Abstract
We investigate single DNA stretching dynamics in a de-wetting flow over micropillars using Brownian dynamics simulation. The Brownian dynamics simulation is coupled with transient flow field computation through a numerical particle tracking algorithm. The droplet formation on the top of the micropillar during the de-wetting process creates a flow pattern that allows DNA to stretch across the micropillars. It is found that DNA nanowire forms if DNA molecules could extend across the stagnation point inside the connecting water filament before its breakup. It also shows that DNA locates closer to the top wall of the micropillar has higher chance to enter the flow pattern of droplet formation and thus has higher chance to be stretched across the micropillars. Our simulation tool has the potential to become a design tool for DNA manipulation in complex biomicrofluidic devices.
INTRODUCTION
To understand the interplay between the flow and large molecules in complex hydrodynamic or electrokinetic systems is important for the design of many polymer and biological processes. Due to the ability to directly visualize individual DNA molecule via fluorescent staining, the single molecular dynamics in micro/nano-fluidic flows has been studied intensively in the past decades.1, 2 DNA stretching is one of the main topics in studying the interplay between DNA and flow field. At the equilibrium state, large DNA molecule such as λ-DNA (48.5 kbp) adopts the convoluted conformation because of entropic elasticity. In order to carry out biological analyses (e.g., gene sequencing or mapping) or use DNA molecules as a building element for bottom-up nanofabrication, stretching or linearization of DNA is a prerequisite process. Basically, the DNA linearization could be classified into four categories depending on the stretching approach and mechanism:3 dynamic stretching, DNA anchoring, confinement, and molecular combing. Dynamic stretching utilizes the dynamic field or field gradient to unravel the DNA molecule in free solution and polymer-enhanced or porous medium. The field could be electric (DC and AC),3, 4, 5, 6, 7, 8, 9, 10 hydrodynamic,2, 11, 12, 13, 14 or combination of both.15 DNA anchoring stretches tethered DNA by using external forces.1, 16, 17 DNA can also be linearized in a confined space with at least one of the dimensions smaller than DNA persistence length.18, 19, 20 Molecular combing utilizes the moving meniscus to stretch and immobilize DNA on the substrate.21, 22 Among these approaches, molecular combing has the potential of permanently stretching the DNA without any sophisticated tools and fabrications.
Molecular combing can be carried out by dipping, spinning, or air-blowing.23 However, the application of this technique is limited by the poor control over the stretching pattern. To overcome this problem, a modified combing strategy was developed in our laboratory to generate highly ordered DNA nanostrands on micropatterned surfaces through a de-wetting process.24, 25 This strategy was also used to produce various nanochannel arrays where the pattern of stretched DNA nanostrands across the micropillar array was used as a precursor to imprint an array of microwell-nanochannel-microwell.26 This facile nanochannel fabrication technique is therefore called DNA combing and imprinting (DCI). We have demonstrated that this simple DCI technique can be applied to fabricate a novel nanochannel electroporation (NEP) device to realize dosage controlled gene, drug, and nanoparticle delivery into individual cells.27
To better understand the mechanism of this approach, we used numerical simulations to study the detailed de-wetting flow patterns over micropillar arrays.28 It was hypothesized that the droplet formation on top of the micropillar after de-wetting creates the flow pattern needed for the stretching of DNA molecules. The effect of the initial micropillar dipping depth was also investigated. Under the over-dipping condition, there would be no droplet formation and we thus assumed no DNA nanostrand was formed. In this study, we use Brownian dynamics (BD) simulation coupled with computational fluid dynamics (CFD) to model the DNA stretching dynamics in the de-wetting flow over micropillars to verify the hypotheses raised in our previous study. The integration of BD simulation with flow/electrical field computation has been carried out to study DNA dynamics in hydrodynamic and electrokinetic flows.3, 5, 7, 29, 30, 31, 32 However, previous studies only focused on 2-D and static/steady-state fields. In this study, we develop a BD-CFD simulation technique for DNA in complex transient physical systems. We apply this technique to study bead-spring modeled DNA molecules in a 3-D transient two-phase flow with complex and dynamic fluid domain. BD simulation is fully integrated with CFD computation so that both DNA and flow dynamics are solved simultaneously. We first study the detailed de-wetting flow pattern by showing both velocity and pressure fields. Then, DNA stretching and immobilization dynamics are investigated through its interplay with the flow field.
SIMULATION METHODS
Typically, simulation of DNA stretching in the receding meniscus flow can be divided into two parts. One is the fluid dynamics computation of the de-wetting two-phase free surface flow, while the other is the DNA molecule stretching dynamics. In the experiments, the DNA nanostrands are composed of multiple stretched DNA molecules.25 In this study, we focus on how the individual DNA molecule is stretched in the de-wetting flow. The effect of multi-chain interactions is not considered in the numerical model and is beyond the scope of this study. We assume that the dilute DNA solution behaves as pure water.
In the experiment, the size of the PDMS stamp is in the order of 1 cm × 1 cm.25 However, the affordable size of computational domain is limited to ∼0.1 mm. Therefore, only a small section of the experimental setup is modeled in the simulation. The de-wetting of the DNA solution between the PDMS stamp and the glass substrate is generated by peeling off the PDMS stamp rotationally. Therefore, the peel-off motion is basically a combination of rotation and translation in the computational domain. In our previous study, the de-wetting generated by pure rotation was investigated.15 Here, we validate the flow pattern by pure translation.
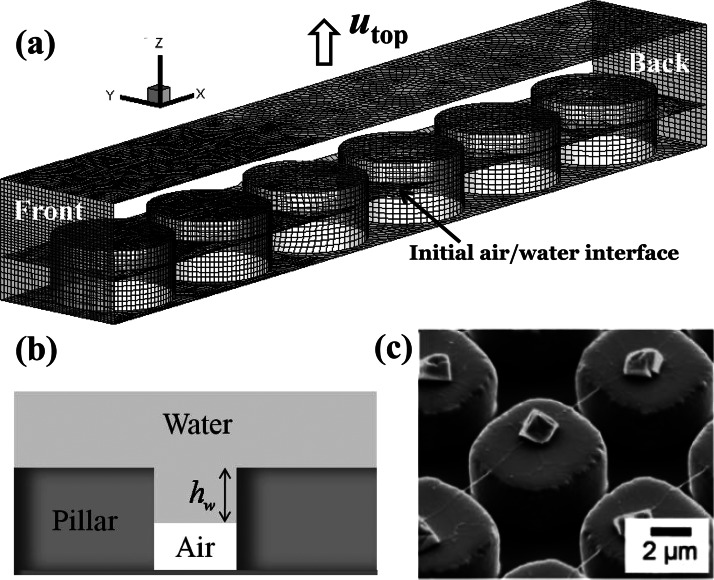
Our simulation setup is depicted in Fig. 1a with meshed boundaries. In the simulation, the de-wetting is generated by the upward moving of the top flat surface. From the front side view, two side boundaries (transparent) are symmetric. With a constant pressure on the front side and a symmetric condition on the back side, the air would flow in from the front side and cause the water to recede backward. The coordinate is set such that the de-wetting is along the x-direction and the top surface moves in the z-direction as shown in Fig. 1a. The diameter, height, and center to center distance of the micropillar are 7, 3.5, and 8.3 μm, respectively.25 Initially, the distance between the top surface and the pillar top is the same as the pillar height. The water is filled in the space between the top surface and the interface and a layer of air exits between the interface and the bottom surface as schematically described in Fig. 1b. A parameter is defined that specifies the initial dipping depth of the micropillar. The top surface moves upward with a speed of to start the de-wetting process. Fig. 1c shows the scanning electron microscopy (SEM) picture of the stretched DNA nanostrands across the micropillars with salt crystals formed on the top of the micropillars. We use the simulation to understand why and how the DNA is stretched, which cannot be observed experimentally.
