Abstract
Hypoxia Inducible Factors (HIFs) are heterodimeric transcription factors induced in a variety of pathophysiological settings, including cancer. We describe the first detailed structure-activity-relationship study of small molecules designed to inhibit HIF-2α–ARNT heterodimerization by binding an internal cavity of the HIF-2α PAS-B domain. Through a series of biophysical characterizations of inhibitor/protein interactions (NMR and X-ray crystallography), we have established the structural requirements for artificial inhibitors of the HIF-2α–ARNT PAS-B interaction. These results may serve as a foundation for discovering therapeutic agents that function by a novel mode of action.
Keywords: Hypoxia, protein-ligand interactions, protein-protein interactions, drug discovery, cancer
INTRODUCTION
Mammalian cells employ pathways by which they recognize and respond to metabolic and environmental cues. Though promoting normal developmental and physiological adaptions in response to stress, such pathways are also frequently misappropriated in the context of human disease. For example, mammalian cells express a hypoxic response pathway by which changes in cellular oxygen availability are sensed. When oxygen availability is low, a family of Hypoxia Inducible Factors (HIFs) are upregulated.1 These transcription factors coordinate the expression of hundreds of downstream target genes that promote appropriate responses, such as anaerobic metabolism and increased oxygen delivery.2 However, HIFs also participate in pathophysiological settings, including cancer.1,3 HIFs are often induced within the hypoxic environment of tumors, where they are conscripted to promote changes in tumor metabolism, angiogenesis, metastasis, and many other pro-tumorigenic responses.4 In fact, the impact of HIF on cancer can be so great that there is often selective pressure for mutations that uncouple HIF induction from oxygen availability, resulting in constitutive HIF activity.5
While expression studies and genetic model systems provide compelling evidence that HIF inhibition may impair tumor progression in many settings, small molecule tools offer several advantages for investigating HIF as a therapeutic target.5 Such reagents offer the potential to assess the consequences of in vivo HIF attenuation on therapeutically relevant time scales in a range of model systems, many of which are not amenable to genetic manipulation. Small molecules can also help address the potential consequences of global HIF attenuation on important physiological pathways throughout a therapeutically-relevant window. Although a handful of high throughput screen (HTS)-derived small molecules or natural products have been reported to modulate the pathway, almost all of these directly interfere with targets other than the HIF proteins themselves, giving rise to pleiotropic effects and limiting their utility in testing HIF-centric hypotheses.6 Indeed, HIFs themselves are an example of a traditionally challenging target for pharmacological intervention: it is a large, intracellular multiprotein complex without any catalytic active sites that are typically used for small molecule substrate binding. Moreover, much of the HIF complexes exist in an extended conformation, reducing the availability of potential ligand binding sites.
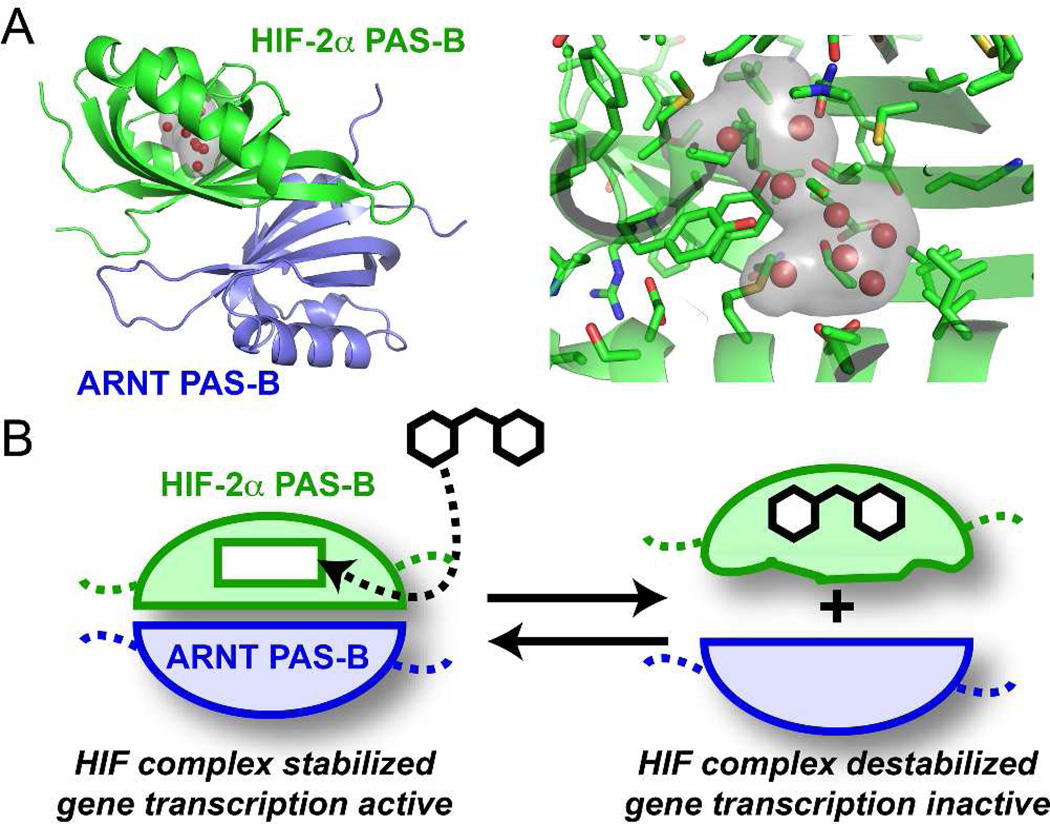
Human HIF transcription factors are heterodimers composed of one of three regulated HIF-α (HIF-1α, HIF-2α/EPAS-1, or HIF-3α) subunits and a constitutive ARNT (also known as HIF-β) subunit, all members of the bHLH–PAS (basic Helix-Loop-Helix–Period-ARNT-Single minded) family.8 HIF PAS domains stabilize the HIF heterodimers via protein-protein interactions across subunits as mutations or deletions in the PAS domains attenuate HIF heterodimer formation and transcriptional activity.9 Though large, hydrophobic protein-protein interfaces are notoriously difficult to disrupt directly with small molecules, PAS domains provide an attractive opportunity. Notably, many PAS-mediated protein-protein interactions are regulated by allosteric conformational changes induced by cofactors that bind within the core of the PAS domain itself.10 We hypothesized that HIF PAS domains might likewise be amenable to binding small molecule antagonists within their cores to induce conformational changes that disrupt HIF dimerization (Figure 1).
Figure 1.
Basis of small molecule regulation of protein-protein interactions in HIF-2. A) Crystal structure of the HIF-2α–ARNT PAS-B heterodimer9b (PDB code: 3F1P), highlighting the internal cavity within HIF-2α PAS-B (grey surface, internal waters represented as red spheres). Sidechains lining the cavity are provided by a mix of hydrophobic and polar residues as shown. B) Schematic for small molecule regulation of HIF-2, with ligand binding to the HIF-2α PAS-B cavity, distorting the adjacent β-sheet that also provides the ARNT PAS-B binding surface.7
Notably, one of the two PAS domains in HIF-2α (PAS-B) is particularly well-suited in this regard. Recently our groups used X-ray crystallography and NMR to identify a large (290 Å3) water-filled cavity in the core of this domain (Figure 1b).9b,11 Cavities of this size are rare and strongly suggestive of a missing cofactor or ligand-binding site. An initial NMR-based small molecule screen identified a number of artificial ligands for the HIF-2α PAS-B domain, most of which were two substituted aromatic rings connected by short (1- or 2-atom) linkers. These initial findings demonstrated that this cavity could accommodate ligand binding to induce conformational changes that weaken the protein-protein interaction between purified PAS domains from the HIF-2α and ARNT subunits. However, these initial lead molecules lacked the efficacy and pharmacological attributes required to modulate HIF-2 gene expression in cells.
