Abstract
Nucleosomes in which histone H3 is replaced by CENP-A direct kinetochore assembly. CENP-A nucleosomes extracted from human and Drosophila cells have been reported to have reduced heights relative to canonical octameric H3 nucleosomes, suggesting a unique tetrameric, hemisomal composition. We demonstrate that even octameric CENP-A nucleosomes assembled in vitro exhibit a reduced height, indicating that they are physically distinct from H3 nucleosomes, and negating the need to invoke the presence of hemisomes.
Conventional nucleosomes wrap 147 base pairs (bp) of DNA 1.65 times around an octameric protein core containing two copies of the histone: H2A, H2B, H3 and H4 (ref. 1). A distinguishing feature of all centromeres is the presence of specialized nucleosomes in which the histone H3 variant, CENP-A (or cenH3), replaces canonical histone H3 (ref. 2). Atomic Force Microscopy (AFM) has shown that CENP-A nucleosomal arrays in chromatin extracted from fly and human cells are reduced in height relative to H3 nucleosomal arrays. This finding underpins the proposal that CENP-A nucleosomes are atypical tetrameric particles containing only a single subunit of H2A, H2B, CENP-A and H4 (hemisomes)3,4. Height measurement has remained the principal assay used to indicate that ex vivo CENP-A nucleosomes in arrays are hemisomal. However, it remains possible that these CENP-A nucleosomes are actually octameric, but that fundamental physical differences between CENP-A and H3 particles makes them appear shorter in height by AFM. To test this we have examined the height of octameric CENP-A and H3 nucleosomes in arrays assembled in vitro from recombinant histones. In vitro assembled CENP-A and H3 nucleosomes were measured in arrays in order to closely emulate ex vivo measurement conditions3–5.
We prepared CENP-A and H3 nucleosomes using untagged recombinant histones from two evolutionarily distant organisms, human and fission yeast (Schizosaccharomyces pombe)6. Nucleosomes were assembled onto arrays of DNA containing 19 × 197 bp repeats of the well-characterized 601 sequence7. Particles assembled by this procedure are consistently octameric1,8,9.
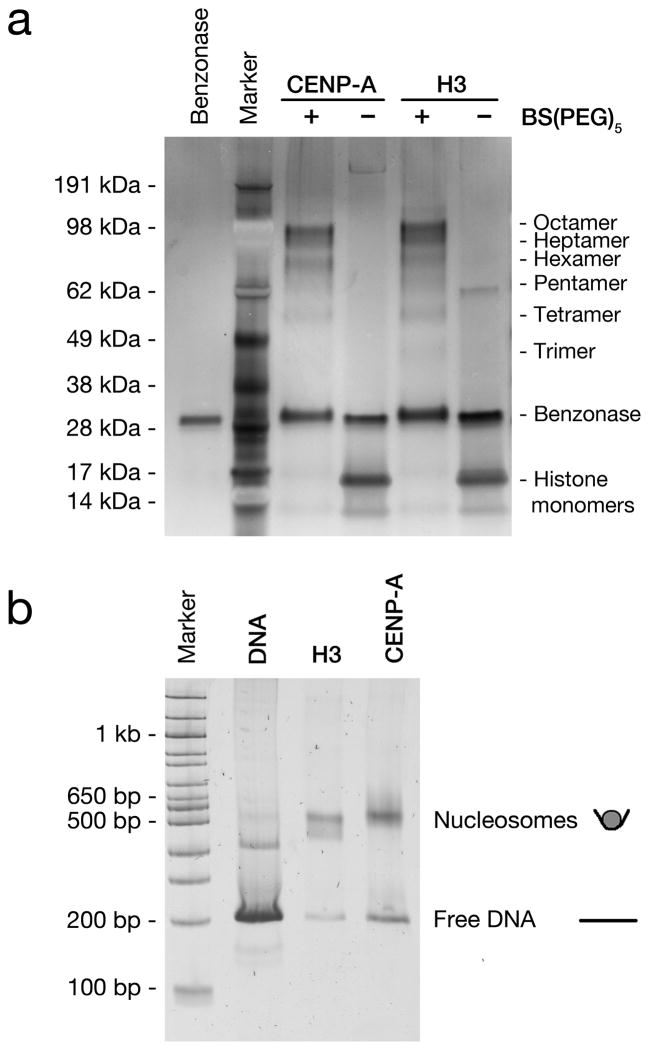
To confirm that the assembled CENP-A and H3 nucleosomes were octameric, we performed cross-linking and gel mobility assays. To approximate their molecular weight we exposed assembled nucleosome arrays to the BS(PEG)5 cross-linker at nanomolar nucleosome concentrations and analyzed them by SDS-PAGE (Fig. 1a, and Supplementary Fig. 1a). Both CENP-A and H3 chromatin yielded the expected molecular weights for octameric nucleosomes and intermediate complexes were observed that are consistent with the progressive fixation of individual histones from monomers up to octameric complexes. This demonstrated that both CENP-A and H3 assemble into similar octameric complexes in vitro. Moreover, increased concentrations of cross-linker did not lead to the formation of complexes with a higher molecular weight than octamers (Supplementary Fig. 1b). The absence of larger complexes indicates that the observed octamers did not result from the progressive cross-linking of tetramers. We also assessed the relative size of recombinant CENP-A and H3 particles by comparing their mobility. Assembled CENP-A and H3 nucleosome arrays were digested to monomers with AvaI, which cuts between each 601 repeat. Analyses by native PAGE showed that CENP-A and H3 nucleosomes have identical motilities (Fig. 1b and Supplementary Fig. 2). Thus, as previously observed, these in vitro assembled nucleosomes consisted of octamers of histones1,8,9. Moreover, mass spectrometry analyses of these same nucleosome bands, extracted from the native PAGE gel, revealed that each nucleosome type contained the full complement of expected histones (Supplementary Fig. 3). To assay the length of DNA wrapped around these H3 and CENP-A nucleosomes, we digested arrays with micrococcal nuclease (MNase) and determined the length of protected DNA by gel electrophoresis or Bioanalyzer measurements. Whilst both particle types protected discrete lengths of DNA with some variability, CENP-A nucleosomes protected approximately 20 bp less than H3 nucleosomes (Supplementary Fig. 4), which is consistent with other in vitro and in vivo analyses of CENP-A nucleosomes10 (B. Black personal communication).
Figure 1. In vitro assembled H3 and CENP-A nucleosomes behave as octamers.
(a) Silver stained SDS-PAGE gel of recombinant human nucleosomes following BS(PEG)5 fixation and digestion with Benzonase. (b) Sybr Green stained native PAGE of in vitro assembled human H3 and CENP-A nucleosome arrays, digested to mono-nucleosomes with AvaI.
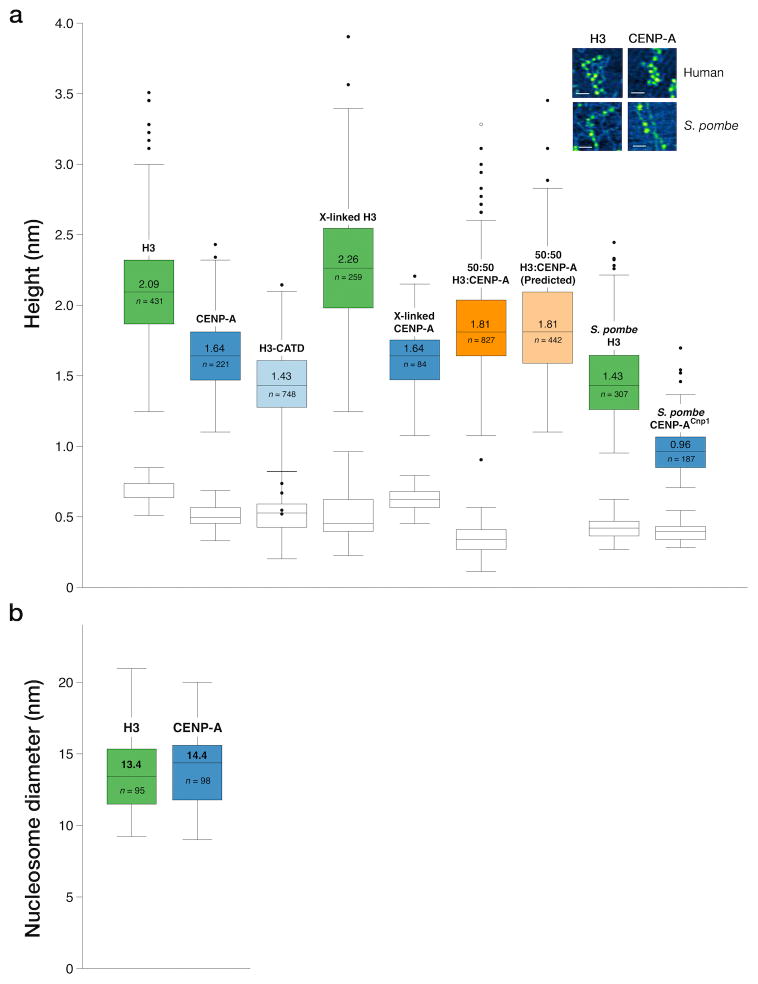
We used these same in vitro assembled nucleosome arrays to determine the height of octameric CENP-A and H3 particles using AFM. At least 180 individual particles of each type, that were clearly part of arrays, were measured. We found that CENP-A nucleosomes assembled in vitro from both human and S. pombe recombinant histones consistently measured lower in height than H3 nucleosomes (Fig. 2a). For human nucleosomes, CENP-A nucleosomes had a median height of 1.64 nm (S.E. ±0.02 nm), compared with 2.09 nm (±0.02 nm) for H3. S. pombe CENP-ACnp1 nucleosomes had a median height of 0.96 nm (±0.01 nm), compared with 1.43 nm (±0.02 nm) for H3 nucleosomes. These nucleosomal heights are less than those observed in crystal structures, however they are typical of AFM images collected in air due to a combination of sample compression and dehydration3,11. As the data is distributed non-parametrically (Shapiro-Wilk test W = 0.969, p = 6.383×10−8), we used two-sample Kolmogorov-Smirnov (KS) tests to compare CENP-A and H3 nucleosome height distributions. The recorded CENP-A particle heights were significantly lower than those of H3 particles; D = 0.5662, p < 2.2×10−16 and D = 0.7723, p < 2.2×10−16 for human and S. pombe CENP-A nucleosomes, respectively.
Figure 2. Octameric CENP-A nucleosomes are lower in height.