Figure 1.
(a) The geometrical setup of the CFD computation with meshed boundaries. (b) The schematic diagram defining the micropillar dipping depth hw. (c) The SEM picture of stretched DNA over micropillars and salt crystals on micropillars.
Two-phase flow computation
The computation of free surface flows is one of the main topics in CFD since they are involved in many engineering problems such as coating, fluidization, and spray atomization.33, 34, 35 At the interface of two fluids, surface tension exists as a result of uneven molecular forces. The prediction of the interface evolution and the treatment of the interface conditions have been a challenging task for numerical simulation. Most existing numerical methods for interfacial flows fall into two main categories: body-fitted grid (front-capturing) and fixed grid (front-tracking) approaches. In the body-fitted grid method, the interface is the boundary that separates different immiscible fluids and coincides with the mesh side. The governing equations for each phase are solved separately and the interfacial forces are imposed directly on the phase boundary. In order to follow the evolution of the interfacial profile, the computational domain must be re-mapped at every time step. In general, the body-fitted grid approach is more accurate, but it is more time-consuming and is difficult to handle severe distortions and complex topological changes. On the other hand, in the fixed grid method, an additional continuous field variable instead of a real boundary is used to identify the interface. This field variable is usually called color function c defining the fraction of volume in each discretized element of one phase, which adopts the concept of volume of fluid (VOF) method.36 The color function is transported convectively in the computational domain. For a two-phase (water/air) flow, only one color function is required. For example, by setting the water as the reference, represents the water domain, represents the air domain, and the free surface is defined as the contour of . The advantage of front-tracking is that a fixed mesh can be used without frequent updates of the mesh with the deformation of interface. Intrinsically, the interface is not the explicit boundary in this approach. Thus, how to impose the boundary conditions at the interface is an issue. This issue could be solved by using a continuum surface force (CSF) model proposed by Brackbill et al.37 The CSF model interprets the surface force as a continuous body force in the domain rather a boundary condition at the interface. The combination of the CSF model and VOF method provides an affordable approach to simulate interfacial flows involving surface tension and complex topological changes. Here, we use this approach to simulate the de-wetting free surface flow.
By assuming constant physical properties, the conservative form of governing equations of an incompressible Newtonian free surface flow can be expressed as
| (1) |
| (2) |
where the control volume V is bounded by a closed surface S with outward normal vector n, u is flow velocity, is the surface velocity of S, ρ is density, t is time, and f is the external force acting on S that can be expressed as
| (3) |
where η the dynamic viscosity, and the velocity and coordinate component in the direction of unit vector of the Cartesian coordinate system, p the pressure, and the surface tension component in the direction of . Using the CSF and VOF models, is determined by
| (4) |
where σ denotes the surface tension and the color function c is governed by
| (5) |
We choose the water phase as the reference and the physical properties can be expressed as
where subscripts w and a represent water and air in the computational domain. We use the fluid properties at 20 °C and contact angles on top (glass) and bottom (PDMS) are 34.3° and 112°, respectively.28
The governing equations are solved using commercial CFD code flunet (ANSYS Inc.) based on the finite volume method. The Euler implicit scheme is used for the time marching, the SIMPLE algorithm is adopted for pressure-velocity coupling, and the convective term is interpolated by first-order upwind scheme.38 The Euler explicit with geometric reconstruction is used for the color function calculation.39
Brownian dynamics simulation
We use the BD simulation with the bead-spring model for a DNA molecule to investigate the individual DNA stretching dynamics. In this study, the DNA chain with a contour length L = 20 μm is simulated, which corresponds to the YOYO-1 labeled λ-DNA molecule used in the experiments.24, 25 The DNA is modeled by N beads connected with N-1 springs. The force balance for the i-th bead could be expressed as
| (7) |
where ξ is the drag coefficient of a bead, is the position of the i-th bead, is the velocity field, is the total spring force acted at the bead, is the Brownian force, is the meniscus force, and denotes the bead-wall interaction. The meniscus force is used to model the surface tension force exerted on the DNA molecule when the DNA is close to the interface. In our approach of simulating the two-phase flow, the meniscus force could be expressed as
| (8) |
where σ is the surface tension and the diameter of the bead, θ the meniscus contact angle on the bead, and is the gradient of the color function at . We also define the unit vector along as . Due to the hydrophilic surface of DNA molecule, we simply use in this study.
When it comes to the dynamic properties of single macromolecule such as DNA, the influence of intra-molecule hydrodynamic interactions (HI) is always a concern. Here, our modeled DNA is λ-DNA with contour length about 20 μm. At this molecule weight and contour length, it shows that the dynamics difference between the HI and free draining (FD) models is negligible40, 41 and FD model can well capture the coil-stretch transition dynamics in both extensional and shear flows.42, 43, 44 HI effect starts to play a role when the contour length is longer than 100 μm.45 Therefore, we assume that the hydrodynamic interactions could be ignored in our simulations.
To carry out the simulation, we non-dimensionalize Eq. 7 by introducing dimensionless parameters
| (9) |
where is the root-mean-square spring length, is the number of Kuhn steps () in one spring, and the thermal energy. Accordingly, the dimensionless force balance equation becomes
| (10) |
where is the bead Peclect number, is the diffusivity of the bead. where is a vector with its components uniformly distributed between −1 and 1 and . The worm-like chain force law46 is used to determine the spring force between bead i and i + 1, denoted as ,
| (11) |
where , the effective persistence length, and . For the stained λ-DNA, the persistence length is = 0.066 μm.47 In the simulation, a higher effective persistence length is used in order to compensate the artificial effect of using multiple beads. To simulate labeled λ-DNA in an extension flow, Larson et al. used = 0.075, 0.082, and 0.096 μm for number of beads N = 5, 10, and 20, respectively.42 An empirical relationship could be concluded based on their parameters48
| (12) |
To better describe the DNA conformation change, we use N = 51 beads in this study and therefore is increased to be 0.139 μm that makes .
The bead-wall interaction is modeled by the modified Heyes-Melrose algorithm where the bead is re-positioned to the nearest wall whenever the bead penetrates through the wall.49, 50 It can be expressed mathematically as50
| (13) |
where is the displacement vector for the penetrated bead, is the vector from the penetrated bead to the nearest wall, is the magnitude of , and H denotes the Heaviside step function that is only a unit value for penetrated beads. The bead is allowed to penetrate into the air in the simulation. When this happens, theoretically the bead drag coefficient should be modified accordingly. However, due to the viscosity difference between the water and air, we simply set in Eq. 10 when the bead is in the air.
