Abstract
The M5 muscarinic acetylcholine receptor is suggested to be a potential pharmacotherapeutic target for the treatment of drug abuse. We describe herein the discovery of a series of M5-preferring orthosteric antagonists based on the scaffold of 1,2,5,6-tetrahydropyridine-3-carboxylic acid. Compound 56, the most selective compound in this series, possesses an 11-fold selectivity for the M5 over M1 receptor, and shows little activity at M2–M4. This compound, although exhibiting modest affinity (Ki = 2.24 μM) for the [3H]N-methylscopolamine binding site on the M5 receptor, is potent (IC50 = 0.45 nM) in inhibiting oxotremorine-evoked [3H]DA release from rat striatal slices. Further, a homology model of human M5 receptor based on the crystal structure of the rat M3 receptor was constructed, and docking studies of compounds 28 and 56 were performed in an attempt to understand the possible binding mode of these novel analogues to the receptor.
INTRODUCTION
Muscarinic acetylcholine receptors (mAcChRsa) are G-protein coupled receptors (GPCRs) activated by the endogenous neurotransmitter acetylcholine (ACh) and the natural alkaloid muscarine. Upon ACh activation, these receptors regulate a variety of central and peripheral physiological functions such as cognition, movement, sleep, cardiovascular function, smooth-muscle contractility, and glandular secretion.1–4 Thus, mAcChRs have emerged as important therapeutic targets for many diseases, such as Alzheimer’s disease, Parkinson’s disease, psychosis, pain, asthma, diabetes, and smooth-muscle disorders.3, 4 Five mAcChR subtypes (M1–M5) have been identified.2 Each of the five mAcChR subtypes has a defined distribution pattern within specific brain regions and peripheral tissues and mediates distinct physiological functions.1–4 To avoid side effects, selectivity at specific mAcChR subtypes is often a major focus of discovery of mAcChR agonists and antagonists as therapeutic agents.
The M5 mAcChR was the last subtype to be cloned.2, 5 A growing body of evidence suggests that this subtype is a potential target for the discovery of treatments for drug abuse.6 The rewarding effects of abused drugs are believed to be mediated by the mesolimbic dopamine (DA) pathway, which projects from the midbrain ventral tegmental area (VTA) to the nucleus accumbens.7–10 M5 mAcChRs are the only muscarinic subtype localized to VTA and that modulate DA release from VTA DA neurons11–17 Consistent with M5 mAcChR modulation of mesolimbic DA transmission, behavioral studies using M5 knockout mice show a reduction in reward and withdrawal responses following repeated morphine or cocaine administration, as well as a reduction in the rate of cocaine self-administration.18–21 Furthermore, microinfusion into VTA of an antisense oligonucleotide targeting M5 receptor mRNA inhibits brain stimulation reward in rats15 and microinfusion into the VTA of the nonselective mAcChR antagonist, scopolamine (1, Figure 1), reduces cocaine-facilitated DA release in nucleus accumbens.17 Microinfusion of the nonselective mAcChR antagonist, atropine (2, Figure 1), into the VTA in rats dose-dependently inhibits morphine-induced conditioned place preference.22 Taken together, these studies suggest that discovery of subtype-selective M5 mAcChR antagonists may provide novel treatments for drug abuse. Importantly, mice lacking M5 receptors exhibit no difference from their wild-type littermates in various behavioral and pharmacologic tests,18,23 suggesting that centrally-active M5 receptor antagonists will be well tolerated.
Figure 1.
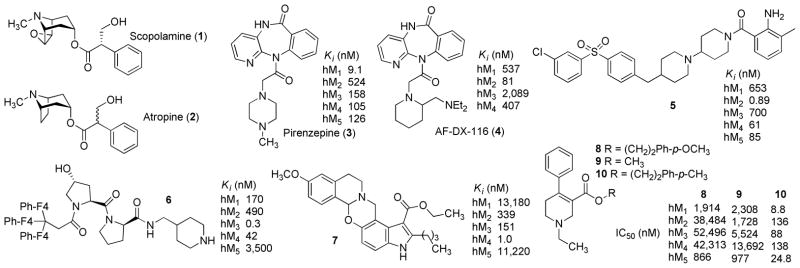
Structures of selected mAcChR antagonists and their literature reported mAcChR affinity data.
Subtype-selective M5 receptor antagonists also will be useful pharmacological tools in defining the physiological functions of this receptor. Currently, little information is available on the effects of selective activation or inhibition of M5 receptors. Recent success in identifying M5-selective positive allosteric modulators may provide new information in this regard.24–26 Nevertheless, no subtype-selective orthosteric M5 antagonists are available to date. Unlike allosteric binding sites, which are usually more divergent across subtypes, orthosteric ACh binding sites across the five mAcChR subtypes have been shown to be highly conserved at the amino acid sequence level (73–83% identity).5 As such, selectivity among the mAcChR subtypes is likely based on conformational dissimilarities, rather than upon single amino acid residues.27–29 In fact, small molecule orthosteric antagonists with preference for an individual subtype have been reported for M1–M4 but not M5 (e.g., compound 3 for M1,30 5 for M2,31 6 for M3,32 and 7 for M4,29 Figure 1), indicating the existence of sufficient differences among the agonist recognition sites on the five mAcChR subtypes allowing targeting of selective compounds to these sites.
Radioligand binding assays using membranes from recombinant cells expressing human mAcChRs (hM1–hM5) show that compound 8 (1-ethyl-4-phenyl-1,2,5,6-tetrahydropyridine-3-carboxylic acid 4-methoxyphenethyl ester, Figure 1) preferentially binds to the M5 receptor (subtype selectivity: M1/M5 = 2.2-fold, M2/M5 = 44-fold, M3/M5 = 60-fold, M4/M5 = 48-fold, Figure 1).33 Thus, the current exploration of the structure activity relationship (SAR) regarding M5 selective orthosteric ligands is based on compound 8. To our best knowledge, compound 8 and its analogue 9 (Figure 1), are the only orthosteric small molecules reported to exhibit preference for M5 receptors.33
The current structural modification strategy is provided in Figure 2. The 1,2,5,6-tetrahydropyridine-3-caboxylate core structure was retained, while modifying each part of the “appending” pharmacophores in a stepwise manner. These “appending” pharmacophores were to be rearranged around the core structure in compound 8 (Figure 2). Herein, we report the synthesis, pharmacological evaluation, and SAR analysis of this new series of mAcChR antagonists. Additionally, a homology model of hM5 mAcChR was constructed based on the crystal structure of the rat M3 mAcChR. Preliminary docking experiments were performed using this model in an attempt to understand the structural basis of the interaction between these ligands and the receptor.
Figure 2.
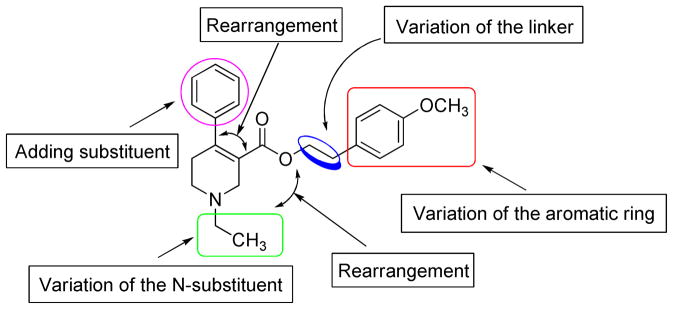
General structural modification strategy.