(a) Box plots of AFM peak heights for individual human and S. pombe H3 and CENP-ACnp1 nucleosomes, fixed nucleosomes and mixed nucleosomes. Naked DNA control for each image (white box plots). Inset (a): AFM image example, bar = 50 nm.
(b) Distribution of nucleosome diameters for 95 H3 and 98 CENP-A human particles. Box Plots: central lines with values = medians; box outer edges = first and third interquartile ranges; whiskers = range; outliers = single dots. n = particle number counted per sample.
One explanation for this height difference is that CENP-A nucleosomes might disassemble during preparation for AFM imaging, whilst H3 nucleosomes remain intact. However, we also measured H3 and CENP-A nucleosomes that were cross-linked with BS(PEG)5 prior to preparation for AFM (Fig. 2a). The level of crosslinking was equivalent to that where mainly octameric complexes for H3 and CENP-A nucleosomes were detected on denaturing gels (Fig. 1a). We found that height measurements of nucleosomes in these samples were very similar to that of uncross-linked material (Fig. 2a). Thus sample preparation does not account for the height difference observed between H3 and CENP-A nucleosomes. We also discounted the possibility that height differences between H3 and CENP-A nucleosomes are due to variable AFM imaging conditions. When we mixed H3 and CENP-A at an equal ratio, prior to imaging, the resulting distribution of observed nucleosome heights lay exactly as predicted for an equal mix of data randomly collected from the individual H3 and CENP-A nucleosomes samples (Fig. 2a). An additional explanation for the height difference is that CENP-A nucleosomes might deform more readily than H3 nucleosomes under the AFM tip. However, we found the diameter of both human CENP-A (14.4 nm S.D. ±2.5 nm, n = 98) and H3 (13.4 ±2.7 nm, n = 95) recombinant particles to be similar (Fig. 2b). To further investigate the observed height difference between CENP-A and H3 particles we assembled nucleosome arrays in vitro that contained a chimeric human H3 (H3CATD), including the CENP-A targeting domain (CATD) region from CENP-A. The CATD region consists of 22 amino acid substitutions from CENP-A that span the loop 1 and α2 helix and are sufficient to target chimeric H3CATD to centromeres12. AFM measurements of these in vitro assembled human H3CATD nucleosomes had a median height of 1.43 nm (S.E. ±0.01 nm), significantly less than the median height of 2.09 nm (±0.02 nm) recorded for H3 (Fig. 2a); KS test D = 0.7676, p < 2.2×10−16. Thus the CATD region is sufficient to account for the reduced height of human CENP-A nucleosomes. Interestingly, the CATD region is known to impart a rigid and compact nature to CENP-A:H4 tetramers in solution12. Whilst these features were not apparent in the CENP-A nucleosome crystal structure9, our data support the conclusion that the CATD region also confers distinct biophysical properties to octameric CENP-A nucleosomes that results in them having a reduced height measurement by AFM.