We use the explicit Euler's method to integrate Eq. 10 to perform BD simulations. In the bulk condition, the simulated radius of gyration is 0.65 ± 0.05 μm, which is close to the experimental value of 0.69 μm.51 We also determine the longest relaxation time of DNA by fitting the time evolution of ensemble averaged mean square end-to-end distance of an initially stretched DNA chain at static condition to an exponential function
| (14) |
where is the dimensionless end-to-end distance of the chain, means the ensemble average, is the equilibrium value of , A is a fitting parameters, and is the dimensionless longest relaxation time. Our relaxation simulation starts with a stretched linear chain with 0.8 extension ratio. Equation 14 is used to fit the simulated curve when the end-to-end distance is less than 30% of its contour length. The dimensionless relaxation time is 32.01 from 100 dependent runs, which agrees with the value in the literature.52
Integration of BD and CFD
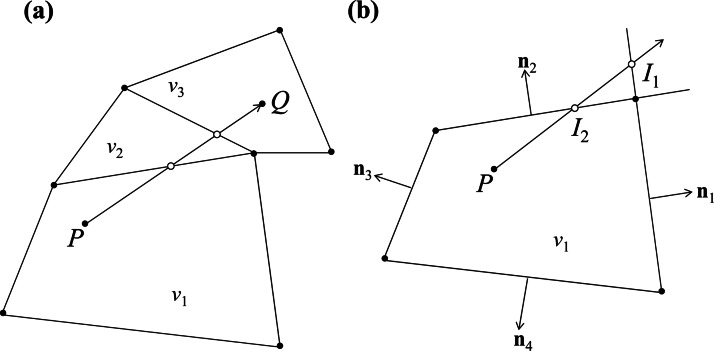
When carrying out the BD simulation for DNA dynamics, DNA beads need to be tracked in the discretized computational fluid domain so that the flow properties at bead coordinates could be interpolated. The DNA tracking is implemented by a particle localization-algorithm that is illustrated in Fig. 2, where the target point Q is tracked from the reference point P as shown in Fig. 2a. To track point Q from point P, we follow the tracking vector and keep tracking the interaction points on the mesh boundaries between points P and Q until we reach the control volume where point Q locates. The searching of the intersection point is further illustrated in Fig. 2b.
Figure 2.
Illustration of the particle tracking algorithm. (a) P is the reference point located in the control volume v1 and Q is the tracking point located in the control volume v3. (b) I1 and I2 are possible interaction points of the tracking vector with the boundaries of v1. n1 to n4 are normal vectors on the boundaries.
If we let and , the possible intersection points will be on boundaries for which , where d is the distance between P and Q, t is the unit vector along the tracking direction, and n is the outward unit normal of the boundary. In this illustration, there are two possible first intersection points on the boundaries of the control volume as shown in Fig. 2b: and . The real intersection point is the one having the smallest distance from point P. The distance between the intersection point and point P could be determined in the procedure as follows.
First, any point on the tracking line can be expressed parametrically as
| (15) |
where α is the parameter that specifies the distance measured from . Then, the intersection point should satisfy
| (16) |
where is the centroid of the boundary. Substituting Eq. 15 into Eq. 16 gives the distance between the intersection points and as
| (17) |
Once the intersection point () is decided, the control volume () adjacent to the intersected boundary could be determined based on the meshing connectivity. Then the particle tracking could be continued by replacing the reference point P by the intersection point and the reference control volume by . Finally, the tracking ends when the control volume where the target point locates is determined.
The coupling between the fluid dynamics and DNA dynamics is one-way coupled based on the assumption that the motion of the DNA molecule will not affect the flow. The simulation procedure is summarized as follows:
-
1.
Given the flow field and locations of DNA beads at the current time level, .
-
2.
Track DNA beads in the discretized computational domain at time level, .
-
3.
Interpolate and at bead locations.
-
4.
Based on and , solve Eq. 10 to update the DNA bead locations at the next time level, .
-
5.
Solve Eqs. 1, 2, 3, 4, 5, 6 to find the flow field at the next time level, .
-
6.
Stop the computation if the prescribed time limit has been reached. Otherwise, set and , then return to step 2 and repeat the computation.
The integration of BD simulation and fluent CFD computation is realized by the use of the user-defined functions of fluent. We perform all of our simulation on a desktop workstation with an Intel® Xeon X5493 processor. The typical run time for studying both the flow and DNA stretching dynamics is about 5–10 h in a fluent serial solver.
RESULTS AND DISCUSSIONS
De-wetting flow pattern over micropillars
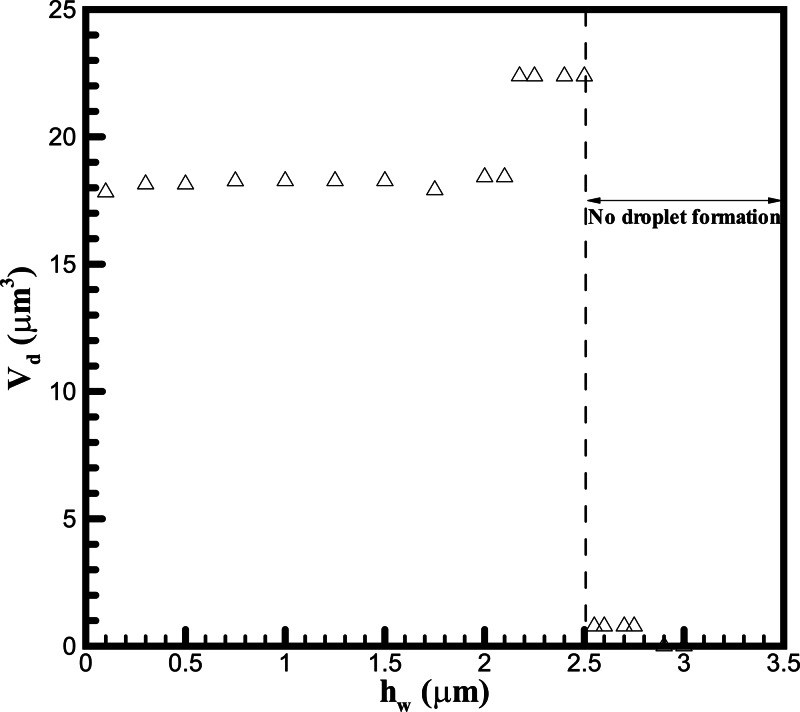
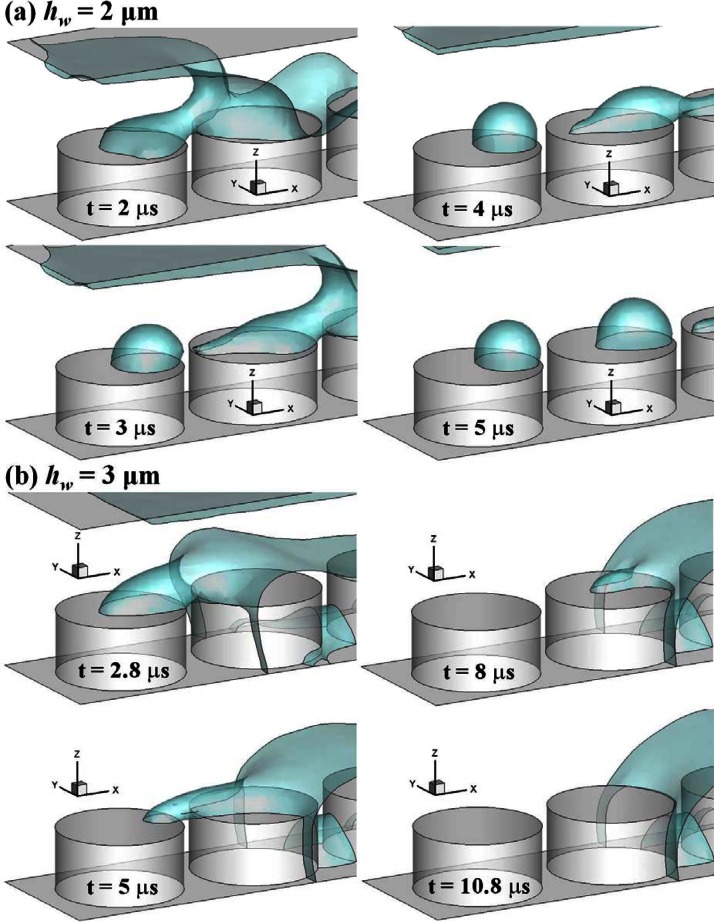
