Abstract
Nicotinic acetylcholine receptors (nAChR) are members of the Cys-loop ligand-gated ion channel superfamily. Muscle nAChR are heteropentamers that assemble from two α, and one each of β, γ, and δ subunits. Each subunit is composed of three domains, extracellular, transmembrane and intracellular. The transmembrane domain consists of four α-helical segments (M1–M4). Pioneering structural information was obtained using electronmicroscopy of Torpedo nAChR. The recently-solved X-ray structure of the first eukaryotic Cys-loop receptor, a truncated (intracellular domain missing) glutamate-gated chloride channel α (GluClα)showed the same overall architecture . However, a significant difference with regard to the vertical alignment between the channel-lining segment M2 and segment M3 was observed. Here we used functional studies utilizing disulfide trapping experiments in muscle nAChR to determine the spatial orientation between M2 and M3. Our results are in agreement with the vertical alignment as obtained when using the GluClα structure as a template to homology model muscle nAChR, however, they cannot be reconciled with the current Torpedo nAChR model. The vertical M2–M3 alignments as observed in X-ray structures of prokaryotic Gloeobacter violaceus ligand-gated ion channel (GLIC) and GluClα are in agreement. Our results further confirm that this alignment in Cys-loop receptors is conserved between prokaryotes and eukaryotes.
Keywords: acetylcholine receptor, nicotinic, ion channel, transmembrane segment, GluCl, GLIC
Introduction
Cys-loop receptors are ligand-gated ion channels that conduct either anions or cations. The individual families are named after the ligand that gates them, γ-amino butyric acid receptors (GABAAR), nicotinic acetylcholine receptors (nAChR), serotonin receptors (5-HT3R), glycine receptors (GlyR). Many drugs in current clinical use target these receptors, and a variety of diseases involve Cys loop receptor dysfunction, for example epilepsy, anxiety, Alzheimer’s and Parkinson’s disease, schizophrenia, depression, attention deficit hyperactivity disorder (ADHD), myasthenia gravis, psychosis, and nicotine addiction.
Pioneering detailed structural knowledge about these receptors was obtained by Nigel Unwin using electronmicroscopy of Torpedo nAChR, which show high sequence-identity with muscle nAChR (Miyazawa et al. 2003, Unwin 2005). The recent identification and subsequent X-ray crystal structure determination of prokaryotic homologues, the Gloeobacter violaceus ligand gated ion channel (GLIC) and Erwinia chrysanthemi LGIC (ELIC), have given high-resolution insights into the receptor structure (Tasneem et al. 2005, Hilf & Dutzler 2008, Hilf & Dutzler 2009). They have also depicted binding sites for numerous allosteric modulators (Nury et al. 2011, Hilf et al. 2010, Pan et al. 2012a, Pan et al. 2012b). The usefulness of these structures for precise mechanistic insights is controversial, as the X-ray structures often do not reflect the functional state that is to be expected (Gonzalez-Gutierrez et al. 2012, Goyal et al. 2011, Parikh et al. 2011). Overall, the same core subunit architecture is found in the structures of metazoan and prokaryotic homologues: a conserved ECD with two anti-parallel β-sheets and a TMD with four α-helical segments (Miyazawa et al. 2003, Unwin 2005, Hilf & Dutzler 2008, Hilf & Dutzler 2009, Hibbs & Gouaux 2011, Bocquet et al. 2009). Compared to eukaryotic Cys-loop receptors, the prokaryotic ones lack the eponymous disulfide-linked cysteines in the Cys-loop, as well as an intracellular domain (Tasneem et al. 2005). The intracellular domain in metazoans is contributed to by a relatively long peptide (ca. 50-270 amino acids) between the transmembrane segments M3 and M4. The M3M4 loop in prokaryotes is barely longer than what is required to link the two transmembrane segments (3–14 amino acids). However, we have previously shown that the long intracellular domain in eukaryotic 5HT3A and GABA-ρ1 receptors can be replaced by a heptapeptide (SQPARAA), the M3–M4 linker in GLIC inferred from multiple-sequence alignment studies, while retaining the ability of the receptors to fold, assemble and function as ligand-gated ion channels upon expression in Xenopus laevis oocytes (Jansen et al. 2008). A comparable approach was utilized for the Caenorhabditis elegans glutamate-gated chloride channel α (GluClα) to obtain a crystallizable construct, in that the intracellular domain was replaced by a tripeptide (Hibbs & Gouaux 2011). This first eukaryotic Cys-loop receptor structure of GluClα surprisingly showed a vertical alignment between the channel-lining transmembrane segment M2 and the transmembrane segment M3 that was distinct from the one observed in the Torpedo nAChR structure (Unwin 2005, Unwin & Fujiyoshi 2012).
Several previous studies have indirectly performed experiments that may be used to assess the question of the vertical alignment between M2 and M3 in nAChR. However, none of them discussed the vertical alignment and/or the discrepancies. The laboratory of Grosman investigated the Cα distances in muscle nAChR of residues along the M1, M2, and M3 segments to the pore’s long axis with a single-channel proton-transfer technique and found that these transmembrane segments only rearrange minimally during gating (Cymes & Grosman 2008, Cymes et al. 2005). While the rates of proton transfer for pre-M2 residues indicated that M2 started closer to the N-terminus than predicted by the Torpedo model, the vertical alignment between M2 and M3 cannot be directly assessed by these measurements. Several recent high-resolution NMR spectroscopy studies investigated the nAChR transmembrane domain. One studied the structure of the sole transmembrane domain (M1 to M4 segments) of the nAChR β2-subunit in and arrived at the conclusion that the resulting four helix bundle “more resembled the structure of the TM domain in GLIC than that in the Torpedoβ1 subunit” (Bondarenko et al. 2010). It was observed that M2 started two residues closer to the N-terminus than in the Torpedo structure. Next, a water-soluble transmembrane domain derived from the nAChR α1-subunit and additionally designed to promote monomeric structure was studied by NMR (Cui et al. 2012). This study specifically discussed the discrepancies in lengths of the various transmembrane segments between the NMR study of isolated individual transmembrane domains and the Torpedo structure. The latest NMR-derived structure resolved the transmembrane domains of α4 and β2 nAChR subunits in lauryldimethylamine-oxide (LDAO) micelles with comparable results. While none of the NMR spectroscopy papers directly addresses the discrepancies in their NMR-derived structures with the Torpedo structure with regard to the vertical alignment between M2 and M3, our analysis of the distances between the residues we investigated in the NMR-structures more closely resembles the distances observed in GluCl over those in Torpedo (Table 3).
TABLE 3.
Distances between engineered Cys based on the X-ray GluCl and NMR nAChR structure.s
| Y277 | M278 | L279 | F280 | T281 | M282 | V283 | |
|---|---|---|---|---|---|---|---|
| GluCl 3RHW | 12.5 | 8.7 | 9.8 | 12.2 | 10.3 | 8.1 | 11.7 |
| Torpedo 2BG9 (A) | 20.4 | 17.5 | 17.5 | 16.8 | 13.3 | 13.6 | 14.2 |
| nAChR α4 2LLY | 14.3 | 10.6 | 12.5 | 15.1 | 13.0 | 11.0 | 14.5 |
| nAChR β2 2LM2 | 13.3 | 10.8 | 13.8 | 14.8 | 11.4 | 11.3 | 14.9 |
| nAChR α1 WSA 2LKG | 12.5 | 8.9 | 10.6 | 13.5 | 12.6 | 11.0 | 14.5 |
| nAChR α1 WSA 2LKH | 12.0 | 8.3 | 9.4 | 12.6 | 11.8 | 10.5 | 13.8 |
Distances (Å) measured between the α-carbons of M2 T254 and M3 positions, or homologous residues, as indicated.
Since the transmembrane domain is of great importance for studies involving a multitude of allosteric modulators like alcohols, general anesthetics, neurosteroids and other synthetic and natural compounds, we sought to address experimentally the vertical alignment of M2 and M3 in nAChR with the present study. Here, we use disulfide trapping in α-subunits from mouse muscle nAChR, that have 77.3 % sequence identity and 88.1 % similarity with α-subunits from Torpedo nAChR (93.4 % identity and 100.0 % similarity, respectively, for the sequences covering M2 and M3), to probe the proximity between engineered Cys in these two transmembrane segments. From our study that includes 13 Cys-pairs we conclude, that the vertical alignment/register of a homology model of muscle nAChR built on the crystallizable engineered GluClα construct is in agreement with our experimental data whereas the Torpedo nAChR structural models conflicts with our results.
