Abstract
Chronic lymphocytic leukemia (CLL) is the most common leukemia in the Western world and remains incurable with conventional chemotherapy treatment approaches. CLL as a disease entity is defined by a relatively parsimonious set of diagnostic criteria and therefore likely constitutes an umbrella term for multiple related illnesses. Of the enduring fundamental biological processes that affect the biology and clinical behavior of CLL, few are as central to the pathogenesis of CLL as recurrent acquired genomic copy number aberrations (aCNA) and recurrent gene mutations. Here, a state-of-the-art overview of the pathological anatomy of the CLL genome is presented, including detailed descriptions of the anatomy of aCNA and gene mutations. Data from SNP array profiling and large-scale sequencing of large CLL cohorts, as well as stimulated karyotyping, are discussed. This review is organized by discussions of the anatomy, underlying pathomechanisms and clinical significance of individual genomic lesions and recurrent gene mutations. Finally, gaps in knowledge regarding the biological and clinical effects of recurrent genomic aberrations or gene mutations on CLL are outlined to provide critical stimuli for future research.
Keywords: CLL, genomic copy number aberrations, gene mutations
INTRODUCTION
The improvements of technologies to accurately measure genomic changes in cancer cells and application of these to chronic lymphocytic leukemia (CLL) have resulted in detailed knowledge of the pathological anatomy of the CLL genome. Knowledge is most complete for acquired genomic copy number aberrations (aCNA) and gene mutations, and it is complemented with karyotypic information, including descriptions of balanced and unbalanced chromosomal translocations.
The knowledge base of aCNA and loss-of-heterozygosity (LOH) in CLL has been greatly expanded through the use of high-resolution single-nucleotide polymorphism (SNP) array-based genomic copy number assessments that have resulted in precise aCNA demarcations at the kilobase to sub-megabase resolution. The knowledge of recurrent gene mutations in CLL has been aided by the revolution in massively parallel sequencing. Gaps in knowledge exist with regard to the types and frequency of genomic changes outside of well-annotated genes, the biological effects of recurrent or rare acquired balanced chromosomal translocations on CLL cells, the stable effects of genomic aberrations on the CLL transcriptome, the mechanistic interplay between aCNA/LOH and gene mutations (and other critical CLL pathways) and the magnitude and importance of epigenetic changes in CLL.
Overall, the CLL genome is characterized by far fewer genomic events than many of the solid tumors and also by fewer events than other B-cell malignancies such as diffuse large B-cell lymphoma, mantle cell lymphoma (MCL) or follicular lymphoma. Within CLL cohorts, however, the range of genomic aberrations is wide, with ~20% of CLL lacking any aCNA (but such cases carry a few gene mutations), whereas others, albeit infrequently, carry >10 such events.
Decade-long research on karyotypic changes in CLL ultimately culminated in the development of a set of fluorescence in situ hybridization (FISH) probes and a CLL-FISH panel that has gained widespread clinical use in CLL. This is because subsets of patients with monoallelic loss of TP53, as detected by TP53-centric FISH probes (as a small part of large deletions on chromosome 17-pcommonly referred to as del17p), or ATM, as detected by ATM-centric FISH probes (as part of heterogeneous interstitial deletions on chromosome 11q- commonly referred to as del11q), when analyzed as cohorts, have shortened survival relative to comparator cohorts lacking these findings.1,2 CLL-FISH is therefore an important test in clinical CLL management and has resulted in the realization of risk-adapted therapies for small subsets of CLL patients, including early use of allogeneic transplantations.3–8
Research on CLL karyotypes has been revolutionized over the last few years through use of innovative CLL cell stimulants, allowing for successful generation of karyotypes for almost all cases. Results of such analyses are complementary to array-based CLL genomics and have identified a substantial subset of CLL (~20%) with complex aberrant karyotypes and short survival. Stimulated karyotypes have also allowed for the detection of sporadic, as well as recurrent, balanced and unbalanced chromosomal translocations, and the presence of such chromosomal translocations has subsequently been associated with shortened patient survival. Overall, however, it is clear that a dominant high-frequency CLL driver in the form of a recurrent balanced translocation, akin to the Philadelphia chromosome in CML or Ph + ALL, does not exist.
Within the last two years, results from large-scale sequencing studies in CLL have been published that in aggregate are notable for (1) the lack of a dominant high-frequency mutated driver gene in CLL;9,10 (2) a low gene mutation load per CLL case; (3) the lack of frequent mutations in the clinically important kinase gene families;11 (4) the identification of novel recurrently mutated genes in CLL, such NOTCH1, SF3B1 and others that, although infrequent, may result in the discovery of novel pathomechanisms in CLL; and, (v) associations of selected gene mutations with specific aCNA, suggesting cooperative effects on the afflicted CLL cell.
In the following paragraphs, a detailed discussion of aCNA, LOH, gene mutations and karyotypes in CLL is presented, and these lesions are reviewed from a biological, diagnostic, prognostic and therapeutic perspective.
ACQUIRED GENOMIC COPY NUMBER ABERRATIONS AND LOSS-OF-HETEROZYGOSITY (LOH) IN CLL
CLL genomic complexity
aCNA and LOH have been measured in paired DNA samples derived from purified CD19 + and CD3 + cells from 255 CLL patients using SNP 6.0 array-based profiling,12 as well as collectively from hundreds of patients (study sizes ranged from 70 to 369 patients) in DNA of varied purity and/or in tumor DNA only.13–17 In addition, measurements using array comparative genomic hybridization (CGH) platforms have been reported.18,19 From this data, a relatively complete view of CLL-associated genomic aCNA has emerged. Overall, CLL is characterized by a relatively stable genome, with the majority of cases displaying between 0–2 aCNA. LOH usually associates with copy loss in CLL, and copy-neutral LOH (cnLOH, also referred to as acquired uniparental disomy (aUPD)) is rare, but when it occurs, for example, in cnLOH-17p, it is of high clinical relevance.20,21
Of clinical importance is a subset of CLL with elevated aCNA counts. For instance, aCNA ≥2 is detected in ~35% of all CLL cases and aCNA ≥3 in ~20% of cases, and as such an elevated aCNA count (referred to as elevated genomic complexity) demarcates a CLL subset with progressive and aggressive disease and short survival.12,14,17,22 In comprehensive bivariate and multivariate analysis, elevated genomic complexity emerged as the dominant predictor of short overall survival (OS), identifying high-risk CLL cases in all CLL cohorts that were previously stratified by other markers. It is thus clear that the inability to maintain genomic stability associates strongly with an aggressive CLL phenotype.
Genomic complexity has also been identified and defined in large-scale CLL karyotyping studies (see below). Approximately 20% of CLL cases were found to carry ≥3 genomic aberrations, and the presence of complex karyotypes in CLL predicted for short OS.23 It is likely, albeit formally unproven, that both SNP array-based and karyotype-based CLL genome analysis identify largely overlapping subsets of patients, with the former, in addition, detecting cases with microdeletions or cnLOH and the latter detecting cases with chromosomal changes that do not result in genomic copy loss (that is, balanced chromosomal translocations, additions or isochromsomes).
With regard to the likely mechanisms of the strong negative prognostic effects of elevated genomic complexity in CLL, one relates to the association with TP53 mutations: most TP53-mutated CLL is genomically complex.12 However, approximately two-thirds of CLL with aCNA ≥2 are TP53 wild type, and despite intense research efforts it is currently unclear as to what molecular aberrations associate with or cause genomic complexity in this CLL subset.
The correlation of known measurable CLL traits with SNP array-based genomic complexity has identified the following traits as independently associated with elevated genomic complexity (in descending order of strength): del17p/TP53 mutations, del11q and large del13q14 inclusive of RB.24 Impaired ATM activation measured following external CLL cell irradiation weakly predicted elevated genomic complexity, but this effect could not be separated from the presence of del11q.24
With regard to the underlying reasons why genomically complex CLL is clinically aggressive, three complementary hypotheses are advanced: (i) frequent association with TP53 mutations/del17p and other defects that impair apoptotic cell responses to genotoxic therapies,12,24 (ii) ongoing clonal evolution, and (iii) associations with telomere attrition and associated genomic instability. The latter case would place genomic instability as a consequence of telomere dysfunction, which remains an attractive but not definitively proven concept.25–29 Furthermore, it remains unclear whether telomere dysfunction can result in interstitial deletions, the most frequent aCNA type in CLL. Ongoing research is directed at the identification of additional molecular aberrations, including gene mutations that are associated with elevated genomic complexity in CLL.
In the following paragraphs, individual high-frequency aCNAs are discussed in detail, followed by a brief summary of novel low-frequency but recurrent aCNAs, for which little detailed information is available at present.
Deletion 13q14
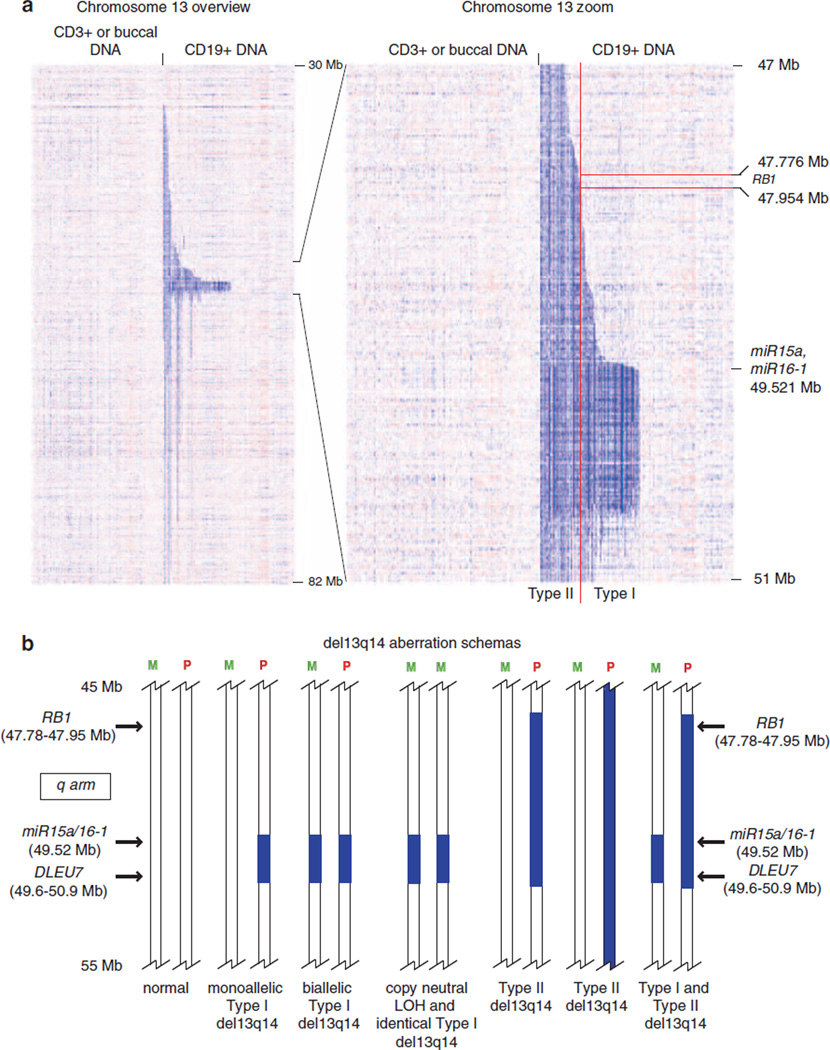
Interstitial deletions of various lengths located at 13q14 are present in ~50% of CLL.2,30–36 These deletions, commonly referred to as del13q14, display substantial anatomic heterogeneity, and a subset extends telomerically and centromerically for many megabases beyond an approximate anchor point at chromosomal physical position 50 Mb. Approximately 15–20% of CLL cases with del13q14 carry a del13q14 on both chromosomes: either the same lesion or lesions of different lengths. Largely as a result of high-resolution SNP array profiling, the exact anatomy and extent of various del13q14 have been defined.21 For illustration purposes, a published heatmap and a schema of del13q14, as detected through SNP 6.0 array profiling of 255 CLL cases, is shown in Figure 1.37 From these studies, a few overriding conclusions can be drawn, including the following: (i) that del13q14 comprises various lesions with likely divergent associated gene deregulations; (ii) previously identified very short lesions used to define a minimal deleted region inclusive of the miR15/16 locus were not identified in large CLL genomic profiling studies, and instead a relatively uniform lesion type (referred to as type I and present in ~60% of all CLL with del13q14) with a deletion length of ~0.8–1 Mb was identified; (iii) the vast majority (~98%) of del13q14 include the miR15/16 locus, as well as DLEU7, and all intervening genes;37–39 (iv) some type I del13q14 are biallelic and exist surrounded by large stretches of (many Mb in length) cnLOH, a lesion type that is otherwise rare in CLL; (v) multiple discrete anatomic del13q14 lesion clusters as defined by centromeric break points can be identified, suggesting divergent but largely unknown biological or clinical consequences.