We first identify the conditions of proper and over-dipping in our fluidic system where the de-wetting is generated by pure translational motion of the top surface. We study the effect of the initial micropillar dipping depth in the simulation. Fig. 3 shows the volume of formed droplet on top of the first micropillar at various initial micropillar dipping depths when m/s. We carry out simulations for from 0.1 toward 3.5 μm which is the height of the micropillar. Higher corresponds to a higher degree of dipping of the micropillar into the water. For simplicity, only the volume of the droplet on the first pillar is calculated. When is increased, first maintains a constant value about 18 μm3. Its value increases to about 23 μm3 when > 2 μm. Then the value of drops largely when is higher than 2.5 μm. Therefore, we define a critical value of hw,critical as 2.5 μm in this study. Above this value, there would be no formation of DNA nanostrands across the micropillars. Thus, we identify the situation of hw < 2.5 μm as “proper-dipping” while hw > 2.5 μm as “over-dipping.” Fig. 4 shows the examples of proper-dipping (= 2 μm) and over-dipping (= 3 μm) where the time evolution of free surface profile is plotted. Under proper-dipping, we see the formation of droplets on the top of micropillars in Fig. 4a. Under the over-dipping, the flow pattern changes and there is no formation of droplet. Since higher , thicker the initial water layer. Therefore, more water could be embraced into the forming droplet when > 2 μm. However, when is too large, the receding meniscus is able to touch the bottom floor surface during the de-wetting. The water does not simply retreat over the micropillar and form the droplet. In this situation, the air would be trapped by water and forms the bubble, for example, the one between the second and third pillar in Fig. 4b. Our simulation result agrees with the experimental process where no DNA nanowires were found when the micropillar stamp was pressed on the solution before the peel-off.28
Figure 3.
The formed droplet volume on the top of the first micropillar after the retreat of water under different initial micropillar dipping depths in the simulation at m/s.
Figure 4.
Time series of free surface profile of (a) proper-dipping (μm) and (b) over-dipping (μm) at m/s.
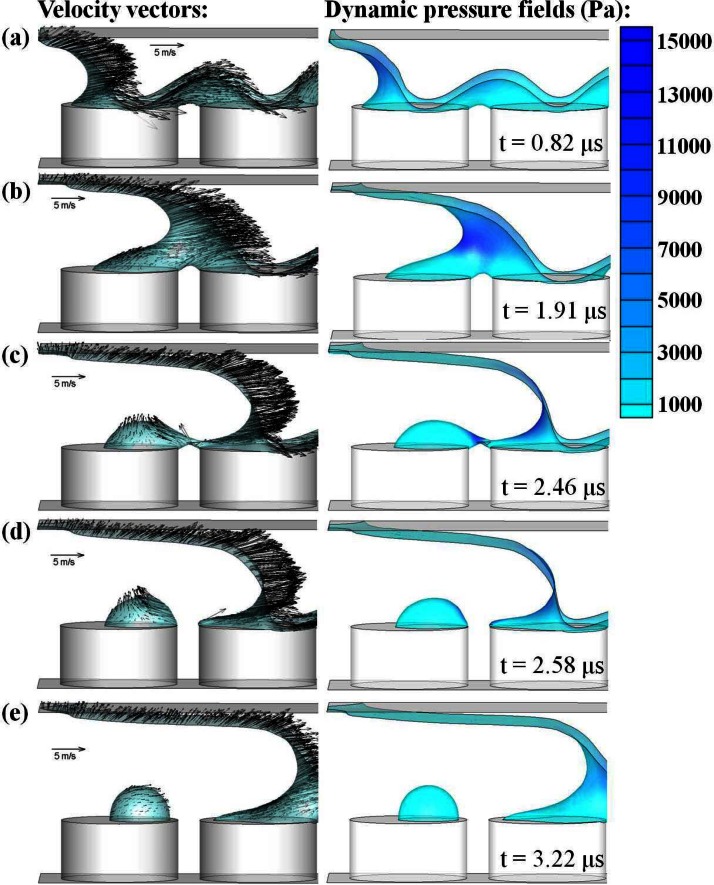
The detailed flow fields of a proper-dipping condition ( = 0.1 μm) are presented in Fig. 5. Five time series of simulated free surface profiles along with velocity vectors and dynamic pressure contours during water de-wetting over the micropillars are depicted. The reference vector is 5 m/s and the scale bar of the dynamic pressure is listed on the right. The dynamic pressure is the pressure associated with the flow motion. The flow-in of air from the left resulting from the upward motion of the top glass substrate pushes the water backward and creates a concave free surface profile in Fig. 5a. The impingement of the air on the water generates a relatively high pressure zone around the vertex of the concave profile as could be seen in Fig. 5b. Then the water starts to form a droplet on the top of the first micropillar and there is a water bridge connecting the forming droplet and the rest of the retreating water in Fig. 5c. At this moment, the forming droplet is about to detach from the main stream and the water filament created between the detaching droplet and the rest of receding water acts like a neck. Interestingly, there is a localized high pressure region generated at the water neck which directly explains the velocity vectors in opposite directions induced across the water filament. Thus, there is a stagnation point existing inside the water filament. This flow pattern is similar to the stagnation flow in a cross-slot that has been used to study single DNA coil-stretch transition dynamics.2, 4 The stagnation flow will shrink the water neck and eventually cause the breakup or pinch-off of the water filament shown at the next frame, Fig. 5d. Right after the pinch-off, some water in the water filament will incorporate into the forming droplet and there is a residual inertia forcing the droplet moving to the left. The droplet will finally settle down on the top of the first pillar and the dynamic pressure on the whole droplet becomes isobaric as shown in Fig. 5e. Since the water will flow downstream through the connecting water bridge during the droplet formation, the time of breakup will determine the final droplet volume. We find that the breakup time reduces when > 2 μm. This could also explain why there is a volume jump in Fig. 3.
Figure 5.
The time series of the flow field for μm and m/s. The velocity vectors and dynamic pressure contours at the de-wetting free surface are plotted.
