutation is extremely highly conserved across eukaryotes, it seems likely that sCSN3c function may be similarly conserved.
Materials and Methods
Plant Materials and Growth Conditions
All Arabidopsis lines used in this study are in the Col-0 ecotype. Seeds were sterilized by 30% bleach + 0.1% Triton-X100 for 10 min and were stratified at 4°C for 1-4 days. Seedlings were grown under sterile conditions on vertically oriented ATS nutrient medium [61] under long-day conditions. Adult plants were grown in soil under long-day conditions at 20°C. The tir1-1, csn1-10, csn3-2 (SALK_106465), eta2-1, axr6-3, and axr1-12 mutants have been described previously [35], [43], [75]. The BA3:GUS [57], DR5:GUS [3], HS:AXR3NT-GUS [4], and PIAA28:IAA28-myc [60] transgenes were introduced into the csn3-3 and csn1-10 backgrounds by crossing. For construction of double mutant and reporter lines, the csn3-3 mutation was genotyped using a CAPS marker for PCR products generated with primers Ex7F (5′-CAACGACGGGAAGATTGGTG-3′) and Ex8R (5′- GCCTCCTTAGCATTACCAAG-3′). When digested with Sty I, the 289 bp CSN3 PCR product is cleaved into 163 and 126 bp fragments, whereas the csn3-3 mutation abolishes the Sty I recognition site. The eta2-1, axr1-12, and axr6-3 mutations were confirmed by sequencing PCR products spanning the mutation sites.
For root growth assays, 5-d.o. seedlings were transferred to ATS medium supplemented with various concentrations of 2,4-D, and root growth was measured after an additional 4 days. Percent inhibition was calculated by dividing the average growth on auxin media by the average growth on ATS control media and subtracting this ratio from 100%. For measuring IAA-induced root growth inhibition, 6-d.o. seedlings were transferred to freshly made IAA plates and were grown under long-day illumination through yellow long-pass filters to slow indolic compound breakdown. Protein extractions for gel filtration and western experiments were made from 7 - 10-d.o. seedlings grown in liquid ATS medium on a shaker at 20°C.
Complementation
The CSN3 genomic DNA construct was composed of the CSN3 coding region together with a 1.3 kb fragment upstream of the start codon plus a 600bp fragment downstream of the stop codon. The whole sequence was amplified from BAC clone F18022 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCCTTTGATGGCGCCATGGTGG-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCGTATGGAAACATGTGATAACC) and Gateway cloned into the vector pEarleyGate301 [76]. The construct was then transformed into csn3-3[BA3:GUS] plants using Agrobacterium tumefaciens strain GV3101 according to standard procedures [77]. Two independent T3 homozygous lines (L2 and L4) were used for the root growth and GUS assays to assess complementation.
GUS Histochemical Staining
BA3:GUS and DR5:GUS assays were conducted as described previously with slight modifications [52]. 6-d.o. seedlings grown vertically on ATS plates were transferred into liquid ATS medium supplemented with 0.5 µM 2,4-D for 12 h (BA3:GUS) and 4 h (DR5:GUS) before GUS staining. For HS:AXR3NT-GUS assays, 6-d.o. Col, csn1-10 and csn3-3 seedlings homozygous for the reporter construct were heat-shocked for 2 h at 37°C to induce expression of the transgene. Seedlings then were transferred to 20°C medium with 1 µM 2,4-D for 20 min before GUS detection. Quantitative measurements of β-glucuronidase activity were conducted as described previously [4] with modifications: after heat shock for 2 h at 37°C, seedlings were transferred to 20°C liquid ATS medium for 15 min. Seedlings were then moved into medium supplemented with 1 µM 2,4-D, sampled at 0, 20, 40 and 60 min thereafter and stored in liquid nitrogen until protein extraction. Frozen seedlings were homogenized by a laboratory vibration mill (Mixer-Mill; Qiagen, cat. no. MM300) in extraction buffer (50 mM KPO4 pH 7, 0.1% triton X-100, 10 mM β-mercaptoethanol, 10 mM EDTA). Samples were then centrifuged to remove debris and the protein level of each sample was measured. Protein extracts were mixed with an equal volume of extraction buffer plus 4 mM MUG (4-methylumbelliferyl-B-D-glucoronide), incubated at 37°C overnight, and the fluorometric signal measured using a BioTek FL600 Fluorescence Microplate Reader (Winooski, VT) as per manufacturer's instructions (excitation at 360 nm, emission at 460 nm). Activity was calculated as fluorometric units per μg protein and were normalized as the percentage of the starting point (0 min).