Figure 1.
Genomic copy number heatmap displays of chromosome 13q of 255 CLL cases ranked by the position of centromeric 13q14 deletion break points. (a) Copy number heatmap displays for paired DNA samples based on SNP 6.0 array profiling were generated using dChipSNP. Left panel: CD3 + or buccal DNA; right panel: CLL CD19 + DNA. Blue indicates copy loss. Each column represents one patient. (b) Schemas of various del13q14 in CLL: M, maternal chromosome; P, paternal chromosome. The approximate locations of selected genes (miR15a/16.1, DLEU7 and RB) are indicated. Deletions are indicated by blue bars.
The high frequency of del13q14 in CLL suggests a driver status for this lesion type in CLL. Importantly, however, del13q14 lesions that are indistinguishable from CLL-associated del13q14 have been identified in other cancers, such as MM, diffuse large B-cell lymphoma, AML, prostate cancer and others, and therefore are unlikely to confer CLL-specific biological traits on CLL cells. Furthermore, the frequency of FISH-detectable del13q14 in CD5 + CLL-like monoclonal B-cell lymphocytosis is 50%, but only 1% of these monoclonal B-cell lymphocytosis cases convert to CLL every year, clearly indicating that del13q14 alone is insufficient for the generation of CLL in humans.40,41
Despite decades of research, a complete understanding of gene deregulations as part of various del13q14 deletions has not emerged, and none of the genes located within type I 13q14 deletions are recurrently somatically mutated. Nonetheless, important progress has been made in elucidating del13q14 biology. One of the contributing genetic elements to del13q14 is the miR15a/16.1 locus.42 Evidence for a role of these miRs in CLL pathogenesis stems from (i) their almost invariate inclusion in all del13q14 lesions;21 (ii) occasional biallelic deletion status, suggestive of a tumor suppressor gene function (albeit with the caveat that in humans a second locus on chromosome 3 at ~chromosomal position 160.122 Mb encodes for the highly related miRs 15b and 16.2 with the potential for compensation of del13q14-resident miR loss; (http://www.mirbase.org/index.shtml); (iii) effects of low miR15a/16.1 levels on anti-apoptotic and cell cycle regulatory molecules that are consistent with a role in CLL pathobiology and cancer in general; (iv) occasional epigenetic miR15a/16.1 downregulation in CLL without del13q14;43 and (v) evidence from spontaneously occurring or artificially engineered mice that develop CLL-like illnesses upon ablation of the mouse homologues of hsa-miR15a/16.1.44–46
Some of the experimental evidence suggesting a less fundamental and possibly just contributory role of 13q14-resident miRs to CLL pathogenesis are (i) that miR expression (in particular miR16.1) levels are not substantially affected by the majority of 13q14 deletions that affect just one chromosome,37 (ii) the lack of recurrent miR15a/16.1 mutations and (iii) the low frequency of CLL-like illnesses that occur in mice engineered to carry miR15a/16.1 deletions alone as opposed to a higher frequency in mice with larger engineered deletions similar to the ones that actually occur in humans.
Recently, interest in del13q14 has shifted to the gene DLEU7, which is expressed at subnormal levels in CLL.47 Effects of DLEU7 on pro-survival cell signaling pathways have been demonstrated in heterologous cell systems, and inherited CNVs spanning this gene have been identified in a family with multiple affected CLL members.48,49 It is therefore possible that ablation of miR15a/16.1 together with DLEU7 and possibly even other genes codetermines 13q14 biology. This research area constitutes work in progress.
Of additional noteworthiness is the fact that an ad-hoc summary analysis of recent large-scale sequencing studies detailed below has not identified a frequently somatically mutated gene located within any of the various 13q14 deletions. It is therefore likely that selected genes located within del13q14 are further inactivated through epigenetic regulation or that selected genes display haploinsufficiency.
With regard to the clinical importance of the presence of del13q14 as detected through FISH in CLL, a few considerations are important: (i) the clinically used CLL FISH panel cannot differentiate del13q14 subtypes, as the probe used is located within all 13q14 deletions; (ii) a monoallelic versus biallelic del13q14 status carries no independent prognostic significance; (iii) the expression levels of the del13q14-resident miR15a/16.1 carry no prognostic significance; (iv) the association of a del13q14 of any type with additional FISH abnormalities indicates a more aggressive disease course than the presence of a sole del13q14; (v) large del13q14 (type II and present in ~40% of all CLL with del13q14) that are inclusive of the Rb gene (approximate physical location at 48 Mb) demarcate a subset of CLL with del13q14 that is clinically more aggressive. In support of this finding, an association of del13q14 type II deletion with elevated genomic lesion load (genomic complexity) has been detected.37–39,50–52 It is thus likely that inclusion of a FISH probe centered on Rb1 will further refine genomic CLL risk stratification.
Deletion 17p
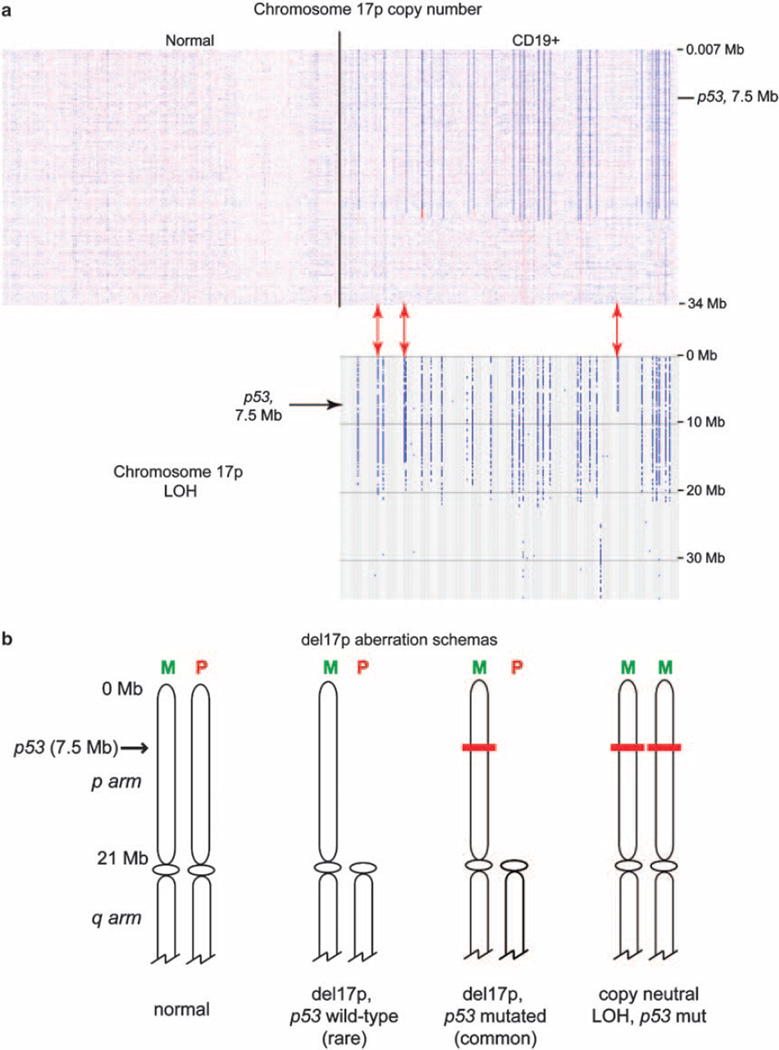
Terminal deletions of the short arm of chromosome 17, commonly referred to as 17p deletions or del17p, are detected by FISH in ~7% of newly diagnosed CLL.2,53 These deletions are almost always large, measure ~18–22 Mb in length, and are the consequence of various structural changes (deletions, unbalanced translocations and isochromosomes). To assist the reader with the visualization of del17p, LOH and cnLOH at 17p in CLL, a copy number heatmap and LOH analysis and a schema of del17p as detected through SNP 6.0 array profiling are displayed in Figure 2. Del17p invariably results in the deletion of one TP53 allele with the second retained TP53 allele reported mutated in 64–100% of cases in various studies.12,20,54–56 Importantly, these studies differ in various technical aspects of TP53 mutation analysis and differ substantially in the frequency of mutations in TP53 exons 4, 9 and 10. Analysis of large unselected CLL cohorts (193, 255 and 400 CLL patients are described in Harferlach et al.,23 Malcikova et al.54 and Ouillette et al.12) identified between 0–6% of del17p CLL without TP53 mutations. It reflects the opinion of the author that the vast majority of CLL with del17p carry TP53 mutations on the retained allele. Small 17p deletions that are centered on TP53 have been described as well, albeit at much lower frequencies than the canonical del17p. Most del17p are present in the majority of the CLL clone as assessed by FISH, indicating a strong competitive advantage for growth or survival.
Figure 2.
(a) Genomic copy number heatmap displays of chromosome 17p of 255 CLL cases: copy number heatmap displays for paired DNA samples based on SNP 6.0 array profiling were generated using dChipSNP. Left panel: CD3 + or buccal DNA; right panel: CLL CD19 + DNA. Blue indicates copy loss, red indicates copy gain. Each column represents one patient. Lower panel: 17p-LOH; red arrows indicate cases with cnLOH-17p. (b) Schemas of del17p in relation to TP53 mutations and LOH at 17p in CLL: M, maternal chromosome; P, paternal chromosome. The approximate location of TP53 is indicated. Red bars indicate mutated TP53.
Almost all research on del17p has been focused on the association with TP53 mutations (see section below). Here the following facts are of clinical importance: (i) all del17p are detectable by the clinically used CLL FISH panel that includes a probe that is centered on the TP53 gene;57 (ii) some 17p deletions convert to cnLOH at 17p (cnLOH-17p; unbiased frequencies of aUPD-17p in untreated or relapsed/ refractory CLL patients are not available; three out of 255 such patients were identified in the published cohort by Ouillette et al.12) that were undetectable by FISH, and as these cases are always associated with homozygous TP53 mutations they constitute one subset of aggressive CLL that may display falsely reassuring FISH findings;20 (iii) sporadic, non-del17p-associated or monoallelic TP53 mutations exist (reported frequencies in large CLL cohorts range from 1–5% and frequencies as high as 18% have been reported in highly selected fludarabine-refractory CLL) independently of del17p (but of unclear relation to cnLOH-17p), and these are also associated with aggressive disease;54–56,58,59 (iv) the incidence of del17p increases with time from diagnosis and in particular with prior therapies for CLL; (v) presence of del17p is particularly deleterious in the setting of genotoxic therapy (purine analogs and alkylators) for CLL, as these therapies rely on functional TP53 protein for apoptosis induction;60 (vi) presence of del17p serves as a basis for risk-adapted therapies in CLL, increasingly involving non-genotoxic therapy approaches and reduced-intensity allogeneic stem cell transplantation as consolidation;3,5,6 (vii) presence of del17p alone should not be used as an indication for initiation of therapy; and (viii) CLL with del17p is clinically heterogeneous and if associated with mutated IgVH loci or early Rai stage may be clinically stable for years.61,62 For instance, patients with CLL and del17p that were further analyzed for IgVH-status and Rai stage demonstrated 3-year OS of 96, 74 and 22% with ≤1, 2 or 3 such factors present.61
Somewhat unclear at present are the exact mechanisms contributing to the observed clinical aggressiveness of CLL with del17p. Although it is likely that apoptosis resistance and therefore lack of complete cell kill underlies del17p effects, it is also likely that del17p and associated TP53 inactivation creates a permissive environment for persistence of new and additional genomic changes, and that some of these changes result in more aggressive CLL subclones. The analysis of clonal evolution in CLL and effects on clinical outcome are a research opportunity in CLL.1,63–65
Given the anatomic size of 17p deletions in CLL, it is likely that the full biological impact of this lesion type on CLL cells involves additional gene deregulations. However, very little additional information is available. Genome-wide analysis of gene mutations in CLL did not identify a frequently mutated gene other than TP53 that is located within del17p. With regard to del17p-associated transcriptome deregulation, unpublished data have identified hundreds of statistically deregulated genes, complicating further in-depth analysis. Associations of del17p with aberrant expression of microRNAs have been reported, and in particular, aberrantly high expression of miR-21 has been proposed to further increase the odds of short survival in CLL.66 This is an area of future research opportunities.