To identify superior HIF-2 antagonist candidates, we screened a collection of > 200,000 structurally diverse small molecules from an in-house compound library using a commercially-available luminescence proximity (AlphaScreen, from Perkin Elmer) assay format. AlphaScreen is a homogenous, bead-based luminescence proximity assay12, which monitors the formation of a complex between two tagged proteins (i.e. GST-HIF-2α PAS-B* and ARNT PAS-B*-FLAG domains, where PAS-B* designates the HIF-2α E247R and ARNT PAS-B R362E variants used to crystallize complexes with small molecule ligands)9b to bring cognate donor and acceptor beads into close proximity (Figure S1). The beads constitute a pair that can be detected by a luminescent signal triggered by diffusion of singlet oxygen from a donor bead and subsequent detection on an acceptor. In the presence of a small molecule that disrupts the protein-protein interaction, this luminescent signal is extinguished. A screen of the 203,520 compounds in this library provided us with approximately 20 candidates suitable for further study by analog synthesis or purchase. Recently, our laboratories characterized one such synthetic small molecule that binds the HIF-2α PAS-B internal cavity and exhibits an AlphaScreen IC50 value of approximately 0.1 µM.7 Isothermal titration calorimetry measurements confirm in vitro binding in the same range (KD = 80–90 nM). In cell culture, these compounds interfere with HIF-2 driven transcription with low-µM potency.
Herein, we describe a full account of the synthesis and evaluation of a series of compounds that culminated in the identification of our most active small molecule binder of the HIF-2α PAS-B domain. In the process, we elucidate the structural features of the most active compound that lead to a disruption of the heterodimeric structure of the HIF transcription factor. We discuss a focused subset of the hundreds of small molecule inhibitors of HIF-2α that were synthesized and evaluated by our laboratories. This work represents the first detailed SAR study of small molecules designed to inhibit HIF-2α–ARNT heterodimerization, which may serve as a foundation for discovering therapeutic agents that function by a novel mode of action.
RESULTS
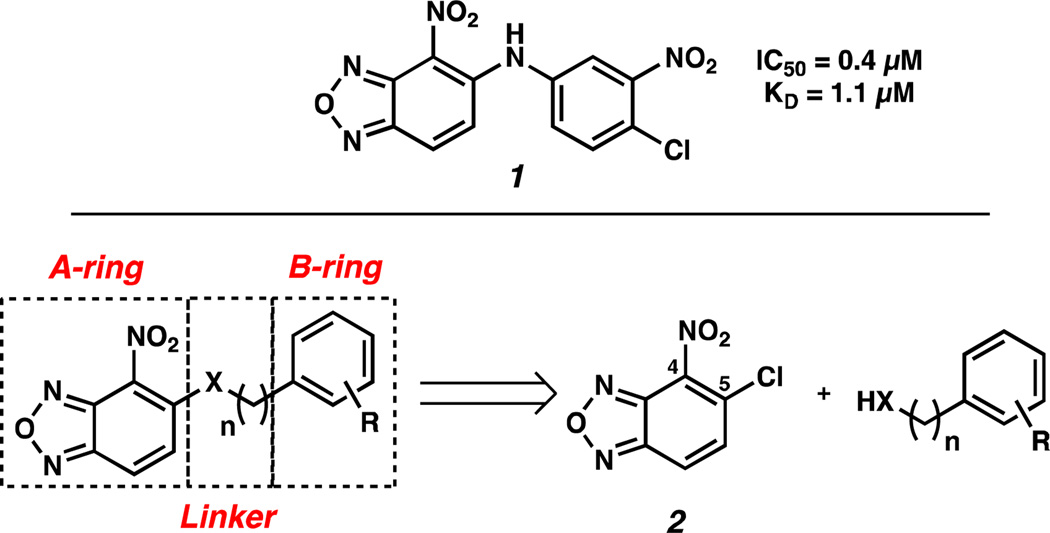
Compound 1 was chosen as the starting point for our SAR campaign because of its superior activity over other promising scaffolds identified in our initial screen of > 200,000 structurally diverse small molecules (Figure 2), exhibiting an IC50 of 0.4 µM in the AlphaScreen and a KD of 1.1 µM by ITC. This compound contains three general structural units that can be modified: the A-ring, the B-ring, and the 1- or 2-atom linker.
Figure 2.
Compound identified from the initial high-throughput screen and Strategic Molecular Subsections for SAR Studies.
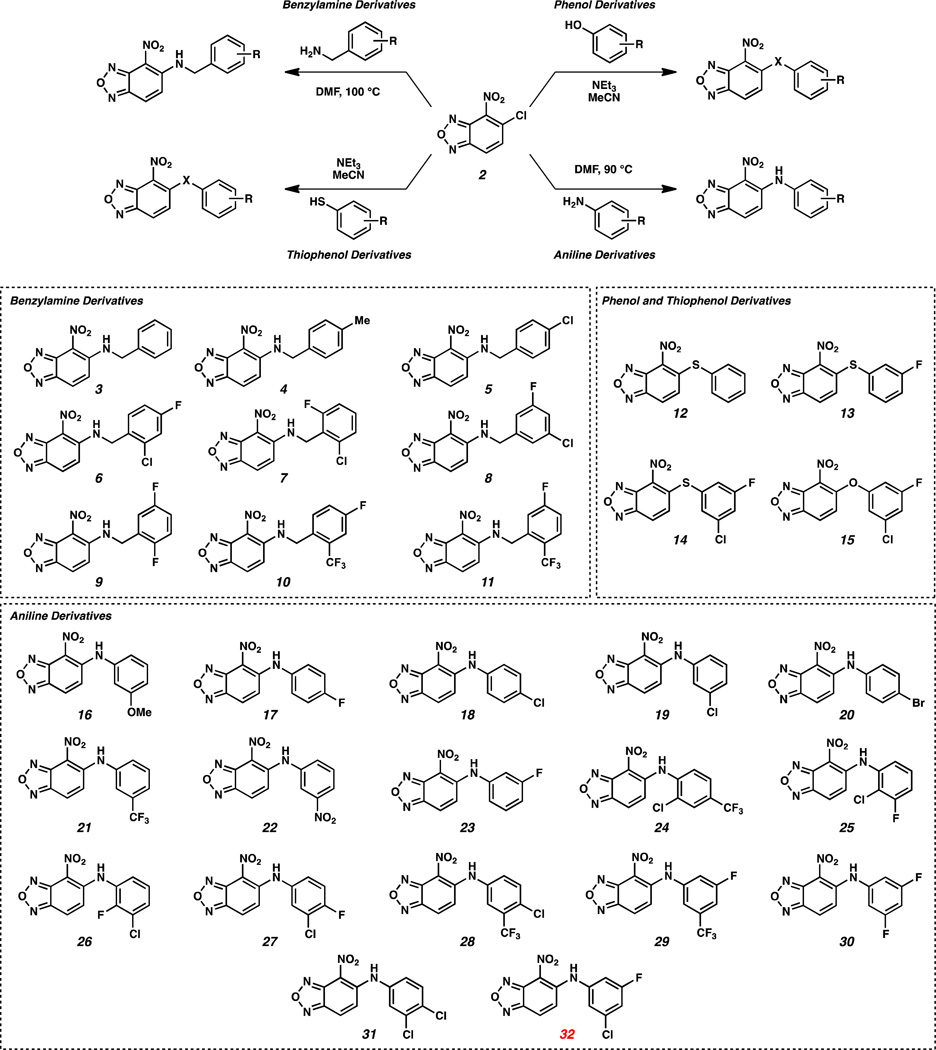
We chose 4-nitrobenzoxadiazole as the initial A-ring for our studies, and we systematically optimized the B-ring and the linker to identify analogs with improved activity. Synthetically, analogs of 1 were accessed from the coupling of commercially available 5-chloro-4-nitrobenzoxadiazole 2 and heteroatom nucleophiles. This convergent strategy facilitated the generation of a diverse array of structures that were evaluated in the AlphaScreen.