Materials and Methods
Plasmids and Mutagenesis
Plasmid pSP64T carrying the gene for the mouse muscle nAChR α-subunit was used to generate mutations in the M2 and M3 transmembrane segments (Akabas et al. 1992). Two endogenous Cys residues, αC192 and αC193, that form a disulfide-linked vicinal cystine at the tip of loop C the turn between antiparallel β strands 9 and 10 of the extracellular domain of all nAChR α-subunits, were replaced with serines to prevent interference from reducing or oxidizing these Cys during the conditions required for our cross-linking experiments (Stewart et al. 2006). The M2 residue αT254 (12’) was mutated to Cys in the αC192S/C193S background and this construct was used to introduce Cys residues in different positions of the M3 segment. We determined α-subunit M3 residues in close proximity (<13 Å) to M2 αT254C based on the structural model of Torpedo nAChR (αS288C to αV293C) and according to a homology model of mouse muscle nAChR that we built on the GluCl structure (αY277C to αV283C)(Hibbs & Gouaux 2011, Unwin 2005). Since α-subunits of mouse muscle nAChR and Torpedo marmorata only differ by 4 out of the 61 amino acids covering M2 and M3, distances calculated for the Torpedo structure or a homology model of muscle nAChR α-subnits based on the Torpedo structure are virtually identical. Mutations were introduced by polymerase chain reaction using the QuikChange II XL mutagenesis kit (Stratagene). DNA isolation was performed by using WizardR Plus Minipreps or WizardR Plus Midipreps DNA purification systems from Promega (Madison, WI). All mutant plasmids were sequenced to confirm the mutations and ascertain the absence of undesired mutations by automated DNA sequencing at Genewiz Inc. Plasmids were linearized (α− and γ-subunits with XbaI, β with SacI, and δ with BamHI) prior to in vitro mRNA transcription with SP6 RNA polymerase (SP6 mMessage mMachine kit, Ambion, Austin, TX). mRNA was purified with the mMega Clear kit (Ambion), precipitated with ammonium acetate, dissolved in diethylpyrocarbonate-treated water, and stored at −80 °C.
Reagents
Acetylcholine (ACh) (Sigma) was prepared as 1M stock solution in water. Dithiothreitol (DTT) (Sigma) was dissolved in water to obtain a 1M stock solution and diluted into recording buffer, OR2 (in mM: 82.5 NaCl, 2 KCl, 1 MgCl2, and 5 HEPES; pH adjusted to 7.5 with NaOH), before each experiment. o-Phenanthroline (Sigma) was prepared as a 1M stock solution in DMSO on the day of the experiment. CuSO4 was made as a 100 mM stock solution in water. CuSO4 and o-phenanthroline were mixed in OR2 before use to a final concentration of 100:200 μM Cu:Phen. Ethylene glycol tetraacetic acid (EGTA) (Sigma) was prepared as 0.5 M solution in water with a pH of 7.4 and was diluted to a concentration of 2 mM in OR2 before application. Collagenase (Type 1A, Sigma) was prepared in Ca2+-free OR2 at a concentration of 2 mg/ml.
Expression in X. laevis oocytes
Stage V–VI oocytes were defolliculated with a 75 min incubation in 2 mg/ml Type 1A collagenase (Sigma) in OR2 (in mM: 82.5 NaCl, 2 KCl, 1 MgCl2, and 5 HEPES; pH adjusted to 7.5 with NaOH). Oocytes were washed thoroughly in OR2 and kept in standard oocyte saline (SOS) medium (in mM: 100 NaCl, 2 KCl, 1.8 CaCl2, 1 MgCl2, and 5 mM HEPES, pH 7.5, supplemented with 1% antibioticantimycotic (100×) liquid (10,000 IU/ml penicillin, 10,000 μg/ml streptomycin, and 25 μg/ml amphotericin B; Invitrogen, Carlsbad, CA) and 5% horse serum (Sigma). Oocytes were injected 24 h after isolation with 50 nl (10 ng) of a 2:1:1:1 mixture of a mouse α:β:γ:δ subunit mRNA and kept at 16°C. Mutant subunit mRNA was substituted for wt α-subunit where necessary.
Two-electrode Voltage Clamp (TEVC) Experiments
Electrophysiological recordings were conducted 3–5 d after injection at room temperature in a ~250 μl chamber continuously perfused at a rate of 5–6 ml/min with Ca2+ -free OR2. Currents were recorded from individual oocytes using two-electrode voltage-clamp at a holding potential of −60 mV. The ground electrode was connected to the bath via a 3 M KCl/agar bridge. Glass microelectrode resistance was <2 MΩ when filled with 3M KCl. Data were acquired at 200 Hz and analyzed using a TEV-200 amplifier (Dagan Instruments), a Digidata 1440 series data interface (analog-digital converter) and pClamp 10.3 software (Molecular Devices). Currents (I) elicited by ACh applications were separated by at least 6 min of wash period with the running buffer for the oocytes to be able to completely recover from desensitization. Currents were considered to be stable if the variation between consecutive IAch varied by ≤ 10–15%. All experiments were performed on at least 3 oocytes from two different batches of oocytes.
Concentration–response analysis. EC50 Determination
Progressively increasing concentrations of ACh were applied to oocytes expressing wild-type or mutant receptors, after obtaining a stable IACh with an approximately EC50 ACh concentration. Currents were normalized to the maximal ACh-induced current (Imax). The ACh concentration–response relationship was determined for wild-type and each mutant. It is calculated by least-squares minimization (GraphPad Prism version 5.0 for Windows, GraphPad Software) of the currents to a logistic equation of the form: where nH is the Hill coefficient and EC50 is the ACh concentration that gives rise to 50% of the maximal current. Parameters from several oocytes were averaged to obtain the mean EC50 and Hill coefficient. Data are presented as mean ± SEM. For determining the effect of reducing and oxidizing agents we subsequently used a concentration of agonist between the EC20 and EC50. A submaximal concentration increases the chances of observing an effect, since receptor modifications can have effects on gating kinetics as well as conductance (Zhang & Karlin 1997, Karlin & Akabas 1998).
DTT effect
10 mM DTT was applied for 2 min, once stable ACh EC20–50 test currents (I) were achieved for each oocyte. The current was recorded again (IDTT) after washing the oocyte for 6 min. Changes in ACh-induced current amplitude upon application of DTT were calculated as follows: .
Cu:Phen effect
After DTT treatment as described above, a stable ACh response was recorded, and subsequently, oocytes were super-fused with the redox catalyst Cu:Phen (100:200 μM) (Kobashi, 1968) solution for 2 min and two more ACh test pulses (ICu:Phen) were applied after a 6 min washout period. To confirm that the Cu:Phen effect was due to the formation of disulfide bonds and not to the binding of Cu2+ to the receptor, 2 mM EGTA was applied for 2 min to chelate possibly bound heavy metals from the receptor. Two ACh pulses were used after EGTA treatment. Inhibition of the current due to disulfide bond formation by Cu:Phen application was normalized to wild-type and calculated as: .
A second application of 10 mM DTT for 2 min was used to test for the reversibility of the Cu:Phen effect.
Statistics
The effect of each reagent on mutant receptors was compared with that obtained with wild-type receptors using one-way ANOVA and Dunnett’s multiple comparison test (Prism 5.0; GraphPad Software, San Diego, CA).
Homology modeling and alignments
Sequence identities and similarities were calculated with EMBOSS Needle - Pairwise Sequence Alignment (http://www.ebi.ac.uk/Tools/psa/emboss_needle/). Homology models for all subunits of mouse muscle nAChR were built based on the recently published X-ray crystal structure of Caenorhabditis elegans glutamate-gated chloride channelα (GluClα)(Hibbs and Gouaux, 2011). Sequences were obtained from the ligand-gated ion channel database (http://www.ebi.ac.uk/compneur-srv/LGICdb/LGICdb.php) or RCSB PDB (Research Collaboratory for Structural Bioinformatics; http://www.rcsb.org/pdb/). Sequences were aligned by using ClustalW (http://www.ebi.ac.uk/clustalw/index.html). Mouse muscle nAChR was aligned as a raw sequence and submitted as a modeling request in Swiss Model (Swiss-Pdb Viewer 4.0.4, http://spdbv.vital-it.ch) (Guex & Peitsch 1997, Schwede et al. 2003). Figures were rendered with PyMOL 1.3 (http://www.pymol.org).