IAA28-myc degradation assay
Experiments were done as described previously [60] with modifications. 7-d.o. light-grown seedlings of Col, csn1-10 and csn3-3 carrying the IAA28-myc construct were removed from ATS plates and floated in liquid ATS supplemented with 0.5 µM IAA. At the indicated time points, roots were excised and homogenized for protein extraction.
Antibodies and Immunoblot Analysis
The CUL1 antibody has been described previously [9] and the CUL4 antibody was obtained from Dr. Xing Wang Deng (Yale U.). The quantification of CUL1-NEDD8:CUL1 ratios was performed in Image J. Antibodies against the CSN1, CSN3, and CSN8 subunits were raised by immunizing New Zealand white rabbits (Cocalico Biological, Reamstown, PA) with recombinant 6xHis-CSNx protein purified from E. coli using standard protocols [9]. Crude sera were subsequently affinity purified against nitrocellulose-bound antigens [78]. The CSN4, CSN5 and Rpt5 antibodies were purchased from BIOMOL Int'l, L.P./Enzo Life Sciences. Monoclonal α-myc 9E10 antibody was purchased from Covance and used as recommended. For IAA28-myc, cullins and CSN subunit immunoblotting, protein extracts were prepared from 7- to 10-d.o. seedlings (or seedling roots for IAA28-myc assay) in protein extraction buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5% NP40, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1X Protease Inhibitor Cocktail Kit (Thermo)). 50 µg of protein were loaded in each lane and separated by 10% SDS-PAGE (cullins and CSN subunits) or 4∼12% NuPAGE® Bis-Tris Gel (Invitrogen). For gel filtration, proteins of each fraction were concentrated with StrataClean™ Resin (Stratagene) and separated on 4∼12% NuPAGE® Bis-Tris Gel (Invitrogen), blotted, and used for western detection.
Gel Filtration Chromatography
Experiments were conducted as described previously [43]. In brief, 7-d.o. seedlings grown in liquid ATS medium were homogenized in extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 10 mM MgCl2, 0.5 mM NaN3, 1 mM DTT, 1 mM PMSF, and 1X Protease Inhibitor Cocktail Kit (Thermo)). Homogenates were centrifuged twice for 10 min at 4°C to remove debris. Supernatants were then filtered though a 0.45 µm HT Tuffryn® Membrane (Pall). 600 µg of total protein was fractionated through a Superdex 200 10/300 GL column (Amersham/GE Healthcare). After loading the sample, proteins were eluted in filtered and degassed extraction buffer at a flow rate of 0.2 mL/min. 0.5 mL fractions were collected after the 6 mL void volume was reached. All procedures were carried out at 4°C.
Supporting Information
IAA induced BA3:GUS expression is reduced by the csn1-10 and csn3-3 mutations to a similar extent. 6-d.o. transgenic Col, csn1-10 and csn3-3 seedlings carrying the BA3:GUS reporter were treated with 1 µM IAA for 3 h before histochemical staining for β-glucuronidase activity.
(TIF)
α-myc western detection of the IAA28-myc fusion protein. The PIAA28:IAA28-myc reporter was introduced into the csn1-10 and csn3-3 backgrounds by crossing. Protein extracts were made from 7-d.o. seedling roots treated with IAA for the indicated time. RPT5 was used as a loading control.
(TIF)
sCSN3c is not a breakdown product of the CSN holocomplex during the in vitro fractionation. Fractions (#5∼9) of the first gel filtration run using Col seedling protein extracts were isolated and injected into the column for a second round of gel filtration. CSN3 western detection was conducted using fractions from the 2nd gel filtration. No CSN3 was detected in the sCSN3c fractions.
(TIF)
Acknowledgments
We would like to thank the Arabidopsis Biological Resource Center and Drs. Bonnie Bartel and Xing Wang Deng for providing reagents used in this study.