Deletion 11q
Interstitial loss of chromosomal material located on chromosome 11q, commonly referred to as del11q, is present at diagnosis in ~10% of CLL patients. Most 11q deletions in CLL are large and measure many Mb to tens of Mb in length.67–69 All 11q deletions that include the ATM gene are classical 11q deletions and all involve one chromosome only. Homozygous 11q deletions do not exist, but rare cnLOH at 11q has been described. Atypical small 11q deletions located closer to the centromere with a minimal deleted region distinct from classical 11q deletions have been identified as well, but nothing is known about their biology or clinical relevance.12,14
Despite the substantial size heterogeneity and length of 11q deletions in CLL, much of the research on del11q has focused on ATM. Mutations in ATM (see section below) in the retained allele have been identified in a minority of 11q-deleted CLL at various frequencies, with the published literature not clearly separating somatically acquired from germline mutations, or even SNPs. In addition, mutations in BIRC3 have recently been identified, albeit only in a small subset of CLL that had relapsed quickly following fludarabine-based therapy and not in de novo CLL.70 At present, data from large-scale CLL gene sequencing studies do not support the existence of other frequently mutated 11q-resident genes.
Given the absence of high-frequency mutated genes in CLL that are located within the boundaries of del11q, the question arises as to what drives clonal selection of CLL cells with del11q? A second question that may or may not be related to the first centers on the almost complete association of del11q status with unmutated IgVH status (defined as ≥98% homology to germline), an association that remains fully unexplained at present.
Recently, aberrantly high insulin receptor (INSR) expression was identified in 60–70% of CLL with del11q.69 Insulin stimulation of CLL with high INSR expression provides anti-apoptotic and pro-growth stimuli to CLL cells, and high INSR expression associates with early CLL disease progression. Currently, it is assumed that one or a few genes located within 11q deletions are negative regulators of INSR expression and that monoallelic deletions of these genes, together with unidentified defects, result in aberrant INSR upregulation. Experiments to identify such INSR regulators are being conducted and should result in greater mechanistic clarity regarding the regulation of INSR expression in CLL.
Summarizing the best available data on the pathophysiology of del11q in CLL, it appears that defects in DNA-double-strand (ds) break and repair responses contribute to the phenotype of CLL with del11q, and such defects are due to (i) occasional ATM mutations; (ii) non-mutational ATM dysfunction/hypoactivation owing to unidentified mechanisms; (iii) compound gene deletions involving other genes that are involved in the DNA-ds break and repair responses (for instance, Mre11a and H2AX together with ATM);24 and (iv) unidentified 11q-associated mechanisms.71 In addition, pro-survival and pro-growth pathways are activated in CLL cells with del11q, either through aberrant overexpression of the INSR or TCL1, providing credible mechanisms for clonal selective pressure that enriches for the presence of del11q.72 Finally, the strong association of del11q with elevated genomic complexity in CLL may underlie clonal evolution/diversification in CLL, which is followed ultimately by outgrowth or regrowth of more aggressive clonal variants. It is likely that future research into 11q biology will uncover additional molecular aberrations that are important to 11q subsets.
With regard to the clinical implications of the presence of del11q in CLL, the following conclusions can be supported by current evidence: (i) essentially all CLL with del11q is progressive: long-term disease stability without the need for initial therapy does not exist at appreciable frequencies; (ii) remission durations after frontline therapy for CLL with del11q are shorter than for patients without del11q (and absence of del17p), although recent improvements following the inclusion of cyclophosphamide into the treatment regimens have been noted;73 (iii) the OS of CLL patient cohorts with del11q is modestly shorter than that for CLL without del11q (and absence of del17p);2,53 and (iv) CLL with del11q can be associated with disproportionate increases in internal lymph node sizes, a phenomenon that may be related to aberrant high INSR expression.74
Trisomy 12
A gain of the entire chromosome 12 (referred to as trisomy 12) is present in ~15–18% of CLL at diagnosis. A few additional CLL cases carry partial gains of chromosome 12, but few, if any, of these gains are recurrent. Despite the substantial fraction of CLL that carries trisomy 12, very little concrete information is available on the molecular mechanisms that are underlying this chromosomal aberration and their possible effects on CLL cells, and patients with trisomy 12 are the least studied of all FISH-defined CLL subsets.
Nonetheless, two recent interesting observations have been reported: (i) the frequent (50%) association of NOTCH1 exon 34 mutations with trisomy 12 (compared with a frequency of NOTCH1 exon 34 mutations in unselected de novo CLL of ~4–6%);75–78 and (ii) the heightened dependence of CLL cells with trisomy 12 on hedgehog pathway activation.79 The latter findings, although still somewhat preliminary, resulted from ex vivo studies of inhibitors of the hedgehog pathway in CLL and findings of elevated levels of Desert hedgehog (DHH) and Gli1 in patients with trisomy 12.
Clinically, an isolated trisomy 12 carries no adverse prognostic information. Importantly, patients with isolated trisomy 12 treated with modern chemoimmunotherapy approaches are almost always sensitive to therapy (possibly because of a strong negative association of trisomy 12 with TP53 mutations or elevated genomic complexity) and often enjoy above-median remission durations.12,80
Recurrent aCNAs in CLL with frequencies of 1–5%
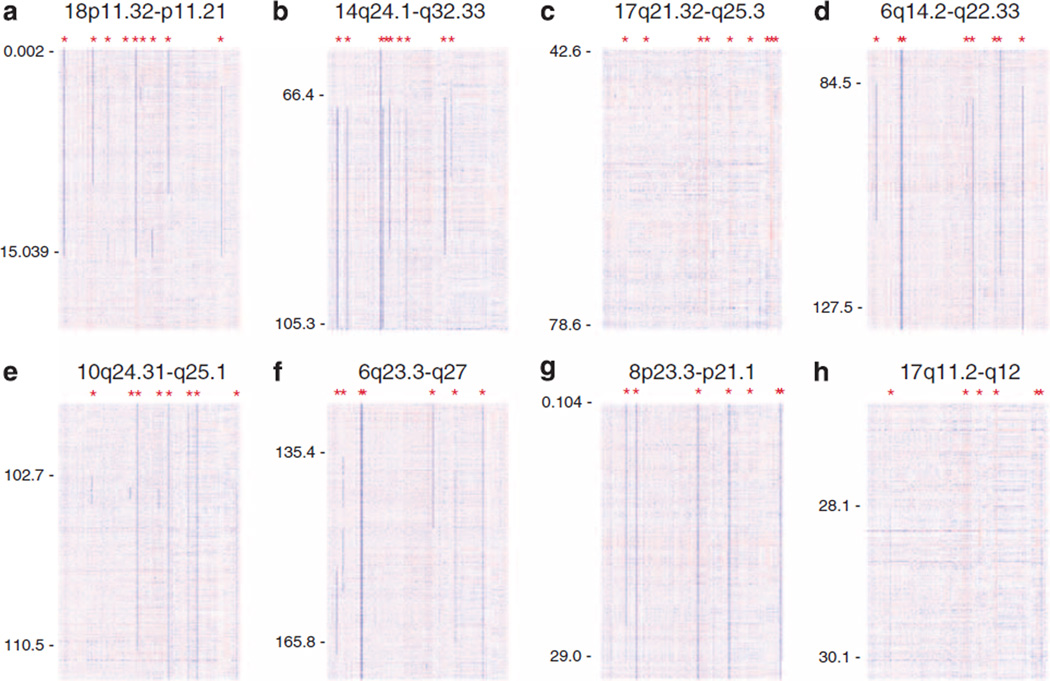
Various aCNAs with frequencies of 1–5% have been identified in CLL, largely as a result of high-resolution SNP array-based genomic assessments. Complicating a complete description of the frequency and identity of these second tier aCNAs are various differences in methodologies used for detection and at times lack of analysis of paired normal DNA. The aCNAs as described by Ouillette et al.,12 which are based on direct comparisons of paired DNA samples from 255 CLL patients isolated from FACS-sorted cells, have been tabulated in Table 1 and have been graphically summarized in Figure 3. In addition, recurrent low-frequency aCNAs can be found in various recent publications.13,15,17,38 Some of these include gains at 3q26 and 8q24 and losses of varying length at 8p. Very little firm knowledge is available about any of these rare recurrent aCNAs. In addition, aCNAs listed in Table 1 often co-occur in the setting of other aCNAs or TP53 mutations, thus complicating reductionistic research approaches and conclusions.
Table 1.
Acquired genomic copy number aberrations detected in 255 CLL cases using SNP 6.0 profiling. MAR: minimally altered regions (inclusive of gains and losses). Based on highly purified DNA from sorted CD19 + and CD3 + cells of 255 CLL cases as described in Ouillette et al. (2011)12
| Chromosome cytoband |
Incidence | MAR start rsID# |
MAR start physical position |
MAR stop rsID# |
MAR stop physical position |
MAR length (Mb) |
Predominant Lesion Type |
|---|---|---|---|---|---|---|---|
| 13q13.2–q31.1 | 51.37% | 2740536 | 49.503 | 201778 | 49.928 | 0.425 | Loss |
| 11q13.5–q23.3 | 10.12% | 10789636 | 107.077 | 4754348 | 108.087 | 1.01 | Loss |
| 17p13.3–p11.1 | 9.80% | 6565733 | 0.007 | 2472708 | 18.858 | 18.851 | Loss |
| 18p11.32–p11.21 | 3.92% | 646699 | 2.657 | 4797120 | 3.715 | 1.058 | Loss > gain |
| 14q24.1–q32.33 | 3.53% | 1954119 | 68.841 | 1955619 | 79.853 | 11.012 | Loss |
| 17q21.32–q25.3 | 3.53% | MAR1: 8077375 | MAR1: 42.691 | MAR1: 16949418 | MAR1: 43.346 | 0.655 | Gain |
| MAR2: 1486751 | MAR2: 47.695 | MAR2: 7226166 | MAR2: 50.795 | 3.1 | Gain > loss | ||
| MAR3: 9944529 | MAR3: 70.793 | MAR3: 8067774 | MAR3: 71.991 | 1.198 | Gain > loss | ||
| MAR4: 4789911 | MAR4: 74.554 | MAR4: 12452661 | MAR4: 74.852 | 0.298 | Gain > loss | ||
| 6q14.2–q22.33 | 3.14% | 1474620 | 97.752 | 10782162 | 109.913 | 12.161 | Loss |
| 10q24.31–q25.1 | 3.14% | 2296586 | 104.236 | 17113122 | 104.596 | 0.36 | Loss |
| 6q23.3–q27 | 2.75% | 11966730 | 138.104 | 17078283 | 148.739 | 10.635 | Loss |
| 8p23.3–p21.1 | 2.75% | 13278518 | 18.508 | 17468779 | 18.836 | 0.328 | Loss |
| 17q11.2–q12 | 2.35% | 28921 | 28.29 | 16968252 | 28.717 | 0.427 | Gain > Loss |
| 18q21.33–q23 | 2.35% | 2732226 | 71.744 | 4891158 | 72.18 | 0.436 | Gain > Loss |
| 8q24.21 | 1.96% | 7824785 | 128.184 | 16902149 | 128.476 | 0.292 | Gain |
| 13q31.1 | 1.96% | 4143260 | 84.332 | 9555564 | 85.938 | 1.606 | Loss > gain |
| 17q12 | 1.96% | 2411182 | 32.134 | 12449696 | 32.526 | 0.392 | Gain > loss |
| 17q21.31 | 1.96% | 4793124 | 40.003 | 2664008 | 41.239 | 1.236 | Gain > loss |
| 19p13.3 | 1.96% | 8100066 | 0.212 | 10420193 | 3.478 | 3.266 | Loss > gain |
| 20p13 | 1.96% | 6081944 | 2.027 | 7272003 | 4.261 | 2.234 | Loss |
| 20p12.3–p12.1 | 1.96% | 3897510 | 6.077 | 201928 | 17.473 | 11.396 | Loss |
| Xp11.4–p11.3 | 1.96% | 5963946 | 41.042 | 17214797 | 42.773 | 1.731 | Loss |
Figure 3.