RESULTS AND DISCUSSION
Synthesis
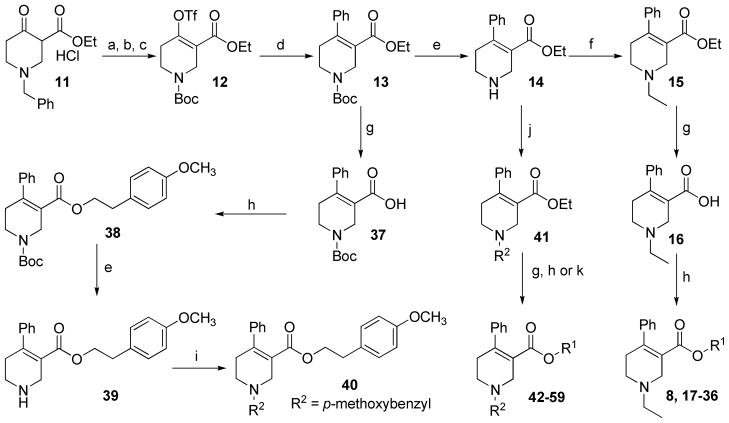
The general synthetic methods for analogues 17–36 (Table 1) with modifications on the p-methoxyphenethyl moiety of compound 8 are depicted in Scheme 1. Transformation of commercially available ethyl 1-benzyl-4-oxo-3-piperidinecarboxylate (11) into compound 12 was achieved by initial conversion of 11 to N-BOC protected compound via a two-step process,34 followed by triflation under diisopropylamine and trifluoromethanesulfonic anhydride.35 Suzuki coupling of 12 with phenylboronic acid afforded compound 13. Removal of BOC in 13 by TFA afforded compound 14, which was then N-ethylated to form compound 15. Compound 15 was subjected to ester hydrolysis under basic condition to afford carboxylic acid 16. Treatment of 16 under a Steglich esterification condition with appropriate alcohols afforded 8 and its analogues 17–36. The synthetic route for analogues with variations on the N-ethyl moiety of compound 8 was initially devised to introduce the N-substituent at the last step. Accordingly, compound 13 was converted to compound 38 in a sequence similar to that for compound 15 to 8 (Scheme 1). However, deprotection of 38 to form 39 using various de-BOC methods (TFA/CH2Cl2, HCl/EtOAc, AcCl/MeOH, or TsOH/CH2Cl2) resulted in complex mixtures. Attempts to purify compound 39 by silica gel column chromatography failed. Impure intermediate 39 (obtained from TFA treatment) was employed to provide N-Me, N-n-Pr, N-n-Bu, or N-p-methoxybenzyl analogues via reductive amination, but only N-p-methoxybenzyl analogue (compound 40) was obtained in pure form after preparative TLC purification. Alternatively, a similar route to that for analogues 17–36 was applied to prepare analogues 42–59 from compound 14 (Scheme 1, Table 2).
Table 1.
Structures and Binding Affinity for Scopolamine (1), Compounds 8, and 17–36 at the hM1 and hM5 mAcChRsa
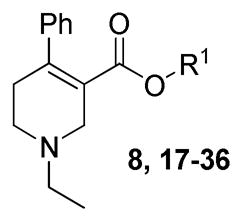
| ||||
|---|---|---|---|---|
| compd | R1 |
Ki, nM
|
hM1/hM5 | |
| hM1 | hM5 | |||
| 1 | - | 7.5 | 17.6 | 0.4 |
| 8 |
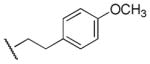
|
170 | 420 | 0.4 |
| 17 |
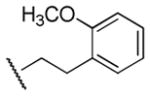
|
1,730 | 1,250 | 1.4 |
| 18 |
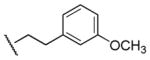
|
470 | 150 | 3.1 |
| 19 |
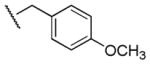
|
3,060 | 3,610 | 0.8 |
| 20 |
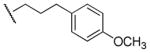
|
3,290 | 470 | 7.0 |
| 21 |
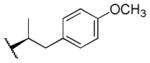
|
>100,000 | >100,000 | - |
| 22 |
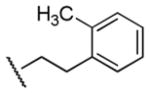
|
200 | 290 | 0.7 |
| 23 |
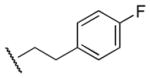
|
100 | 190 | 0.5 |
| 24 |
|
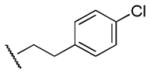
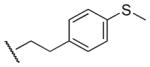
560 | 410 | 1.4 |
| 25 |
|
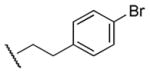
240 | 370 | 0.6 |
| 26 |
|
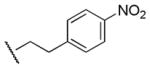
810 | 440 | 1.8 |
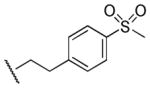
| 27 |
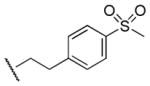
|
>100,000 | >100,000 | - |
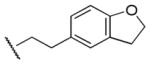
| 28 |
|
1,390 | 230 | 6.0 |
| 29 |
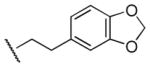
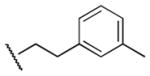
|
200 | 280 | 0.7 |
| 30 |
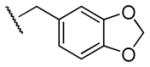
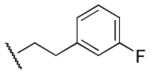
|
60 | 120 | 0.5 |
| 31 |
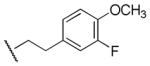
|
11,700 | 2,460 | 4.8 |
| 32 |
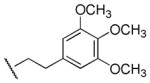
|
>100,000 | >100,000 | - |
| 33 |
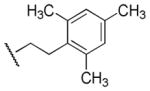
|
>100,000 | >100,000 | - |
| 34 |
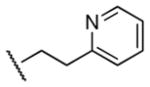
|
410 | 760 | 0.5 |
| 35 |
|
370 | 140 | 2.6 |
| 36 |
|
140 | 190 | 0.7 |
At least three independent experiments with samples evaluated in duplicate were performed to obtain the Ki value.
Scheme 1. Synthesis of Compounds 8, 17–36, 40, and 42–59a.
aReagents and conditions: (a) H2, 10% Pd/C (5 w/w%), EtOH; (b) (BOC)2O, Et3N, CH2Cl2; (c) Tf2O, iPr2NH, CH2Cl2, −78 °C; (d) PhB(OH)2, Pd(PPh4)3, Na2CO3 (2.0 M), THF, 65 °C; (e) TFA/CH2Cl2 (1:1); (f) EtI, K2CO3, EtOH; (g) 10% KOH (aq.)/EtOH (1:1); (h) alcohols, EDCI, DMAP, CH2Cl2; (i) p-methoxybezaldehyde, NaBH(OAc)3, HOAc, THF; (j) aldehydes, NaBH3CN, EtOH; (k) alcohols, 2,4,6-trichlorobenzoyl chloride, DMAP, Et3N, THF.
Table 2.
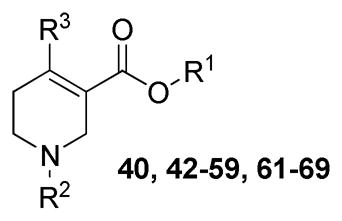
Structures and Binding Affinity for Analogues 40, 42–59, and 61–69 at the hM1 and hM5 mAcChRsa
| ||||||
|---|---|---|---|---|---|---|
| compd | R1 | R2 | R3 |
Ki, nM
|
||
| hM1 | hM5 | hM1/hM5 | ||||
| 40 |
|
4-MeO-Bn | Ph | >100,000 | >100,000 | - |
| 42 | ibid. | Me | Ph | 20 | 30 | 0.7 |
| 43 | ibid. | nPr | Ph | >100,000 | >100,000 | - |
| 44 | ibid. | nBu | Ph | >100,000 | >100,000 | - |
| 45 |
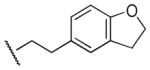
|
Me | Ph | 290 | 60 | 4.8 |
| 46 | ibid. | nPr | Ph | >100,000 | >100,000 | - |
| 47 | ibid. | nBu | Ph | >100,000 | >100,000 | - |
| 48 |
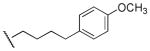
|
Me | Ph | 5,400 | 1,160 | 4.7 |
| 49 |
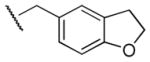
|
Me | Ph | >100,000 | >100,000 | - |
| 50 |
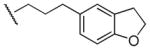
|
Me | Ph | 920 | 630 | 1.5 |
| 51 |
|
Me | Ph | 340 | 190 | 1.8 |
| 52 |
|
Me | Ph | 510 | 110 | 4.6 |
| 53 |
|
Me | Ph | 880 | 280 | 3.1 |
| 54 |
|
Me | Ph | 330 | 80 | 4.1 |
| 55 |
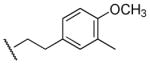
|
Me | Ph | 2,700 | 740 | 3.6 |
| 56 |
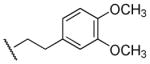
|
Me | Ph | 25,300 | 2,240 | 11.3 |
| 57 |
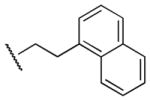
|
Me | Ph | 24,400 | 5,040 | 4.8 |
| 58 |
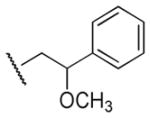
|
Me | Ph | >100,000 | >100,000 | - |
| 59 |
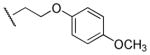
|
Me | Ph | 6,190 | 3,070 | 2.0 |
| 61 |
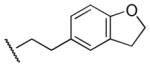
|
Et | 4-F-Ph | 7,900 | 2,350 | 3.4 |
| 62 | ibid. | Me | 3-F-Ph | >100,000 | >100,000 | - |
| 63 | ibid. | Me | 2-F-Ph | 5,270 | 740 | 7.0 |
| 64 | ibid. | Me | 3-MeO-Ph | 2,430 | 2,680 | 0.9 |
| 65 | ibid. | Me | 2-MeO-Ph | 8,350 | 2,410 | 3.5 |
| 66 |
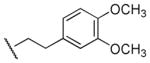
|
Me | 3-F-Ph | 27,000 | 3,340 | 8.1 |
| 67 | ibid. | Me | 2-F-Ph | >100,000 | >100,000 | - |
| 68 | ibid. | Me | 3-MeO-Ph | >100,000 | >100,000 | - |
| 69 | ibid. | Me | 2-MeO-Ph | >100,000 | >100,000 | - |
At least three independent experiments with samples evaluated in duplicate were performed to obtain the Ki value.