The in vitro assembled CENP-A nucleosomes utilized here migrated through native PAGE gels similarly to octameric H3 nucleosomes and cross-linked as octameric complexes (Fig. 1, Supplementary Fig. 1 and 2). However, when CENP-A and H3 nucleosome heights were compared using AFM, CENP-A particles registered a significantly lower height. This difference was apparent whether using independently produced human or S. pombe nucleosomal arrays. Previously, the observed difference in height between CENP-A and H3 nucleosomes in ex vivo arrays was considered to support the conclusion that CENP-A particles are hemisomal complexes. In contrast, our analyses demonstrate that AFM actually detects an intrinsic difference in the biophysical properties of octameric CENP-A nucleosomes that causes them to appear lower in height than their H3 counterparts when assembled as nucleosomal arrays in vitro. Moreover, the CATD region that confers specific biological properties to CENP-A is sufficient to account for this difference.
The shorter length of DNA protected by human CENP-A nucleosomes in vivo is similar to that of in vitro assembled CENP-A nucleosomes (Supplementary Fig. 4), indicating that both sources of CENP-A nucleosomes have similar properties13. This reduced protection most likely results from the slacker association of DNA at the entry and exit points of CENP-A nucleosomes due to the less extensive αN-helix9. Both the length of wrapped DNA and the integrity of the αN-helix of H3 have previously been observed to alter nucleosome height11,14. Our analyses show that the CATD domain also influences nucleosome height (Fig. 2a). It remains to be determined whether reduced DNA wrapping, or some other structural property conferred by the CATD region (for example increased rigidity) results in decreased particle height15.
The heights reported here for octameric in vitro assembled human nucleosome arrays are consistent with previously observed heights for CENP-A and H3 nucleosomes on arrays extracted from human cells3,5. Furthermore, most CENP-A residing in mononucleosomes extracted from Drosophila cells can be cross-linked as dimers, consistent with it forming mainly octameric particles16. Thus our analyses suggest that CENP-A nucleosomes extracted from human and S. pombe cells are also likely to be octameric, and that they are unlikely to be hemisomes as has been proposed3–5.
Online Methods
Nucleosome fixation with BS(PEG)5
Nucleosomes were dialyzed to a fixation buffer of 20 mM Hepes pH 7, 2 mM EDTA. Then the primary amine cross-linker BS(PEG)5 (Thermo Scientific) was added at the required molar excess (1000 – 5000 x for full fixation of the histone octamer). Samples were fixed for 2 h at 37 C with gentle shaking before addition of Tris pH 7 to a final concentration of 200 mM to quench the fixative. To check the extent of fixation, 15 pmol of nucleosomes were digested with 0.5 μl of Benzonase (Novagen) at RT for 10 minutes then boiled in SDS-PAGE loading buffer (Life Technologies), run on a 4–12% NuPAGE SDS-PAGE gel in MES buffer (Life Technologies) alongside an unfixed control sample and the gel stained with a silver staining kit (Life Technologies).