DNA stretching dynamics
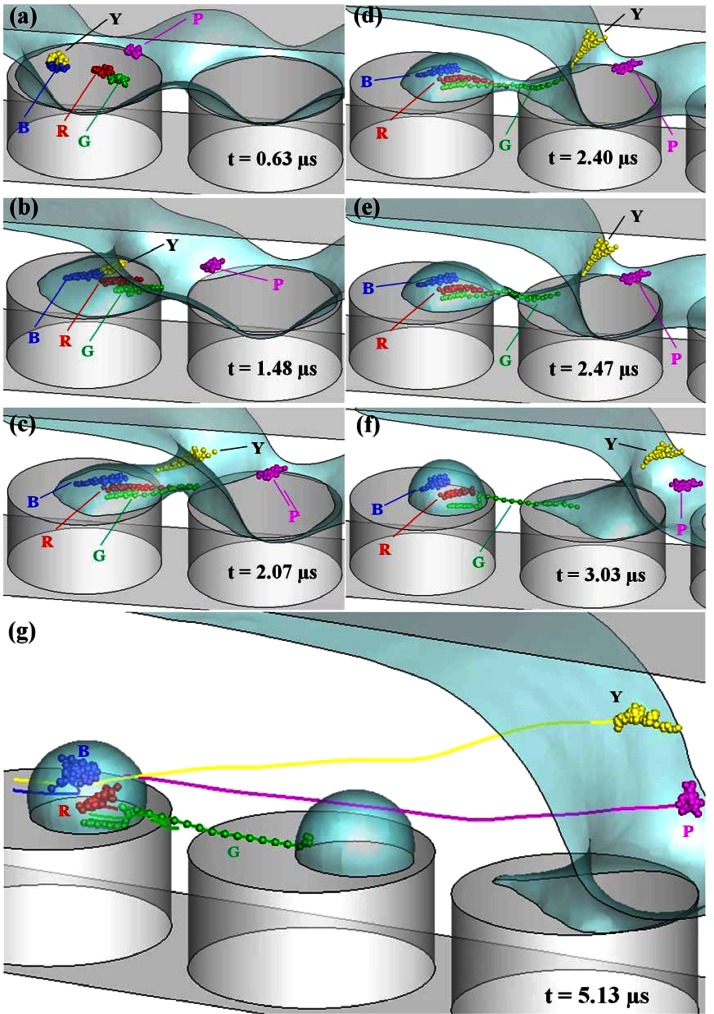
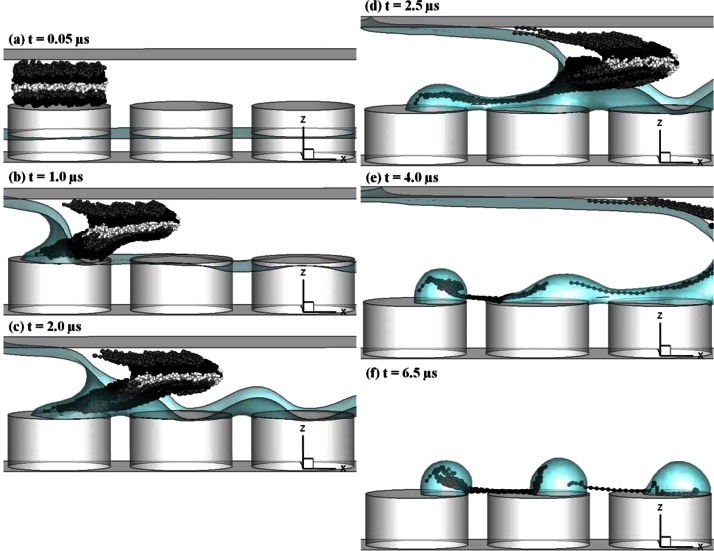
We then integrate the BD simulation with CFD computation to study the interplay between the DNA and the flow. Fig. 6 shows the time series of DNA dynamics in the described de-wetting flow. The simulated free surface profiles and conformations of five DNA molecules are presented. Five DNA chains marked in colors of blue (B), green (G), purple (P), red (R), and yellow (Y) initially locate above on the top of left micropillar in Fig. 6a. The initial DNA conformations are obtained by running the random walk for a period of relaxation time from randomly generated conformations. Due to the flow and velocity gradients, DNAs start to move with the fluid and deform. Compared to other DNA molecules, the purple DNA locates further away from the micropillar top wall. Because of the no-slip boundary condition on the wall, other four DNAs experience a lower flow velocity but a higher velocity gradient. Thus, the purple DNA moves further, while the other four begin to unravel as seen in Fig. 6b. In Fig. 6c, the droplet is forming on the top of the left pillar and the water neck develops. At this moment, the yellow DNA catches up with the green one and moves into the water neck. The red and green DNAs stretch further and reach the water neck region, while the blue DNA stays inside the forming droplet.
Figure 6.
The time series of DNA dynamics in the de-wetting flow (μm and m/s). (a)-(f) Five instantaneous DNA conformations in yellow (Y), blue (B), red (R), green (G), and purple (P). (g) Instantaneous DNA conformations after the formation of the second droplet along with DNA center-of-mass trajectories starting from (a).
When the droplet develops further, the water neck shrinks as shown in Fig. 6d. The yellow DNA is squeezed out during the shrinkage of the water neck. It slips over and rotates counterclockwise along the concave free surface profile and catches up with the purple DNA. The green DNA is stretched across the micropillars inside the water neck. The red DNA heads to the smallest section of the water neck with its tail trailed by the forming droplet. The blue DNA inside the forming droplet recoils back slightly. At the next stage, the water neck is about to break up in Fig. 6d. As depicted in Fig. 5c, the flows in opposite directions are generated along the water neck. This flow pattern pushes the red DNA back to the forming droplet if we compare its locations in Figs. 6d, 6e. Meanwhile, the green DNA still stays inside the water neck since it extends across the water neck. Clearly, the dynamics of each DNA is very different. In the beginning, all DNAs migrate from top of the left pillar. After their interplays with the flow field, the yellow and purple ones move forward with the fluid, the blue and red ones are trapped in the forming droplet, while the green one is stretched across two micropillars inside the shrinking water neck. Once the water neck is pinched off, the stretched green DNA strand is immobilized across the two micropillars as shown in Fig. 6f.