Analogues 61–69 (Table 2) were synthesized in a similar manner to analogues 42–59 from triflate 12. Phenyl ring substituted phenylboronic acids were used instead of phenylboronic acid as the Suzuki coupling partners (Scheme 2).
Scheme 2. Synthesis of Compounds 61–69a.
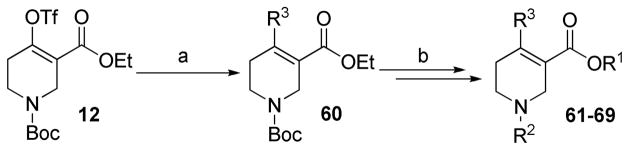
aReagents and conditions: (a) substituted phenylboronic acids, Pd(PPh4)3, Na2CO3 (2.0 M), THF, 65 °C; (b) same as from compound 13 to 42 in Scheme 1.
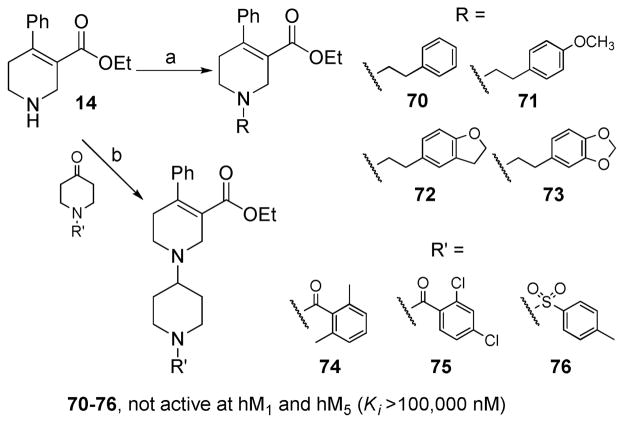
Analogues 70–73 were prepared by N-alkylation of compound 14 with appropriate alkyl halides (Scheme 3). Analogues 74–76 were prepared by reductive amination of compound 14 with an N-sulfonated or N-acylated 4-piperidinone using sodium triacetoxyborohydride (Scheme 3).
Scheme 3. Synthesis of Compounds 70–76a.
aReagents and conditions: (a) alkylbromides, KI, K2CO3, CH3CN, reflux; (b) NaBH(OAc)3, HOAc, THF, 65 °C for 74, rt for 75 and 76.
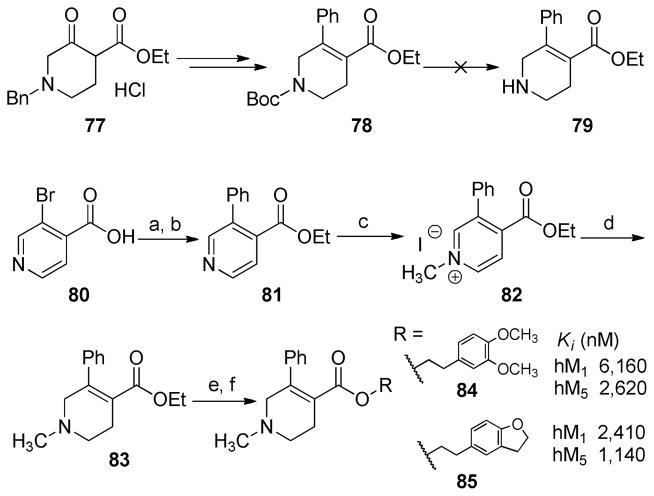
The synthesis of analogues 84 and 85 were initially attempted by applying a similar route to that used for the synthesis of their regioisomers in Scheme 1 (analogues 56 and 45, respectively), starting from ethyl 1-benzyl-3-oxo-4-piperidinecarboxylate hydrochloride (77) (Scheme 4). However, de-BOC reaction of compound 78 failed to yield the desired intermediate 79. As an alternative, analogues 84 and 85 were synthesized starting from 3-bromoisonicotinic acid (80). Ethyl esterification of 80 followed by Suzuki coupling with phenylboronic acid afforded compound 81, which was treated with methyl iodide to yield the corresponding quaternary pyridinium salt 82. Reduction of 82 with sodium borohydride gave 1,2,5,6-tetrahydropyridine derivative 83, which underwent ester hydrolysis and subsequent esterification with appropriate alcohols to afford analogues 84 and 85.
Scheme 4. Synthesis of Compounds 84 and 85a.
aReagents and conditions: (a) EtOH, H2SO4 (con.), reflux; (b) PhB(OH)2, Pd(PPh4)3, Na2CO3 (2.0 M), THF, 65 °C; (c) MeI, acetone; (d) NaBH4, EtOH; (e) 10% KOH (H2O)/EtOH (1:1); (f) alcohols, EDCI, DMAP, CH2Cl2.
Synthesis of analogue 89 was achieved by initial Suzuki coupling of 4-bromopyridine with 2-(ethoxycarbonyl)phenylboronic acid, followed by a similar route to that for analogues 84 and 85 from compound 81 (Scheme 5).
Scheme 5. Synthesis of Compound 89a.
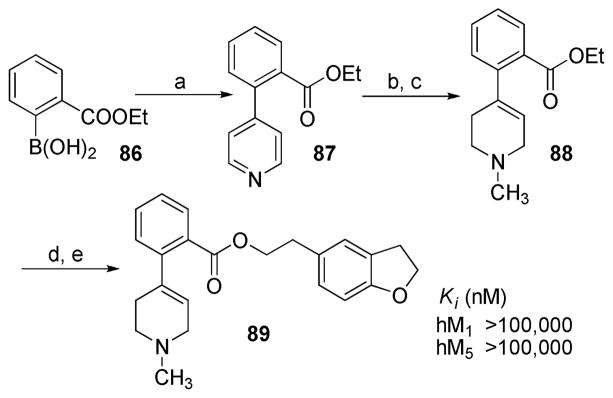
aReagents and conditions: (a) 4-bromopyridine, Pd(PPh3)4, Na2CO3 (2.0 M), THF, 65 °C; (b) MeI, acetone; (c) NaBH4, EtOH; (d) 10% KOH (H2O)/EtOH (1:1); (e) 2-(2,3-dihydrobenzofuran-5-yl)ethanol, EDCI, DMAP, CH2Cl2.
Radioligand Binding Assay
Receptor affinities were determined by measuring inhibition of the binding of [3H]N-methylscopolamine (NMS), an orthosteric antagonist probe, to membranes from Chinese hamster ovarian (CHO) cells individually expressing human hM1–hM5 mAcChR receptors. Compound 1 (scopolamine) and compound 8 were used as reference compounds. Methods for these binding assays are described briefly in the Experimental Section. All synthesized analogues were evaluated first at hM1 and hM5 subtypes (Table 1 and 2, Scheme 3–5). Selectivity for M5 over M1 is important, because antagonism of CNS M1 receptors has been suggested to result in cognitive deficits36 and increase DA release in striatum.37 Binding affinity for analogues which exhibited binding preference for M5 over M1 were determined at each of the five mAcChR subtypes (Table 3).
Table 3.
Binding Affinity and Selectivity for Selected Analogues at the hM1-hM5mAcChRsa
| Compd |
Ki, nM
|
||||
|---|---|---|---|---|---|
| hM5 | hM1 (M1/M5)b | hM2(M2/M5)b | hM3(M3/M5)b | hM4 (M4/M5)b | |
| 1 | 17.6 | 7.5 (0.4) | 9.5 (0.5) | 6.5 (0.4) | 36.9 (2.1) |
| 8 | 420 | 170 (0.4) | 1,100 (2.6) | 1,370 (3.3) | 1,730 (4.1) |
| 20 | 470 | 3,290 (7.0) | 2,030 (4.3) | 4,070 (8.7) | 11,500 (24) |
| 28 | 230 | 1,390 (6.0) | >100,000 (>435) | 31,100 (135) | 10,500 (46) |
| 45 | 60 | 290 (4.8) | 1,170 (20) | 2,840 (47) | 1,890 (32) |
| 56 | 2,240 | 25,300 (11) | >100,000 (>45) | >100,000 (>45) | >100,000 (>45) |
| 57 | 5,040 | 24,400 (4.8) | >100,000 (>20) | >100,000 (>20) | >100,000 (>20) |
| 63 | 740 | 5,270 (7.1) | 540 (0.7) | 4,890 (6.6) | 5,740 (7.8) |
| 66 | 3,340 | 27,000 (8.0) | >100,000 (>30) | >100,000 (>30) | >100,000 (>30) |
At least three independent experiments with samples evaluated in duplicate were performed to obtain the Ki value;
Numbers in the parentheses are the ratios of binding affinity between M5 and the respective subtype.