(a–h) Genomic copy number heatmap display of eight distinct recurrent aCNAs based on 255 CLL cases: Blue indicates copy loss, red indicates copy gain. Asterisks indicate an aCNA. Based on highly purified DNA from sorted CD19 + and CD3 + cells as described in Ouillette et al. (2011).12
With regard to the clinical importance of rare but recurrent aCNA, the existing data have to be interpreted with caution, as none are based on large enough CLL cohorts to allow for multivariate analyses and assessments of the independence of effects. Although it is plausible that individual lesions directly affect CLL clinical progression or even chemotherapy resistance, no direct evidence is available to confirm such a hypothesis.
RECURRENTLY MUTATED GENES IN CLL
At present, large-scale sequencing data on > 200 CLL cases have been published.9–11,81,82 Overall it appears that CLL-associated gene mutations are relatively infrequent and mostly private. One of the important findings from this data in aggregate is the fact that a high-frequency mutated driver gene in CLL has not been identified and likely does not exist. However, CLL subsets that carry recurrent gene mutations in the 1–10% range exist, and knowledge is evolving with regards to the biological and clinical implications of these findings.
Proposals have been advanced to categorize CLL-associated mutations based on gene functions into pathways. Complicating such synthetic approaches is the lack of detailed studies that measure effects of mutations of genes on afflicted CLL pathways. Nonetheless, in addition to the well-known mutations affecting the DNA-ds break response and repair pathway (see below), mutations recurrently affecting innate immune system molecules and RNA splicing have been identified.
In the following paragraphs, selected, recurrently mutated genes in CLL and their biological and clinical effects are discussed in detail. This research area is advancing rapidly and the reader is referred to ongoing research findings for up-to-date information.
TP53 mutations
Somatically acquired mutations in TP53 are detectable using direct sequencing of PCR amplicons that are templated on genomic DNA in ~10% of newly diagnosed CLL.54,83,84 The frequency of CLL-associated TP53 mutations in community-based assessments, however, may be substantially less than 10%.85 Functional assays for detection of TP53 mutations have been described as well.20,71 In CLL, the TP53 mutation frequency increases slightly with disease duration and is higher in the relapse setting, following the use of genotoxic therapy. Most TP53 mutations in CLL are missense mutations targeting the DNA-binding domain and most mutations occur in TP53 exons 5–9. A few additional mutations occur in TP53 exon 4 and rarely in exon 10.20,56,58,59 Most missense-mutated TP53 proteins accumulate to high levels in the CLL cells, the pathophysiological relevance of which is unknown. A subset of TP53 mutations are frameshift mutations because of either indels, nonsense or splice-site mutations, resulting in severely truncated TP53 proteins. It is currently assumed that these are null mutations.
CLL cells with TP53 mutations are completely resistant to radiation-induced apoptotic cell death, demonstrating the central and absolute requirement for functional TP53 protein in the DNA-ds break-induced apoptotic responses.24,86 Unclear at present is what other TP53 functions are important contributors to the severe clinical phenotype inflicted by TP53 mutants on CLL cells. It is furthermore unclear if some of the CLL-associated TP53 mutants have additional gain-of-function phenotypes and what the exact phenotype is.
Clinically, CLL with TP53 mutations is aggressive.87,88 The OS of patients receiving conventional therapies when assessed from the time of detection of a TP53 mutation is measured in years, but substantial interpatient variability exists.89,90 Most of the negative survival impact conferred by TP53 mutations appears owing to incomplete responses and short response durations to standard therapies, with a subset of patients displaying frank therapy resistance. The latter phenomenon is clinically catastrophic and not well understood in molecular terms, in particular given that the spectrum of clinical responses in TP53-mutated CLL can range from CR (albeit rare) to progressive disease.
CLL with TP53 mutations is currently treated with a variety of risk-adapted therapies and provides the best-studied indication for use of reduced-intensity allogeneic stem cell transplantation (RIC-allo-TX) as consolidation.91,92
ATM mutations
The frequency of somatically acquired mutations in ATM in unselected CLL cohorts is unknown.93–95 If extrapolations from large-scale CLL sequencing studies are used, and assuming that all ATM exons were adequately covered, it is substantially less than 10%. Within the context of del11q, which invariably results in the removal of one ATM allele, the ATM mutation frequency estimates range from 8–30%.96,97 ATM mutations comprise missense, nonsense and frameshift mutations, the latter type suggestive of gene product inactivation. Little information on the function of ATM proteins carrying single amino-acid substitutions is available and it is unclear what fraction of these constitutes complete or partial loss-of-function mutations.
The analysis of ATM protein levels and radiation-induced ATM autophosphorylation in a large CLL cohort of 250 cases has further qualified the incidence of ATM aberrations in CLL.24,97 From this data, it has become clear that ATM aberrations are infrequent and of modest effect on CLL clinical outcome. Although remission durations of ATM-aberrant CLL are shorter than for various comparator cohorts, effects of ATM aberrations on OS were not detected, suggesting effective salvage of ATM-aberrant CLL in the relapse setting.
Interestingly, while various cell types with ATM inactivation demonstrate a radiation-sensitivity phenotype in clonogeneic plating assays, ATM-aberrant CLL cells are neither resistant nor supersensitive to radiation-induced apoptosis (unpublished observation on the basis of the comparative quantitative analysis of radiation-induced CLL apoptosis and radiation-induced ATM autophosphorylation in purified cells from 210 CLL patients with wild-type TP53). This is clearly very distinct from the findings in TP53-mutated CLL. Even though ATM functions upstream of TP53 in the DNA-ds break repair and response pathway, a defect in ATM, phenotypically or clinically, does not equal a defect in TP53, suggesting redundant TP53 activation pathways within the CLL cell.
With regard to the clinical importance of ATM mutations in CLL, the existing literature is conflicted.96,98–100 Most importantly, effects of ATM mutations and effects of del11q are not separated in published work, and germ-line mutations in ATM are not clearly distinguished from somatically acquired mutations. Finally, prospective assessments of the effect of ATM mutations on CLL survival in well-defined CLL cohorts using multivariate analysis are not available.
NOTCH1 mutations
The application of massively parallel sequencing to CLL genomes and exomes, and candidate gene resequencing has resulted in the identification of novel recurrent mutations in NOTCH1 in CLL.81,82 These mutations cluster in the very large exon 34, disrupt the PEST domain, and have the net effect of stabilization of the NOTCH1 protein. The most commonly identified mutation is a recurrent dinucleotide deletion (NOTCH1 c.7541_7542het_delCT). Very little biological information of the effect of stabilized NOTCH1 protein on afflicted CLL cells is available, but it is likely that mutated-NOTCH1 protein is still ligand-dependent for full pathway activation and not autonomously activated.
The frequency of NOTCH1 mutations in large unselected and untreated CLL validation cohorts is ~6%, with higher estimates derived from the original discovery cohorts (mixtures of untreated and relapsed CLL) and higher frequencies in relapsed CLL. It is presently unclear but under active investigation at what frequency NOTCH1 mutations are acquired during CLL disease evolution. Interestingly, NOTCH1 mutations are not equally distributed across all CLL but are highly enriched for CLL cases that are ZAP70 + and IgVH unmutated, therefore displaying strong associations with proliferative and progressive CLL. No information is available that would mechanistically explain this association. In addition, ~50% of CLL with NOTCH1 mutations also carry trisomy 12, a frequency substantially higher than expected by chance; this again suggests cooperation between these genomic aberrations, but specific information is unavailable.75,101
With regard to the clinical implications of NOTCH1 mutations in CLL, the existing evidence is conflicted.78 One report, based on large patient numbers and multivariate modeling, suggested a very adverse effect on survival by NOTCH1 mutations and equated the negative prognostic impact of the presence of NOTCH1 mutations with the presence of TP53 mutations.102 Data from an independent large CLL cohort employing comprehensive multivariate analyses did not detect such effects.75 It is thus clear that only carefully conducted research on very large, prospectively collected CLL cohorts can provide definitive answers. In the meantime, various data are somewhat at odds with the postulated strong negative prognostic effects of NOTCH1 mutations in CLL, including: (i) the strong association with trisomy 12, which usually does not identify aggressive CLL; and, (ii) the lack of associated high-risk genomic features like del17p, del11q, elevated genomic complexity or TP53 mutations.
Of additional interest is the reported elevated frequency of CLL transformation to large cell lymphoma (Richter’s transformation) in CLL with NOTCH1 mutations.81,82 Here again, prospectively analyzed large CLL cohorts are needed to prove this hypothesis. If confirmed, CLL with NOTCH1 mutations may be eligible for novel intervention trials aimed at prevention of such lethal transformation events.
Future work in CLL carrying stabilizing NOTCH1 mutations will focus on testing the suitability of mutated NOTCH1 as a target for therapy. Such targeted therapy is contingent upon the demonstration that CLL cells rely on NOTCH1 for either survival or proliferation or alternatively, that NOTCH1 mutations confer chemotherapy resistance on afflicted CLL cells.
SF3B1 mutations
Massively parallel sequencing of CLL exomes and candidate gene resequencing approaches resulted in the unexpected finding of novel missense mutations in the splicing cofactor SF3B1.9,10,103 Mutation frequency estimates based on the original discovery cohorts combined are ~10%, whereas estimates based on large-scale validation studies are closer to 6% (unpublished). Associations with fludarabine resistance, as well as the presence of del11q, have been reported but are in need of validation, especially given the fact that many biomarkers in CLL are enriched in relapsed/refractory CLL without causal link to therapy resistance.
Mutations in SF3B1 are mono-allelic missense mutations involving recurrently affected codons. Given that SF3B1 is involved in mRNA 3′-splice-site recognition, follow-up studies using RNAseq to detect aberrant mRNA species in cells with SF3B1 mutations were performed. Indeed, some mRNA species demonstrated aberrant intron retention.10 It is, however, unclear at present whether aberrant mRNA splicing is indeed the critical pathomechanistic effect of SF3B1 mutations and if so, what critical mRNA are affected.
With regard to the clinical impact of SF3B1 mutations in CLL, the data are sparse. In other hematological malignancies which also carry SF3B1 mutations, effects on prognosis are usually favorable, providing a framework from which to view the effect of SF3B1 mutations in CLL. Large-scale CLL studies are needed to advance our understanding in this area.
Other recurrently mutated genes in CLL
A number of genes have been found to be recurrently mutated in CLL, albeit at low and largely unknown frequencies in validation cohorts, and the data in this research area are still actively evolving. The best current information can be found in two published large CLL exome sequencing studies and large-scale sequence analysis of kinase genes.9–11,104 Largely unknown at present is the functional consequence of the known mutations on affected genes, unless genes are mutated through frameshift or nonsense changes, which usually indicate loss of function. A few recurrently mutated genes not discussed above are briefly mentioned here:
Activating BRAF mutations occur in 2% of CLL and may lead to the clinical use of BRAF inhibitors for selected cases.11
Mutations in MYD88, similar to the mutations described in NHL, have been identified in CLL and appear over-represented in CLL with del13q14. MYD88 is involved in TLR signaling and the CLL/NHL-associated mutations are activating and result in NFκB activation.9,81,105
XPO1/Exportin mutations (a protein involved in nuclear protein export) have been identified in a few percent of CLL patients.81
FBXW7 (a ubiquitin ligase) mutations have been found in sporadic CLL cases and may provide an alternative, albeit infrequent, mechanism for NOTCH1 activation.