Funding Statement
This work was supported by the National Institutes of Health (GM067203 to WMG) and a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft (to MQ). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1. Woodward AW, Bartel B (2005) Auxin: Regulation, action, and interaction. Ann. Bot 95: 707–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tiwari SB, Hagen G, Guilfoyle TJ (2004) Aux/IAA Proteins Contain a Potent Transcriptional Repression Domain. Plant Cell 16: 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA Proteins Repress Expression of Reporter Genes Containing Natural and Highly Active Synthetic Auxin Response Elements. Plant Cell 9: 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276. [DOI] [PubMed] [Google Scholar]
- 5. Maraschin FD, Memelink J, Offringa R (2009) Auxin-induced, SCFTIR1-mediated poly-ubiquitination marks AUX/IAA proteins for degradation. Plant J 59: 100–109. [DOI] [PubMed] [Google Scholar]
- 6. Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, et al. (2005) Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9: 109–119. [DOI] [PubMed] [Google Scholar]
- 7. Pickart CM (2001) Mechanisms underlying ubiquitination. Ann Rev Biochem 70: 503–533. [DOI] [PubMed] [Google Scholar]
- 8. Petroski MD, Deshaies RJ (2005) Function and regulation of Cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6: 9–20. [DOI] [PubMed] [Google Scholar]
- 9. Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, et al. (1999) Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev 13: 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Patton EE, Willems AR, Tyers M (1998) Combinatorial control in ubiquitin-dependent proteolysis: don't Skp the F-box hypothesis. Trends Genet 14: 236–243. [DOI] [PubMed] [Google Scholar]
- 11. Dharmasiri N, Dharmasiri S, Estelle M (2005) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445. [DOI] [PubMed] [Google Scholar]
- 12. Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451. [DOI] [PubMed] [Google Scholar]
- 13. Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, et al. (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645. [DOI] [PubMed] [Google Scholar]
- 14. Hobbie L, McGovern M, Hurwitz LR, Pierro A, Liu NY, et al. (2000) The axr6 mutants of Arabidopsis thaliana define a gene involved in auxin response and early development. Development 127: 23–32. [DOI] [PubMed] [Google Scholar]
- 15. Quint M, Ito H, Zhang W, Gray WM (2005) Characterization of a novel temperature-sensitive allele of the CUL1/AXR6 subunit of SCF ubiquitin-ligases. Plant J 43: 371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. del Pozo JC, Dharmasiri S, Hellmann H, Walker L, Gray WM, et al. (2002) AXR1-ECR1-dependent conjugation of RUB1 to the Arabidopsis cullin AtCUL1 is required for auxin response. Plant Cell 14: 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. del Pozo JC, Estelle M (1999) The Arabidopsis cullin AtCUL1 is modified by the ubiquitin-related protein RUB1. Proc Natl Acad Sci USA 96: 15342–15347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dharmasiri S, Dharmasiri N, Hellmann H, Estelle M (2003) The RUB/Nedd8 conjugation pathway is required for early development in Arabidopsis. EMBO J 22: 1762–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dharmasiri N, Dharmasiri S, Weijers D, Karunarathna N, Jurgens G, et al. (2007) AXL and AXR1 have redundant functions in RUB conjugation and growth and development in Arabidopsis. Plant J 52: 114–123. [DOI] [PubMed] [Google Scholar]
- 20. Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, et al. (2009) An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458: 732–736. [DOI] [PubMed] [Google Scholar]
- 21. Yang X, Menon S, Lykke-Andersen K, Tsuge T, Di X, et al. (2002) The COP9 Signalosome Inhibits p27kip1 Degradation and Impedes G1-S Phase Progression via Deneddylation of SCF Cul1. Curr Biol 12: 667–672. [DOI] [PubMed] [Google Scholar]
- 22. Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, et al. (2001) Promotion of NEDD8-CUL1 Conjugate Cleavage by COP9 Signalosome. Science 292: 1382–1385. [DOI] [PubMed] [Google Scholar]