Identification of compound 8 was the starting point. Compound 8 was an analogue of the 1-ethyl-4-phenyl-tetrahydropyridine-3-carboxylic acid scaffold aimed at identifying M1 selective antagonists. Compound 8 was reported to have a 2.2-fold preference for M5 over M1, and to be more than 40-fold selective for M5 over M2–M4 mAcChR subtypes (Figure 1).33 However, in the current study, compound 8 showed no selectivity for M5 over M1–M4 (Table 3). Preliminary SAR from the previous report33 suggested that small changes on the p-methoxyphenyl ring of 8 afforded remarkably different binding affinity and selectivity profiles at mAcChR subtypes. For example, when the methoxy group on the phenyl ring of 8 was replaced by a methyl group, the resulting analogue 10 bound preferentially to the M1 receptor, while binding affinity at the M5 receptor increased 35-fold, when compared to compound 8 (Figure 1).33
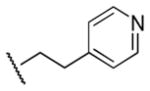
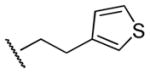
Results in Table 1 summarize the modifications to the p-methoxyphenethyl moiety of compound 8 evaluated in the current study and the influence of these modifications on binding affinity and selectivity for M1 and M5 receptors. Results from para-, meta-, and ortho-methoxy analogues, 8, 17, and 18, respectively, indicate that the meta-substitution was favorable to M5 selectivity. Compared to 8 (Ki = 420 nM) and 17 (Ki = 1,250 nM), meta-analogue (18, Ki = 150 nM) exhibited a small 3- and 8-fold, respectively, increase in affinity at M5 over M1. Replacement of the ethylene link in the p-methoxyphenethyl moiety by a methylene (compound 19, Ki = 3,610 nM) or a methyl ethylene link (compound 21, Ki > 100 μM) significantly decreased binding potency at M5 receptor, whereas replacement with a propylene link (compound 20, Ki = 470 nM) retained binding affinity at M5 and markedly increased selectivity (7-fold) over M1 receptors. Interestingly, there were no marked differences in M5 affinities when the 4-methoxy was replaced with another para-substitution group, including halogens (compounds 23–25) and nitro group (compound 26), indicating modification at this position is somewhat tolerated. However, compound 27, in which a 4-methylsulfonyl group was attached to the phenyl ring, exhibited little affinity for either M1 or M5 receptors. Compounds 28 and 31, wherein both 3- and 4-positions of the phenyl ring were substituted, not only retained affinity at the M5 receptor (Ki = 230 and 2,460 nM, respectively), but also exhibited preference for this subtype (6.0- and 4.8-fold M5/M1, respectively). Tri-substitution of the phenyl ring in compounds 32 and 33 revealed that this modification was detrimental to binding. Finally, replacement of the 4-methoxyphenyl ring with a heterocyclic ring, including pyridyl (compounds 34 and 35) and thiophenyl (compound 36) rings, resulted in retention of binding affinity at both M1 and M5 receptors.
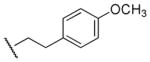
In parallel with the modification on the p-methoxyphenethyl group, the N-ethyl moiety of compound 8 (Table 2) also was altered. Replacement of the ethyl group by a methyl group (compound 42) resulted in an 8-fold and 14-fold increase in binding affinity at M1 and M5 receptors, respectively. Increasing the size of the substituted group from ethyl to n-propyl (compound 43), n-butyl (compound 44), or 4-methoxybenzyl group (compound 40) eliminated affinity for these two receptors. Similar results were observed for N-methyl, N-n-propyl, and N-n-butyl analogues (compounds 45, 46, and 47, respectively) of compound 28. Interestingly, the M1/M5 selectivity of two N-methyl containing analogues, compound 42 (0.7-fold) or 45 (4.8-fold), were similar to their corresponding N-ethyl analogues, 8 (0.4-fold) and 28 (6.0-fold), respectively.
Based on the above SAR data, our focus changed to analogues bearing an N-methyl group in lieu of N-ethyl group, and the SAR was extended regarding the substitution group of the carboxylic ester moiety (Table 2). Replacement of the ethyl link in compound 45 by a methylene (compound 49) or propylene (compound 50) link resulted in a decrease in binding affinity at both M1 and M5 receptors, and also resulted in a decrease in selectivity for M5 over M1 receptors. SAR among compounds 45, 49, and 50 was inconsistent with previous SAR among compounds 8, 19 and 20, indicating an unpredictable nature of this component of the structure. On the other hand, analogues containing substituents at the meta-position or both the meta- and para-position of the ester phenyl ring (compounds 53–56) consistently exhibited a binding preference at M5 over M1 receptor. Compound 56 was identified as the most selective (11-fold for M1/M5) M5 compound.
Based on compounds 45 and 56, SAR was further extended by introduction of an electron-withdrawing (fluoro) or electron-donating (methoxy) group onto the phenyl ring on the C4 of the tetrahydropyridine core (compounds 61–69, Table 2). In general, these analogues exhibited decreased or completely abolished activity at either M1 or M5 receptor when compared to compound 45 and 56, suggesting that this component of the molecule is less tolerant to structural modification.
Further SAR exploration was focused on previous hypotheses that spatial rearrangement or reorientation of the pharmacophore elements in mAcChR ligands would alter affinity and selectivity profiles.38 For example, rearrangement of the amino group in M1 preferring antagonist 3 (pirenzepine) afforded M2 preferring antagonist 4 (Figure 1).39 To test this hypothesis with respect to the current analogues, we conducted three types of pharmacophoric “rearrangements”. First, transposition of the substituted group on the ester functionality and the ethyl group on the N atom in compound 8 and analogues afforded compounds 70–73 (Scheme 3). An N-acylated or N-sulfonated piperidine-4-yl group also was introduced to the N atom of the tetrahydropyridine ring (compounds 74–76, Scheme 3). The N-substituents were selected because they are present in potent mAcChR orthosteric antagonists, such as darifenacin (90), zamifenacin (91), and compounds 92 and 9340 (Figure 3). Surprisingly, none of these new analogues displayed any activity at either M1 or M5 receptors, indicating a different receptor binding mode for the basic N atom in compound 8 and its analogues compared with compounds 90–93.
Figure 3.
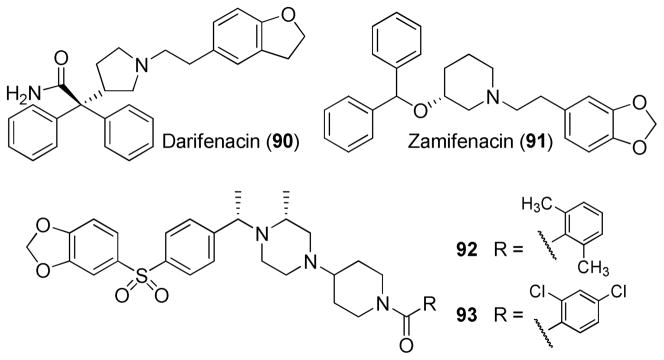
Structures of Darifenacin (90), Zamifenacin (91), and compounds 92 and 93.
Second, the ester group on C3 and the phenyl group on C4 of the 1,2,5,6-tetrahydropyridine ring in compounds 56 and 45 were transposed to produce compounds 84 and 85 (Scheme 4), respectively. Both compounds 84 (M1/M5 = 2.4) and 85 (M1/M5 = 2.1) displayed reduced selectivity for M5 over M1 receptors, when compared to their corresponding position isomers, 56 and 45, respectively. Last, the ester functionality in compound 45 was moved from C3 of the tetrahydropyridine ring to the phenyl ring on C4 (compound 89, Scheme 5). This “parallel” shift resulted in a complete loss of binding affinity at both M1 and M5 receptors.
At saturation concentrations, all analogues, except for those with Ki > 100,000 nM, exhibited complete inhibition (maximal inhibition, Imax = 100%, data not show) of the binding of the orthosteric antagonist [3H]NMS at mAcChRs, which is consistent with an orthosteric mechanism of action. Selected compounds, including 20, 28, 45, 56, 57, 63, and 66, which exhibited binding preference for hM5 over hM1 mAcChRs, also were evaluated for affinity at M2, M3, and M4 mAcChR subtypes (Table 3). Results showed that 5 of these 7 compounds exhibited good selectivity for M5 over M2, M3, and M4 receptors. The most selective compound, 56 (11-fold for M5 over M1 receptor), had no affinity (Ki > 100 μM) at the M2, M3, and M4 mAcChR subtypes. Compound 28 exhibited 6-fold, >435-fold, 135-fold, and 46-fold for M5 over M1, M2, M3, and M4, respectively. Although slightly less selective for the M5 over the M1 receptor, it exhibited higher affinity at M5 when compared to compound 56.