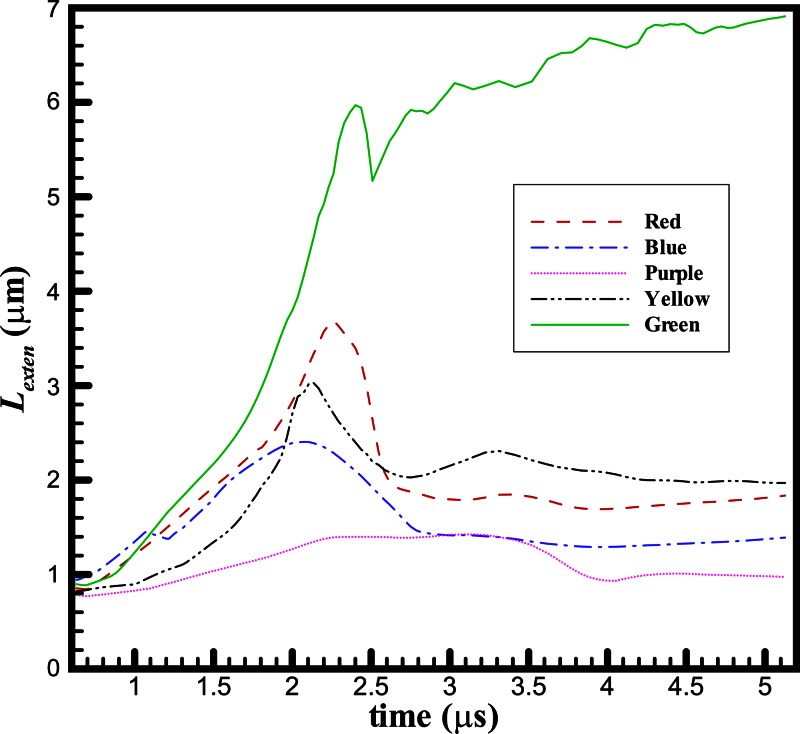
Finally, Fig. 6g shows the dynamics of DNA and the flow after another droplet is formed on the top of the next micropillar. In addition, the center-of-mass trajectory of each DNA from its initial location is plotted. The yellow and purple DNAs avoid the trapping and migrate much longer distances than the other three DNAs. Reversed trajectories are found for the blue and red DNAs, while the trajectory of the green DNA ends at the gap between two micropillars due to stretching. In reality, the formed droplet will evaporate and the DNA molecules and other solutes trapped inside the droplet will form condensed dot-like microstructures as shown in Fig. 1c. In addition, the portion of green DNA inside the droplet would be unwound because of the movement of the meniscus in evaporation and would attach onto the wall. We also summarize temporal profiles of chain extension Lexten for these five DNAs in Fig. 7. Clearly, the purple DNA has smallest extension because it simply migrates with the flow. Compared to red, blue, and green ones, the yellow DNA has delayed extension. It starts to extend when getting into the water neck region as shown in Fig. 6c. Once it slides over the shrinking water neck, it recoils back. Initially, the red and blue DNAs have similar extension profiles until t = 1.8 μs. After that moment, the blue DNA cannot extend further due to being trapped in the forming droplet. On the other hand, the red DNA extends further until t = 2.2 μs. Then, the flows in opposite directions form and it is pushed back to the forming droplet. The green DNA is the one that can extend further even after the formation of flows in opposite directions since it moves across the whole water filament. The slight drop of extension before the breakup is caused by the chain recoiling inside the droplet. When the next droplet starts to form, the green DNA is stretched further.
Figure 7.
Temporal profiles of chain extension for five DNAs shown in Fig. 6.
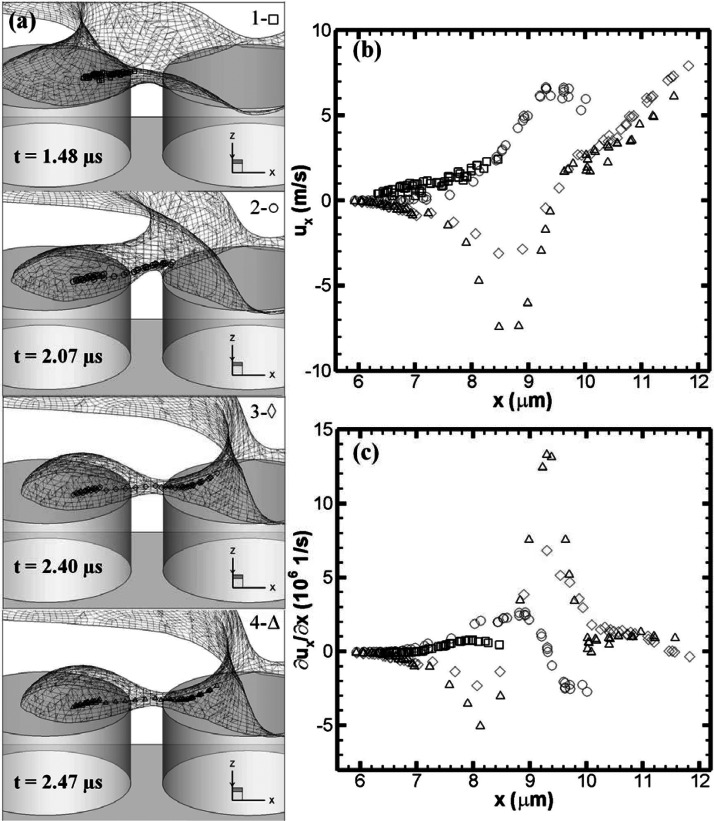
Fig. 8 highlights the DNA stretching dynamics using the stretched green DNA in Fig. 6 as the model. The DNA configurations are marked by different symbols using square, circle, delta, and diamond from top to bottom as emphasized on the top-right corner of each frame in Fig. 8a. For simplicity, we call the time series of the DNA stretching process in Fig. 8a stage 1 to stage 4. Accordingly, the flow velocity along the x-component velocity (the de-wetting direction) and its gradient with respect to the x-coordinate at DNA beads are marked using the same symbols as plotted in Figs. 8b, 8c along with bead x-coordinates. From the velocity profiles in Fig. 8b, we understand that the forming droplet tries to seize the trail of the DNA. The flow is nearly static when it is close to the central area of the forming droplet. At stages 1 and 2, the DNA only experiences flows in the positive x-direction. The increase of the flow from stage 1 to 2 is caused by the decrease of the cross section area of water. On the other hand, the velocity profiles at stages 3 and 4 are characterized by the V-shaped velocity profiles. The velocity profiles bounce back at around μm. They are across approximately at μm where the flow changes direction, which is exactly the location of the stagnation point. Thus, the stagnation point could be seen as a flow divider. To the left, the water moves to the left; to the right, the water moves to the right. As a result, the water filament will eventually be pinched off. The closer to the pinch-off point, the higher the speed of the velocity is. Fig. 8c shows the corresponding velocity gradients. The value of the velocity gradient can generally be linked to the meniscus profile in this study. The concave meniscus profile of the water neck contributes to the positive velocity gradient while the convex meniscus profile of the forming droplet contributes to the negative velocity gradient. In addition, the magnitude of the velocity gradient is proportional to the curvature of the free surface profile. The positive velocity gradients are able to stretch DNA chains as we see in Fig. 8a. The localized positive velocity gradients at stages 3 and 4 force the DNA to be straightened out across the micropillars.
Figure 8.
The interplay between the flow and the stretching DNA (μm and m/s). (a) Free surface profiles and DNA instantaneous configurations at four specific moments. DNA conformations are marked by square (□), circle (○), diamond (⋄), and delta (Δ). (b) Extracted x-component flow velocities at DNA beads versus bead x-coordinates. (c) Extracted x-component velocity gradients in the x-direction versus bead x-coordinates.
From previous results, we know that the flow pattern of droplet formation allows the DNA molecule to be anchored by the forming droplet and be stretched inside the shrinking water filament. However, as we find in Fig. 6 only one out of five DNA molecules could be stretched. The yellow, red, and green DNAs have the chance to reach the forming water neck as shown in Fig. 6c. However, only the green DNA could be stretched in the flow and its interplay with the flow field is further investigated in Fig. 8. On the other hand, the yellow DNA migrates through the shrinking water filament in Fig. 6d, while the red one is able to stay further inside the water filament. However, the red one does not reach the right-hand side of the stagnation point and is pushed back to the droplet after the breakup of the water filament. We also study the effect of initial DNA conformation by assign different DNAs at the same center-of-mass location of the green DNA in Fig. 6. However, we do not find that the correlation between the initial configuration and combing. For one DNA to be stretched in current dynamic de-wetting flow, it has to enter the water filament at a right moment with a proper position in order to be stretched. We therefore carry out a parametric study to understand which initial location of DNA has a higher chance to be stretched.