POT1 (a member of the shelterin complex) mutations have been detected in CLL and may affect telomere maintenance.10
LRP1B (a cell surface receptor) mutations occur in CLL and may affect a generic cancer-relevant pathway, as this gene is one of the most frequently altered genes in all cancer.10
Sporadic inactivating RB1 mutations have been identified in the setting of large 13q14 deletions, resulting in a small CLL subset that is Rb null.37
THE LANDSCAPE OF aCNA AND CHROMOSOMAL TRANSLOCATIONS IN CLL AS DEFINED THROUGH CLL CELL KARYOTYPING
CLL karyotyping using basic cell culture conditions has been performed for decades and has resulted in valuable information on gross structural aberrations.106–109 The major limitations of this tool are the inability to generate karyotypes for the majority of patients, as well as the unreliable detection of smaller genomic lesions (sub-megabase to 5 Mb range). Over the last few years, novel CLL cell culture conditions have been established, which now allow for the generation of so-called ‘stimulated karyotypes’ in the vast majority of patients110 and results from karyotyping efforts of large CLL patient collections have been reported.111–114 The largest study of CLL genomics based on stimulated karyotypes by Haferlach et al.,23 very nicely summarizes CLL genomics on the basis of karyotyping of 500 CLL; therefore only a few pertinent points are highlighted in the following bullets:23
Eighty-three percent of analyzed CLL cases carried an aCNA; the median was 1.7 aCNA per case; 2 or more aCNA were detected in ~44% of cases, whereas 3 or more aCNA were detected in ~21%.
Reciprocal translocations, which are understudied in CLL, were detected in ~20% of cases. Of these, translocations involving the Ig loci (heavy- and light-chain loci), as well as cytoband 13q14 were the most frequent. In addition, studies on translocations involving cytoband 14q32 identified BCL2 t(14;18)(q32q21) and BCL3 t(14;19)(q32;q13) as recurrent translocation partners; t(14;19)(q32;q13)/(Ig-BCL3) may be associated with more aggressive CLL, as well as atypical CLL presentations.115–118
As expected, karyotyping detected more aCNA than the commonly used CLL-FISH test
aCNAs as detected by karyotyping in CLL with normal FISH are prognostically adverse.119,120
A complex aberrant karyotype identifies a high-risk subgroup of CLL with short survival; the overlap of CLL cases with a complex aberrant karyotype and presence of either del17p or del11q is partial, and,
The mean number of aCNAs in TP53-mutated CLL was five, as compared with a mean of 1.5 in the TP53 wild-type group.
Some of the caveats regarding the routine use of stimulated karyotypes in CLL management include the following: (i) prospective evaluations of the prognostic value of stimulated karyotyping results in CLL cohorts have not been reported; (ii) as expected, even stimulated karyotyping misses smaller genomic lesions; for instance, short del13q14 as detected through CLL FISH is often not detected and therefore CLL-FISH results are still necessary for proper genomic assessments; (iii) interlaboratory differences in karyotyping results exist; and (iv) differences in the detection rates (frequencies) and types of abnormal karyotypes exist, which is dependent on what specific culture conditions are used.
FUTURE PERSPECTIVES
Despite substantial advances in our knowledge about the biological and clinical implications of aCNA and recurrent gene mutations in CLL, many questions remain. To assist the reader with understanding the limits of current knowledge, as well as with the planning and design of future experimental approaches aimed at improving the understanding of CLL biology and clinical behavior, selected questions have been tabulated in Table 2.
Table 2.
Selected questions in CLL genomics
| A comprehensive molecular understanding of determinants of genomic instability and complexity in CLL. |
| The relative contributions of individual acquired genomic copy number aberrations and recurrent gene mutations on (i) drug-mediated CLL cell kill versus resistance; (ii) in vivo CLL growth kinetics; (iii) CLL clonal diversification; and, (iv) CLL patient survival. |
| The frequency of all acquired genomic copy number aberrations and recurrent gene mutations in CLL at diagnosis, with focus on minor subclones and possible effects of therapy on clonal selection.53,63,65 |
| The direct mutagenic effects of CLL therapy: does CLL therapy cause genomic aberrations, or alternatively, facilitate acquisition of acquired genomic copy number aberrations or gene mutations in CLL through clonal selection? |
| A complete characterization of acquired genomic copy number aberrations and recurrent gene mutations in relapsed CLL, including CLL with short remission durations following chemoimmunotherapy. |
| The molecular aberrations associated with del17p other than TP53 mutations that influence CLL prognosis. Understanding why CLL with TP53 mutations/del17p and mutated IgVH status are clinically less aggressive. |
| The molecular aberrations associated with del11q subtypes that influence CLL prognosis. |
| The critical genes and pathways deregulated or altered as part of various 13q14 deletions and del13q14 subtypes. |
| The genomic aberrations associated with (i) CD5 + monoclonal B-lymphocytosis (MBL) and (ii) with progression (if any) of MBL to CLL. |
| The effects of second-tier acquired genomic copy number aberrations on CLL biology and clinical outcome: which second-tier acquired genomic copy number aberrations in CLL directly affect CLL prognosis? |
| The molecular pathways and networks that are stably perturbed by recurrent acquired genomic copy number aberrations, gene mutations or both in CLL subsets.69,121 |
| The identification of therapies that are particularly effective in CLL genomic subsets or CLL subsets identified by specific recurrent gene mutations. |
| The landscape and importance of epigenetic aberrations in CLL.122 |
| The effects of NOTCH1 mutations on CLL biology and patient outcome. |
| The effects of SF3B1 mutations on CLL biology and patient outcome. |
| The effects of individual balanced chromosomal translocations on CLL biology and outcome. |
| Understanding the clinical importance of acquired genomic copy number aberrations and recurrent gene mutations in the era of targeted therapies.123–125 For instance, how do CLL patients with elevated genomic complexity, TP53 mutations, del17p or del11q respond to treatment with novel small molecules (inhibitors of BTK, SYK or PI3Kdelta)? |
Summarizing briefly the important deficiencies in current knowledge as outlined in Table 2, these center on (1) a more complete molecular characterization of the effects of aCNA on the biology of afflicted CLL cells, including the effects on defined signal transduction pathways and the interplay with pathways known to be central to CLL biology (for instance, the B-cell receptor complex signaling pathway); (ii) an understanding of the effects of recurrent gene mutations on CLL subset biology; (iii) the role of stable transcriptome changes in CLL subsets and the role and effects of epigenetic gene modulation; and (iv) the clinical importance and prognostic effects of rare aCNA and gene mutations—a question that can only be addressed in large-scale CLL collaborations possibly involving thousands of patients.
Given the central importance of aCNA and gene mutations in CLL, a deeper understanding of their mechanistic functions will inform better prognostication and better therapy selection and will ultimately result in further improvements in the outcome for patients with CLL.
ACKNOWLEDGEMENTS
This research is supported by the National Institutes of Health through R01 CA136537-01 (SM), the Translational Research Program of the Leukemia and Lymphoma Society of America (SM), the Scholars in Clinical Research Program of the Leukemia and Lymphoma Society of America (SM) and a CLL collaborative grant from the Lymphoma Research Foundation. This research is supported (in part) by the National Institutes of Health through the University of Michigan’s Cancer Center Support Grant (5 P30 CA46592) and Oncology Research Training Grant (T32 CA 009357-30). I am grateful for services provided by the microarray core of the University of Michigan Comprehensive Cancer Center.
CONFLICT OF INTEREST
Dr Malek’s work has been funded by the NIH, the Leukemia and Lymphoma Society of America and the Lymphoma Research Foundation. He has received honoraria from Roche and Teva.
Footnotes
Author contributions: Sami N Malek wrote the paper.
REFERENCES
- 1.Shanafelt TD, Hanson C, Dewald GW, Witzig TE, LaPlant B, Abrahamzon J, et al. Karyotype evolution on fluorescent in situ hybridization analysis is associated with short survival in patients with chronic lymphocytic leukemia and is related to CD49d expression. J Clin Oncol. 2008;26:e5–e6. doi: 10.1200/JCO.2008.16.7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 3.Pettitt AR, Jackson R, Carruthers S, Dodd J, Dodd S, Oates M, et al. Alemtuzumab in combination with methylprednisolone is a highly effective induction regimen for patients with chronic lymphocytic leukemia and deletion of TP53: Final results of the National Cancer Research Institute CLL206 Trial. J Clin Oncol. 2012;30:1647–1655. doi: 10.1200/JCO.2011.35.9695. [DOI] [PubMed] [Google Scholar]
- 4.Spaner DE. Oral high-dose glucocorticoids and ofatumumab in fludarabine-resistant chronic lymphocytic leukemia. Leukemia. 2012;26:1144–1145. doi: 10.1038/leu.2011.329. [DOI] [PubMed] [Google Scholar]
- 5.Stilgenbauer S, Zenz T, Winkler D, Buhler A, Schlenk RF, Groner S, et al. Subcutaneous alemtuzumab in fludarabine-refractory chronic lymphocytic leukemia: clinical results and prognostic marker analyses from the CLL2H study of the German Chronic Lymphocytic Leukemia Study Group. J Clin Oncol. 2009;27:3994–4001. doi: 10.1200/JCO.2008.21.1128. [DOI] [PubMed] [Google Scholar]
- 6.Castro JE, James DF, Sandoval-Sus JD, Jain S, Bole J, Rassenti L, et al. Rituximab in combination with high-dose methylprednisolone for the treatment of chronic lymphocytic leukemia. Leukemia. 2009;23:1779–1789. doi: 10.1038/leu.2009.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parikh SA, Keating MJ, O’Brien S, Wang X, Ferrajoli A, Faderl S, et al. Frontline chemoimmunotherapy with fludarabine, cyclophosphamide, alemtuzumab, and rituximab for high-risk chronic lymphocytic leukemia. Blood. 2011;118:2062–2068. doi: 10.1182/blood-2011-01-329177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dreger P, Dohner H, Ritgen M, Bottcher S, Busch R, Dietrich S, et al. Allogeneic stem cell transplantation provides durable disease control in poor-risk chronic lymphocytic leukemia: long-term clinical and MRD results of the German CLL Study Group CLL3× trial. Blood. 2010;116:2438–2447. doi: 10.1182/blood-2010-03-275420. [DOI] [PubMed] [Google Scholar]