Inhibition of Oxotremorine-Evoked Striatal [3H]DA Release
In vitro functional assays for mAcChR antagonists measure the ability of molecules to block mAcChR agonist-induced receptor activation at recombinant mAcChR subtypes expressed in cells.41 Pharmacological studies of M5 receptors using mouse basilar artery also have been reported.42 However, these recombinant and native M5 receptors functional assays are far removed from a potential role for M5 receptors in cocaine and opiate addiction. Studies have shown that oxotremorine, a non-selective mAcChR agonist, concentration dependently increases [3H]DA release from striatal slices prepared from wild-type mice and that oxotremorine-evoked striatal [3H]DA release was reduced significantly in M5 receptor knockout mice.18, 43 We hypothesized that an M5 receptor selective antagonist would also reduce oxotremorine-mediated rat striatal [3H]DA release. Current results show that oxotremorine evokes [3H]DA release from rat striatal slices and that scopolamine inhibits this effect in a concentration-dependent manner (Figure 4). These results support the contention that this functional assay probes native M5 receptors. Furthermore, this functional assay is highly relevant to the underlying dopaminergic mechanisms involved in drug reward and abuse.
Figure 4.
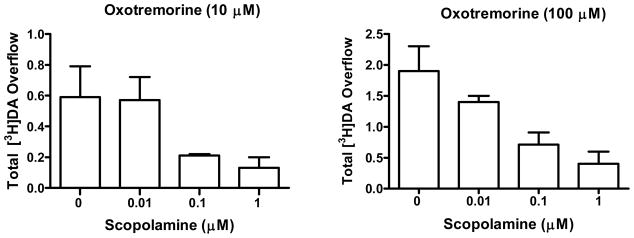
Scopolamine (0.01–1 μM) inhibits oxotremorine (10 μM and 100 μM) evoked [3H]DA release from rat striatal slices (data are expressed as Mean ± SEM, n = 3).
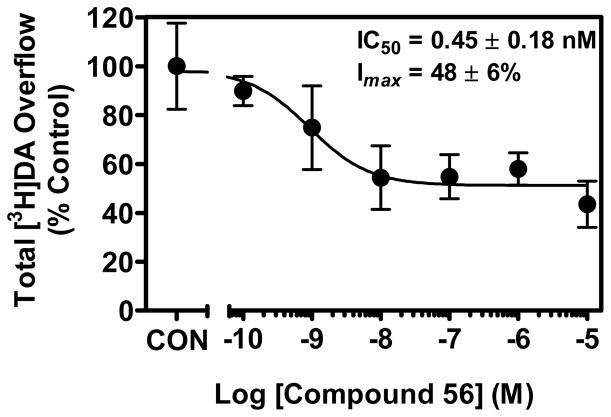
Results revealed that compound 56 inhibited (IC50 = 0.45 nM) oxotremorine (100 μM) evoked [3H]DA release from rat striatal slices (Figure 5). Unlike scopolamine (1 μM), which completely inhibits oxotremorine-mediated [3H]DA release from rat striatal slices (Figure 4), compound 56 produced maximal inhibition (Imax) of only 48% of the oxotremorine-evoked [3H]DA release (Figure 5). These current results are consistent with previous reports that ~50% of oxotremorine-evoked [3H]DA release from striatal slices was eliminated in M5 knock-out mice compared to wild-type mice,18 indicating that other mAcChR subtype(s) also mediate oxotremorine-evoked striatal DA release. In agreement with this hypothesis, studies using mAcChR knock-out mice suggested that M3 and M4 receptors also were involved in mediating striatal DA release.43 The observations that both compound 56 and the deletion of the M5 receptor resulted in similar effects on oxotremorine-evoked striatal [3H]DA release, together with the selective binding of 56 to M5 over M3 and M4 receptors, strongly suggest that 56 interacts with M5 receptors to inhibit muscarinic agonist-induced striatal DA release.
Figure 5.
Compound 56 inhibits oxotremorine (100 μM) evoked [3H]DA release from rat striatal slices (data are expressed as Mean ± SEM, n = 4).
It is noteworthy that compound 56 (IC50 = 0.45 nM) appears more potent than scopolamine in inhibiting oxotremorine-evoked [3H]DA release from rat striatal slices although its [3H]NMS binding affinity on the M5 receptor is 127-fold less than scopolamine. One explanation on the lack of correlation between binding and function is that the [3H]NMS binding assay was performed on a single receptor in a recombinant system but the [3H]DA release assay was on heterogeneous brain slices. Compound 56 may potently act at other sites that also inhibit [3H]DA release. Alternatively, recent studies on the crystal structure of the rat M3 receptor with antagonist tiotropium bound to the orthosteric binding site suggested that tiotropium binds transiently to an allosteric site en route to the orthosteric binding pocket.44 Compound 56 could have a similar allosteric interaction with the receptor, causing the inhibition of oxotremorine-evoked striatal [3H]DA release. However, further pharmacological studies are needed to elucidate the mechanism.
Binding Mode for M5 mAcChRs for Lead Compounds 28 and 56
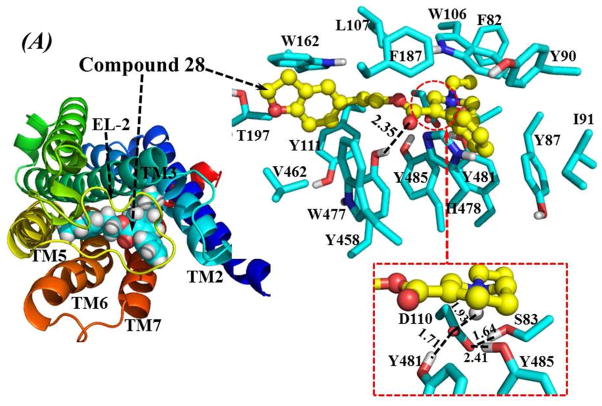
To study the interaction of our compounds with M5 mAcChR in atomic detail, homology modeling and molecular docking operations were performed. Compounds 28 and 56 were selected for these studies, since both analogues preferencially bind to M5 mAcChR, and thus, can be considered current lead compounds. Importantly, compounds 28 and 56 have an order of magnitude difference in binding affinities at M5 receptor (Ki = 230 nM vs 2240 nM, respectively). This moderate difference in affinity between 28 and 56 is ideal for testing the reliability of our homology models. The structural model of the human M5 mAcChR was constructed based on the newly available X-ray crystal structure of the rat M3 mAcChR with antagonist tiotropium bound to the orthosteric binding site.44 Compounds 28 and 56 were docked into possible binding sites among the transmembrane (TM) helices of the M5 mAcChR. Binding structures were selected from the docking results and subjected to energy minimization. In accordance with the minimized binding structures, the binding site for compounds 28 and 56 at the M5 mAcChR is the orthosteric site located near the extracellular end of TM3, TM5, TM6, and TM7. As shown in Figure 6, TM2 and TM4 are also partially involved in the formation of the antagonist-binding site. In addition, the antagonist binding site is partially covered by extracellular loop 2 (EL-2). As depicted in Figure 6A, compound 28 is orientated horizontally inside the binding pocket. The cationic head of compound 28 is anchored around the negatively charged side chain of residue D110 of TM3, interacting through electrostatic attraction and strong hydrogen bonding. Meanwhile, residue D110 is also hydrogen-bonded with the side chain of S83 from TM2, and the side chains of Y481 and Y485 from TM7. The cationic head of compound 28 is in close contact with residues W106 and L107 from TM3. The phenyl group on C4 of tetrahydropyridine ring is closely packed with Y87, Y90 and I91 from TM2, W106 from TM3, F187 from EL-2, and H478 from TM7. The ethyl group at the cationic head of compound 28 contacts with the aromatic side chain of residue F82 from TM2. The carbonyl oxygen of compound 28 is weakly hydrogen-bonded with the hydroxyl group at the side chain of Y458 from TM6. The tail group (2,3-dihydrobenzofuran-5-ethyl) of compound 28 is packed in parallel with the underneath Y111 from TM3, and packed perpendicularly with the side chain of W162 from TM4. The tail group of compound 28 is also surrounded by T197 from TM5 and V462 from TM6.