Ava1 digests of nucleosome arrays
Nucleosome arrays were digested with Ava1 (NEB) in buffer containing 50 mM KOAc, 20 mM Tris-OAc pH 7.9, 1 mM DTT, 0.1 mg mL−1 BSA, 0.5 mM MgCl2. Digests were left to proceed overnight at room temperature then run on 5% non-denaturing acrylamide gels (29:1 acrylamide:bis-acrylamide) in 0.5 x TBE (45 mM Tris base, 45 mM boric acid, 1 mM EDTA), with 0.5 x TBE as the running buffer and standard DNA-gel glycerol loading buffer.
Micrococcal nuclease digests of nucleosome arrays
Nucleosome arrays were digested on ice for 1 minute with micrococcal nuclease (Worthington Biochemical) in a buffer containing 10 mM Tris pH 8, 50 mM NaCl2, 1 mM CaCl2. Reactions were quenched with the addition of 0.5 volumes of 600 mM NaCl, 0.3% SDS, 30 mM EGTA and 10 mM Tris pH8. Proteins were digested by incubation with proteinase K (Fermentas) at 37 C, 15 minutes and the DNA was extracted with phenol:chloroform. DNA extracted from digested and undigested control samples of both H3 and CENP-A nucleosome arrays were run on 5% non-denaturing acrylamide gels (29:1 acrylamide:bis-acrylamide) in 0.5 x TBE (45 mM Tris base, 45 mM boric acid, 1 mM EDTA), with 0.5 x TBE as the running buffer and standard DNA-gel glycerol loading buffer. DNA was visualized by staining with SyBr Gold and scanned on a versadoc gel imager (Bio-Rad).
Analysis of nucleosome components by Mass spectrometry
Gel bands were excised, cut into small pieces and digested as described previously17. The resulting peptides were desalted using StageTips and analysed using a nanoLC (Dionex UltiMate 3000 system) coupled to a Q-Exactive mass spectrometer (Thermo Fisher Scientific)18. Full MS scans were acquired in the Orbitrap mass analyser over the range m/z 300–1750, and the ten most intense peaks were fragmented in the HCD collision cell. The MS data were analysed using Mascot version 2.2.0 (Matrix Science Ltd). MS/MS data were searched against the UniProtKB human database, containing 807454 protein sequences (released February 2013).
Preparation of samples and surfaces for Atomic force microscopy (AFM)
Freshly cleaved V1 grade Mica (SPI supplies) was functionalized with 3-aminopropyltriethoxysilane (APTES – Sigma) as described in Lyubchenko et al (2009)19. Samples were pipetted onto the functionalized surface at a titration of nucleosome concentrations centered on 0.1 nM. Deposited samples were left for 5 minutes to adhere at RT then rinsed twice with molecular biology grade water (Sigma). A stream of argon was used to gently dry the surfaces and they were imaged immediately. At least two biological replicates were imaged for each sample.
AFM imaging
AFM imaging was performed in air at minimal force in intermittent contact mode using either a Veeco Explorer or a Veeco Nanoman VS with a Dimension 3100 controller (Bruker). In our hands both machines gave images of comparable quality and the nucleosome height data collected was essentially identical from either machine. Images were collected over an area of between 1–5 μm at a typical scan rate of ~ 1.2 Hz. The DLC-10 probes used (Bruker) had a nominal resonance of 160 kHz, stiffness of 5 N m−1, and a nominal tip radius of 1 nm.
AFM image processing
AFM images were first leveled using the NanoScope Analysis software (Veeco) then exported for further analysis using ImageJ (NIH). The background height was subtracted from the image and a mask layer used to remove particles above 5 nm in height. All nucleosome-like particles that could be clearly distinguished as round “bead-on-a-string” particles were selected manually. Manual selection of nucleosomes was preferred as in our hands this was found to include fewer non-nucleosomal particles in the analysis than an automated, filter-based approach. Particles were classed as non-nucleosomal or excluded from analysis if they were located within regions compacted such that individual nucleosomes could not be easily distinguished, if they were potentially deposited on top of DNA or other particles or if the particle diameter was above 25 nm. The maximum height and diameter of selected particles was recorded from the original, background-subtracted image. The height of DNA was recorded from at least 10 points within each image to be used as an internal control of DNA height. The median height of DNA across all images was 0.49 nm (S.E. ±0.01 nm, n = 323), which is typical of dehydrated DNA under pressure from the AFM probe and absorbed on a surface4.