- 23. Gusmaroli G, Figueroa P, Serino G, Deng XW (2007) Role of the MPN subunits in COP9 signalosome assembly and activity, and their regulatory interaction with Arabidopsis Cullin3-based E3 ligases. Plant Cell 19: 564–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Menon S, Chi H, Zhang H, Deng XW, Flavell RA, et al. (2007) COP9 signalosome subunit 8 is essential for peripheral T cell homeostasis and antigen receptor-induced entry into the cell cycle from quiescence. Nat Immunol 8: 1236–1245. [DOI] [PubMed] [Google Scholar]
- 25. Mundt KE, Liu C, Carr AM (2002) Deletion Mutants in COP9/Signalosome Subunits in Fission Yeast Schizosaccharomyces pombe Display Distinct Phenotypes. Mol Biol Cell 13: 493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chamovitz DA, Wei N, Osterlund MT, von Arnim AG, Staub JM, et al. (1996) The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 86: 115–121. [DOI] [PubMed] [Google Scholar]
- 27. Wei N, Chamovitz DA, Deng X-W (1994) Arabidopsis COP9 is a component of a novel signaling complex mediating light control of development. Cell 78: 117–124. [DOI] [PubMed] [Google Scholar]
- 28. Schwechheimer C, Serino G, Callis J, Crosby WL, Lyapina S, et al. (2001) Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science 292: 1379–1382. [DOI] [PubMed] [Google Scholar]
- 29. Chen H, Shen Y, Tang X, Yu L, Wang J, et al. (2006) Arabidopsis CULLIN4 Forms an E3 Ubiquitin Ligase with RBX1 and the CDD Complex in Mediating Light Control of Development. Plant Cell 18: 1991–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Olma MH, Roy M, Le Bihan T, Sumara I, Maerki S, et al. (2009) An interaction network of the mammalian COP9 signalosome identifies Dda1 as a core subunit of multiple Cul4-based E3 ligases. J Cell Sci 122: 1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dohmann EMN, Nill C, Schwechheimer C (2010) DELLA proteins restrain germination and elongation growth in Arabidopsis thaliana COP9 signalosome mutants. Eur J Cell Biol 89: 163–168. [DOI] [PubMed] [Google Scholar]
- 32. Feng S, Ma L, Wang X, Xie D, Dinesh-Kumar SP, et al. (2003) The COP9 Signalosome Interacts Physically with SCFCOI1 and Modulates Jasmonate Responses. Plant Cell 15: 1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cope G, Suh GB, Aravind L, Schwarz S, Zipursky SL, et al. (2002) Role of Predicted Metalloprotease Motif of Jab1/Csn5 in Cleavage of Nedd8 from Cul1. Science 298: 608–611. [DOI] [PubMed] [Google Scholar]
- 34. Dohmann EM, Kuhnle C, Schwechheimer C (2005) Loss of the CONSTITUTIVE PHOTOMORPHOGENIC9 signalosome subunit 5 is sufficient to cause the cop/det/fus mutant phenotype in Arabidopsis. Plant Cell 17: 1967–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dohmann EM, Levesque MP, Isono E, Schmid M, Schwechheimer C (2008) Auxin responses in mutants of the Arabidopsis CONSTITUTIVE PHOTOMORPHOGENIC9 signalosome. Plant Physiol 147: 1369–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, et al. (2008) Structural insights into NEDD8 activation of Cullin-RING ligases: Conformational control of conjugation. Cell 134: 995–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Saha A, Deshaies RJ (2008) Multimodal Activation of the Ubiquitin Ligase SCF by Nedd8 Conjugation. Mol Cell 32: 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu K, Chen A, Pan Z-Q (2000) Conjugation of Nedd8 to CUL1 Enhances the Ability of the ROC1-CUL1 Complex to Promote Ubiquitin Polymerization. J Biol Chem 275: 32317–32324. [DOI] [PubMed] [Google Scholar]
- 39. Lykke-Andersen K, Schaefer L, Menon S, Deng XW, Miller JB, et al. (2003) Disruption of the COP2 signalosome Csn2 subunit in mice causes deficient cell proliferation, accumulation of p53 and cyclin E, and early embryonic death. Mol Cell Biol 23: 6790–6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stuttmann J, Lechner E, Guerois R, Parker JE, Nussaume L, et al. (2009) COP9 signalosome- and 26S proteasome-dependent regulation of SCFTIR1 accumulation in Arabidopsis. J Biol Chem 284: 7920–7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wee S, Geyer RK, Toda T, Wolf DA (2005) CSN facilitates Cullin-RING ubiquitin ligase function by counteracting autocatalytic adapter instability. Nat Cell Biol 7: 387–391. [DOI] [PubMed] [Google Scholar]
- 42. Wu JT, Lin HC, Hu YC, Chien CT (2005) Neddylation and deneddylation regulate Cul1 and Cul3 protein accumulation. Nat Cell Biol 7: 1014–1020. [DOI] [PubMed] [Google Scholar]