Results
Single Cys mutant and all double Cys mutants are functional
To create the double Cys mutants required for intra-subunit Cys cross-linking studies between the transmembrane segments M2 and M3 in muscle nAChR, we decided to utilize the α-subunit. This subunit appears twice in the hetero-pentameric receptor assembled in a clock-wise fashion when viewed from the extracellular side αγαβδ, whereas the other three subunits are only present once (Fig. 1). Therefore, the effect of cross-linking may be observed more readily in αsubunits. We chose to mutate position M2 αT254 to Cys. This position corresponds to the 12’ position, based on the prime numbering system for M2 that denotes an absolutely conserved cationic residue in eukaryotic Cys-loop receptors towards the cytoplasmic N-terminal end of M2 with 0’, and gives increasing numbers moving in C-terminal direction towards the extracellular side. The numbering of equivalent positions in M2 was introduced by Miller (Miller 1989). For the nAChR α-subunit 0’ corresponds to αK241 and 20’, the conserved extracellular ring of charge position in M2, to αE262. Based on the Torpedo nAChR model the 12’ position is in the cytoplasmic half of the M2 segment, facing away from the channel lumen towards the M3 segment of the same subunit (Fig. 1(b)). We determined the α-subunit M3 residues in close proximity to M2 T254 (<13 Å between α-carbons) based on the Torpedo nAChR model (αS288 to αV293, SIIITV) and according to a homology model of mouse muscle nAChR that we built on the GluCl structure (αY277 to αV283, YMLFTMV) (Table 1, Fig. 2). Two sets of double Cys mutants, covering six or seven consecutive positions in M3, respectively, were generated, αT254C+αY277C to αT254C+αV283C based on our homology model built on the GluCl structure, and αT254C+αS288C to αT254C+αV293C based on the Torpedo structure. All constructs including wild-type had the background mutation αC192S-C193S to avoid interference from reducing the extracellular disulfide bridge between the vicinal Cys that form a cystine in loop C under our experimental conditions (Stewart et al. 2006). The single Cys mutant (αT254C) and all double Cys mutants were functional after expression in X. laevis oocytes as determined by two-electrode voltage-clamp experiments (Table 2, Fig. 3). The ACh EC50, the concentration of ACh that induces a half-maximal effect, was 150 ± 30μM for wild-type (with αC192S-C193S background mutation as for all constructs), and increased 3-fold in the M2 single mutant αT254C (515 ± 23 μM). The EC50 ranged between 105 and 699 μM for the double mutants of the GluCl set and between 624 and 1,058 μM for the double mutants based on the Torpedo structure. One mutant, αT254C+αM282C of the GluCl set had a significantly different EC50 from wild-type as determined by one-way ANOVA with Dunnett’s multiple comparison test (p<0.001). Of the Torpedo set all mutants except αT254C+αI291C had a significantly different EC50 from wild-type as determined by one-way ANOVA with Dunnett’s multiple comparison test (p<0.001). The Hill coefficient, nH, was not significantly different from wild-type for any of the single or double Cys mutants (Table 2). Two mutants of the GluCl set, αT254C+αY277C and αT254C+αM282C, and three mutants from the Torpedo set, αT254C+αS288C, αT254C+αI289C and αT254C+αT292C, had significantly different maximum current amplitudes as compared to wild-type upon ACh application (Table 2).
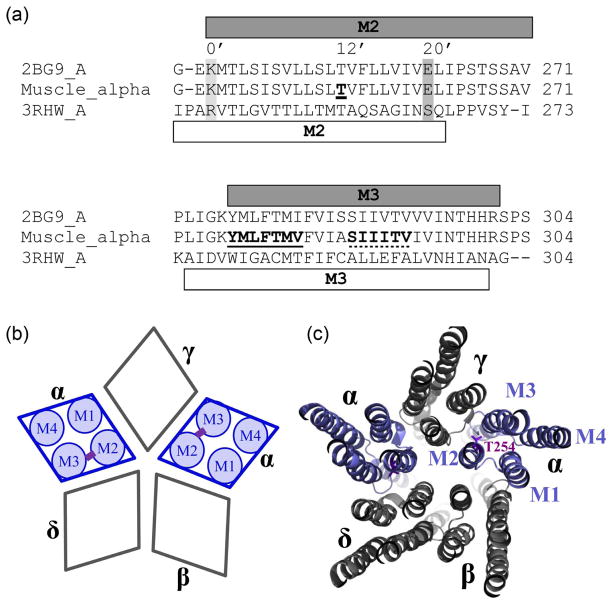
Fig 1.
Multiple sequence alignments of Torpedo, muscle nAChR and GluCl α-subunits as obtained with ClustalW, and schematic nAChR representation. (a) Only the alignment comprising the transmembrane segments M2 and M3 is shown. Note that α-subunits of mouse nAChR and Torpedo are identical for 61 out of the shown 65 residues. The α-helical content as in the Torpedo structural model (PDB# 2BG9) is depicted with a grey bar on top of, and as in the GluCl structure (PDB# 3RHW) with a white bar below the sequences. For the M2 segment the position we mutated to Cys, 12’ corresponding to αT254 is indicated by bold font and underlined, and for orientation also 0’ and 20’ are marked by a light and dark grey box, respectively, and additionally the respective numbering. Positions mutated to Cys in M3 are indicated by bold font and by a continuous line for the GluCl-based set (277-YMLFTMV) and by a dotted line for the Torpedo-based set (288-SIIITV). (b) Subunit arrangement of muscle nAChR with a schematic representation of transmembrane segments (M1–M4) for the two α-subunits, and the potential cross-link schematically indicated in purple for the studied α-subunits. Subunits are labeled with greek letters, α, β, γ, andδ. (c) Muscle nAChR modeled on the Torpedo structure as viewed from the extracellular side in a slab-representation of the transmembrane domain. Subunits are labeled with greek letters and transmembrane segments M1 to M4 are indicated for one of the two α-subunits. The 12’ M2 residue, T254, is in purple stick representation.
TABLE 1.
Distances between engineered Cys based on the GluCl and Torpedo structures
| GluCl-based set | |||||||
|---|---|---|---|---|---|---|---|
| Y277 | M278 | L279 | F280 | T281 | M282 | V283 | |
| GluCl 3RHW | 12.5 | 8.7 | 9.8 | 12.2 | 10.3 | 8.1 | 11.7 |
| Torpedo 2BG9 (A) | 20.4 | 17.5 | 17.5 | 16.8 | 13.3 | 13.6 | 14.2 |
| Torpedo-based set | ||||||
|---|---|---|---|---|---|---|
| S288 | I289 | I290 | I291 | T292 | V293 | |
| GluCl 3RHW | 15.6 | 15.2 | 17.5 | 19.9 | 20.1 | 21.2 |
| Torpedo 2BG9 (A) | 8.2 | 8.0 | 11.7 | 12 | 9.6 | 11.2 |
Distances (Å) measured between the α-carbons of M2 T254 and M3 positions, or homologous positions, as indicated.
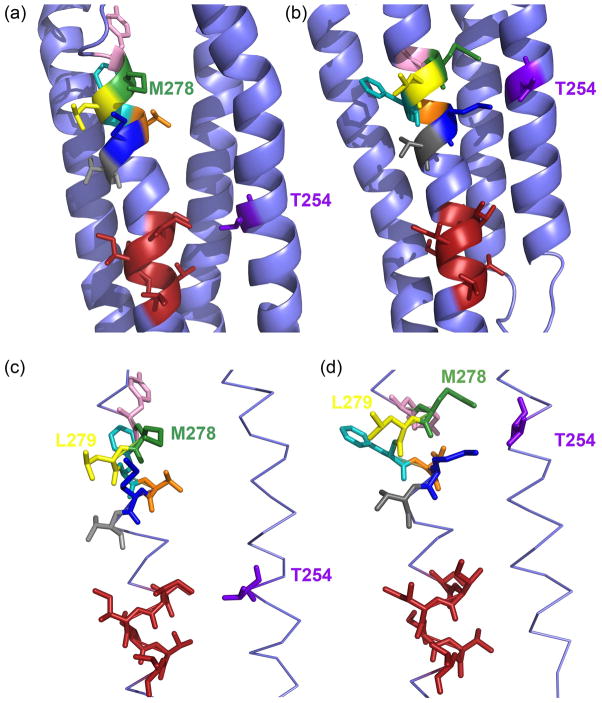
Fig 2.
Relative Orientation of the M2 and M3 transmembrane segments of the muscle nAChR αsubunit. (a) Orientation of the muscle nAChR homology modelM278 based on the Torpedo nAChR structure (PDB# 2BG9), and (b) based on the Caenorhabditis elegans glutamate-gated chloride channel (GluCl) α-subunit structure (PDB# 3RHW) as a template. Residue M2 αT254 is colored purple. M3 residues, which are selected and replaced to Cys according to the GluCl structure are color-coded as follows: Y277, pink; M278, green; L279, yellow; F280, cyan; T281, orange; M282, dark blue; V283, grey. Side chains in red color are shown for the M3 residues S288 to V293, which are in close proximity to M2 αT254 based on the Torpedo structure. (c) and (d) Same as in (a) and (b) except that only M2 and M3 are shown for clarity in stick representation. Note that in (a) and (c)αT254C is in close proximity to the red-colored side-chains and in (b) and (d) it is close to the differently-colored side-chains.