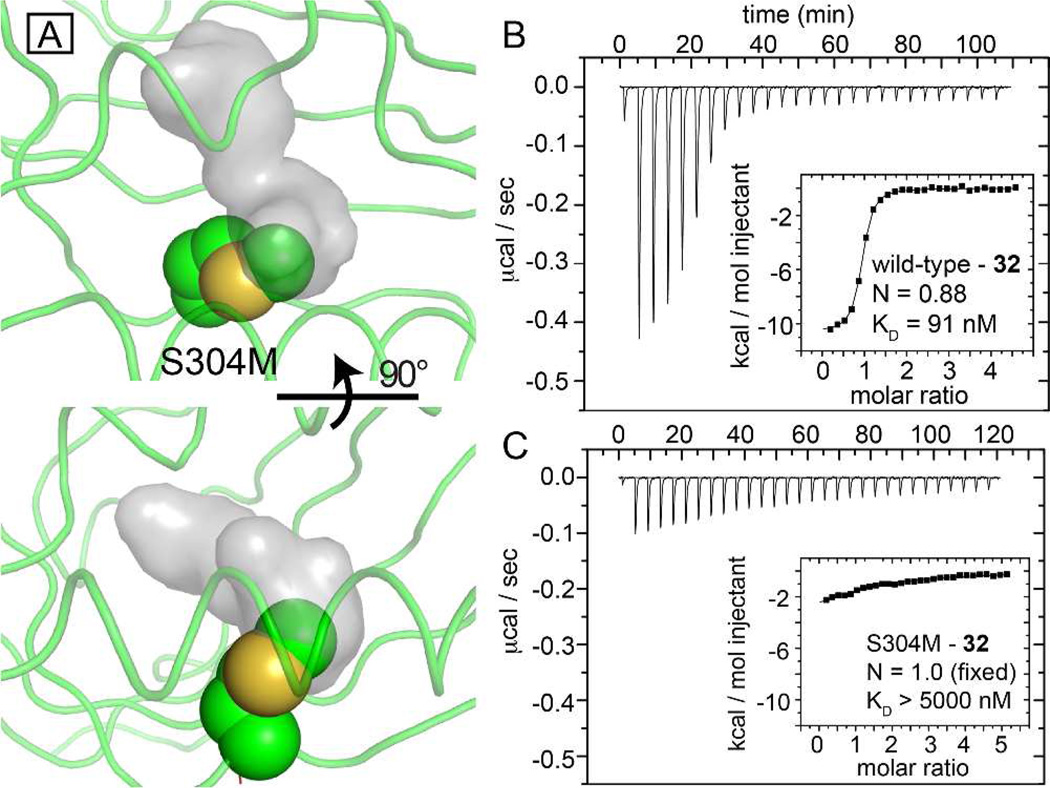
Initially, benzoxadiazole 2 was coupled with a series of benzyl amines in DMF at 100 °C to yield N-linked analogs 3–11 (Scheme 1). Thiophenols and phenol also reacted with benzoxadiazole 2 in the presence of triethylamine in MeCN at ambient temperature to furnish S-linked analogs 12–14 and O-linked analog 15. The coupling of benzoxadiazole 2 with sterically and electronically diverse anilines at elevated temperatures generated diarylamines 16–32. Compounds 3–32 were evaluated for their ability to interrupt HIF-2α–ARNT heterodimerization using the AlphaScreen described above (Table 1). To better use this assay to identify allosteric inhibitors that work by binding to the HIF-2α PAS-B cavity, we sought to develop negative control proteins which would be unable to bind ligands to the internal cavity while still folding properly and retaining the ability to bind ARNT PAS-B. We generated approximately 25 HIF-2α PAS-B variants for this purpose, each of which contained point mutations in one or more residues lining the internal cavity. Of this group, the S304M mutation best met our design criteria, placing the newly-introduced long aliphatic methionine sidechain into the internal cavity (Figure 3A). This change does not grossly affect the HIF-2α PAS-B protein fold, as judged by similarities in backbone NMR chemical shifts (Figure S2A), but very effectively interferes with binding of small molecules into the hydrophobic core (Figure 3B,C). Critically, HIF-2α PAS-B* (S304M) retains the ability to bind ARNT PAS-B* (Figure S2B), essential to its use as a control reagent with the AlphaScreen assay.
Scheme 1.
Synthesis of Analogs of 1.
Table 1.
AlphaScreen IC50 for 3 – 32 (values in µM)
| Compound | Alpha | Compound | Alpha |
|---|---|---|---|
| 3 | > 5 | 18 | NA |
| 4 | NA | 19 | 0.18 |
| 5 | ND | 20 | NA |
| 6 | 0.33 | 21 | 0.46 |
| 7 | NA | 22 | 2.8 |
| 8 | 2 | 23 | 2.1 |
| 9 | >10 | 24 | NA |
| 10 | 0.5 | 25 | ND |
| 11 | NA | 26 | 0.76 |
| 12 | ND | 27 | 0.17 |
| 13 | ND | 28 | 0.09 |
| 14 | ND | 29 | 0.43 |
| 15 | ND | 30 | 2 |
| 16 | NA | 31 | 0.12 |
| 17 | NA | 32 | 0.1 |
ND – compound disrupts the control. NA – no activity at the highest concentration tested.
Figure 3.
A single internal mutation within the HIF-2α PAS-B cavity attenuates ligand binding. A) Two views of a model of the S304M mutation (spheres) suggests that the new sidechain will intrude upon the apo- protein cavity (grey surface, from PDB code: 3F1P).9b (B, C) ITC of wild-type (B) and S304M (C) complexes with compound 32, demonstrating that the mutation reduces the affinity of the protein over 50-fold, validating the biophysically-characterized ligand binding site.
A number of molecules, designated as ND in Table 1, typically inhibited heterodimerization of ARNT with the WT and S304 mutant HIF-2α PAS-B, suggesting they are either interfering with the assay conditions or cause disruption of heterodimerization through an alternative pathway that may not involve binding to the internal cavity. From the initial SAR studies, it was apparent that modifications of the linker had deleterious effects on the activity. Of the nine benzylamine derivatives, only 6 (IC50 = 0.33 µM) and 10 (IC50 = 0.5 µM) demonstrated appreciable activity in the AlphaScreen. The common feature of these two analogs is the ortho, para substitution pattern with electron withdrawing substituents. Further discussion of the activity of 6 and 10 will be included below. Changing from N-linked to O- and S-linked analogs diminished all activity, regardless of the substitution pattern (compounds 12–15). Based on our initial success with the aniline series 16–32 described below, we abandoned further efforts with the O- and S-linked analogs. We believe there may be a role for hydrogen bonding in the –NH of the linker, which is not possible for the O- and S-linked analogs. This is further demonstrated below, where N-methylation of the linker reduces activity.
The series of analogs most exhaustively explored was the diarylamines, where a clear pattern of activity emerged. Generally, mono-substitution of the B-ring resulted in inactive compounds, with the exception of 19 (IC50 = 0.18 µM) and 21 (IC50 = 0.46 µM), which had m-Cl and m-CF3 substituents, respectively. No analogs with mono-substitution in the para-position showed inhibition of heterodimerization. The most potent compounds to emerge from this series were the di-substituted diarylamines, with analogs containing at least one meta-substituent providing the most potent disruption in the AlphaScreen (IC50 values ranging from 0.09 µM to 2.8 µM).
To further explore the most potent inhibitors, compounds that had significant activity by AlphaScreen (i.e. IC50 < 2.0 µM) were tested by ITC to obtain KD values for binding of small molecules to the HIF-2α PAS-B domain (Table 2). Overall, we found good correlation between the IC50 values measured by AlphaScreen and KD values measured by ITC. Ultimately, compound 32 emerged as the most potent compound, both in terms of the AlphaScreen assay and by ITC.
Table 2.