- 9.Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011;365:2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quesada V, Conde L, Villamor N, Ordonez GR, Jares P, Bassaganyas L, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2011;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, Reis M, Khoriaty R, Li Y, Ouillette P, Samayoa J, et al. Sequence analysis of 515 kinase genes in chronic lymphocytic leukemia. Leukemia. 2011;25:1908–1910. doi: 10.1038/leu.2011.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ouillette P, Collins R, Shakhan S, Li J, Peres E, Kujawski L, et al. Acquired genomic copy number aberrations and survival in chronic lymphocytic leukemia. Blood. 2011;118:3051–3061. doi: 10.1182/blood-2010-12-327858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gunnarsson R, Mansouri L, Isaksson A, Goransson H, Cahill N, Jansson M, et al. Array-based genomic screening at diagnosis and during follow-up in chronic lymphocytic leukemia. Haematologica. 2011;96:1161–1169. doi: 10.3324/haematol.2010.039768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gunnarsson R, Isaksson A, Mansouri M, Goransson H, Jansson M, Cahill N, et al. Large but not small copy-number alterations correlate to high-risk genomic aberrations and survival in chronic lymphocytic leukemia: a high-resolution genomic screening of newly diagnosed patients. Leukemia. 2010;24:211–215. doi: 10.1038/leu.2009.187. [DOI] [PubMed] [Google Scholar]
- 15.Lehmann S, Ogawa S, Raynaud SD, Sanada M, Nannya Y, Ticchioni M, et al. Molecular allelokaryotyping of early-stage, untreated chronic lymphocytic leukemia. Cancer. 2008;112:1296–1305. doi: 10.1002/cncr.23270. [DOI] [PubMed] [Google Scholar]
- 16.Pfeifer D, Pantic M, Skatulla I, Rawluk J, Kreutz C, Martens UM, et al. Genome-wide analysis of DNA copy number changes and LOH in CLL using high-density SNP arrays. Blood. 2007;109:1202–1210. doi: 10.1182/blood-2006-07-034256. [DOI] [PubMed] [Google Scholar]
- 17.Brown JR, Hanna M, Tesar B, Werner L, Pochet N, Asara JM, et al. Integrative genomic analysis implicates gain of PIK3CA at 3q26 and MYC at 8q24 in chronic lymphocytic leukemia. Clin Cancer Res. 2012;18:3791–3802. doi: 10.1158/1078-0432.CCR-11-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grubor V, Krasnitz A, Troge JE, Meth JL, Lakshmi B, Kendall JT, et al. Novel genomic alterations and clonal evolution in chronic lymphocytic leukemia revealed by representational oligonucleotide microarray analysis (ROMA) Blood. 2009;113:1294–1303. doi: 10.1182/blood-2008-05-158865. [DOI] [PubMed] [Google Scholar]
- 19.Kay NE, Eckel-Passow JE, Braggio E, Vanwier S, Shanafelt TD, Van Dyke DL, et al. Progressive but previously untreated CLL patients with greater array CGH complexity exhibit a less durable response to chemoimmunotherapy. Cancer Genet Cytogenet. 2010;203:161–168. doi: 10.1016/j.cancergencyto.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saddler C, Ouillette P, Kujawski L, Shangary S, Talpaz M, Kaminski M, et al. Comprehensive biomarker and genomic analysis identifies p53 status as the major determinant of response to MDM2 inhibitors in chronic lymphocytic leukemia. Blood. 2008;111:1584–1593. doi: 10.1182/blood-2007-09-112698. [DOI] [PubMed] [Google Scholar]
- 21.Ouillette P, Erba H, Kujawski L, Kaminski M, Shedden K, Malek SN. Integrated genomic profiling of chronic lymphocytic leukemia identifies subtypes of deletion 13q14. Cancer Res. 2008;68:1012–1021. doi: 10.1158/0008-5472.CAN-07-3105. [DOI] [PubMed] [Google Scholar]
- 22.Kujawski L, Ouillette P, Erba H, Saddler C, Jakubowiak A, Kaminski M, et al. Genomic complexity identifies patients with aggressive chronic lymphocytic leukemia. Blood. 2008;112:1993–2003. doi: 10.1182/blood-2007-07-099432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haferlach C, Dicker F, Schnittger S, Kern W, Haferlach T. Comprehensive genetic characterization of CLL: a study on 506 cases analysed with chromosome banding analysis, interphase FISH, IgV(H) status and immunophenotyping. Leukemia. 2007;21:2442–2451. doi: 10.1038/sj.leu.2404935. [DOI] [PubMed] [Google Scholar]
- 24.Ouillette P, Fossum S, Parkin B, Ding L, Bockenstedt P, Al-Zoubi A, et al. Aggressive chronic lymphocytic leukemia with elevated genomic complexity is associated with multiple gene defects in the response to DNA double-strand breaks. Clin Cancer Res. 2010;16:835–847. doi: 10.1158/1078-0432.CCR-09-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Britt-Compton B, Lin TT, Ahmed G, Weston V, Jones RE, Fegan C, et al. Extreme telomere erosion in ATM-mutated and 11q-deleted CLL patients is independent of disease stage. Leukemia. 2012;26:826–830. doi: 10.1038/leu.2011.281. [DOI] [PubMed] [Google Scholar]
- 26.Augereau A, T’Kint de Roodenbeke C, Simonet T, Bauwens S, Horard B, Callanan M, et al. Telomeric damage in early stage of chronic lymphocytic leukemia correlates with shelterin dysregulation. Blood. 2011;118:1316–1322. doi: 10.1182/blood-2010-07-295774. [DOI] [PubMed] [Google Scholar]
- 27.Lin TT, Letsolo BT, Jones RE, Rowson J, Pratt G, Hewamana S, et al. Telomere dysfunction and fusion during the progression of chronic lymphocytic leukemia: evidence for a telomere crisis. Blood. 2010;116:1899–1907. doi: 10.1182/blood-2010-02-272104. [DOI] [PubMed] [Google Scholar]
- 28.Brugat T, Nguyen-Khac F, Grelier A, Merle-Beral H, Delic J. Telomere dysfunction-induced foci arise with the onset of telomeric deletions and complex chromosomal aberrations in resistant chronic lymphocytic leukemia cells. Blood. 2010;116:239–249. doi: 10.1182/blood-2009-12-257618. [DOI] [PubMed] [Google Scholar]
- 29.Roos G, Krober A, Grabowski P, Kienle D, Buhler A, Dohner H, et al. Short telomeres are associated with genetic complexity, high-risk genomic aberrations, and short survival in chronic lymphocytic leukemia. Blood. 2008;111:2246–2252. doi: 10.1182/blood-2007-05-092759. [DOI] [PubMed] [Google Scholar]
- 30.Bullrich F, Veronese ML, Kitada S, Jurlander J, Caligiuri MA, Reed JC, et al. Minimal region of loss at 13q14 in B-cell chronic lymphocytic leukemia. Blood. 1996;88:3109–3115. [PubMed] [Google Scholar]
- 31.Liu Y, Hermanson M, Grander D, Merup M, Wu X, Heyman M, et al. 13q deletions in lymphoid malignancies. Blood. 1995;86:1911–1915. [PubMed] [Google Scholar]
- 32.Kalachikov S, Migliazza A, Cayanis E, Fracchiolla NS, Bonaldo MF, Lawton L, et al. Cloning and gene mapping of the chromosome 13q14 region deleted in chronic lymphocytic leukemia. Genomics. 1997;42:369–377. doi: 10.1006/geno.1997.4747. [DOI] [PubMed] [Google Scholar]
- 33.Kitamura E, Su G, Sossey-Alaoui K, Malaj E, Lewis J, Pan HQ, et al. A transcription map of the minimally deleted region from 13q14 in B-cell chronic lymphocytic leukemia as defined by large scale sequencing of the 650 kb critical region. Oncogene. 2000;19:5772–5780. doi: 10.1038/sj.onc.1203978. [DOI] [PubMed] [Google Scholar]
- 34.Kapanadze B, Makeeva N, Corcoran M, Jareborg N, Hammarsund M, Baranova A, et al. Comparative sequence analysis of a region on human chromosome 13q14, frequently deleted in B-cell chronic lymphocytic leukemia, and its homologous region on mouse chromosome 14. Genomics. 2000;70:327–334. doi: 10.1006/geno.2000.6386. [DOI] [PubMed] [Google Scholar]
- 35.Migliazza A, Bosch F, Komatsu H, Cayanis E, Martinotti S, Toniato E, et al. Nucleotide sequence, transcription map, mutation analysis of the 13q14 chromosomal region deleted in B-cell chronic lymphocytic leukemia. Blood. 2001;97:2098–2104. doi: 10.1182/blood.v97.7.2098. [DOI] [PubMed] [Google Scholar]
- 36.Mabuchi H, Fujii H, Calin G, Alder H, Negrini M, Rassenti L, et al. Cloning and characterization of CLLD6, CLLD7, and CLLD8, novel candidate genes for leukemogenesis at chromosome 13q14, a region commonly deleted in B-cell chronic lymphocytic leukemia. Cancer Res. 2001;61:2870–2877. [PubMed] [Google Scholar]
- 37.Ouillette P, Collins R, Shakhan S, Li J, Li C, Shedden K, et al. The prognostic significance of various 13q14 deletions in chronic lymphocytic leukemia. Clin Cancer Res. 2011;17:6778–6790. doi: 10.1158/1078-0432.CCR-11-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mosca L, Fabris S, Lionetti M, Todoerti K, Agnelli L, Morabito F, et al. Integrative genomics analyses reveal molecularly distinct subgroups of B-cell chronic lymphocytic leukemia patients with 13q14 deletion. Clin Cancer Res. 2010;16:5641–5653. doi: 10.1158/1078-0432.CCR-10-0151. [DOI] [PubMed] [Google Scholar]
- 39.Parker H, Rose-Zerilli MJ, Parker A, Chaplin T, Wade R, Gardiner A, et al. 13q deletion anatomy and disease progression in patients with chronic lymphocytic leukemia. Leukemia. 2011;25:489–497. doi: 10.1038/leu.2010.288. [DOI] [PubMed] [Google Scholar]
- 40.Fazi C, Scarfo L, Pecciarini L, Cottini F, Dagklis A, Janus A, et al. General population low-count CLL-like MBL persists over time without clinical progression, although carrying the same cytogenetic abnormalities of CLL. Blood. 2011;118:6618–6625. doi: 10.1182/blood-2011-05-357251. [DOI] [PubMed] [Google Scholar]
- 41.Lanasa MC, Allgood SD, Slager SL, Dave SS, Love C, Marti GE, et al. Immunophenotypic and gene expression analysis of monoclonal B-cell lymphocytosis shows biologic characteristics associated with good prognosis CLL. Leukemia. 2011;25:1459–1466. doi: 10.1038/leu.2011.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sampath D, Liu C, Vasan K, Sulda M, Puduvalli VK, Wierda WG, et al. Histone deacetylases mediate the silencing of miR-15a, miR-16 and miR-29b in chronic lymphocytic leukemia. Blood. 2012;119:1162–1172. doi: 10.1182/blood-2011-05-351510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T, et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer cell. 2010;17:28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 45.Lia M, Carette A, Tang H, Shen Q, Mo T, Bhagat G, et al. Functional dissection of the chromosome 13q14 tumor-suppressor locus using transgenic mouse lines. Blood. 2012;119:2981–2990. doi: 10.1182/blood-2011-09-381814. [DOI] [PubMed] [Google Scholar]
- 46.Raveche ES, Salerno E, Scaglione BJ, Manohar V, Abbasi F, Lin YC, et al. Abnormal microRNA-16 locus with synteny to human 13q14 linked to CLL in NZB mice. Blood. 2007;109:5079–5086. doi: 10.1182/blood-2007-02-071225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hammarsund M, Corcoran MM, Wilson W, Zhu C, Einhorn S, Sangfelt O, et al. Characterization of a novel B-CLL candidate gene--DLEU7--located in the 13q14 tumor suppressor locus. FEBS Lett. 2004;556:75–80. doi: 10.1016/s0014-5793(03)01371-1. [DOI] [PubMed] [Google Scholar]
- 48.Brown JR, Hanna M, Tesar B, Pochet N, Vartanov A, Fernandes SM, et al. Germline copy number variation associated with Mendelian inheritance of CLL in two families. Leukemia. 2012;26:1710–1713. doi: 10.1038/leu.2012.33. [DOI] [PubMed] [Google Scholar]
- 49.Palamarchuk A, Efanov A, Nazaryan N, Santanam U, Alder H, Rassenti L, et al. 13q14 deletions in CLL involve cooperating tumor suppressors. Blood. 2010;115:3916–3922. doi: 10.1182/blood-2009-10-249367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dal Bo M, Rossi FM, Rossi D, Deambrogi C, Bertoni F, Del Giudice I, et al. 13q14 deletion size and number of deleted cells both influence prognosis in chronic lymphocytic leukemia. Genes Chromosomes Cancer. 2011;50:633–643. doi: 10.1002/gcc.20885. [DOI] [PubMed] [Google Scholar]