TABLE 2.
Dose-response results for all constructs. ACh EC50, Hill coefficient and maximal currents for wild-type and all Cys-mutants.
| Mutants | EC50, mM | nH | Imax | n |
|---|---|---|---|---|
| Wild-type | 0.15 ± 0.03 | 1.06 ± 0.04 | −3804 | 5 |
| T254C | 0.51 ± 0.06 | 2.0 ± 0.03 | −2782 | 4 |
| T254C+Y277C | 0.5 ± 0.2 | 1.72 ± 0.4 | −987*** | 3 |
| T254C+M278C | 0.16 ± 0.03 | 1.49 ± 0.19 | −4245 | 4 |
| T254C+L279C | 0.15 ± 0.02 | 1.61 ± 0.1 | −3812 | 3 |
| T254C+F280C | 0.35 ± 0.07 | 1.3 ± 0.2 | −3027 | 4 |
| T254C+T281C | 0.1 ± 0.01 | 1.46 ± 0.14 | −3647 | 4 |
| T254C+M282C | 0.7 ± 0.2*** | 1.82 ± 0.4 | −1996*** | 4 |
| T254C+V283C | 0.18 ± 0.02 | 1.49 ± 0.22 | −3656 | 4 |
| T254C+S288C | 0.94 ± 0.12*** | 1.3 ± 0.11 | −116*** | 4 |
| T254C+I289C | 1.06 ± 0.03*** | 1.59 ± 0.1 | −530*** | 3 |
| T254C+I290C | 0.85 ± 0.16*** | 1.63 ± 0.24 | −2916 | 3 |
| T254C+I291C | 0.62 ± 0.14 | 1.5 ± 0.03 | −3209 | 3 |
| T254C+T292C | 0.96 ± 0.14*** | 1.84 ± 0.24 | −871*** | 3 |
| T254C+V293C | 1.01 ± 0.12*** | 1.69 ± 0.04 | −3292 | 3 |
For all mutants the concentration that induces a half-maximal response, EC50, was determined by applying successively higher ACh concentrations in two-electrode voltage clamp experiments, and using the current amplitude as readout. Currents were normalized to the highest obtained current (Imax) for each oocyte and fitted to obtain EC50 and nH. The numbers of independent experiments are given with n.
indicates significantly different from wild-type (p<0.001) using one-way ANOVA and Dunnett’s multiple comparison test.
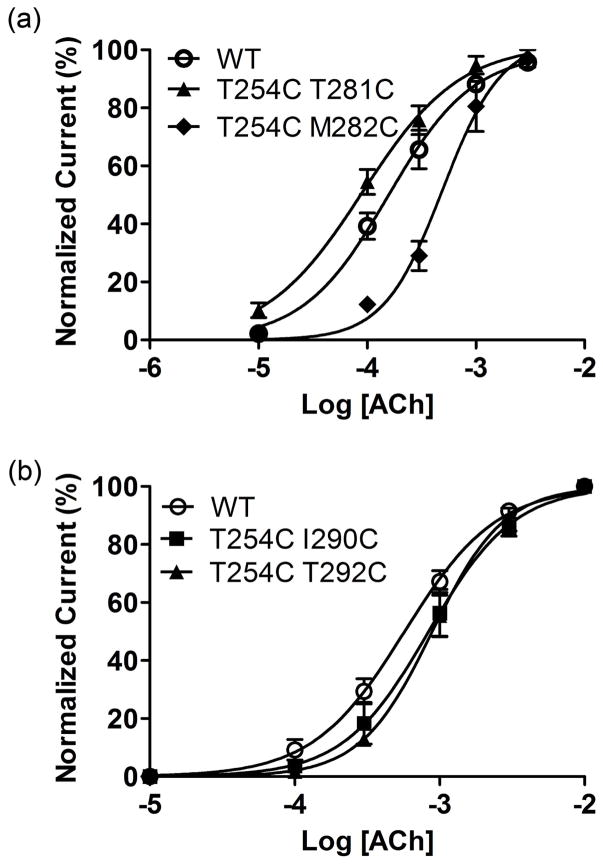
Fig 3.
Concentration response curves for oocytes expressing wild-type and mutant mouse muscle nACh receptors. (a) Data for wild-type (wt) and the mutant with the lowest and highest EC50 for the GluCl set are shown. (b) Data for wild-type and the mutants with the lowest and highest EC50 for the Torpedo set are shown. Currents were normalized to the ACh maximum response for individual oocytes. The data were fit to a sigmoidal dose-response curve shown by the solid line. Points represent the averages (±SEM) from at least 3 oocytes.
Absence of preformed disulfide bonds in Cys mutants
To investigate intrasubunit disulfide bond formation between engineered Cys pairs, one Cys in M2 and one in M3, in the muscle nAChR α-subunit, we measured the effect of reducing and oxidizing treatments on ACh-induced current amplitudes. We compared the current amplitude before and after reducing and oxidizing treatments. It is possible that disulfide bonds between the engineered Cys-pairs are pre-formed during folding of subunits, assembly of subunits, trafficking of pentameric receptors to the plasma membrane, or while the receptors are residing on the plasma membrane, and that such an oxidation does not require the presence of exogenous oxidizing agents. Several previous studies of Cys-loop receptors have found such pre-formed disulfide-linked Cys or Cys-pairs that cross-link under very mild oxidizing conditions like the ambient oxygen dissolved in buffers (Horenstein et al. 2001, Bera & Akabas 2005, Jansen & Akabas 2006). These are usually referred to as “spontaneously formed” disulfide bonds, meaning they do not require oxidizing agent, as opposed to “induced” disulfide bonds that do require stronger oxidizing conditions in the form of exogenous application of oxidizing agent (hydrogen peroxide, iodine, Cu:Phen) to form. Fig. 4 and Fig. 5(a) illustrate the experimental results obtained for wild-type and mutants. After obtaining a stable ACh response with two or more subsequent ACh applications, oocytes were superfused with the reducing agent DTT (10 mM, 2 min), before recording the ACh-induced current amplitude with the same ACh concentration as before again. Compared to the initial current before DTT application (100%), the current amplitude after DTT was not significantly altered in wild-type, the single-Cys mutant T254C, or any of the double-Cys mutants of either the GluCl-based (277–283) or Torpedo-based mutant sets (288–293). For the GluCl-based mutants (277–283) the currents after DTT were within 94 ± 1 and 97 ± 3% of the initial current before DTT. For the Torpedo-based double mutants the response after reducing agent was between 94 ± 8 and 106 ± 2%. These percentages correspond to an effect between −11 ± 4 to −1 ± 2% in the case of GluCl-based mutants (277–283), and −6 ± 8% and +6 ± 2% effect for the Torpedo-based constructs (288–293). We infer that spontaneous formation of disulfide bonds between Cys did not occur in any of our constructs.
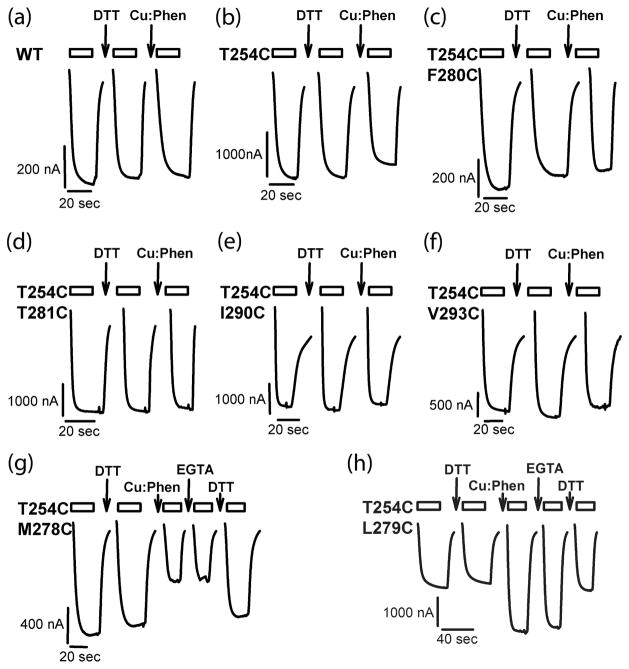
Fig 4.