- 43. Zhang W, Ito H, Quint M, Huang H, Noel LD, et al. (2008) Genetic analysis of CAND1-CUL1 interactions in Arabidopsis supports a role for CAND1-mediated cycling of the SCFTIR1 complex. Proc Natl Acad Sci USA 105: 8470–8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cope GA, Deshaies RJ (2003) COP9 Signalosome: A Multifunctional Regulator of SCF and Other Cullin-Based Ubiquitin Ligases. Cell 114: 663–671. [DOI] [PubMed] [Google Scholar]
- 45. Wu JT, Chan YR, Chien CT (2006) Protection of cullin-RING E3 ligases by CSN-UBP12. Trends Cell Biol 16: 362–369. [DOI] [PubMed] [Google Scholar]
- 46. Enchev RI, Scott DC, da Fonseca PC, Schreiber A, Monda JK, et al. (2012) Structural basis for a reciprocal regulation between SCF and CSN. Cell Rep 2: 616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Emberley ED, Mosadeghi R, Deshaies RJ (2012) Deconjugation of Nedd8 from Cul1 is directly regulated by Skp1-F-box and substrate, and the COP9 signalosome inhibits deneddylated SCF by a noncatalytic mechanism. J Biol Chem 287: 29679–29689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Serino G, Su H, Peng Z, Tsuge T, Wei N, et al. (2003) Characterization of the last subunit of the Arabidopsis COP9 signalosome: implications for the overall structure and origin of the complex. Plant Cell 15: 719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Serino G, Tsuge T, Kwok S, Matsui M, Wei N, et al. (1999) Arabidopsis cop8 and fus4 mutations define the same gene that encodes subunit 4 of the COP9 signalosome. Plant Cell 11: 1967–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Peng Z, Serino G, Deng XW (2001) Molecular characterization of subunit 6 of the COP9 signalosome and its role in multifaceted developmental processes in Arabidopsis. Plant Cell 13: 2393–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Quint M, Barkawi LS, Fan K-T, Cohen JD, Gray WM (2009) Arabidopsis IAR4 Modulates Auxin Response by Regulating Auxin Homeostasis. Plant Physiol 150: 748–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chuang HW, Zhang W, Gray WM (2004) Arabidopsis ETA2, an apparent ortholog of the human cullin-interacting protein CAND1, is required for auxin responses mediated by the SCF(TIR1) ubiquitin ligase. Plant Cell 16: 1883–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gray WM, Muskett PR, Chuang HW, Parker JE (2003) Arabidopsis SGT1b is required for SCF(TIR1)-mediated auxin response. Plant Cell 15: 1310–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ito H, Gray WM (2006) A gain-of-function mutation in the Arabidopsis pleiotropic drug resistance transporter PDR9 confers resistance to auxinic herbicides. Plant Physiol 142: 63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Peng Z, Serino G, Deng XW (2001) A role of Arabidopsis COP9 signalosome in multifaceted developmental processes revealed by the characterization of its subunit 3. Development 128: 4277–4288. [DOI] [PubMed] [Google Scholar]
- 56. Tsuge T, Matsui M, Wei N (2001) The subunit 1 of the COP9 signalosome suppresses gene expression through its N-terminal domain and incorporates into the complex through the PCI domain. J Mol Biol 305: 1–9. [DOI] [PubMed] [Google Scholar]
- 57. Oono Y, Chen QG, Overvoorde PJ, Kohler C, Theologis A (1998) age Mutants of Arabidopsis exhibit altered auxin-regulated gene expression. Plant Cell 10: 1649–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gusmaroli G, Feng S, Deng XW (2004) The Arabidopsis CSN5A and CSN5B subunits are present in distinct COP9 signalosome complexes, and mutations in their JAMM domains exhibit differential dominant negative effects on development. Plant Cell 16: 2984–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kwok SF, Solano R, Tsuge T, Chamovitz DA, Ecker JR, et al. (1998) Arabidopsis homologs of a c-Jun coactivator are present both in monomeric form and in the COP9 complex, and their abundance is differentially affected by the pleiotropic cop/det/fus mutations. Plant Cell 10: 1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Strader LC, Monroe-Augustus M, Bartel B (2008) The IBR5 phosphatase promotes Arabidopsis auxin responses through a novel mechanism distinct from TIR1-mediated repressor degradation. BMC Plant Biol 8: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lincoln C, Britton JH, Estelle M (1990) Growth and Development of the axr1 Mutants of Arabidopsis. Plant Cell 2: 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hardtke CS, Berleth T (1998) The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J 17: 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Oron E, Mannervik M, Rencus S, Harari-Steinberg O, Neuman-Silberberg S, et al. (2002) COP9 signalosome subunits 4 and 5 regulate multiple pleiotropic pathways in Drosophila melanogaster. Development 129: 4399–4409. [DOI] [PubMed] [Google Scholar]