We generate five layers of DNA molecule where each layer is marked by the same color in gray scale. The layers are named based on their color and order by black-1 (B-1), gray-2 (G-2), white-3 (W-3), black-4 (B-4), and gray-5 (G-5) from bottom to top as shown in Fig. 9a. In this case, another proper-dipping condition is used where μm. The center-of-mass of DNA in each layer is located at the plane of z = constant. The distance between each layer is 0.5 μm, which is also the distance between the bottom B-1 layer and the top surface of the pillar. In each layer, 36 DNA molecules are assigned. Again, we do not consider the chain interaction in this parametric study. Thus, the simulation result presented in Fig. 9 is the superposition of multiple 180 chains. From Figs. 9b, 9c, 9d, we see how these five layers of DNA interact with the flow. Clearly, DNAs from B-1, G-2, and G-5 experience a higher degree of deformation than DNAs in W-3 and B-4. Among these DNAs, only B-1 DNAs have the chance to be trapped into the region of the forming droplet and water neck. As a result, B-1 DNAs have the highest degree of deformation. Due to the shear induced by the walls, the DNA molecule distribution displays a parabolic or bullet-like profile. Since only the B-1 DNAs get into the forming droplet region, finally only B-1 DNAs are stretched and immobilized across the micropillars. Among 36 B-1 chains, 7 chains are stretched with 6 chains across the first and second pillar and 1 chain across the second and third pillar in Fig. 9f. For the DNA to be stretched, it has to be embraced by the forming droplet. Therefore, the closer the DNA chain is to the top of the pillar, the higher chance the DNA has for being stretched.
Figure 9.
Time series of simulated DNA configurations (μm and m/s). Five layers of DNAs initially locate over the first pillar from the left that are plotted in gray scale. From bottom to top, each layer is named as black-1 (B-1), gray-2 (G-2), white-3 (W-3), black-4 (B-4), and gray-5 (G-5), respectively. In each layer, there are 36 DNA chains with a constant z-coordinate. From the pillar top wall, each layer of DNA is assigned every 0.5 μm.
CONCLUSION
We carry out Brownian dynamics simulations to study the single DNA stretching dynamics in a de-wetting flow over a micropillar-patterned surface. The Brownian dynamics simulation is integrated with the transient flow field computation through a particle tracking algorithm so that DNA and fluid dynamics are computed simultaneously. The de-wetting is generated by the translational upward motion of the top flat surface in the simulation. The effect of the initial micropillar dipping depth on the droplet is verified in this de-wetting process. Detailed velocity and pressure fields during the droplet formation under proper-dipping are analyzed. Through the interplay with the flow, DNA molecules could either transport over the micropillar, be trapped into the formed droplet, or be stretched and immobilized across micropillars. The DNA has to migrate into the shrinking water filament and extend across the stagnation point of the water filament before the breakup in order to form the stretched nanostrands across the micropillars. Our simulation technique can be used to investigate the DNA dynamics in not only two-phase flows52, 53, 54 but also other complex transient flows55, 56, 57 in microfluidic systems. The computational time in this approach could be further reduced when being integrated with high performance parallel computing. More importantly, it provides the platform for two-way coupled direct numerical simulation due to the coupling with flow field. We expect that this developed simulation approach has the potential to become a design tool for DNA manipulation in complex biomicrofluidic devices.
ACKNOWLEDGMENTS
This work was supported from NSF under Grant No. NSEC EEC-0914790. Authors acknowledge Professor Shiu-Wu Chau and Dr. Chung-Hsun Lin for their valuable comments on CFD computation.
References
- Perkins T. T., Smith D. E., Larson R. G., and Chu S., Science 268(5207), 83 (1995). 10.1126/science.7701345 [DOI] [PubMed] [Google Scholar]
- Perkins T. T., Smith D. E., and Chu S., Science 276(5321), 2016 (1997). 10.1126/science.276.5321.2016 [DOI] [PubMed] [Google Scholar]
- Hsieh C.-C. and Lin T.-H., Biomicrofluidics 5(4), 044106 (2011). 10.1063/1.3655565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juang Y.-J., Wang S., Hu X., and Lee L. J., Phys. Rev. Lett. 93(26), 268105 (2004). 10.1103/PhysRevLett.93.268105 [DOI] [PubMed] [Google Scholar]
- Trahan D. W. and Dolye P. S., Biomicrofluidics 3(1), 012803 (2009). 10.1063/1.3055275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W.-C., Watari N., Wang S., Hu X., Larson R. G., and Lee L. J., Electrophoresis 31(16), 2813 (2010). 10.1002/elps.201000081 [DOI] [PubMed] [Google Scholar]
- Hsieh C.-C., Lin T.-H., and Huang C.-D., Biomicrofluidics 6(4), 0440105 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji N., Ueda M., and Baba Y., Biophys. J. 82(1), 335–344 (2002). 10.1016/S0006-3495(02)75398-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namasivayam V., Larson R. G., Burke D. T., and Burns M. A., Anal. Chem. 74(14), 3378–3385 (2002). 10.1021/ac025551h [DOI] [PubMed] [Google Scholar]
- Lin H.-Y., Tsai L.-C., Chi P.-Y., and Chen C.-D., Nanotechnology 16(11), 2738–2742 (2005). 10.1088/0957-4484/16/11/046 [DOI] [Google Scholar]
- Smith D. E. and Chu S., Science 281(5381), 1335–1340 (1998). 10.1126/science.281.5381.1335 [DOI] [PubMed] [Google Scholar]
- Smith D. E., Babcock H. P., and Chu S., Science 283(5408), 1724–1727 (1999). 10.1126/science.283.5408.1724 [DOI] [PubMed] [Google Scholar]
- Wong P. K., Lee Y.-K., and Ho C.-M., J. Fluid Mech. 497, 55–65 (2003). 10.1017/S002211200300658X [DOI] [Google Scholar]
- Lee C.-H. and Hsieh C.-C., Biomicrofluidics 7(1), 014109 (2013). 10.1063/1.4790821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren F., Zu Y., Rajagopalan K. K., and Wang S., Biomicrofluidics 6(4), 044103 (2012). 10.1063/1.4762852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M. D., Yin H., Landick R., Gelles J., and Block S. M., Biophys. J. 72(3), 1335 (1997). 10.1016/S0006-3495(97)78780-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante C., Byrant Z., and Smith S. B., Nature 421(6921), 423–427 (2003). 10.1038/nature01405 [DOI] [PubMed] [Google Scholar]