Current traces for effect of oxidation and reduction. Representative ACh-induced current traces recorded before and after DTT (10 mM, 2 min), Cu:Phen (100:200 uM, 2 min) and EGTA (2mM, 2 min) applications for wild-type and mutant receptors. Current traces during reagent application are not shown. The bars above the traces indicate the duration of application of ACh; the arrows indicate the application of the given reagent. Holding potential was −60 mV.
Fig 5.
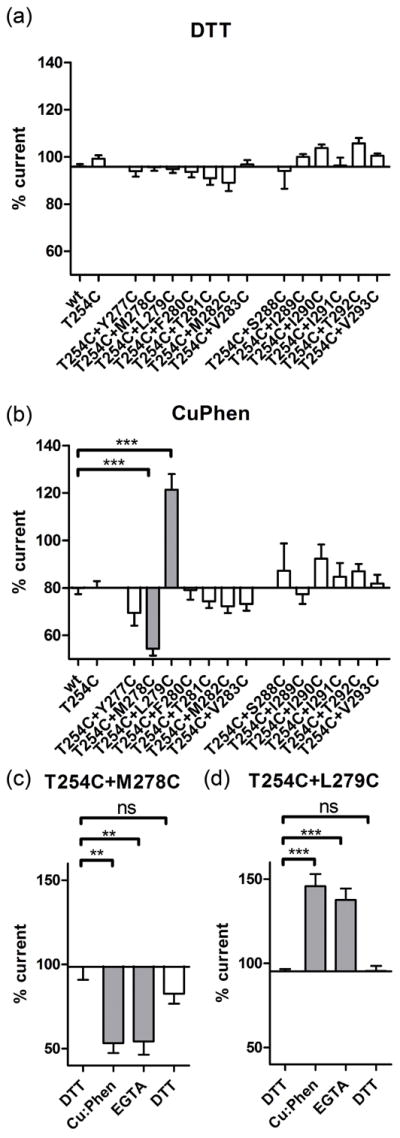
DTT and Cu:Phen effects on ACh-induced current amplitudes. (a) Effect of DTT (10 mM, 2 min) and (b) Cu:Phen (100:200 μM, 2 min) on ACh-induced EC20–50 current amplitudes for wild-type (wt), single and double Cys mutants. GluCl-based and Torpedo-based mutants are indicated in panel A. Holding potential was −60 mV. Percentage ACh-induced current amplitude after DTT or Cu:Phen treatment is shown. One-way ANOVA with Dunnett's post-test vs. wild-type shown as *** for p < 0.0001. (c) and (d) For the double mutants with significant effect of Cu:Phen application as shown in (b) the reversal of this effect was investigated by recording the effect of application of EGTA (2 mM, 2 min), and DTT (10 mM, 2 min) on current amplitudes. One-way ANOVA with Dunnett's post-test vs. the initial DTT application shown as ** for p < 0.05, and *** for p < 0.001.
Disulfide bonds form upon treatment with Cu:Phen in two double mutants
To investigate whether disulfide bond formation could be induced under more oxidizing conditions, we applied the redox catalyst Cu:Phen (100:200 μM, 2 min). Cu(II)(1,10 phenanthroline) complexes lead to the formation of oxyradicals (hydroxyl radical and superoxide anion) out of ambient oxygen dissolved in buffers. The oxyradicals reach the vicinity of Cys sulfhydryls by diffusion and generate Cys sulfhydryl radicals that can subsequently yield disulfides upon collision with a second thiol(-radical)(Careaga & Falke 1992b). After stable ACh responses were obtained, we measured the effect of Cu:Phen application on ACh-EC50-induced current amplitudes. Fig.4 and Fig. 5(b) illustrate the experimental results for wild-type, single mutant and double mutants. Current amplitudes after Cu:Phen application for wild-type and the single mutant T254C were 80 ± 3 and 80 ± 3%, respectively. For the double mutants based on the Torpedo-model they ranged between 77 ± 4 and 92 ± 6% of the initial response, indicating no significant effect of oxidizing agent on these mutants. The effect of Cu:Phen was a reduction between −3 ± 4 and +7 ± 11% for the double mutants after normalization towards wild-type. On the contrary, double mutants from the GluCl-based constructs showed 54 ± 3% to 121 ± 6% of the initial response, corresponding to an effect of −26 ± 3 to 40 ± 7%. Upon statistical analysis using one-way ANOVA with Dunnett’s multiple comparison test comparing all 13 double mutants and the single T254C mutant with wild-type, two double mutants, namely αT254C+M278C (−26 ± 3%) and αT254C+L279C (+40 ± 7 %) were different from wild-type. We infer that for the two double mutants αT254C+M278C and αT254C+L279C Cu:Phen application induced the formation of a disulfide bond with the M2 Cys T254C. To rule out that chelation of Cu2+ by the Cys pairs was responsible for the change in ACh-induced current amplitudes after oxidizing agent application, we applied the chelator EGTA (2 mM, 2 min). The EGTA effect on ACh-induced current amplitudes was 1 ± 2 % for αT254C+M278C and −9 ± 3 % for αT254C+L279C, indicating that EGTA was not able to recover the current amplitudes. On the contrary, DTT, that is a reducing agent in addition to being a weak chelator, increased the current amplitudes by 29 ± 1 % for αT254C+M278C and decreased them by 47 ± 11 % for αT254C+L279C. For both double mutants DTT application was able to restore the current amplitudes to initial levels. The redox chemistry of the Cys likely first leads to the intermediate oxidation state of sulfenic acid (CYS-SOH), from which disulfides that can be reduced with DTT or additional oxidation products like for example sulfinic acid (CYS-SO2H) and sulfonic acid (CYS-SO3H) that cannot be reduced back to Cys-SH with DTT are formed (Rehder & Borges 2010). Therefore, the reversibility by DTT clearly indicates that the effect of oxidation is mainly due to disulfide bond formation.
Discussion
The aim of the present study was to experimentally probe the vertical alignment between the transmembrane segments M2 and M3 in nAChR to address a discrepancy between the Torpedo nAChR model and the recently published GluClα structure in this area (Unwin 2005, Hibbs & Gouaux 2011). The vertical alignment/register between M2 and M3 is crucial, since the transmembrane segments M1–M4 come together to contribute to intra- and inter-subunit binding sites for allosteric ligands. The antiparasitic agent ivermectin binds to an inter-subunit crevice lined by M1, M2 and M3 in GluClα, and likely in a similar fashion to other Cys-loop receptors (Krause et al. 1998, Adelsberger et al. 2000, Shan et al. 2001, Hibbs & Gouaux 2011). General anesthetic and neurosteroid binding site(s) in GABAAR have been mapped to inter- and intra- subunit sites contributed to by M1, M2 and M3, using mutagenesis, Cys-scanning techniques, and photolabeling (Belelli et al. 1997, Bali & Akabas 2004, Hosie et al. 2006, Li et al. 2006, Bali et al. 2009, Chiara et al. 2012) and in the prokaryotic Gloeobacter violaceus ligand-gated ion channel (GLIC) the general anesthetic site was identified by X-ray crystallography to an intra-subunit site that is linked to an inter-subunit site by a so-called “linking channel” (Nury et al. 2011). Similarly, other allosteric modulators like alcohols and PNU-120596 have been shown to interact with positions in the transmembrane domain (Mihic et al. 1997, Lobo et al. 2004, Young et al. 2008, daCosta et al. 2011). These studies investigating the location of binding site(s) for allosteric ligands to Cys-loop receptors that interact with the transmembrane domain exemplify the importance of determining the vertical alignment and orientation between individual transmembrane segments.
The Torpedo nAChR atomic model was obtained by electronmicroscopy of tubular crystals grown from the postsynaptic membranes of the electric organ of Torpedo marmorata (Unwin 2005, Miyazawa et al. 2003). It is generally expected, that several factors that may be problematic with X-ray crystallographic structure determination, like genetic engineering, overexpression in a heterologous host, nonnative detergent solubilization, lipid reconstitution, and other artificial crystallization conditions are not relevant for the Torpedo structure. To obtain the GluCl α-subunit structure several experimental hurdles had to be overcome (Hibbs & Gouaux 2011). As all eukaryotic Cys-loop receptors GluCl α-subunits contain a long intracellular domain between M3 and M4 that only consists out of three to 14 amino acids in prokaryotic members. This 58 amino acid long intracellular domain in GluClα was replaced with an Ala-Gly-Thr tripeptide in the crystallized GluClα construct. In addition, 41 amino acids of the N-terminus and 6 of the C-terminus were removed. Besides this extensive genetic engineering, the overexpression in insect cells (Sf9), extraction from membranes with n-dodecyl-B-D-maltopyranoside (C12M), and artificial crystallization conditions - extensive contacts with the Fab fragments, addition of nonnative lipids as well as PEG, the acidic pH of 4.5, and the temperature of 4°C - may distort physiologically relevant structures. In particular, it is not clear whether the drastic replacement of the intracellular domain with a tripeptide lead to changes in the length of M3 and/or M4 and also their orientation and spatial alignment. It has been shown previously, that M4 of Torpedo nAChR can be replaced by a different unrelated α-helical peptide, a hydrophobic transmembrane segment from stomatitis virus glycoprotein or human interleukin-2 receptor, while retaining the ability of the whole receptor to function as a ligand-gated ion channel upon expression in X. laevis oocytes (Tobimatsu et al. 1987).