KD values for most active compounds (values in µM)
| # | ITC KD | # | ITC KD |
|---|---|---|---|
| 6 | 0.5 | 26 | 1.1 |
| 8 | 2 | 27 | 0.4 |
| 10 | 0.54 | 28 | 0.17 |
| 19 | 0.16 | 29 | 0.37 |
| 21 | 0.64 | 30 | 0.73 |
| 22 | 3 | 31 | 0.17 |
| 23 | 2.2 | 32 | 0.09 |
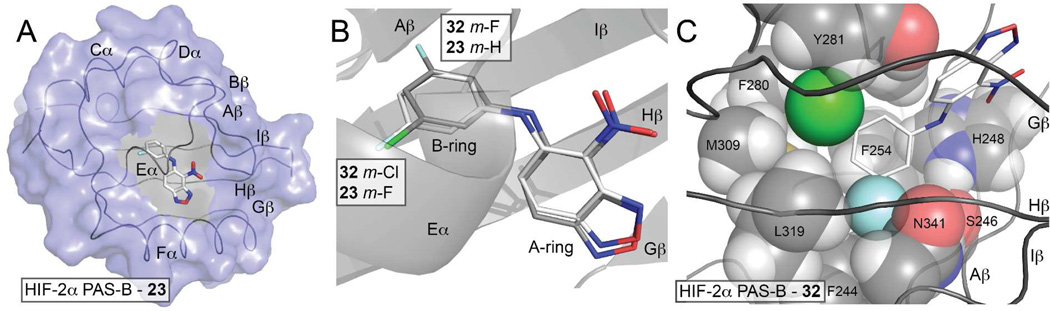
To explore the structural basis of the SAR trends we observed in Tables 1 and 2, we performed NMR-based structural analysis and X-ray crystallographic studies with the HIF-2α PAS-B domain and our synthetic small molecules. For the co-crystal structures we utilized the engineered, high affinity HIF2 PAS-B* heterodimer of HIF-2α E247R and ARNT PAS-B R362E domains employed in the AlphaScreen assay.9b A comparison of the co-crystal structures of the mildly disrupting mono-fluorinated analog 23 (IC50 = 2.1 µM) and the potently disrupting disubstituted analog 32 reveals that both inhibitors bind in the HIF-2α PAS-B internal cavity, fully sequestered from bulk solvent (Figure 4A). The A-ring of both 23 and 32 is oriented to accommodate a weak electrostatic interaction between the nitro-group and the H248 imidazole sidechain. The B-rings occupy similar binding sites, however they are oriented in slightly different conformations (Figure 4B). The m-F of the disubstituted B-ring of 32 packs against the PAS-B β-sheet, and the m-Cl of 32 is positioned near the E helix. In contrast, the m-F in compound 23 points away from the β-sheet and occupies the same site near the E helix as the m-Cl in compound 32 (Figure 4C).
Figure 4.
Binding modes of HIF-2 antagonists. A) The HIF-2 PAS-B* - 23 ternary complex (PDB code: 4GS9) confirms that this inactive HTS-lead analog binds into the HIF-2α PAS-B internal cavity. To better view the internally-bound ligand, secondary structural elements are indicated along the ribbon diagram and the HIF-2α PAS-B surface (blue) has been cut away for residues that separate the binding site from bulk solvent (from this perspective). Although present in these structures, the ARNT PAS-B domain has been omitted for clarity. B) Comparison of the HIF-2 PAS-B complexes with compounds 23 and 32 shows that the mono-substituted m-fluorinated B-ring of 23 flips roughly 180 degrees relative to 32 (PDB code: 4GHI7). C) Close protein - 32 contacts at the m-fluorine site suggests that bound 23 adopts a lower energy bound conformation by placing its single halogen where 32 positions its m-chlorine.
The different orientations of the m-F in the B-ring of a series of analogs may account for differences in activity, which is supported by several observations. Notably, m-F of 32 packs very tightly against the β-sheet surface of the binding site, but makes a modest contribution to the binding energy. A comparison of the ITC data from 32 and 19, differing by a m-F, demonstrates a two fold reduction in affinity (Table 2). Extrapolating from the experimental structure of 32 bound within HIF-2a, we believe the ortho, para substitution pattern of the active benzylamine analogs 6 and 10 allow them to pack their B-rings similarly to 32, placing one substituent against the PAS-B beta-sheet and one near the Ea helix.
Alternatively, the loss of the m-Cl (23) or the replacement of the m-Cl with a m-F (30) weaken binding by 25- and 8-fold, respectively. These observations suggest that the m-F binding site of 32 is sterically strained in the HIF-2α PAS-B* ternary complex and, in the absence of a m-Cl, adopts a lower energy binding pose by flipping into the m-Cl binding site of 32. Together, these observations suggest a mechanism where the m-Cl of 32 pays the energetic cost of forcing the m-F into a suboptimal pocket juxtaposed to the HIF-2α PAS-B β-sheet, effecting conformational changes to the PAS-B heterodimerization interface and weakening the important interaction between the HIF-2α and ARNT PAS-B domains.
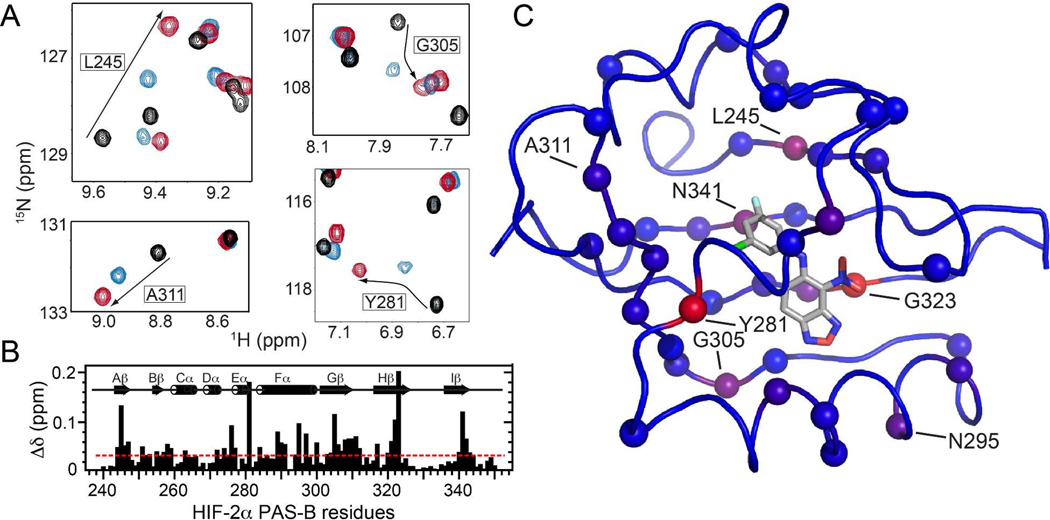
To further characterize the consequences of ligand binding in solution, we examined the wild-type HIF-2α PAS-B domain by solution NMR spectroscopy. 15N/1H HSQC NMR spectra collected from 15N-enriched HIF-2α PAS-B reveal a well-ordered protein with significant spectral changes induced by complexation with either 32 or 23 (Figure 5). At many sites we can assign the 23-bound state by inference from the experimentally determined resonance assignments for both ligand-free9a and 32-bound states.7 At several such sites, we observe crosspeaks for 23-bound HIF-2α PAS-B located between the corresponding peaks for apo-protein or the 32 complex, indicating that binding to 23 causes smaller protein structural changes than binding to 32 (Figure 5A,B). Notably, a number of the larger chemical shift differences localize to the HIF-2α PAS-B β-sheet (Figure 5B,C), which constitutes the ARNT PAS-B binding site.
Figure 5.
Solution NMR characterization of the 23 and 32 complexes with HIF-2α PAS-B. A) 15N/1H HSQC spectra of apo HIF-2α PAS-B (black) and its complexes with 23 (blue) and 32 (red) reveal sites differentially perturbed by the two analogs. B) Chemical shift differences in HIF-2α PAS-B observed between the complexes (with ligands 23 and 32) are plotted along the protein sequence. The red line at 0.033 ppm denotes the average difference observed across all sites. C) Chemical shift differences mapped onto the HIF-2α PAS-B – 32 complex structure (PDB code: 4GHI) as a blue to red gradient, with spheres marking sites experiencing a greater than average difference.
Once the 3-chloro-5-fluoro-aniline 32 emerged as the most active compound and we could rationalize the necessity for the B-ring substitutions, we examined modifications to the A-ring to ascertain the role of the 4-nitrobenzoxadiazole functionality. The co-crystal structures of 32 and 23 indicated there was space for additional steric bulk around the A-ring. In particular, the A-ring of 32 was not filling the pocket para to the nitro group and in the vicinity of the benzoxadiazole functionality. Moreover, in all co-crystals with analogs that contain a nitro-group in the A-ring (e.g., 32), the molecule is oriented to potentially accommodate weak electrostatic interactions between its nitro-group and the H248 imidazole sidechain. We examined a series of A-ring analogs of 32 that probed these observations.