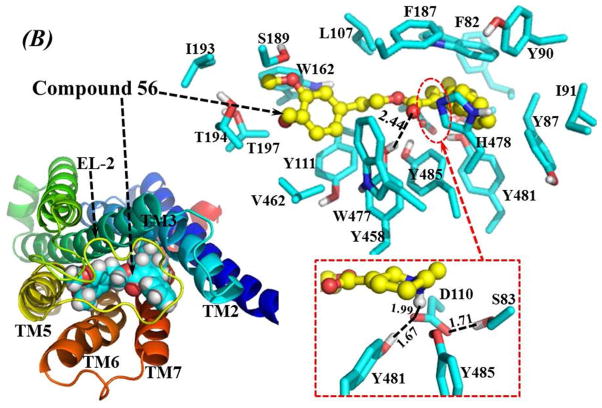
Figure 6.
Homology model of human M5 mAcChR based on the X-ray crystal structure of rat M3 mAcChR (PDB entry of 4DAJ at 3.4 Å resolution, A chain). (A) Top view on the energy-minimized binding structure of M5 mAcChR-Compound 28 complex. (B) Top view on the energy-minimized binding structure of M5 mAcChR-Compound 56 complex. The receptor proteins in (A) and (B) are represented as ribbon in rainbow color, and compound 28 and 56 are shown as spheres (left panel) or ball-and-stick (right panel). Residues within 5 Å of compound 28 and 56 are labeled and shown in stick. 28 and 56 have very similar hydrogen bonding interactions with the protein, including interactions between the D110 side chain and the cationic heads of the compounds, interactions between the carbonyl oxygen atoms of the compounds and the Y458 side chain, and the interactions among side chains of D110, S83, Y481 and Y485. These hydrogen bonding interactions are shown in dashed lines along with the labeled distances.
As depicted in Figure 6B, the binding mode of the M5 mAcChR with compound 56 is essentially the same as that with compound 28. One difference between the binding of 56 and 28 to the M5 mAcChR is that residues I193 and T194 from TM5 are both within 5 Å of compound 56 (Figure 6B), while these two residues have no contacts with compound 28 (Figure 6A). Another difference is the distance of hydrogen bonding formed by the side chain of residue D110 with the cationic head group of the antagonist. As shown in Figure 6, the distance between the hydrogen bonding between residue D110 and the cationic head of compound 28 is shorter than that of the hydrogen bonding between residue D110 and the cationic head of compound 56 (1.93 Å vs 1.99 Å, respectively), indicating a stronger bond with 28 than with 56 to the protein. These structural differences, especially the difference in hydrogen bonding distance, may contribute to the difference in binding affinity as represented by the experimentally measured values of Ki (230 nM for 28 vs. 2240 nM for 56) for these two lead compounds (Table 3). The modeled binding structures helped to qualitatively understand the observed SAR and provided clues to design a valuable virtual library of new analogues for computational screening.
SUMMARY
Starting from compound 8 as a lead structure, we have identified the first M5-preferring antagonists through a systematic structural modification strategy. The greatest mAcChR selectivity and potency shifts came from the modification of the substituents on the ester group. Replacing the N-Et group on the tetrahydropyridine ring with an N-Me group generally resulted in a significant increase in mAcChR binding affinity, while maintaining mAcChR subtype-selectivity profile. This preliminary SAR study provides a basis for further discovery of potent and selective M5 ligands. In addition, we have successfully established a functional assay for M5 receptors using oxotremorine-evoked DA release from superfused rat striatal slices. One of our lead compound, 56, demonstrated inhibition of oxotremorine-mediated striatal [3H]DA release, with a maximal inhibition of ~50%. This result is similar to the effects of M5 knock-out on striatal [3H]DA release, providing validation for the current assay. Further, we have constructed a homology model of human M5 based on the newly available crystal structure of the rat M3 receptor. Docking studies performed on compounds 28 and 56 revealed that both possibly interact with the orthosteric binding site on the M5 receptor, which is consistent with the current results from the [3H]NMS binding assay, indicating an orthosteric mechanism of action for these new analogues. We are building and validating homology models for the other four mAcChR subtypes, and these will be used for virtual library screening. The predictability of these models regarding mAcChR subtype selectivity remains to be tested.
EXPERIMENTAL SECTION
Chemistry
All purchased reagents and solvents were used without further purification unless otherwise noted. All reaction sensitive to air and/or moisture were carried out under argon atmosphere in oven-dried glassware. Flash column chromatography was carried out using 32–63 μm, 60 Ǻ (230–400 mesh) silica gel. Analytical thin layer chromatography was carried out on glass plates precoated with 250 μm silica gel 60 F254. NMR spectra were recorded in CDCl3 on a Varian 300 MHz or 500 MHz spectrometer and chemical shifts are reported in ppm relative to tetramethylsilane as the internal standard. Coupling constants are reported in Hertz (Hz). Mass spectra were recorded on a JEOL JMS-700T MStation. GC-Mass spectra were recorded on an Agilent 6890 GC incorporating an Agilent 7683 autosampler and an Agilent 5973 MSD. Elemental analyses were carried out on a COSTECH elemental combustion system and are within ± 0.4% of theory. All final compounds for biological testing were prepared as salts in ≥ 95% purity, in accord with results from combustion analysis. A detailed description of synthetic methodologies as well as analytical and spectroscopic data for all described compounds is included in the Supporting Information.
Binding Assay
Analogue binding affinities for the five mAcChR subtypes were determined in assays evaluating inhibition of [3H]NMS binding to membranes from CHO-K1 cells expressing one of the recombinant hM1–hM5 mAcChRs. The CHO-K1 cell lines expressing the five subtypes of muscarinic receptors were obtained as a gift from Dr. Tom Bonner of National Institute of Mental Health (NIMH). Cells were grown at 37 °C with 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 4 mM glutamine, 50 ng/mL geneticin G418 and 1% Pen-Strep. Cells were harvested at 70–90% confluency. To obtain cell membranes, cells were scraped into ice cold 50 mM Tris-HCl (pH 7.4), sonicated for 30 s and centrifuged (48,000 g for 30 min). Pellets were re-suspended in 1.5 ml ice-cold 50 mM Tris-HCl buffer, sonicated for 30 s and stored at −80 °C until assay. Assays were performed using 96-well plates. Membrane aliquots (100 μL) containing 10–40 μg protein were added to wells containing 1 nM to100 μM of test compound (25 μL), 0.3 nM [3H]NMS (25 μL; scopolamine methyl chloride [N-methyl-3H]; specific activity 82 Ci/mmol; Perkin Elmer/NEN, Boston, MA) and buffer (50 mM Tris-HCl, pH 7.4; 125 μL) for a total volume of 250 μL. Atropine (1 mM) was used to determine nonspecific binding. Samples were incubated for 120 min at 25 °C with constant agitation. Reactions were terminated by rapid filtration onto GF/B filters using a Filtermate Harvester (Perkin Elmer Life and Analytical Sciences, Boston, MA). Samples were washed three times with 350 μL of ice-cold 50 mM Tris-HCl buffer, and dried for 60 min at 50 °C. Subsequently, 40 μL MicroScint 20 (PerkinElmer Life and Analytical Sciences, Waltham, MA) was added to each well and radioactivity bound determined using liquid scintillation spectrometry. IC50 values were obtained and Ki values calculated using the equation of Cheng and Prusoff.45
Inhibition of Oxotremorine-Evoked [3H]DA Release Assay
Assays were performed according to previously published methods,47 with minor modifications. Striata were dissected, and coronal slices (500 μm, 4–6 mg) obtained with a McIlwain chopper. Slices were incubated for 30 min in Krebs’ buffer (in mM: 108 NaCl, 4.7 KCl, 1.2 MgCl2, 1 NaH2PO4, 1.3 CaCl2, 11.1 glucose, 25 NaHCO3, 0.11 L-ascorbic acid and 0.004 disodium EDTA, pH 7.4, saturated with 95% O2/5% CO2) in a metabolic shaker at 34°C. Slices were incubated with 0.1 μM [3H]DA during the latter 30 min of a 60-min incubation period. Each slice was transferred to a superfusion chamber and superfused (0.6 mL/min at 34 °C) for 60 min with Krebs’ buffer containing nomifensine (10 μM) and pargyline (10 μM) to inhibit reuptake and prevent metabolism, respectively, ensuring that [3H]overflow primarily represents [3H]DA, rather than [3H]metabolites.47 Sample collection began after 60 min of superfusion, when the rate of release was stable. Consecutive 4-min (2.4 mL) samples were collected to determine basal [3H]outflow. Superfusion continued in the absence or presence of a range of analogue concentrations (0.1 nM – 1 mM) for 40 min, followed by 40 min with oxotremorine (100 μM) added to the superfusion buffer. Radioactivity in slices and superfusate samples were determined via liquid scintillation spectroscopy. Data were analyzed by weighted, least squares regression analysis of the sigmoidal concentration-effect curves to obtain IC50 values.