Disulfide trapping has been widely applied to investigate proximity and mobility of engineered Cys-pairs in both soluble and membrane proteins (Careaga & Falke 1992b, Falke & Koshland 1987, Matsumura et al. 1989, Pakula & Simon 1992, Zhan et al. 1994, Gruber & Capaldi 1996, Danielson et al. 1997, Krovetz et al. 1997, Yu et al. 1999, Elling et al. 2000, Shapovalov et al. 2003, Horenstein et al. 2005, Horenstein et al. 2001, Bera & Akabas 2005, Chen et al. 2006, Yang et al. 2007). Here, we use disulfide bond formation as a measure for proximity. The maximum Cβ -Cβ distance observed in protein disulfide bonds is 4.6 Å (Sowdhamini et al. 1989). Therefore, for a disulfide bond to form, the respective Cys β-carbons have to come within 4.6 Å of one another. Additional constraints arise from factors like side-chain bonding, local steric barriers, and orientational constraints resulting from the transition state (Careaga & Falke 1992b, Careaga & Falke 1992a). Generally, the separation distance of the α-carbons is assessed, and expected to be <5.6 Å for disulfides to form. In the present study, we constructed pairs of engineered Cys residues with site-directed mutagenesis, with one Cys in the muscle nAChR αsubunit transmembrane segment M2 at position T254C, and 13 different positions in M3 of the same subunit. All α-subunit constructs expressed, folded, assembled, and trafficked to the plasma membrane and formed functional ACh-gated ion-channels after heterologous expression in X. laevis oocytes, as evidenced by outward currents recorded upon application of ACh in two-electrode voltage clamp experiments. For none of the Cys-pairs disulfide bond formation was observed in the absence of the redox catalyst Cu:Phen, since 1) repetitive ACh-applications produced stable current responses, and 2) the application of the reducing agent DTT to oocytes from which a stable ACh-response had been recorded did not produce a significant change in current amplitude for wild-type, single or double mutants. Here, we provide evidence for disulfide cross-linking between engineered Cys pairs, one at position T254C and the other in M3. The Cys pairs that could be cross-linked by application of the redox catalyst Cu:Phen, M2 T254C and M3 M278C, as well as M2 T254C and M3 L279C, clearly favor the vertical alignment as observed in the GluClα structure. Additional evidence for cross-linking of Cys pairs is provided by the fact, that the reducing agent DTT could reverse the effect of Cu:Phen on the ACh-induced current amplitudes. The chelator EGTA, on the contrary, could not reverse Cu2+:Phen effects, indicating that chelation of Cu by one or more engineered Cys and/or other amino acids did not significantly contribute to the Cu:Phen effect on ACh-induced current amplitudes. Importantly, double Cys mutants that should have had their Cys in a favorable distance if the Torpedo structure were to be correct could not be cross-linked.
When the initial coordinates based on the electronmicrocopic studies of two-dimensional crystals of Torpedo nAChR were released (PDB# 1OED), an extended remark was included in the coordinates file “The link between M1 and M2 was poorly resolved and the trace here is almost certainly wrong in detail. … Users should bear in mind that because of the limited resolution the conformations of the side chains and their atomic coordinates are not individually reliable. Also the ends of the helices are uncertain by at least one residue” (Miyazawa et al. 2003). The location that was described as the most problematic one, the M1 and M2 loop, is exactly where the discrepancy between the Torpedo atomic model and the GluCl structure originates. Hibbs and Gouaux noted “that in the α-subunit M2 pore-lining helix and the M3 α-helix, the nAChR amino acid assignment is off in register by 4 residues or ~1 turn of an α-helix beginning with the M1–M2 loop”(Hibbs & Gouaux 2011). Importanty, a similar discrepancy between amino acid and structure-based alignment had been previously observed and described for the comparison between GLIC and nAChR (Corringer et al. 2010).
As stated earlier, the interface between the extracellular domain and the transmembrane domain is crucial for coupling ligand binding to gating. Coupling involves the M2–M3 linker, the β1-β2 loop, the β8-β9 loop, the Cys loop and the pre-M1 linker (Lee & Sine 2005, Reeves et al. 2005, Mercado & Czajkowski 2006). In nAChR mutant cycle analysis has shown functional coupling between Glu45 and Val46, located in the β1-β2 linker, to Ser269 and Pro272, part of the M2–M3 loop (Lee & Sine 2005). Pro272 has also been implicated as a cis-trans isomerization switch for channel activation (Lummis et al. 2005). Based on the proximity of the residues in the 2BG9 cryo-EM structure, where Glu45 and Val46 straddle Pro272, it was inferred that the coupling was direct as opposed to allosteric. However, distant allosteric coupling has been observed in nAChR by mutant cycle analysis for positions separated by 50–60 Å (Gleitsman et al. 2008). This demonstrates that mutant cycle analysis does not necessarily require physical proximity of mutant cycle interacting residues. Functional coupling between residues that are far apart has been commonly seen (Alexiev et al. 2000). Our results imply a shift in the vertical alignment between M2 and M3 as compared to the 2BG9 Torpedo nAChR model. This will change the M2–M3 linker interactions in the GluClα-based nAChR homology model. An additional complication for the homology model is that sequence alignments suggest that the GluClα M2–M3 linker is one amino acid shorter than in the nAChR. In the mouse nAChR model based on GluClα, Glu45 and Val46 straddle Pro264. In this model Glu45 forms a salt bridge to Arg209 in the pre-M1 linker; this links together the inner and outer β-sheets of the extracellular domain (Lee & Sine 2005). These residues are conserved in the Cys-loop superfamily (Mercado & Czajkowski 2006) and are crucial for gating in both models.
In summary, our experimental data are in agreement with the vertical alignment between M2 and M3 as obtained in a homology model of muscle nAChR based on the recently published GluClα structure, but not as in the Torpedo nAChR structural model. Since the vertical alignment between the prokaryotic GLIC and GluClα is in agreement, our current study implies the same alignment between all Cys-loop receptor homologues, both prokaryotic and eukaryotic. Overall, amino acids are shifted by four positions for the M2 and M3 segments, each, leading to the register between the two segments to be different by two α-helical turns when Torpedo nAChR and GluCl are compared. If the muscle nAChR is homology-modeled on the recently published GluClα structure this leads to all M2 residues moving up the α-helix by four residues towards the extracellular side and consequently all M3 residues moving down by four residues towards the intracellular side. Due to the low resolution of the 2BG9 Torpedo nAChR electron density, it is reasonable to model the relative orientation between M2 and M3 as described in the present study. Our study is limited in that we focused on the vertical alignment between M2 and M3. However, our observations can be used to interpret previous studies and it provides a model on which to base additional investigations. Further studies utilizing the mutants with engineered Cys pairs that could be cross-linked in this study or other double Cys mutants may be useful in investigating relative motions of the M2 and M3 segments during channel gating as well as the points of contact between loops of the transmembrane and extracellular domain. At present, no available structure of Cys-loop receptors is perfect. They are either hampered by low resolution, prokaryotic origin or genetic engineering, indicating the clear need for atomic resolution eukaryotic structures.
Acknowledgments
The authors have no conflict of interest to declare. Research reported in this publication was supported by the National Institute of Neurological Disorders and Stroke (NINDS) of the National Institutes of Health (NIH) under award number R00 NS059841 (to MJ). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank Phaneendra Kumar Duddempudi, Nirupama S. Nishtala, and Prachi Nakashe, for helpful discussions, Dane Langsjoen, Jarrett Linder, and Camille Perot for initial experiments, and Gautham Brahmamudi and Raman Goyal for expert technical support.