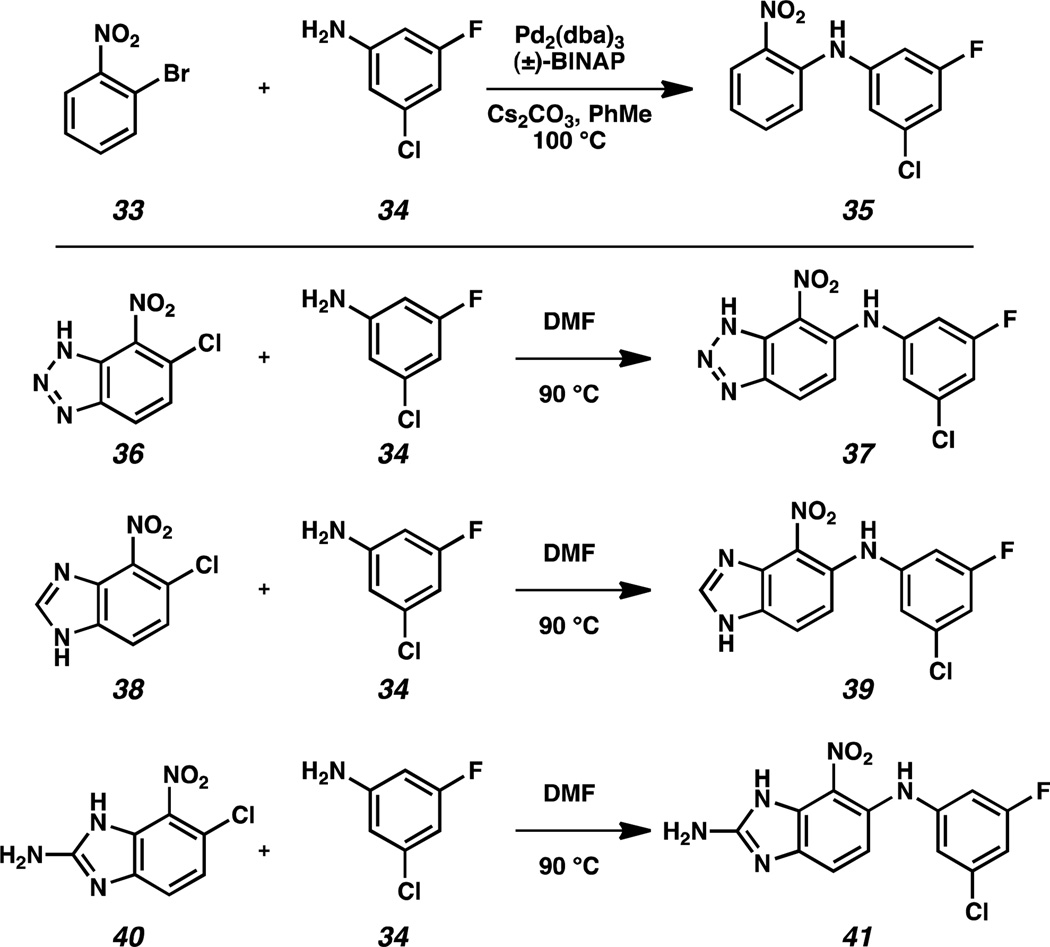
The first set of compounds we targeted involved replacement of the benzoxadiazole with other aromatic and heteroaromatic rings. For example, ortho-nitrophenyl analog 35 was generated by Buchwald-Hartwig coupling between ortho-nitrochlorobenzene 33 and 3-chloro-5-fluoro-aniline 34 (Scheme 2).13 Nitrobenzotriazole 37 was accessed from chloride precursor 36 via nucleophilic aromatic substitution. Benzimidazole 39 and 2-aminobenzimidazole 41 were similarly synthesized from 6-chloro-7-nitro-benzimidazoles 38 and 40, respectively.
Scheme 2.
A-ring Modifications of Compound 32.
Evaluation of the four alternative A-ring heterocycles with AlphaScreen revealed dramatically reduced activity for all of the analogs, with IC50 values of ~2 to 20 µM (Table 3). A few of these analogs were evaluated by ITC, further supporting attenuated binding to the HIF-2α PAS-B cavity. These results suggest that the unique distribution of electron density in the benzoxadiazole ring may play a key role in the optimal binding and disrupting activity of analog 32.
Table 3.
AlphaScreen IC50 values and KD values for A-ring analogs (values in µM).
| # | Alpha | ITC KD | # | Alpha | ITC KD |
|---|---|---|---|---|---|
| 35 | ND | 4.3 | 47 | > 30 | ~6 |
| 37 | >10 | >20 | 48 | NA | – |
| 39 | >5 | – | 49 | NA | – |
| 41 | >2 | – | 50 | NA | – |
| 44 | ND | >10 | 54 | NA | 5 |
| 46 | NA | ~3 | 55 | NA | 5 |
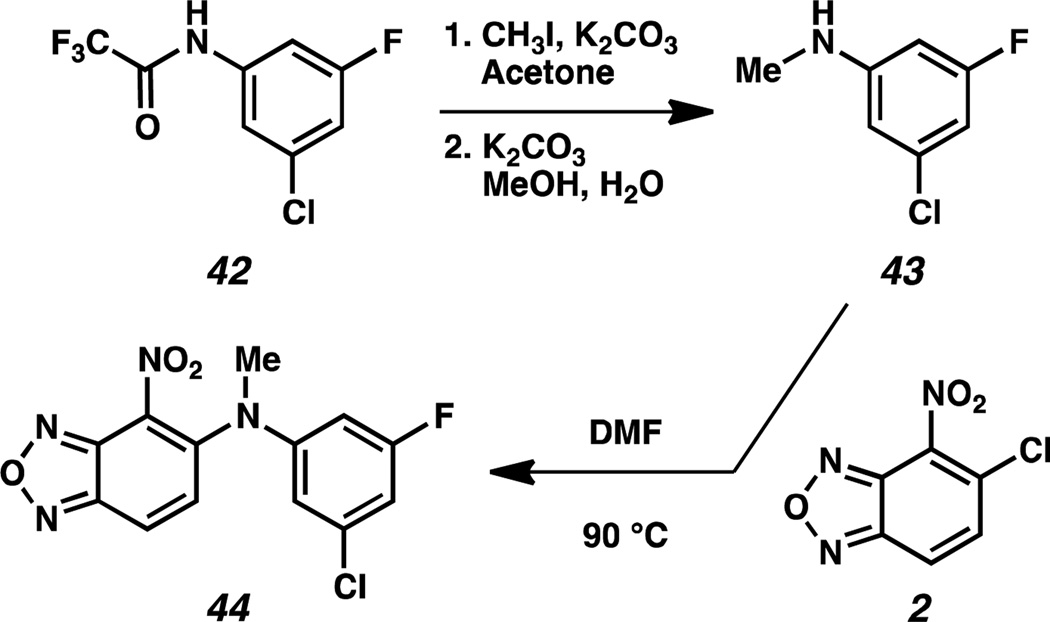
Since initial studies revealed the superiority of the NH-linked analogs 16–32 over S-linked analogs 12–14 and O-linked analog 15, we synthesized N-methylated analog 44 to determine the importance of the bis-anilinic proton in compound 32 (Scheme 3). Trifluoromethyl(3-chloro-5-fluorophenyl)carbamate 42 was converted to N-methyl-3-chloro-5-fluoro-aniline 43 in a two-step sequence. This intermediate was then coupled with benzoxadiazole 2 to furnish N-methylated analog 44. Compound 44 has an IC50 value > 3 µM by AlphaScreen and was further shown by ITC to have a KD > 10 µM.
Scheme 3.
Synthesis of N-methylated analog 45.