- 51.Liu Y, Szekely L, Grander D, Soderhall S, Juliusson G, Gahrton G, et al. Chronic lymphocytic leukemia cells with allelic deletions at 13q14 commonly have one intact RB1 gene: evidence for a role of an adjacent locus. Proc Natl Acad Sci USA. 1993;90:8697–8701. doi: 10.1073/pnas.90.18.8697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stilgenbauer S, Dohner H, Bulgay-Morschel M, Weitz S, Bentz M, Lichter P. High frequency of monoallelic retinoblastoma gene deletion in B-cell chronic lymphoid leukemia shown by interphase cytogenetics. Blood. 1993;81:2118–2124. [PubMed] [Google Scholar]
- 53.Shanafelt TD, Witzig TE, Fink SR, Jenkins RB, Paternoster SF, Smoley SA, et al. Prospective evaluation of clonal evolution during long-term follow-up of patients with untreated early-stage chronic lymphocytic leukemia. J Clin Oncol. 2006;24:4634–4641. doi: 10.1200/JCO.2006.06.9492. [DOI] [PubMed] [Google Scholar]
- 54.Malcikova J, Smardova J, Rocnova L, Tichy B, Kuglik P, Vranova V, et al. Monoallelic and biallelic inactivation of TP53 gene in chronic lymphocytic leukemia: selection, impact on survival and response to DNA damage. Blood. 2009;114:5307–5314. doi: 10.1182/blood-2009-07-234708. [DOI] [PubMed] [Google Scholar]
- 55.Zenz T, Habe S, Denzel T, Mohr J, Winkler D, Buhler A, et al. Detailed analysis of p53 pathway defects in fludarabine-refractory chronic lymphocytic leukemia (CLL): dissecting the contribution of 17p deletion, TP53 mutation, p53-p21 dysfunction, and miR34a in a prospective clinical trial. Blood. 2009;114:2589–2597. doi: 10.1182/blood-2009-05-224071. [DOI] [PubMed] [Google Scholar]
- 56.Rossi D, Cerri M, Deambrogi C, Sozzi E, Cresta S, Rasi S, et al. The prognostic value of TP53 mutations in chronic lymphocytic leukemia is independent of Del17p13: implications for overall survival and chemorefractoriness. Clin Cancer Res. 2009;15:995–1004. doi: 10.1158/1078-0432.CCR-08-1630. [DOI] [PubMed] [Google Scholar]
- 57.Dohner H, Fischer K, Bentz M, Hansen K, Benner A, Cabot G, et al. p53 gene deletion predicts for poor survival and non-response to therapy with purine analogs in chronic B-cell leukemias. Blood. 1995;85:1580–1589. [PubMed] [Google Scholar]
- 58.Zenz T, Krober A, Scherer K, Habe S, Buhler A, Benner A, et al. Monoallelic TP53 inactivation is associated with poor prognosis in chronic lymphocytic leukemia: results from a detailed genetic characterization with long-term follow-up. Blood. 2008;112:3322–3329. doi: 10.1182/blood-2008-04-154070. [DOI] [PubMed] [Google Scholar]
- 59.Dicker F, Herholz H, Schnittger S, Nakao A, Patten N, Wu L, et al. The detection of TP53 mutations in chronic lymphocytic leukemia independently predicts rapid disease progression and is highly correlated with a complex aberrant karyotype. Leukemia. 2009;23:117–124. doi: 10.1038/leu.2008.274. [DOI] [PubMed] [Google Scholar]
- 60.Rosenwald A, Chuang EY, Davis RE, Wiestner A, Alizadeh AA, Arthur DC, et al. Fludarabine treatment of patients with chronic lymphocytic leukemia induces a p53-dependent gene expression response. Blood. 2004;104:1428–1434. doi: 10.1182/blood-2003-09-3236. [DOI] [PubMed] [Google Scholar]
- 61.Tam CS, Shanafelt TD, Wierda WG, Abruzzo LV, Van Dyke DL, O’Brien S, et al. De novo deletion 17p13.1 chronic lymphocytic leukemia shows significant clinical heterogeneity: the M. D. Anderson and Mayo Clinic experience. Blood. 2009;114:957–964. doi: 10.1182/blood-2009-03-210591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Best OG, Gardiner AC, Davis ZA, Tracy I, Ibbotson RE, Majid A, et al. A subset of Binet stage A CLL patients with TP53 abnormalities and mutated IGHV genes have stable disease. Leukemia. 2009;23:212–214. doi: 10.1038/leu.2008.260. [DOI] [PubMed] [Google Scholar]
- 63.Knight SJ, Yau C, Clifford R, Timbs AT, Sadighi Akha E, Dreau HM, et al. Quantification of subclonal distributions of recurrent genomic aberrations in paired pre-treatment and relapse samples from patients with B-cell chronic lymphocytic leukemia. Leukemia. 2012;26:1564–1575. doi: 10.1038/leu.2012.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Braggio E, Kay NE, Vanwier S, Tschumper RC, Smoley S, Eckel-Passow JE, et al. Longitudinal genome-wide analysis of patients with chronic lymphocytic leukemia reveals complex evolution of clonal architecture at disease progression and at the time of relapse. Leukemia. 2012;26:1698–1701. doi: 10.1038/leu.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stilgenbauer S, Sander S, Bullinger L, Benner A, Leupolt E, Winkler D, et al. Clonal evolution in chronic lymphocytic leukemia: acquisition of high-risk genomic aberrations associated with unmutated VH, resistance to therapy, and short survival. Haematologica. 2007;92:1242–1245. doi: 10.3324/haematol.10720. [DOI] [PubMed] [Google Scholar]
- 66.Rossi S, Shimizu M, Barbarotto E, Nicoloso MS, Dimitri F, Sampath D, et al. microRNA fingerprinting of CLL patients with chromosome 17p deletion identify a miR-21 score that stratifies early survival. Blood. 2010;116:945–952. doi: 10.1182/blood-2010-01-263889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fegan C, Robinson H, Thompson P, Whittaker JA, White D. Karyotypic evolution in CLL: identification of a new sub-group of patients with deletions of 11q and advanced or progressive disease. Leukemia. 1995;9:2003–2008. [PubMed] [Google Scholar]
- 68.Neilson JR, Auer R, White D, Bienz N, Waters JJ, Whittaker JA, et al. Deletions at 11q identify a subset of patients with typical CLL who show consistent disease progression and reduced survival. Leukemia. 1997;11:1929–1932. doi: 10.1038/sj.leu.2400819. [DOI] [PubMed] [Google Scholar]
- 69.Saiya-Cork K, Collins R, Parkin B, Ouillette P, Kuizon E, Kujawski L, et al. A pathobiological role of the insulin receptor in chronic lymphocytic leukemia. Clin Cancer Res. 2011;17:2679–2692. doi: 10.1158/1078-0432.CCR-10-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rossi D, Fangazio M, Rasi S, Vaisitti T, Monti S, Cresta S, et al. Disruption of BIRC3 associates with fludarabine chemorefractoriness in TP53 wild-type chronic lymphocytic leukemia. Blood. 2012;119:2854–2862. doi: 10.1182/blood-2011-12-395673. [DOI] [PubMed] [Google Scholar]
- 71.Mohr J, Helfrich H, Fuge M, Eldering E, Buhler A, Winkler D, et al. DNA damage-induced transcriptional program in CLL: biological and diagnostic implications for functional p53 testing. Blood. 2011;117:1622–1632. doi: 10.1182/blood-2010-08-300160. [DOI] [PubMed] [Google Scholar]
- 72.Herling M, Patel KA, Weit N, Lilienthal N, Hallek M, Keating MJ, et al. High TCL1 levels are a marker of B-cell receptor pathway responsiveness and adverse outcome in chronic lymphocytic leukemia. Blood. 2009;114:4675–4686. doi: 10.1182/blood-2009-03-208256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tsimberidou AM, Tam C, Abruzzo LV, O’Brien S, Wierda WG, Lerner S, et al. Chemoimmunotherapy may overcome the adverse prognostic significance of 11q deletion in previously untreated patients with chronic lymphocytic leukemia. Cancer. 2009;115:373–380. doi: 10.1002/cncr.23993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dohner H, Stilgenbauer S, James MR, Benner A, Weilguni T, Bentz M, et al. 11q deletions identify a new subset of B-cell chronic lymphocytic leukemia characterized by extensive nodal involvement and inferior prognosis. Blood. 1997;89:2516–2522. [PubMed] [Google Scholar]
- 75.Shedden K, Li Y, Ouillette P, Malek SN. Characteristics of chronic lymphocytic leukemia with somatically acquired mutations in NOTCH1 exon 34. Leukemia. 2012;26:1108–1110. doi: 10.1038/leu.2011.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Balatti V, Bottoni A, Palamarchuk A, Alder H, Rassenti LZ, Kipps TJ, et al. NOTCH1 mutations in CLL associated with trisomy 12. Blood. 2012;119:329–331. doi: 10.1182/blood-2011-10-386144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lopez C, Delgado J, Costa D, Conde L, Ghita G, Villamor N, et al. Different distribution of NOTCH1 mutations in chronic lymphocytic leukemia with isolated trisomy 12 or associated with other chromosomal alterations. Genes Chromosomes Cancer. 2012;51:881–889. doi: 10.1002/gcc.21972. [DOI] [PubMed] [Google Scholar]
- 78.Del Giudice I, Rossi D, Chiaretti S, Marinelli M, Tavolaro S, Gabrielli S, et al. NOTCH1 mutations in +12 chronic lymphocytic leukemia (CLL) confer an unfavorable prognosis, induce a distinctive transcriptional profiling and refine the intermediate prognosis of +12 CLL. Haematologica. 2012;97:437–441. doi: 10.3324/haematol.2011.060129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Decker S, Zirlik K, Djebatchie L, Hartmann D, Ihorst G, Schmitt-Graeff A, et al. Trisomy 12 and elevated GLI1 and PTCH1 transcript levels are biomarkers for Hedgehog-inhibitor responsiveness in CLL. Blood. 2012;119:997–1007. doi: 10.1182/blood-2011-06-359075. [DOI] [PubMed] [Google Scholar]
- 80.Zenz T, Vollmer D, Trbusek M, Smardova J, Benner A, Soussi T, et al. TP53 mutation profile in chronic lymphocytic leukemia: evidence for a disease specific profile from a comprehensive analysis of 268 mutations. Leukemia. 2010;24:2072–2079. doi: 10.1038/leu.2010.208. [DOI] [PubMed] [Google Scholar]
- 81.Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fabbri G, Rasi S, Rossi D, Trifonov V, Khiabanian H, Ma J, et al. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J Exp Med. 2011;208:1389–1401. doi: 10.1084/jem.20110921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Trbusek M, Smardova J, Malcikova J, Sebejova L, Dobes P, Svitakova M, et al. Missense mutations located in structural p53 DNA-binding motifs are associated with extremely poor survival in chronic lymphocytic leukemia. J Clin Oncol. 2011;29:2703–2708. doi: 10.1200/JCO.2011.34.7872. [DOI] [PubMed] [Google Scholar]
- 84.Gaidano G, Ballerini P, Gong JZ, Inghirami G, Neri A, Newcomb EW, et al. p53 mutations in human lymphoid malignancies: association with Burkitt lymphoma and chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 1991;88:5413–5417. doi: 10.1073/pnas.88.12.5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zainuddin N, Murray F, Kanduri M, Gunnarsson R, Smedby KE, Enblad G, et al. TP53 mutations are infrequent in newly diagnosed chronic lymphocytic leukemia. Leuk Res. 2011;35:272–274. doi: 10.1016/j.leukres.2010.08.023. [DOI] [PubMed] [Google Scholar]
- 86.Johnson GG, Sherrington PD, Carter A, Lin K, Liloglou T, Field JK, et al. A novel type of p53 pathway dysfunction in chronic lymphocytic leukemia resulting from two interacting single nucleotide polymorphisms within the p21 gene. Cancer Res. 2009;69:5210–5217. doi: 10.1158/0008-5472.CAN-09-0627. [DOI] [PubMed] [Google Scholar]
- 87.el Rouby S, Thomas A, Costin D, Rosenberg CR, Potmesil M, Silber R, et al. p53 gene mutation in B-cell chronic lymphocytic leukemia is associated with drug resistance and is independent of MDR1/MDR3 gene expression. Blood. 1993;82:3452–3459. [PubMed] [Google Scholar]