- Douville N., Huh D., and Takayama S., Anal. Bioanal. Chem. 391(7), 2395 (2008). 10.1007/s00216-008-1995-y [DOI] [PubMed] [Google Scholar]
- Tang J., Trahan D. W., and Doyle P. S., Macromolecules 43(6), 3081 (2010). 10.1021/ma902689c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S. F., Karpusenko A., Sakon J. J., Hook J. A., Lamar T. A., and Riehn R., Biomicrofluidics 5(3), 034106 (2011). 10.1063/1.3613671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalet X., Ekong R., Fougerousse F., Rousseaux S., Schurra C., Hornigold N., vanSlegtenhorst M., Wolfe J., Povey S., Beckmann J. S., and Benison A., Science 277(5331), 1518 (1997). 10.1126/science.277.5331.1518 [DOI] [PubMed] [Google Scholar]
- Petit C. A. P. and Carbeck J. D., Nano Lett. 3(8), 1141–1146 (2003). 10.1021/nl034341x [DOI] [Google Scholar]
- Kim J. H., Shi W.-X., and Larson R. G., Langmuir 23(2), 755 (2007). 10.1021/la062505u [DOI] [PubMed] [Google Scholar]
- Guan J. and Lee L. J., Proc. Natl. Acad. Sci. U.S.A. 102(51), 18321 (2005). 10.1073/pnas.0506902102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan J., Yu B., and Lee L. J., Adv. Mater. 19(9), 1212 (2007). 10.1002/adma.200602466 [DOI] [Google Scholar]
- Guan J., Pouyan B., Hemminger O., Chiou N.-R., Zha W., Cavanaugh M., and Lee L. J., Adv. Mater. 22(36), 3997 (2010). 10.1002/adma.201000136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouyan B., Moss A., Liao W.-C., Hanslee B., Jung H. C., Zhang X., Yu B., Wang X., Wu Y., Li L., Gao K., Hu X., Zhao X., Hemminger O., Lu W., Lafyatis G. P., and Lee L. J., Nature Nanotech. 6(11), 747 (2011). 10.1038/nnano.2011.164 [DOI] [PubMed] [Google Scholar]
- Lin C. H., Guan J., Chau S. W., Chen S. C., and Lee L. J., Biomicrofluidics 4(3), 034103 (2010). 10.1063/1.3474639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panwar A. S. and Kumar S., J. Chem. Phys. 118(2), 925 (2003). 10.1063/1.1523912 [DOI] [Google Scholar]
- Kim J. M. and Doyle P. S., J. Chem. Phys. 125(7), 074906 (2006). 10.1063/1.2222374 [DOI] [PubMed] [Google Scholar]
- Duggal R., Sunthar P., and Prakash J. R., J. Rheol. 52(6), 1405 (2008). 10.1122/1.2994729 [DOI] [Google Scholar]
- Xin H., Wang S., and Lee L. J., Phys. Rev. E 79(4), 041911 (2009). 10.1103/PhysRevE.79.041911 [DOI] [PubMed] [Google Scholar]
- Lee S.-L. and Liao W.-C., Int. J. Heat Mass Transfer 51(9-10), 2433 (2008). 10.1016/j.ijheatmasstransfer.2007.08.030 [DOI] [Google Scholar]
- Lee S.-L. and Tien W.-B., Int. J. Heat Mass Transfer 52(13-14), 3000 (2009). 10.1016/j.ijheatmasstransfer.2009.01.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.-L. and Yang C.-F., Can. J. Chem. Eng. 90(3), 612 (2012). 10.1002/cjce.20632 [DOI] [Google Scholar]
- Hirt C. W. and Nichols B. D., J. Comput. Phys. 39(1), 201 (1981). 10.1016/0021-9991(81)90145-5 [DOI] [Google Scholar]
- Brackbill J. U., Kothe D. B., and Zemach C., J. Comput. Phys. 100(2), 335 (1992). 10.1016/0021-9991(92)90240-Y [DOI] [Google Scholar]
- Ferziger J. H. and Perić M., Computational Methods for Fluid Dynamics, 3rd ed. (Springer, 2001). [Google Scholar]
- Morton K. W. and Baines M. J., Numerical Methods for Fluid Dynamics, 1st ed. (Academic Press, 1983). [Google Scholar]
- Jendrejack R. M., de Pablo J. J., and Graham M. D., J. Chem. Phys. 116(17), 7752–7759 (2002). 10.1063/1.1466831 [DOI] [Google Scholar]
- Hsieh C.-C., Li L., and Larson R. G., J. Non-Newtonian Fluid Mech. 113(2-3), 147–191 (2003). 10.1016/S0377-0257(03)00107-1 [DOI] [Google Scholar]
- Larson R. G., Hu H., Smith D. E., and Chu S., J. Rheology 43(2), 267–304 (1999). 10.1122/1.550991 [DOI] [Google Scholar]
- Hur J. S., Shaqfeh E. S. G., and Larson R. G., J. Rheology 44(4), 713–742 (2000). 10.1122/1.551115 [DOI] [Google Scholar]
- Hur J. S., Shaqfeh E. S. G., Babcock H. P., Smith D. E., and Chu S., J. Rheol. 45(2), 421–450 (2001). 10.1122/1.1339246 [DOI] [Google Scholar]
- Schroeder C. M., Shaqfeh E. S. G., and Chu S., Macromolecules 37(24), 9242–9256 (2004). 10.1021/ma049461l [DOI] [Google Scholar]
- Marko J. F. and Sigga E. D., Macromolecules 28(26), 8759 (1995). 10.1021/ma00130a008 [DOI] [Google Scholar]
- Schroeder C. M., Teixeira R. E., Shaqfeh E. S. G., and Chu S., Macromolecules 38(5), 1967 (2005). 10.1021/ma0480796 [DOI] [Google Scholar]
- Hu X., Ph.D. dissertation, The Ohio State University, 2006. [Google Scholar]
- Heyes D. and Melrose J., J. Non-Newtonian Fluid Mech. 46(1), 1 (1993). 10.1016/0377-0257(93)80001-R [DOI] [Google Scholar]
- Kim J. M. and Doyle P. S., Lab Chip 7(2), 213–225 (2007). 10.1039/b612021k [DOI] [PubMed] [Google Scholar]
- Chen Y.-L., Graham M. D., de Pablo J. J., Randall G. C., Gupta M., and Doyle P. S., Phys. Rev. E 70(6), 060901 (2004). 10.1103/PhysRevE.70.060901 [DOI] [PubMed] [Google Scholar]
- Our dimensionless relaxation will be 0.48 after transformed to the bead-spring model used in Ref. 3 where 19 beads were used for a 20.5 μm long DNA and their dimensionless relaxation time is 0.52. During the transformation, we use ξDNA = Nbξ where ξDNA the DNA drag coefficient in bulk solution and Nb the number of beads. The difference may result from the difference in contour length (19.8 μm in this study).
- Hsieh S.-F., Chang C.-P., Juang Y.-J., and Wei H.-H., Appl. Phys. Lett. 93(8), 084103 (2008). 10.1063/1.2969047 [DOI] [Google Scholar]
- Juarez G. and Arratia P. E., Soft Matter 7(19), 9444–9452 (2011). 10.1039/c1sm05347g [DOI] [Google Scholar]
- Hu S.-W., Sheng Y.-J., and Tsao H.-K., Biomicrofluidics 6(2), 024130(2012). 10.1063/1.4729129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M., Kurita H., Komatsu J., Mizuno A., and Katsura S., Appl. Phys. Lett. 89(13), 133901 (2006). 10.1063/1.2357885 [DOI] [Google Scholar]
- Ichikawa M., Ichikawa H., Yoshikawa K., and Kimura Y., Phys. Rev. Lett. 99(14), 148104 (2007). 10.1103/PhysRevLett.99.148104 [DOI] [PubMed] [Google Scholar]