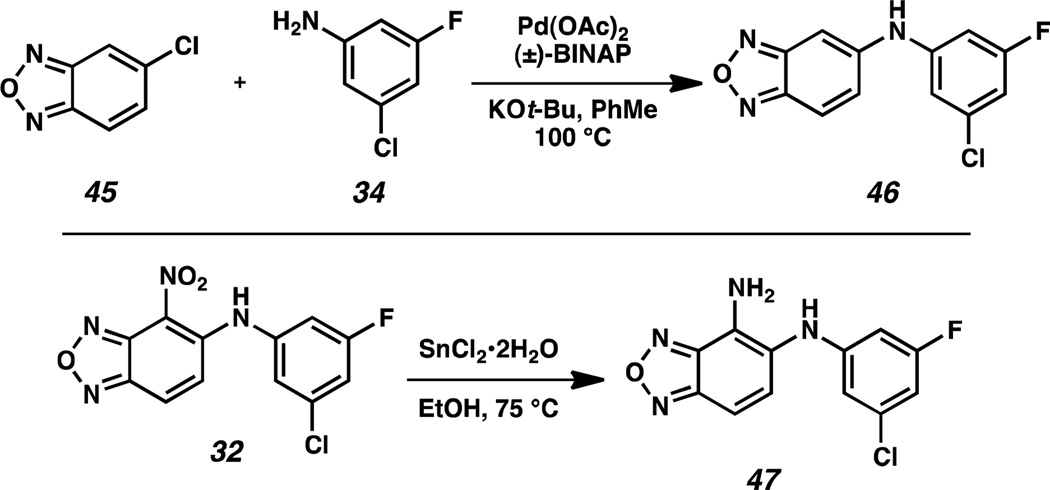
We were also interested in studying the role of the nitro functional group in compound 32. Because of the potentially deleterious pharmacokinetic properties of a nitro group as well as its weak interaction with the H248 imidazole sidechain in the HIF-2α cavity, we examined suitable replacements. Omission of the nitro group in analog 46 was achieved by coupling 5-chlorobenzoxadiazole 45 and aniline 34 with a slightly modified Buchwald-Hartwig amination protocol (Scheme 4). The nitro functional group in 32 was also directly converted under reductive conditions into an amino group, yielding 4-aminobenzoxadiazole 47. These changes led to relatively insoluble compounds. Both 46 and 47 exhibited weak activity in the AlphaScreen, although 46 was also active against the S304M control. We further characterized these analogs by ITC and NMR, with both methods demonstrating weak binding (KD = 3 µM for 46, 6 µM for 47).
Scheme 4.
Conversion of 4-nitro functional group.
Primary aniline 47 was treated with a series of electrophiles, including acetic anhydride, chloroacetylchloride, and maleic anhydride (Scheme 5) to generate N-acyl analogs. These transformations generated 4-amidobenzoxadiazoles 48–50. Analogs 49 and 50 were especially compelling because of their potential as irreversible disruptors of HIF-2α–ARNT heterodimerzation through covalent interactions with the C339 sidechain, which is proximal to the nitro group of compound 32 in the co-crystal structure. However none of these analogs exhibited activity.
Scheme 5.
Synthesis of 4-amidobenzoxadiazoles.
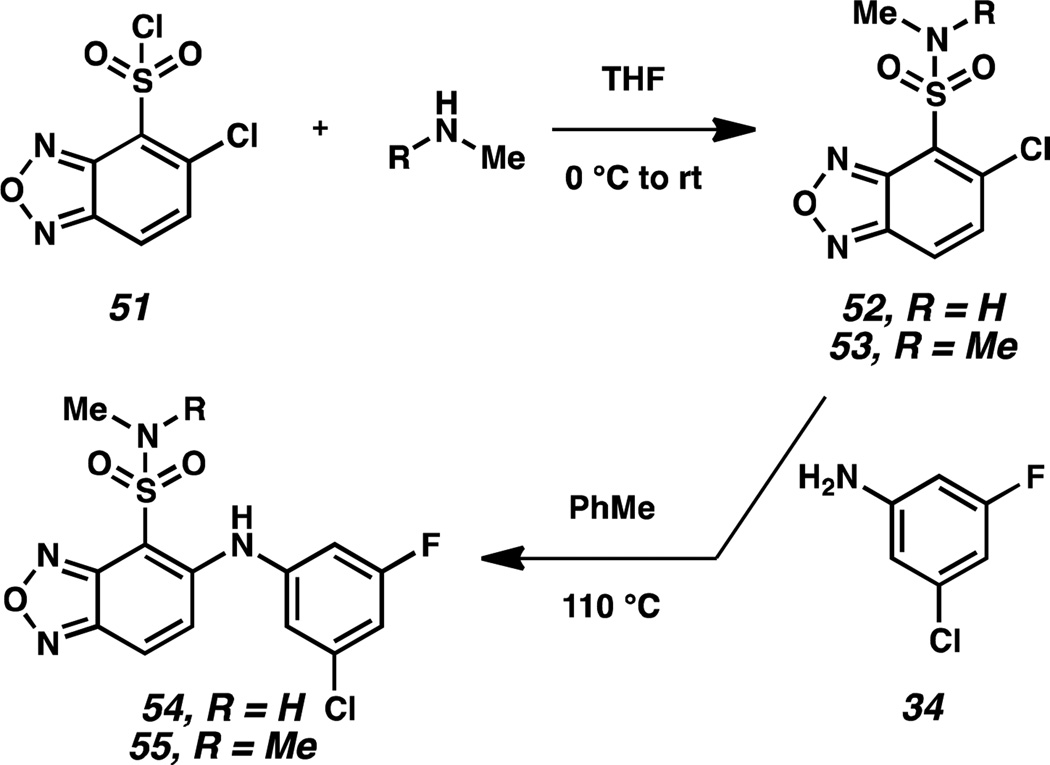
To determine whether the 4-nitro group in compound 32 was beneficial because of its electron withdrawing properties, we synthesized electronically analogous 4-sulfonamidobenzoxadiazoles 54 and 55 (Scheme 6). These compounds were also examined because of the metabolic stability of sulfonamides as electron withdrawing substituents. Synthetically, 5-chlorobenzoxadiazole-4-sulfonyl chloride 51 was converted to 5-chlorobenzoxadiazole-4-sulfonamides 52 and 53 in the presence of methylamine and dimethylamine, respectively. These intermediates were then coupled with aniline 34 to furnish the desired 4-sulfonamidobenzoxadiazoles 54 and 55. Replacement of the nitro group with the electronically similar sulfonamides failed to result in compounds with equal activity. Both 54 and 55 were inactive as disruptors, but had weak affinity for the HIF-2α cavity, with KD values of 5 µM.
Scheme 6.
Synthesis of 4-sulfonamidobenzoxadiazoles.