Homology Modeling and Molecular Docking
The homology model of human M5 mAcChR was built based on the X-ray crystal structure of rat M3 mAcChR (PDB entry of 4DAJ at 3.4 Å resolution, A chain)44 by using the Protein Modeling module of Discovery Studio (version 2.5.5, Accelrys, Inc. San diego, CA). The model of human M5 mAcChR structure was constructed and refined in a very similar manner as in our previous study,48 i.e. the best sequence alignment was selected based on both the alignment score and the reciprocal positions of the conserved residues among all mAcChR subtypes. The coordinates of the conserved regions were directly transformed from the template structure, while the non-conserved residues were mutated from the template to the corresponding ones in M5 mAcChR. Structural optimization and energy minimization for the M5 mAcChR structue were performed using the Amber 11 program suite. The convergence criterion for the energy minization was set to 0.001 kcal mol−1 Å−1. Based on the optimized M5 mAcChR structure, the binding mode of the receptor with two lead compounds 28 and 56 was explored through molecular docking using AutoDock 3.0.5 program.48 This molecular docking approach was similar to that described in our previous study.49 A reasonable binding structure of M5 mAcChR in complex with either compound 28 or 56 was obtained after energy minimization on each complex structure.
Supplementary Material
Acknowledgments
This work was supported financially by a Pilot Project Grant from Center for Drug Abuse Research Translation at the University of Kentucky (DA05312) and grants from National Institute of Drug Abuse, National Institute of Health (DA025948, DA030667 and TR000117).
ABBREVIATIONS
- VTA
ventral tegmental area
- NMS
N-methylscopolamine
- CHO
Chinese hamster ovarian
- TM
transmembrane
- EL
extracellular loop
Footnotes
The authors declare no conflict or financial interest.
Synthetic procedures and analytical data of all compounds prepared. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Caulfield MP. Muscarinic receptors - characterization, coupling and function. Pharmacol Therapeut. 1993;58:319–379. doi: 10.1016/0163-7258(93)90027-b. [DOI] [PubMed] [Google Scholar]
- 2.Caulfield MP, Birdsall NJ International Union of Pharmacology. XVII. Classification of muscarinic acetylcholine receptors. Pharmacol Rev. 1998;50:279–290. [PubMed] [Google Scholar]
- 3.Wess J. Muscarinic acetylcholine receptor knockout mice: novel phenotypes and clinical implications. Annu Rev Pharmacol Toxicol. 2004;44:423–450. doi: 10.1146/annurev.pharmtox.44.101802.121622. [DOI] [PubMed] [Google Scholar]
- 4.Wess J, Eglen RM, Gautam D. Muscarinic acetylcholine receptors: mutant mice provide new insights for drug development. Nat Rev Drug Discov. 2007;6:721–733. doi: 10.1038/nrd2379. [DOI] [PubMed] [Google Scholar]
- 5.Bonner TI, Young AC, Brann MR, Buckley NJ. Cloning and expression of the human and rat M5 muscarinic acetylcholine-receptor genes. Neuron. 1988;1:403–410. doi: 10.1016/0896-6273(88)90190-0. [DOI] [PubMed] [Google Scholar]
- 6.Raffa RB. The M5 muscarinic receptor as possible target for treatment of drug abuse. J Clin Pharm Ther. 2009;34:623–629. doi: 10.1111/j.1365-2710.2009.01059.x. [DOI] [PubMed] [Google Scholar]
- 7.Di Chiara G, Imperato A. Drugs abused by humans preferentially increase synaptic dopamine concentrations in the mesolimbic system of freely moving rats. Proc Natl Acad Sci U S A. 1988;85:5274–5278. doi: 10.1073/pnas.85.14.5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koob GF, Swerdlow NR. The functional output of the mesolimbic dopamine system. Ann N Y Acad Sci. 1988;537:216–227. doi: 10.1111/j.1749-6632.1988.tb42108.x. [DOI] [PubMed] [Google Scholar]
- 9.Koob GF, Nestler EJ. The neurobiology of drug addiction. J Neuropsychiatry Clin Neurosci. 1997;9:482–497. doi: 10.1176/jnp.9.3.482. [DOI] [PubMed] [Google Scholar]
- 10.Cami J, Farre M. Drug addiction. N Engl J Med. 2003;349:975–986. doi: 10.1056/NEJMra023160. [DOI] [PubMed] [Google Scholar]
- 11.Vilaro MT, Palacios JM, Mengod G. Localization of M5 muscarinic receptor messenger-RNA in rat-brain examined by in situ hybridization histochemistry. Neurosci Lett. 1990;114:154–159. doi: 10.1016/0304-3940(90)90064-g. [DOI] [PubMed] [Google Scholar]
- 12.Yasuda RP, Ciesla W, Flores LR, Wall SJ, Li M, Satkus SA, Weisstein JS, Spagnola BV, Wolfe BB. Development of antisera selective for M4 and M5 muscarinic cholinergic receptors-distribution of M4 and M5 receptors in rat-brain. Mol Pharmacol. 1993;43:149–157. [PubMed] [Google Scholar]
- 13.Weiner DM, Brann MR. Distribution of M1-M5 muscarinic receptor messenger-RNAs in rat-brain. Trends Pharmacol Sci. 1989:115–115. [Google Scholar]
- 14.Reever CM, FerrariDiLeo G, Flynn DD. The M5 (m5) receptor subtype: fact or fiction? Life Sci. 1997;60:1105–1112. doi: 10.1016/s0024-3205(97)00054-4. [DOI] [PubMed] [Google Scholar]
- 15.Yeomans JS, Takeuchi J, Baptista M, Flynn DD, Lepik K, Nobrega J, Fulton J, Ralph MR. Brain-stimulation reward thresholds raised by an antisense oligonucleotide for the M5 muscarinic receptor infused near dopamine cells. J Neurosci. 2000;20:8861–8867. doi: 10.1523/JNEUROSCI.20-23-08861.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller AD, Blaha CD. Midbrain muscarinic receptor mechanisms underlying regulation of mesoaccumbens and nigrostriatal dopaminergic transmission in the rat. Eur J Neurosci. 2005;21:1837–1846. doi: 10.1111/j.1460-9568.2005.04017.x. [DOI] [PubMed] [Google Scholar]
- 17.Lester DB, Miller AD, Blaha CD. Muscarinic receptor blockade in the ventral tegmental area attenuates cocaine enhancement of laterodorsal tegmentum stimulation-evoked accumbens dopamine efflux in the mouse. Synapse. 2010;64:216–223. doi: 10.1002/syn.20717. [DOI] [PubMed] [Google Scholar]
- 18.Yamada M, Lamping KG, Duttaroy A, Zhang WL, Cui YH, Bymaster FP, McKinzie DL, Felder CC, Deng CX, Faraci FM, Wess J. Cholinergic dilation of cerebral blood vessels is abolished in M5 muscarinic acetylcholine receptor knockout mice. Proc Natl Acad Sci U S A. 2001;98:14096–14101. doi: 10.1073/pnas.251542998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basile AS, Fedorova I, Zapata A, Liu X, Shippenberg T, Duttaroy A, Yamada M, Wess J. Deletion of the M5 muscarinic acetylcholine receptor attenuates morphine reinforcement and withdrawal but not morphine analgesia. Proc Natl Acad Sci U S A. 2002;99:11452–11457. doi: 10.1073/pnas.162371899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fink-Jensen A, Sorensen L, Bay-Richter C, Frikke-Schmidt H, Wess J, Woldbye DP, Wortwein G. Involvement of the M5 muscarinic receptor in cocaine and amphetamine induced behaviour: Studies in M5 knockout mice backcrossed to the C57BL/6NTac strain. Schizophrenia Research. 2006;81:301–302. [Google Scholar]
- 21.Thomsen M, Woldbye DPD, Wortwein G, Fink-Jensen A, Wess J, Caine SB. Reduced cocaine self-administration in muscarinic M-5 acetylcholine receptor-deficient mice. J Neurosci. 2005;25:8141–8149. doi: 10.1523/JNEUROSCI.2077-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rezayof A, Nazari-Serenjeh F, Zarrindast MR, Sepehri H, Delphi L. Morphine-induced place preference: involvement of cholinergic receptors of the ventral tegmental area. Eur J Pharmacol. 2007;562:92–102. doi: 10.1016/j.ejphar.2007.01.081. [DOI] [PubMed] [Google Scholar]
- 23.Yamada M, Basile AS, Fedorova I, Zhang WL, Duttaroy A, Cui YH, Lamping KG, Faraci FM, Deng CX, Wess J. Novel insights into M-5 muscarinic acetylcholine receptor function by the use of gene targeting technology. Life Sci. 2003;74:345–353. doi: 10.1016/j.lfs.2003.09.022. [DOI] [PubMed] [Google Scholar]