Supplementary Material
Acknowledgments
The authors acknowledge A. Kerr for statistical analysis and M. Barrios-Llerena for mass-spectrometry analyses. AFM was enabled by the Bioimaging Small Research Facility, School of Engineering, University of Edinburgh. The Wellcome Trust supported the work of R.C.A. [095021], [065061]; J.R [084229]; T.O-H. [095062] and the Edinburgh Protein Production Facility [081287]. Work in the Wellcome Trust Centre for Cell Biology is supported by the Wellcome Trust core funding [092076]. M.M. was supported by a Scottish University Life Sciences Alliance (SULSA) Prize studentship. A.F.S. was supported by the US National Institutes of Health (R01 GM074728), C.J.F. by the Stanford Lieberman Fellowship and A.G. by the German Research Foundation (D.F.G.). R.C.A. is a Wellcome Trust Principal Fellow.
Footnotes
Author contributions
M.M. jointly conceived the study with R.C.A.. M.M. produced the S. pombe nucleosome arrays with assistance from T.O-H. and performed the AFM experiments with assistance from A.D.. C.J.F. and A.G. produced the human nucleosome arrays with assistance from A.F.S.. A.G. and M.M. jointly performed AvaI digestion experiments, C.J.F. and M.M jointly performed MNase digestion experiments and H.M.B. and M.M jointly performed the fixation experiments. M.B-L. performed the mass spectrometry analysis with assistance from J.R.. A.K. performed statistical analysis on AFM data. M.M. prepared the manuscript, which was edited by A.F.S. and R.C.A.. All authors discussed the results and implications and commented on the manuscript at all stages.
References
- 1.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Voullaire LE, Slater HR, Petrovic V, Choo KH. Am J Hum Genet. 1993;52:1153–1163. [PMC free article] [PubMed] [Google Scholar]
- 3.Dimitriadis EK, Weber C, Gill RK, Diekmann S, Dalal Y. PNAS. 2010;107:20317–20322. doi: 10.1073/pnas.1009563107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dalal Y, Wang H, Lindsay S, Henikoff S. PLoS Biol. 2007;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bui M, et al. Cell. 2012;150:317–326. doi: 10.1016/j.cell.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luger K, Rechsteiner TJ, Richmond TJ. Methods Mol Biol. 1999;119:1–16. doi: 10.1385/1-59259-681-9:1. [DOI] [PubMed] [Google Scholar]
- 7.Huynh VAT, Robinson PJJ, Rhodes D. J Mol Biol. 2005;345:957–968. doi: 10.1016/j.jmb.2004.10.075. [DOI] [PubMed] [Google Scholar]
- 8.Camahort R, et al. Mol Cell. 2009;35:794–805. doi: 10.1016/j.molcel.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tachiwana H, et al. Nature. 2011;476:1–6. [Google Scholar]
- 10.Panchenko T, et al. PNAS. 2011;108:16588–16593. doi: 10.1073/pnas.1113621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bussiek M, Müller G, Waldeck W, Diekmann S, Langowski J. Eur Biophys J. 2007;37:81–93. doi: 10.1007/s00249-007-0166-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Black B, et al. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 13.Conde e Silva N, et al. J Mol Biol. 2007;370:555–573. doi: 10.1016/j.jmb.2007.04.064. [DOI] [PubMed] [Google Scholar]
- 14.Leuba SH, Bustamante C, Zlatanova J, van Holde K. Biophysj. 1998;74:2823–2829. doi: 10.1016/S0006-3495(98)77989-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Black B, Brock MA, Bédard S, Woods VL, Cleveland DW. PNAS. 2007;104:5008–5013. doi: 10.1073/pnas.0700390104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang W, Colmenares SU, Karpen GH. Mol Cell. 2012;45:263–269. doi: 10.1016/j.molcel.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. Nat Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 18.Rappsilber J, Ishihama Y, Mann M. Anal Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 19.Lyubchenko YL, Shlyakhtenko LS. Methods. 2009;47:206–213. doi: 10.1016/j.ymeth.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.