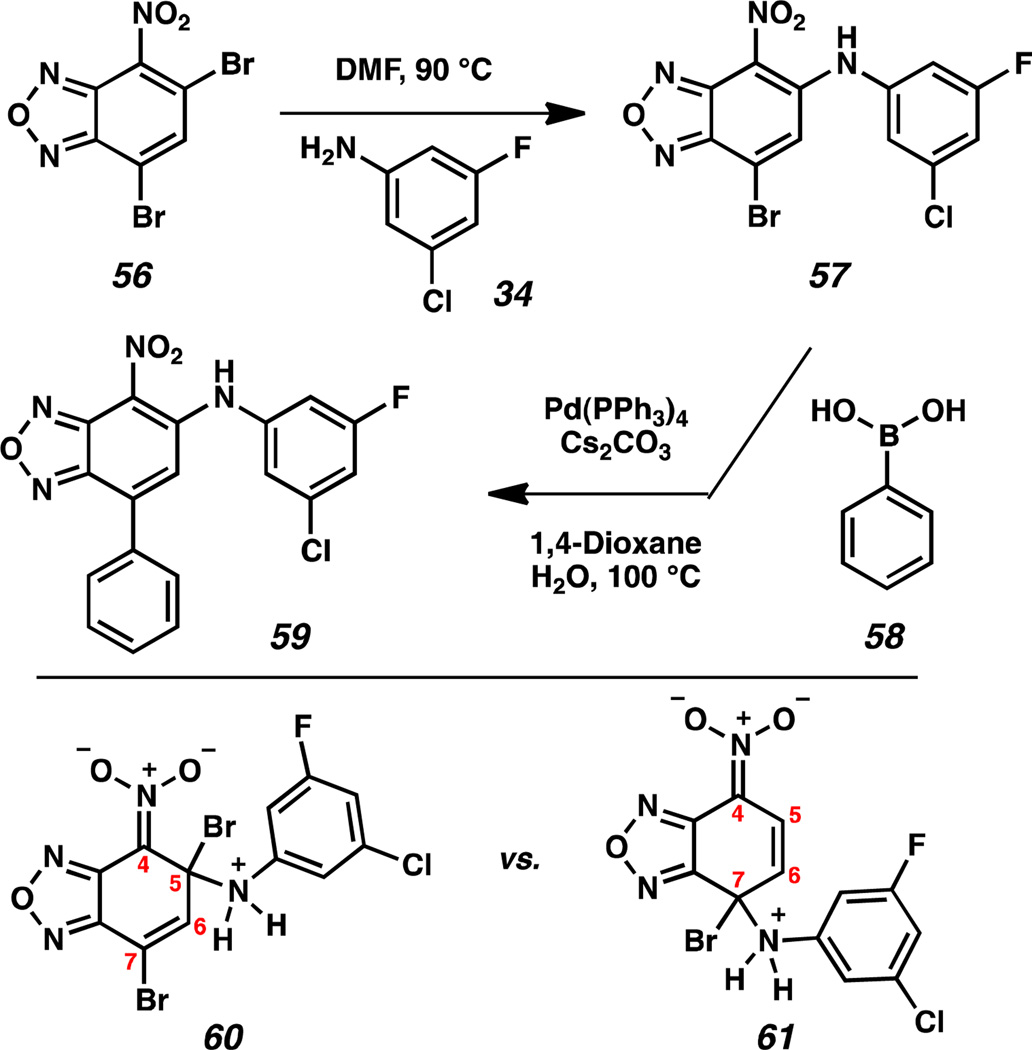
Finally, we functionalized the 7-position of the benzoxadiazole ring system (Scheme 7), because X-ray co-crystal structures of several small molecules and the HIF-2α PAS-B domain indicated room for expansion in this chemical space of our analogs. Dibromo-4-nitrobenzoxadiazole 56 was coupled with aniline 34 under standard thermal conditions to yield 7-bromo-4-nitrobenzoxadiazole 57. We anticipated the displacement of the 5-bromo substituent in preference to displacement of the 7-bromo substituent for two reasons: (1) the enhanced stability of linear conjugation with the benzoxadiazole ring in intermediate 60 over 61; and (2) the larger Coulombic term at the 5-position.14 This regioselectivity was confirmed by examining crystals of benzoxadiazole 57 that were suitable for X-ray diffraction. 7-Bromobenzoxadiazole 57 was then subjected to Suzuki-Miyaura cross-coupling conditions with phenylboronic acid 58 to furnish 7-phenylbenzoxadiazole 59.15 Evaluation of 57 and 59 by AlphaScreen and ITC revealed complete loss of activity by the addition of the –Br and –Ph substituents. This result suggests the importance of the electronic and steric environment of the A-ring for activity.
Scheme 7.
Functionalization of the 7-position of the benzoxadiazole.
DISCUSSION
Taken together, our data provide a useful assessment of the structural requirements for artificial inhibitors of the HIF-2α–ARNT PAS-B interaction essential for assembling the HIF transcription factor. Our compounds work by triggering allosteric changes in the HIF-2α PAS-B β-sheet, initiated by ligand binding to a completely buried internal cavity. This binding site places certain structural restraints on the inhibitors, as demonstrated by integrating these structure-activity-relationships with our prior structural work.7,9b,11 These restraints can be satisfied by a variety of compounds with two aromatic/heterocyclic rings coupled by short linkers, including the nitrobenzoxadiazoles reported here, along with modified trifluorotoluenes.9b,11 Both types of compounds can utilize 1- or 2-atom linkers, with secondary amines being most thoroughly explored.
Within the B-ring, we find that substitutions are most tolerated at the positions where the apo-form of the cavity is largest, corresponding to the meta-position of the ligand B-ring. Crystal structures of the apo- and ligand-bound protein show that the cavity approaches close to the para-position of the B-ring, with a corresponding decrease in binding affinity and disruption potency for para- vs. meta- substituted compounds (e.g., compare 17 vs. 23; 18 vs. 19). While explored in less detail here, ortho-substitutions also appear to slightly decrease affinity and potency.
In contrast, the protein cavity near the A-ring appears to accommodate a smaller range of groups, as exhibited by the diversity of compounds reported here and previously.9b,11 These share a requirement for an electron-deficient aromatic ring, here achieved by an NO2-substituted benzoxadiazole and previously by a NO2, CF3-disubstituted phenyl ring. This ring interacts with residues surrounding the cavity, including Y281.9b Notably, the nitro group that is common among these compounds, often avoided in medicinal chemistry applications,16 appears to be essential for high affinity and potency. Notably, its replacement by an NH2 worsens binding and disruption by 30–60 fold (compare 32 vs. 47), underscoring its importance. This has been attributed to favorable interactions with H2489b or involvement in an intramolecular H-bond,11 the latter of which is also supported by the deleterious effects of N-methylation (compare 32 vs. 44).
In addition to improving binding affinity and potency, these diverse compounds enable studies that show the importance of protein dynamics at all stages of ligand binding and action. Entry of ligands from solvent into the pocket buried over 6 Å within the protein requires substantial flexibility of several helices.9a,b All of our compounds require some change in the shape of the apo- cavity to accommodate chemical groups, as perhaps best exemplified by the A-ring nitro group described above, which displaces the M252 sidechain. Indeed, ligand-induced structural changes are essential for triggering the allosteric changes required to displace ARNT PAS-B from the other side of the HIF-2α PAS-B β-sheet. This is clearly demonstrated with binding of compound 32 to the HIF-2α PAS-B domain, which pushes on several β-sheet residues (predominantly C339 and N341 in the Iβ strand and F244 in Aβ), and bulges the sheet in such a way that distorts the ARNT binding surface.7 SAR comparisons from compounds that bind similarly, but with lower potency (e.g. 32 vs. 23; Figures 4 and 5) provide a critical (and practical) way to assess the relative flexibility of the pocket for different ligands.
More broadly, it is becoming readily apparent that a wide range of natural and artificial ligands are used by PAS domains to disrupt protein-protein interactions via allosteric mechanisms. Best studied among these are a group of photosensitive flavin-binding PAS domains, known as LOV domains, which harness the photochemically-driven formation of a protein-ligand bond to initiate similar conformational changes as we observe with HIF-2α PAS-B.7,17 Studies of the aryl hydrocarbon receptor (AhR), a transcription factor which responds to a wide range of xenobiotic compounds and natural metabolites10a suggest that AhR may bind compounds analogously to HIF-2α, folding the surrounding domain.18 Based on these data and the presence of the large, hydrated cavity in the HIF-2α PAS-B domain – most typical of apo- forms of ligand binding proteins – we postulate that there is an endogenous ligand involved in HIF-2 regulation. If so, future elucidation of such a natural ligand(s) may provide novel insights into binding of small molecules in the HIF-2α PAS-B pocket as well as insights into the metabolic regulation of HIF in physiological settings.
Supplementary Material
ACKNOWLEDGMENT
We acknowledge the following grants for funding this project: NIH P01 CA095471, CPRIT RP100846, Cancer Center Support Grant 5P30 CA142543 and the Welch Foundation I-1689 (JBM) and I-1568 (RKB). JBM is a Chilton/Bell Foundation Endowed Scholar. UKT and KHG are W. W. Caruth, Jr. Scholars in Biomedical Research. RKB is the Michael L. Rosenberg Scholar in Medical Research and was supported by a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund. This investigation was conducted in a facility constructed with support from the Research Facilities Improvement Program (Grant # C06 RR 15437-01) from the National Center for Research Resources, National Institutes of Health. Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by U. Chicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357
ABBREVIATIONS USED
- HIFs
hypoxia inducible factors
- ARNT
aryl hydrocarbon receptor nuclear translocator
- PAS
Period-ARNT-single minded
- bHLH
basic Helix-Loop-Helix
- HTS
high-throughput screen
- ITC
isothermal calorimetry
- AHR
aryl hydrocarbon receptor
Footnotes
ASSOCIATED CONTENT
Supporting Information. General procedures, compound characterization, assay procedures, data tables, biophysical characterization. This material is available free of charge via the Internet at http://pubs.acs.org
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
PDB ID Codes: The atomic coordinates and structure factor amplitudes associated with the HIF-2α PAS-B* - ARNT-B* heterodimer crystal structure (3F1P) and its ternary complex with compound 23 (4GS9) has been deposited in the PDB macromolecular database.