- 24.Bridges TM, Marlo JE, Niswender CM, Jones CK, Jadhav SB, Gentry PR, Plumley HC, Weaver CD, Conn PJ, Lindsley CW. Discovery of the First Highly M5-Preferring Muscarinic Acetylcholine Receptor Ligand, an M5 Positive Allosteric Modulator Derived from a Series of 5-Trifluoromethoxy N-Benzyl Isatins. J Med Chem. 2009;52:3445–3448. doi: 10.1021/jm900286j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bridges TM, Kennedy JP, Cho HP, Breininger ML, Gentry PR, Hopkins CR, Conn PJ, Lindsley CW. Chemical lead optimization of a pan G(q) mAChR M-1, M-3, M-5 positive allosteric modulator (PAM) lead. Part I: Development of the first highly selective M-5 PAM. Bioorg Med Chem Lett. 2010;20:558–562. doi: 10.1016/j.bmcl.2009.11.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bridges TM, Kennedy JP, Hopkins CR, Conn PJ, Lindsley CW. Heterobiaryl and heterobiaryl ether derived M5 positive allosteric modulators. Bioorg Med Chem Lett. 2010;20:5617–5622. doi: 10.1016/j.bmcl.2010.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J, Schoneberg T, van Rhee M, Wess J. Mutational analysis of the relative orientation of transmembrane helices I and VII in G protein-coupled receptors. J Biol Chem. 1995;270:19532–19539. doi: 10.1074/jbc.270.33.19532. [DOI] [PubMed] [Google Scholar]
- 28.Bohme TM, Keim C, Dannhardt G, Mutschler E, Lambrecht G. Design and pharmacology of quinuclidine derivatives as M2-selective muscarinic receptor ligands. Bioorg Med Chem Lett. 2001;11:1241–1243. doi: 10.1016/s0960-894x(01)00186-x. [DOI] [PubMed] [Google Scholar]
- 29.Bohme TM, Augelli-Szafran CE, Hallak H, Pugsley T, Serpa K, Schwarz RD. Synthesis and pharmacology of benzoxazines as highly selective antagonists at M(4) muscarinic receptors. J Med Chem. 2002;45:3094–3102. doi: 10.1021/jm011116o. [DOI] [PubMed] [Google Scholar]
- 30.Choppin A, Stepan GJ, Loury DN, Watson N, Eglen RM. Characterization of the muscarinic receptor in isolated uterus of sham operated and ovariectomized rats. Br J Pharmacol. 1999;127:1551–1558. doi: 10.1038/sj.bjp.0702696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Chackalamannil S, Hu Z, Greenlee WJ, Clader J, Boyle CD, Kaminski JJ, Billard W, Binch H, 3rd, Crosby G, Ruperto V, Duffy RA, Cohen-Williams M, Coffin VL, Cox KA, Grotz DE, Lachowicz JE. Improving the oral efficacy of CNS drug candidates: discovery of highly orally efficacious piperidinyl piperidine M2 muscarinic receptor antagonists. J Med Chem. 2002;45:5415–5418. doi: 10.1021/jm0255163. [DOI] [PubMed] [Google Scholar]
- 32.Sagara Y, Sagara T, Uchiyama M, Otsuki S, Kimura T, Fujikawa T, Noguchi K, Ohtake N. Identification of a novel 4-aminomethylpiperidine class of M3 muscarinic receptor antagonists and structural insight into their M3 selectivity. J Med Chem. 2006;49:5653–5663. doi: 10.1021/jm051205r. [DOI] [PubMed] [Google Scholar]
- 33.Augelli-Szafran CE, Blankley CJ, Jaen JC, Moreland DW, Nelson CB, Penvose-Yi JR, Schwarz RD, Thomas AJ. Identification and characterization of m1 selective muscarinic receptor antagonists. J Med Chem. 1999;42:356–363. doi: 10.1021/jm980067l. [DOI] [PubMed] [Google Scholar]
- 34.Solymar MFE, Fulop F. Enzyme-catalyzed kinetic resolution of piperidine hydroxy esters. Tetrahedron: Asymmetry. 2004;15:3281–3287. [Google Scholar]
- 35.Inokuchi E, Narumi T, Niida A, Kobayashi K, Tomita K, Oishi S, Ohno H, Fujii N. Efficient synthesis of trifluoromethyl and related trisubstituted alkene dipeptide isosteres by palladium-catalyzed carbonylation of amino acid derived allylic carbonates. J Org Chem. 2008;73:3942–3945. doi: 10.1021/jo702318d. [DOI] [PubMed] [Google Scholar]
- 36.Anagnostaras SG, Murphy GG, Hamilton SE, Mitchell SL, Rahnama NP, Nathanson NM, Silva AJ. Selective cognitive dysfunction in acetylcholine M1 muscarinic receptor mutant mice. Nat Neurosci. 2003;6:51–58. doi: 10.1038/nn992. [DOI] [PubMed] [Google Scholar]
- 37.Gerber DJ, Sotnikova TD, Gainetdinov RR, Huang SY, Caron MG, Tonegawa S. Hyperactivity, elevated dopaminergic transmission, and response to amphetamine in M1 muscarinic acetylcholine receptor-deficient mice. Proc Natl Acad Sci U S A. 2001;98:15312–15317. doi: 10.1073/pnas.261583798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naito R, Yonetoku Y, Okamoto Y, Toyoshima A, Ikeda K, Takeuchi M. Synthesis and antimuscarinic properties of quinuclidin-3-yl 1,2,3,4-tetrahydroisoquinoline-2-carboxylate derivatives as novel muscarinic receptor antagonists. J Med Chem. 2005;48:6597–6606. doi: 10.1021/jm050099q. [DOI] [PubMed] [Google Scholar]
- 39.Choppin A, Eglen RM, Hegde SS. Pharmacological characterization of muscarinic receptors in rabbit isolated iris sphincter muscle and urinary bladder smooth muscle. Br J Pharmacol. 1998;124:883–888. doi: 10.1038/sj.bjp.0701920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCombie SW, Lin SI, Tagat JR, Nazareno D, Vice S, Ford J, Asberom T, Leone D, Kozlowski JA, Zhou G, Ruperto VB, Duffy RA, Lachowicz JE. Synthesis and structure-activity relationships of M(2)-selective muscarinic receptor ligands in the 1-[4-(4-arylsulfonyl)-phenylmethyl]-4-(4-piperidinyl)-piperazine family. Bioorg Med Chem Lett. 2002;12:795–798. doi: 10.1016/s0960-894x(02)00024-0. [DOI] [PubMed] [Google Scholar]
- 41.Watson N, Daniels DV, Ford APDW, Eglen RM, Hegde SS. Comparative pharmacology of human muscarinic M3 and M5 cholino-ceptors expressed in Chinese hamster ovary (CHO) cells. Br J Pharmacol. 1998;123:U153–U153. [Google Scholar]
- 42.Pulido-Rios MT, Steinfeld T, Armstrong S, Watson N, Choppin A, Eglen R, Hegde SS. In vitro isolated tissue functional muscarinic receptor assays. Curr Protoc Pharmacol. 2010;Chapter 4(Unit 4):15. doi: 10.1002/0471141755.ph0415s48. [DOI] [PubMed] [Google Scholar]
- 43.Zhang W, Yamada M, Gomeza J, Basile AS, Wess J. Multiple muscarinic acetylcholine receptor subtypes modulate striatal dopamine release, as studied with M1-M5 muscarinic receptor knock-out mice. J Neurosci. 2002;22:6347–6352. doi: 10.1523/JNEUROSCI.22-15-06347.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kruse AC, Hu J, Pan AC, Arlow DH, Rosenbaum DM, Rosemond E, Green HF, Liu T, Chae PS, Dror RO, Shaw DE, Weis WI, Wess J, Kobilka BK. Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature. 2012;482:552–556. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng Y, Prusoff WH. Relationship between the inhibition constant (Ki) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 46.Dwoskin LP, Zahniser NR. Robust modulation of [3H]dopamine release from rat striatal slices by D-2 dopamine receptors. J Pharmacol Exp Ther. 1986;239:442–453. [PubMed] [Google Scholar]
- 47.Zumstein A, Karduck W, Starke K. Pathways of dopamine metabolism in the rabbit caudate nucleus in vitro. Naunyn Schmiedebergs Arch Pharmacol. 1981;316:205–217. doi: 10.1007/BF00505651. [DOI] [PubMed] [Google Scholar]
- 48.Huang X, Zheng G, Zhan CG. Microscopic binding of M5 muscarinic acetylcholine receptor with antagonists by homology modeling, molecular docking, and molecular dynamics simulation. J Phys Chem B. 2012;116:532–541. doi: 10.1021/jp210579b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morris GMG, Halliday DS, Huey RS, Hart RWE, Belew RK, Olson AJ. Automated docking using a lamarckian genetic algorithm and empirical binding free energy function. J Comput Chem. 1998;19:1639–1662. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.