Abbreviations
- nAChr
nicotinic actylcholine receptor
- GluClα
glutamate-gated chloride channel α
References
- Adelsberger H, Lepier A, Dudel J. Activation of rat recombinant alpha(1)beta(2)gamma(2S) GABA(A) receptor by the insecticide ivermectin. Eur J Pharmacol. 2000;394:163–170. doi: 10.1016/s0014-2999(00)00164-3. [DOI] [PubMed] [Google Scholar]
- Akabas MH, Stauffer DA, Xu M, Karlin A. Acetylcholine receptor channel structure probed in cysteine-substitution mutants. Science. 1992;258:307–310. doi: 10.1126/science.1384130. [DOI] [PubMed] [Google Scholar]
- Alexiev U, Mollaaghababa R, Khorana HG, Heyn MP. Evidence for long range allosteric interactions between the extracellular and cytoplasmic parts of bacteriorhodopsin from the mutant R82A and its second site revertant R82A/G231C. J Biol Chem. 2000;275:13431–13440. doi: 10.1074/jbc.275.18.13431. [DOI] [PubMed] [Google Scholar]
- Bali M, Akabas MH. Defining the propofol binding site location on the GABAA receptor. Mol Pharmacol. 2004;65:68–76. doi: 10.1124/mol.65.1.68. [DOI] [PubMed] [Google Scholar]
- Bali M, Jansen M, Akabas MH. GABA-induced intersubunit conformational movement in the GABAA receptor alpha 1M1-beta 2M3 transmembrane subunit interface: experimental basis for homology modeling of an intravenous anesthetic binding site. J Neurosci. 2009;29:3083–3092. doi: 10.1523/JNEUROSCI.6090-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belelli D, Lambert JJ, Peters JA, Wafford K, Whiting PJ. The interaction of the general anesthetic etomidate with the gamma-aminobutyric acid type A receptor is influenced by a single amino acid. Proc Natl Acad Sci USA. 1997;94:11031–11036. doi: 10.1073/pnas.94.20.11031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bera AK, Akabas MH. Spontaneous thermal motion of the GABA(A) receptor M2 channel-lining segments. J Biol Chem. 2005;280:35506–35512. doi: 10.1074/jbc.M504645200. [DOI] [PubMed] [Google Scholar]
- Bocquet N, Nury H, Baaden M, Le Poupon C, Changeux JP, Delarue M, Corringer PJ. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 2009;457:111–114. doi: 10.1038/nature07462. [DOI] [PubMed] [Google Scholar]
- Bondarenko V, Tillman T, Xu Y, Tang P. NMR structure of the transmembrane domain of the n-acetylcholine receptor beta2 subunit. Biochim Biophys Acta. 2010;1798:1608–1614. doi: 10.1016/j.bbamem.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Careaga CL, Falke JJ. Structure and dynamics of Escherichia coli chemosensory receptors. Engineered sulfhydryl studies. Biophys J. 1992a;62:209–216. doi: 10.1016/S0006-3495(92)81806-4. discussion 217–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Careaga CL, Falke JJ. Thermal motions of surface alpha-helices in the D-galactose chemosensory receptor. Detection by disulfide trapping. J Mol Biol. 1992b;226:1219–1235. doi: 10.1016/0022-2836(92)91063-u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Durkin KA, Casida JE. Spontaneous mobility of GABAA receptor M2 extracellular half relative to noncompetitive antagonist action. J Biol Chem. 2006;281:38871–38878. doi: 10.1074/jbc.M608301200. [DOI] [PubMed] [Google Scholar]
- Chiara DC, Dostalova Z, Jayakar SS, Zhou X, Miller KW, Cohen JB. Mapping general anesthetic binding site(s) in human alpha1beta3 gamma-aminobutyric acid type A receptors with [(3)H]TDBzl-etomidate, a photoreactive etomidate analogue. Biochemistry. 2012;51:836–847. doi: 10.1021/bi201772m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corringer PJ, Baaden M, Bocquet N, Delarue M, Dufresne V, Nury H, Prevost M, Van Renterghem C. Atomic structure and dynamics of pentameric ligand-gated ion channels: new insight from bacterial homologues. J Physiol. 2010;588:565–572. doi: 10.1113/jphysiol.2009.183160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui T, Mowrey D, Bondarenko V, et al. NMR structure and dynamics of a designed water-soluble transmembrane domain of nicotinic acetylcholine receptor. Biochim Biophys Acta. 2012;1818:617–626. doi: 10.1016/j.bbamem.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cymes GD, Grosman C. Pore-opening mechanism of the nicotinic acetylcholine receptor evinced by proton transfer. Nat Struct Mol Biol. 2008;15:389–396. doi: 10.1038/nsmb.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cymes GD, Ni Y, Grosman C. Probing ion-channel pores one proton at a time. Nature. 2005;438:975–980. doi: 10.1038/nature04293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- daCosta CJ, Free CR, Corradi J, Bouzat C, Sine SM. Single-channel and structural foundations of neuronal alpha7 acetylcholine receptor potentiation. J Neurosci. 2011;31:13870–13879. doi: 10.1523/JNEUROSCI.2652-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielson MA, Bass RB, Falke JJ. Cysteine and disulfide scanning reveals a regulatory alpha-helix in the cytoplasmic domain of the aspartate receptor. J Biol Chem. 1997;272:32878–32888. doi: 10.1074/jbc.272.52.32878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elling CE, Raffetseder U, Nielsen SM, Schwartz TW. Disulfide bridge engineering in the tachykinin NK1 receptor. Biochemistry. 2000;39:667–675. doi: 10.1021/bi991777b. [DOI] [PubMed] [Google Scholar]
- Falke JJ, Koshland DE., Jr Global flexibility in a sensory receptor: a site-directed cross-linking approach. Science. 1987;237:1596–1600. doi: 10.1126/science.2820061. [DOI] [PubMed] [Google Scholar]
- Gleitsman KR, Kedrowski SM, Lester HA, Dougherty DA. An intersubunit hydrogen bond in the nicotinic acetylcholine receptor that contributes to channel gating. J Biol Chem. 2008;283:35638–35643. doi: 10.1074/jbc.M807226200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Gutierrez G, Lukk T, Agarwal V, Papke D, Nair SK, Grosman C. Mutations that stabilize the open state of the Erwinia chrisanthemi ligand-gated ion channel fail to change the conformation of the pore domain in crystals. Proc Natl Acad Sci U S A. 2012;109:6331–6336. doi: 10.1073/pnas.1119268109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal R, Salahudeen AA, Jansen M. Engineering a prokaryotic Cys-loop receptor with a third functional domain. J Biol Chem. 2011;286:34635–34642. doi: 10.1074/jbc.M111.269647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber G, Capaldi RA. The trapping of different conformations of the Escherichia coli F1 ATPase by disulfide bond formation. Effect on nucleotide binding affinities of the catalytic sites. J Biol Chem. 1996;271:32623–32628. doi: 10.1074/jbc.271.51.32623. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Hibbs RE, Gouaux E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature. 2011;474:54–60. doi: 10.1038/nature10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilf RJ, Bertozzi C, Zimmermann I, Reiter A, Trauner D, Dutzler R. Structural basis of open channel block in a prokaryotic pentameric ligand-gated ion channel. Nat Struct Mol Biol. 2010;17:1330–1336. doi: 10.1038/nsmb.1933. [DOI] [PubMed] [Google Scholar]
- Hilf RJ, Dutzler R. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature. 2008;452:375–379. doi: 10.1038/nature06717. [DOI] [PubMed] [Google Scholar]
- Hilf RJ, Dutzler R. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature. 2009;457:115–118. doi: 10.1038/nature07461. [DOI] [PubMed] [Google Scholar]
- Horenstein J, Riegelhaupt P, Akabas MH. Differential protein mobility of the gamma-aminobutyric acid, type A, receptor alpha and beta subunit channel-lining segments. J Biol Chem. 2005;280:1573–1581. doi: 10.1074/jbc.M410881200. [DOI] [PubMed] [Google Scholar]
- Horenstein J, Wagner DA, Czajkowski C, Akabas MH. Protein mobility and GABA-induced conformational changes in GABAA receptor pore-lining M2 segment. Nat Neurosci. 2001;4:477–485. doi: 10.1038/87425. [DOI] [PubMed] [Google Scholar]
- Hosie AM, Wilkins ME, da Silva HM, Smart TG. Endogenous neurosteroids regulate GABAA receptors through two discrete transmembrane sites. Nature. 2006;444:486–489. doi: 10.1038/nature05324. [DOI] [PubMed] [Google Scholar]