- 88.Wattel E, Preudhomme C, Hecquet B, Vanrumbeke M, Quesnel B, Dervite I, et al. p53 mutations are associated with resistance to chemotherapy and short survival in hematologic malignancies. Blood. 1994;84:3148–3157. [PubMed] [Google Scholar]
- 89.Gonzalez D, Martinez P, Wade R, Hockley S, Oscier D, Matutes E, et al. Mutational status of the TP53 gene as a predictor of response and survival in patients with chronic lymphocytic leukemia: results from the LRF CLL4 trial. J Clin Oncol. 2011;29:2223–2229. doi: 10.1200/JCO.2010.32.0838. [DOI] [PubMed] [Google Scholar]
- 90.Zenz T, Eichhorst B, Busch R, Denzel T, Habe S, Winkler D, et al. TP53 mutation and survival in chronic lymphocytic leukemia. J Clin Oncol. 2010;28:4473–4479. doi: 10.1200/JCO.2009.27.8762. [DOI] [PubMed] [Google Scholar]
- 91.Sorror ML, Storer BE, Sandmaier BM, Maris M, Shizuru J, Maziarz R, et al. Five-year follow-up of patients with advanced chronic lymphocytic leukemia treated with allogeneic hematopoietic cell transplantation after nonmyeloablative conditioning. J Clin Oncol. 2008;26:4912–4920. doi: 10.1200/JCO.2007.15.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Malek S. Clinical utility of prognostic markers in chronic lymphocytic leukemia. ASCO Education Book. 2010:263–267. [review] [Google Scholar]
- 93.Bullrich F, Rasio D, Kitada S, Starostik P, Kipps T, Keating M, et al. ATM mutations in B-cell chronic lymphocytic leukemia. Cancer Res. 1999;59:24–27. [PubMed] [Google Scholar]
- 94.Schaffner C, Stilgenbauer S, Rappold GA, Dohner H, Lichter P. Somatic ATM mutations indicate a pathogenic role of ATM in B-cell chronic lymphocytic leukemia. Blood. 1999;94:748–753. [PubMed] [Google Scholar]
- 95.Stankovic T, Weber P, Stewart G, Bedenham T, Murray J, Byrd PJ, et al. Inactivation of ataxia telangiectasia mutated gene in B-cell chronic lymphocytic leukaemia. Lancet. 1999;353:26–29. doi: 10.1016/S0140-6736(98)10117-4. [DOI] [PubMed] [Google Scholar]
- 96.Austen B, Skowronska A, Baker C, Powell JE, Gardiner A, Oscier D, et al. Mutation status of the residual ATM allele is an important determinant of the cellular response to chemotherapy and survival in patients with chronic lymphocytic leukemia containing an 11q deletion. J Clin Oncol. 2007;25:5448–5457. doi: 10.1200/JCO.2007.11.2649. [DOI] [PubMed] [Google Scholar]
- 97.Ouillette P, Li J, Shaknovich R, Li Y, Melnick A, Shedden K, et al. Incidence and clinical implications of ATM aberrations in chronic lymphocytic leukemia. Genes Chromosomes Cancer. 2012 doi: 10.1002/gcc.21997. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Austen B, Powell JE, Alvi A, Edwards I, Hooper L, Starczynski J, et al. Mutations in the ATM gene lead to impaired overall and treatment-free survival that is independent of IGVH mutation status in patients with B-CLL. Blood. 2005;106:3175–3182. doi: 10.1182/blood-2004-11-4516. [DOI] [PubMed] [Google Scholar]
- 99.Cejkova S, Rocnova L, Potesil D, Smardova J, Novakova V, Chumchalova J, et al. Presence of heterozygous ATM deletion may not be critical in the primary response of chronic lymphocytic leukemia cells to fludarabine. Eur J Haematol. 2009;82:133–142. doi: 10.1111/j.1600-0609.2008.01177.x. [DOI] [PubMed] [Google Scholar]
- 100.Lozanski G, Ruppert AS, Heerem NA, Lozanski A, Luca DM, Gordon A, et al. Variations of the ATM gene in chronic lymphocytic leukemia patients lack substantial impact on progression-free survival and overall survival: a Cancer and Leukemia Group B Study. Leuk Lymphoma. 2012;53:1743–1748. doi: 10.3109/10428194.2012.668683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Balatti V, Bottoni A, Palamarchuk A, Alder H, Rassenti LZ, Kipps TJ, et al. NOTCH1 mutations in CLL associated with trisomy 12. Blood. 2012;119:329–331. doi: 10.1182/blood-2011-10-386144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rossi D, Rasi S, Fabbri G, Spina V, Fangazio M, Forconi F, et al. Mutations of NOTCH1 are an independent predictor of survival in chronic lymphocytic leukemia. Blood. 2012;119:521–529. doi: 10.1182/blood-2011-09-379966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rossi D, Bruscaggin A, Spina V, Rasi S, Khiabanian H, Messina M, et al. Mutations of the SF3B1 splicing factor in chronic lymphocytic leukemia: association with progression and fludarabine-refractoriness. Blood. 2011;118:6904–6908. doi: 10.1182/blood-2011-08-373159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brown JR, Levine RL, Thompson C, Basile G, Gilliland DG, Freedman AS. Systematic genomic screen for tyrosine kinase mutations in CLL. Leukemia. 2008;22:1966–1969. doi: 10.1038/leu.2008.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ngo VN, Young RM, Schmitz R, Jhavar S, Xiao W, Lim KH, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470:115–119. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gahrton G, Robert KH, Friberg K, Zech L, Bird AG. Nonrandom chromosomal aberrations in chronic lymphocytic leukemia revealed by polyclonal B-cell-mitogen stimulation. Blood. 1980;56:640–647. [PubMed] [Google Scholar]
- 107.Juliusson G, Oscier DG, Fitchett M, Ross FM, Stockdill G, Mackie MJ, et al. Prognostic subgroups in B-cell chronic lymphocytic leukemia defined by specific chromosomal abnormalities. N Engl J Med. 1990;323:720–724. doi: 10.1056/NEJM199009133231105. [DOI] [PubMed] [Google Scholar]
- 108.Peterson LC, Lindquist LL, Church S, Kay NE. Frequent clonal abnormalities of chromosome band 13q14 in B-cell chronic lymphocytic leukemia: multiple clones, subclones, and nonclonal alterations in 82 midwestern patients. Genes Chromosomes Cancer. 1992;4:273–280. doi: 10.1002/gcc.2870040402. [DOI] [PubMed] [Google Scholar]
- 109.Dohner H, Stilgenbauer S, Fischer K, Bentz M, Lichter P. Cytogenetic and molecular cytogenetic analysis of B cell chronic lymphocytic leukemia: specific chromosome aberrations identify prognostic subgroups of patients and point to loci of candidate genes. Leukemia. 1997;11:S19–S24. [PubMed] [Google Scholar]
- 110.Dicker F, Schnittger S, Haferlach T, Kern W, Schoch C. Immunostimulatory oligonucleotide-induced metaphase cytogenetics detect chromosomal aberrations in 80% of CLL patients: A study of 132 CLL cases with correlation to FISH, IgVH status, and CD38 expression. Blood. 2006;108:3152–3160. doi: 10.1182/blood-2006-02-005322. [DOI] [PubMed] [Google Scholar]
- 111.Put N, Konings P, Rack K, Jamar M, Van Roy N, Libouton JM, et al. Improved detection of chromosomal abnormalities in chronic lymphocytic leukemia by conventional cytogenetics using CpG oligonucleotide and interleukin-2 stimulation: a Belgian multicentric study. Genes Chromosomes Cancer. 2009;48:843–853. doi: 10.1002/gcc.20691. [DOI] [PubMed] [Google Scholar]
- 112.Mayr C, Speicher MR, Kofler DM, Buhmann R, Strehl J, Busch R, et al. Chromosomal translocations are associated with poor prognosis in chronic lymphocytic leukemia. Blood. 2006;107:742–751. doi: 10.1182/blood-2005-05-2093. [DOI] [PubMed] [Google Scholar]
- 113.Heerema NA, Byrd JC, Dal Cin PS, Dell’ Aquila ML, Koduru PR, Aviram A, et al. Stimulation of chronic lymphocytic leukemia cells with CpG oligodeoxynucleotide gives consistent karyotypic results among laboratories: a CLL Research Consortium (CRC) Study. Cancer Genet Cytogenet. 2010;203:134–140. doi: 10.1016/j.cancergencyto.2010.07.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Muthusamy N, Breidenbach H, Andritsos L, Flynn J, Jones J, Ramanunni A, et al. Enhanced detection of chromosomal abnormalities in chronic lymphocytic leukemia by conventional cytogenetics using CpG oligonucleotide in combination with pokeweed mitogen and phorbol myristate acetate. Cancer Genet. 2011;204:77–83. doi: 10.1016/j.cancergen.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Huh YO, Schweighofer CD, Ketterling RP, Knudson RA, Vega F, Kim JE, et al. Chronic lymphocytic leukemia with t(14;19)(q32;q13) is characterized by atypical morphologic and immunophenotypic features and distinctive genetic features. Am J Clin Pathol. 2011;135:686–696. doi: 10.1309/AJCPOEFP3SLX6HXJ. [DOI] [PubMed] [Google Scholar]
- 116.Martin-Subero JI, Ibbotson R, Klapper W, Michaux L, Callet-Bauchu E, Berger F, et al. A comprehensive genetic and histopathologic analysis identifies two subgroups of B-cell malignancies carrying a t(14;19)(q32;q13) or variant BCL3-translocation. Leukemia. 2007;21:1532–1544. doi: 10.1038/sj.leu.2404695. [DOI] [PubMed] [Google Scholar]
- 117.Ueshima Y, Bird ML, Vardiman JW, Rowley JDA. 14;19 translocation in B-cell chronic lymphocytic leukemia: a new recurring chromosome aberration. Int J Cancer. 1985;36:287–290. [PubMed] [Google Scholar]
- 118.Nguyen-Khac F, Chapiro E, Lesty C, Grelier A, Luquet I, Radford-Weiss I, et al. Specific chromosomal IG translocations have different prognoses in chronic lymphocytic leukemia. Am J Blood Res. 2011;1:13–21. [PMC free article] [PubMed] [Google Scholar]
- 119.Van Den Neste E, Robin V, Francart J, Hagemeijer A, Stul M, Vandenberghe P, et al. Chromosomal translocations independently predict treatment failure, treatment-free survival and overall survival in B-cell chronic lymphocytic leukemia patients treated with cladribine. Leukemia. 2007;21:1715–1722. doi: 10.1038/sj.leu.2404764. [DOI] [PubMed] [Google Scholar]
- 120.Rigolin GM, Cibien F, Martinelli S, Formigaro L, Rizzotto L, Tammiso E, et al. Chromosome aberrations detected by conventional karyotyping using novel mitogens in chronic lymphocytic leukemia with ‘normal’ FISH: correlations with clinicobiologic parameters. Blood. 2012;119:2310–2313. doi: 10.1182/blood-2011-11-395269. [DOI] [PubMed] [Google Scholar]
- 121.Visone R, Rassenti LZ, Veronese A, Taccioli C, Costinean S, Aguda BD, et al. Karyotype-specific microRNA signature in chronic lymphocytic leukemia. Blood. 2009;114:3872–3879. doi: 10.1182/blood-2009-06-229211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kanduri M, Cahill N, Göransson H, Enström C, Ryan F, Isaksson A, et al. Differential genome-wide array-based methylation profiles in prognostic subsets of chronic lymphocytic leukemia. Blood. 2010;115:296–305. doi: 10.1182/blood-2009-07-232868. [DOI] [PubMed] [Google Scholar]
- 123.Herman SE, Gordon AL, Hertlein E, Ramanunni A, Zhang X, Jaglowski S, et al. Bruton tyrosine kinase represents a promising therapeutic target for treatment of chronic lymphocytic leukemia and is effectively targeted by PCI-32765. Blood. 2011;117:6287–6296. doi: 10.1182/blood-2011-01-328484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hoellenriegel J, Meadows SA, Sivina M, Wierda WG, Kantarjian H, Keating MJ, et al. The phosphoinositide 3′-kinase delta inhibitor, CAL-101, inhibits B-cell receptor signaling and chemokine networks in chronic lymphocytic leukemia. Blood. 2011;118:3603–3612. doi: 10.1182/blood-2011-05-352492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ponader S, Chen SS, Buggy JJ, Balakrishnan K, Gandhi V, Wierda WG, et al. The Bruton tyrosine kinase inhibitor PCI-32765 thwarts chronic lymphocytic leukemia cell survival and tissue homing in vitro and in vivo. Blood. 2012;119:1182–1189. doi: 10.1182/blood-2011-10-386417. [DOI] [PMC free article] [PubMed] [Google Scholar]