- 64. Sharon M, Mao H, Erba EB, Stephens E, Zheng N, et al. (2009) Symmetrical Modularity of the COP9 Signalosome Complex Suggests its Multifunctionality. Structure 17: 31–40. [DOI] [PubMed] [Google Scholar]
- 65. Uhle S, Medalia O, Waldron R, Dumdey R, Henklein P, et al. (2003) Protein kinase CK2 and protein kinase D are associated with the COP9 signalosome. EMBO J 22: 1302–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhou CS, Wee S, Rhee E, Naumann M, Dubiel W, et al. (2003) Fission yeast COP9/signalosome suppresses cullin acivity through recruitment of the deubiquitylating enzyme Ubp12p. Mol Cell 11: 927–938. [DOI] [PubMed] [Google Scholar]
- 67. Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, et al. (2003) The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113: 357–367. [DOI] [PubMed] [Google Scholar]
- 68. Ullah Z, Buckley MS, Arnosti DN, Henry RW (2007) Retinoblastoma protein regulation by the COP9 signalosome. Mol Biol Cell 18: 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Harari-Steinberg O, Cantera R, Denti S, Bianchi E, Oron E, et al. (2007) COP9 signalosome subunit 5 (CSN5/Jab1) regulates the development of the Drosophila immune system: effects on Cactus, Dorsal and hematopoiesis. Genes To Cells 12: 183–195. [DOI] [PubMed] [Google Scholar]
- 70. Oron E, Tuller T, Li L, Rozovsky N, Yekutieli D, et al. (2007) Genomic analysis of COP9 signalosome function in Drosophila melanogaster reveals a role in temporal regulation of gene expression. Mol Syst Biol 3: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Fukumoto A, Tomoda K, Kubota M, Kato J-y, Yoneda-Kato N (2005) Small Jab1-containing subcomplex is regulated in an anchorage- and cell cycle-dependent manner, which is abrogated by ras transformation. FEBS Lett 579: 1047–1054. [DOI] [PubMed] [Google Scholar]
- 72. Tomoda K, Kubota Y, Arata Y, Mori S, Maeda M, et al. (2002) The Cytoplasmic Shuttling and Subsequent Degradation of p27Kip1 Mediated by Jab1/CSN5 and the COP9 Signalosome Complex. J Biol Chem 277: 2302–2310. [DOI] [PubMed] [Google Scholar]
- 73. Rubio V, Shen Y, Saijo Y, Liu Y, Gusmaroli G, et al. (2005) An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J 41: 767–778. [DOI] [PubMed] [Google Scholar]
- 74. Wei N, Deng XW (2003) The COP9 signalosome. Annu Rev Cell Dev Biol 19: 261–286. [DOI] [PubMed] [Google Scholar]
- 75. Ruegger M, Dewey E, Gray W, Hobbie L, Turner J, et al. (1998) The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev 12: 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Earley KW, Haag JR, Pontes O, Opper K, Juehne T, et al. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629. [DOI] [PubMed] [Google Scholar]
- 77. Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743. [DOI] [PubMed] [Google Scholar]
- 78. Pringle JR, Preston RA, Adams AEM, Stearns T, Drubin DG, et al. (1989) Fluorescence microscopy methods for yeast. Meth Cell Biol 31: 357–435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
IAA induced BA3:GUS expression is reduced by the csn1-10 and csn3-3 mutations to a similar extent. 6-d.o. transgenic Col, csn1-10 and csn3-3 seedlings carrying the BA3:GUS reporter were treated with 1 µM IAA for 3 h before histochemical staining for β-glucuronidase activity.
(TIF)
α-myc western detection of the IAA28-myc fusion protein. The PIAA28:IAA28-myc reporter was introduced into the csn1-10 and csn3-3 backgrounds by crossing. Protein extracts were made from 7-d.o. seedling roots treated with IAA for the indicated time. RPT5 was used as a loading control.
(TIF)
sCSN3c is not a breakdown product of the CSN holocomplex during the in vitro fractionation. Fractions (#5∼9) of the first gel filtration run using Col seedling protein extracts were isolated and injected into the column for a second round of gel filtration. CSN3 western detection was conducted using fractions from the 2nd gel filtration. No CSN3 was detected in the sCSN3c fractions.
(TIF)