- Jansen M, Akabas MH. State-dependent cross-linking of the M2 and M3 segments: functional basis for the alignment of GABAA and acetylcholine receptor M3 segments. J Neurosci. 2006;26:4492–4499. doi: 10.1523/JNEUROSCI.0224-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen M, Bali M, Akabas MH. Modular design of Cys-loop ligand-gated ion channels: functional 5-HT3 and GABA rho1 receptors lacking the large cytoplasmic M3M4 loop. J Gen Physiol. 2008;131:137–146. doi: 10.1085/jgp.200709896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin A, Akabas MH. Substituted-cysteine accessibility method. Methods Enzymol. 1998;293:123–145. doi: 10.1016/s0076-6879(98)93011-7. [DOI] [PubMed] [Google Scholar]
- Krause RM, Buisson B, Bertrand S, Corringer PJ, Galzi JL, Changeux JP, Bertrand D. Ivermectin: a positive allosteric effector of the alpha7 neuronal nicotinic acetylcholine receptor. Mol Pharmacol. 1998;53:283–294. doi: 10.1124/mol.53.2.283. [DOI] [PubMed] [Google Scholar]
- Krovetz HS, VanDongen HM, VanDongen AM. Atomic distance estimates from disulfides and high-affinity metal-binding sites in a K+ channel pore. Biophys J. 1997;72:117–126. doi: 10.1016/S0006-3495(97)78651-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WY, Sine SM. Principal pathway coupling agonist binding to channel gating in nicotinic receptors. Nature. 2005;438:243–247. doi: 10.1038/nature04156. [DOI] [PubMed] [Google Scholar]
- Li GD, Chiara DC, Sawyer GW, Husain SS, Olsen RW, Cohen JB. Identification of a GABAA receptor anesthetic binding site at subunit interfaces by photolabeling with an etomidate analog. J Neurosci. 2006;26:11599–11605. doi: 10.1523/JNEUROSCI.3467-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo IA, Mascia MP, Trudell JR, Harris RA. Channel gating of the glycine receptor changes accessibility to residues implicated in receptor potentiation by alcohols and anesthetics. J Biol Chem. 2004;279:33919–33927. doi: 10.1074/jbc.M313941200. [DOI] [PubMed] [Google Scholar]
- Lummis SC, Beene DL, Lee LW, Lester HA, Broadhurst RW, Dougherty DA. Cis-trans isomerization at a proline opens the pore of a neurotransmitter-gated ion channel. Nature. 2005;438:248–252. doi: 10.1038/nature04130. [DOI] [PubMed] [Google Scholar]
- Matsumura M, Becktel WJ, Levitt M, Matthews BW. Stabilization of phage T4 lysozyme by engineered disulfide bonds. Proc Natl Acad Sci U S A. 1989;86:6562–6566. doi: 10.1073/pnas.86.17.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado J, Czajkowski C. Charged residues in the alpha1 and beta2 pre-M1 regions involved in GABAA receptor activation. J Neurosci. 2006;26:2031–2040. doi: 10.1523/JNEUROSCI.4555-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihic SJ, Ye Q, Wick MJ, et al. Sites of alcohol and volatile anaesthetic action on GABA(A) and glycine receptors. Nature. 1997;389:385–389. doi: 10.1038/38738. [DOI] [PubMed] [Google Scholar]
- Miller C. Genetic manipulation of ion channels: a new approach to structure and mechanism. Neuron. 1989;2:1195–1205. doi: 10.1016/0896-6273(89)90304-8. [DOI] [PubMed] [Google Scholar]
- Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423:949–955. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- Nury H, Van Renterghem C, Weng Y, et al. X-ray structures of general anaesthetics bound to a pentameric ligand-gated ion channel. Nature. 2011;469:428–431. doi: 10.1038/nature09647. [DOI] [PubMed] [Google Scholar]
- Pakula AA, Simon MI. Determination of transmembrane protein structure by disulfide cross- linking: the Escherichia coli Tar receptor. Proc Natl Acad Sci U S A. 1992;89:4144–4148. doi: 10.1073/pnas.89.9.4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Chen Q, Willenbring D, Mowrey D, Kong XP, Cohen A, Divito CB, Xu Y, Tang P. Structure of the pentameric ligand-gated ion channel GLIC bound with anesthetic ketamine. Structure. 2012a;20:1463–1469. doi: 10.1016/j.str.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Chen Q, Willenbring D, et al. Structure of the pentameric ligand-gated ion channel ELIC cocrystallized with its competitive antagonist acetylcholine. Nature communications. 2012b;3:714. doi: 10.1038/ncomms1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikh RB, Bali M, Akabas MH. Structure of the M2 transmembrane segment of GLIC, a prokaryotic Cys loop receptor homologue from Gloeobacter violaceus, probed by substituted cysteine accessibility. J Biol Chem. 2011;286:14098–14109. doi: 10.1074/jbc.M111.221895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves DC, Jansen M, Bali M, Lemster T, Akabas MH. A role for the beta 1- beta 2 loop in the gating of 5-HT3 receptors. J Neurosci. 2005;25:9358–9366. doi: 10.1523/JNEUROSCI.1045-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehder DS, Borges CR. Cysteine sulfenic acid as an intermediate in disulfide bond formation and nonenzymatic protein folding. Biochemistry. 2010;49:7748–7755. doi: 10.1021/bi1008694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q, Haddrill JL, Lynch JW. Ivermectin, an unconventional agonist of the glycine receptor chloride channel. J Biol Chem. 2001;276:12556–12564. doi: 10.1074/jbc.M011264200. [DOI] [PubMed] [Google Scholar]
- Shapovalov G, Bass R, Rees DC, Lester HA. Open-State Disulfide Crosslinking between Mycobacterium tuberculosis Mechanosensitive Channel Subunits. Biophys J. 2003;84:2357–2365. doi: 10.1016/S0006-3495(03)75041-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowdhamini R, Srinivasan N, Shoichet B, Santi DV, Ramakrishnan C, Balaram P. Stereochemical modeling of disulfide bridges. Criteria for introduction into proteins by site-directed mutagenesis. Protein Eng. 1989;3:95–103. doi: 10.1093/protein/3.2.95. [DOI] [PubMed] [Google Scholar]
- Stewart DS, Chiara DC, Cohen JB. Mapping the structural requirements for nicotinic acetylcholine receptor activation by using tethered alkyltrimethylammonium agonists and antagonists. Biochemistry. 2006;45:10641–10653. doi: 10.1021/bi060686t. [DOI] [PubMed] [Google Scholar]
- Tasneem A, Iyer LM, Jakobsson E, Aravind L. Identification of the prokaryotic ligand-gated ion channels and their implications for the mechanisms and origins of animal Cys-loop ion channels. Genome Biol. 2005;6:R4. doi: 10.1186/gb-2004-6-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobimatsu T, Fujita Y, Fukuda K, Tanaka KI, Mori Y, Konno T, Mishina M, Numa S. Effects of substitution of putative transmembrane segments on nicotinic acetylcholine receptor function. FEBS Letters. 1987;222:56–62. doi: 10.1016/0014-5793(87)80191-6. [DOI] [PubMed] [Google Scholar]
- Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J Mol Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Unwin N, Fujiyoshi Y. Gating movement of acetylcholine receptor caught by plunge-freezing. J Mol Biol. 2012;422:617–634. doi: 10.1016/j.jmb.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Webb TI, Lynch JW. Closed-state cross-linking of adjacent beta1 subunits in alpha1beta1 GABAa receptors via introduced 6' cysteines. J Biol Chem. 2007;282:16803–16810. doi: 10.1074/jbc.M611555200. [DOI] [PubMed] [Google Scholar]
- Young GT, Zwart R, Walker AS, Sher E, Millar NS. Potentiation of alpha7 nicotinic acetylcholine receptors via an allosteric transmembrane site. Proc Natl Acad Sci U S A. 2008;105:14686–14691. doi: 10.1073/pnas.0804372105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Kono M, Oprian DD. State-dependent disulfide cross-linking in rhodopsin. Biochemistry. 1999;38:12028–12032. doi: 10.1021/bi990948+. [DOI] [PubMed] [Google Scholar]
- Zhan H, Choe S, Huynh PD, Finkelstein A, Eisenberg D, Collier RJ. Dynamic transitions of the transmembrane domain of diphtheria toxin: disulfide trapping and fluorescence proximity studies. Biochemistry. 1994;33:11254–11263. doi: 10.1021/bi00203a022. [DOI] [PubMed] [Google Scholar]
- Zhang H, Karlin A. Identification of acetylcholine receptor channel-lining residues in the M1 segment of the beta-subunit. Biochemistry. 1997;36:15856–15864. doi: 10.1021/bi972357u. [DOI] [PubMed] [Google Scholar]