REFERENCES
- 1.Semenza GL. Hypoxia-inducible factors: mediators of cancer progression and targets of cancer therapy. Trends Pharm. Sci. 2012;33:207–214. doi: 10.1016/j.tips.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xia X, Lemieux ME, Li W, Carroll JS, Brown M, Liu XS, Kun AL. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc. Natl. Acad. Sci. U.S.A. 2009;106:4260–4265. doi: 10.1073/pnas.0810067106. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Schodel J, Oikonomopoulos S, Ragoussis J, Pugh CW, Ratcliffe PJ, Mole DR. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood. 2011;117:e207–e217. doi: 10.1182/blood-2010-10-314427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bos R, van der Groep P, Greijer AE, Shvarts A, Meijer S, Pinedo HM, Semenza GL, van Diest PJ, van der Wall E. Levels of hypoxia-inducible factor-1alpha independently predict prognosis in patients with lymph node negative breast carcinoma. Cancer. 2003;97:1573–1581. doi: 10.1002/cncr.11246. [DOI] [PubMed] [Google Scholar]; (b) Zagzag D, Zhong H, Scalzitti JM, Laughner E, Simons JW, Semenza GL. Expression of hypoxia-inducible factor 1alpha in brain tumors: association with angiogenesis, invasion, and progression. Cancer. 2000;88:2606–2618. [PubMed] [Google Scholar]
- 4.Semenza GL. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer. 2003;10:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 5.Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol. Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Nagle DG, Zhou Y-D. Natural product-based inhibitors of hypoxia-inducible factor-1 (HIF-1) Curr. Drug Targets. 2006;7:355–369. doi: 10.2174/138945006776054979. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Semenza GL. Evaluation of HIF-1 inhibitors as anticancer agents. Drug Disc. Today. 2007;12:853–859. doi: 10.1016/j.drudis.2007.08.006. [DOI] [PubMed] [Google Scholar]; (c) Onnis B, Rapisarda A, Melillo G. Development of HIF-1 inhibitors for cancer therapy. J. Cell. Mol. Med. 2009;13:2780–2786. doi: 10.1111/j.1582-4934.2009.00876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheuermann TH, Li Q, Ma H-W, Key J, Zhang L, Chen R, Garcia JA, Naidoo J, Longgood J, Frantz DE, Tambar UK, Gardner KH, Bruick RK. Allosteric inhibition of hypoxia inducible factor 2 with small molecules. Nat. Chem. Biol. doi: 10.1038/nchembio.1185. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. U.S.A. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Erbel PJ, Card PB, Karakuzu O, Bruick RK, Gardner KH. Structural basis for PAS domain heterodimerization in the basic helix--loop--helix-PAS transcription factor hypoxia-inducible factor. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15504–15509. doi: 10.1073/pnas.2533374100. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Scheuermann TH, Tomchick DR, Machius M, Guo Y, Bruick RK, Gardner KH. Artificial ligand binding within the HIF2α PAS-B domain of the HIF2 transcription factor. Proc. Natl. Acad. Sci. U.S.A. 2009;106:450–455. doi: 10.1073/pnas.0808092106. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Yang J, Zhang L, Erbel PJ, Gardner KH, Ding K, Garcia JA, Bruick RK. Functions of the Per/ARNT/Sim (PAS) domains of the hypoxia inducible factor (HIF) J. Biol. Chem. 2005;280:36047–36054. doi: 10.1074/jbc.M501755200. [DOI] [PubMed] [Google Scholar]
- 10.(a) Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu. Rev. Pharmacol. Toxicol. 2003;43:309–334. doi: 10.1146/annurev.pharmtox.43.100901.135828. [DOI] [PubMed] [Google Scholar]; (b) Crosson S, Rajagopal S, Moffat K. The LOV domain family: photoresponsive signaling modules coupled to diverse output domains. Biochemistry. 2003;42:2–10. doi: 10.1021/bi026978l. [DOI] [PubMed] [Google Scholar]; (c) Gilles-Gonzalez MA, Gonzalez G. Heme-based sensors: defining characteristics, recent developments, and regulatory hypotheses. J. Inorg. Biochem. 2005;99:1–22. doi: 10.1016/j.jinorgbio.2004.11.006. [DOI] [PubMed] [Google Scholar]; (d) Hoff WD, Dux P, Hard K, Devreese B, Nugteren-Roodzant IM, Crielaard W, Boelens R, Kaptein R, van Beeumen J, Hellingwerf KJ. Thiol ester-linked p-coumaric acid as a new photoactive prosthetic group in a protein with rhodopsin-like photochemistry. Biochemistry. 1994;33:13959–13962. doi: 10.1021/bi00251a001. [DOI] [PubMed] [Google Scholar]
- 11.Key J, Scheuermann TH, Anderson PC, Daggett V, Gardner KH. Principles of ligand binding within a completely buried cavity in HIF2α PAS-B. J. Am. Chem. Soc. 2009;131:17647–17654. doi: 10.1021/ja9073062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ullman EF, Kirakossian H, Switchenko AC, Ishkanian J, Ericson M, Wartchow CA, Pirio M, Pease J, Irvin BR, Singh S, Singh R, Patel R, Dafforn A, Davalian D, Skold C, Kurn N, Wagner DB. Luminescent oxygen channeling assay (LOCI): sensitive, broadly applicable homogeneous immunoassay method. Clin. Chem. 1996;42:1518–1526. [PubMed] [Google Scholar]
- 13.(a) Paul F, Patt J, Hartwig JF. Palladium-catalyzed formation of carbon-nitrogen bonds - Reaction intermediates and catalyst improvements in the hetero cross-coupling of aryl halides and tin amides. J. Am. Chem. Soc. 1994;116:5969–5970. [Google Scholar]; (b) Guram AS, Buchwald SL. Palladium-catalyzed aromatic aminations with in situ generated aminostannanes. J. Am. Chem. Soc. 1994;116:7901–7902. [Google Scholar]; (c) Hartwig JF. Evolution of a fourth generation catalyst for the amination and thioetherification of aryl halides. Acc. Chem. Res. 2008;41:1534–1544. doi: 10.1021/ar800098p. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Surry DS, Buchwald SL. Dialkylbiaryl phosphines in Pd-catalyzed amination: a user's guide. Chem. Sci. 2011;2:27–50. doi: 10.1039/C0SC00331J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleming I. Frontier orbitals and organic chemical reactions. England: John Wiley & Sons; 1976. pp. 68–69. [Google Scholar]
- 15.Miyaura N, Suzuki A. Palladium-catalyzed cross-coupling reactions of organoboron compounds. Chem. Rev. 1995;95:2457–2483. [Google Scholar]
- 16.Wermuth CG, editor. The practice of medicinal chemistry. 2nd Ed. London, UK: Elsevier Academic Press; 2003. [Google Scholar]
- 17.Halavaty AS, Moffat K. N- and C-terminal flanking regions modulate light-induced signal transduction in the LOV2 domain of the blue light sensor phototropin 1 from Avena sativa. Biochemistry. 2007;46:14001–14009. doi: 10.1021/bi701543e. [DOI] [PubMed] [Google Scholar]
- 18.Pandini A, Denison MS, Song Y, Soshilov AA, Bonati L. Structural and functional characterization of the aryl hydrocarbon receptor ligand binding domain by homology modeling and mutational analysis. Biochemistry. 2007;46:696–708. doi: 10.1021/bi061460t. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.