Abstract
In plants, microRNA399 (miR399) is a major regulator of phosphate (Pi) homeostasis by way of post-transcriptional mechanisms including transcript cleavage and transcriptional repression. Although miRNA genomic organization, biogenesis, and mode of action in plants are known, the regulatory mechanisms affecting miRNAs are poorly understood. We have shown that AtMYB2 functions as a transcriptional activator for miR399f expression in the context of phosphate homeostasis. AtMYB2 directly binds to a MYB-binding site in the promoter of the miR399f precursor and regulates miR399f expression. In addition, AtMYB2 transcripts are induced under Pi deficiency. The overexpression of AtMYB2 affects root system architecture (RSA), indicated by suppression of primary root growth and enhanced development of root hairs. AtMYB2 and miR399f are expressed and localized in the same tissues under Pi limitation. This study establishes that AtMYB2 regulates Pi-starvation responses (PSR) by activating of miR399f transcript, suggesting that an analysis of this miRNA promoter could reveal the existence and extent of crosstalk with other signaling mechanisms.
Keywords: Arabidopsis MYB2, microRNA399, phosphate, phytohormones, signaling
Phosphorus (P), a macronutrient, is an essential ingredient of plant growth and propagation, and is taken up by plant roots from the soil. A deficiency of phosphorus constitutes a severely limiting factor for crop productivity all over the world.1 Plants experiencing Pi-deficient conditions employ systemic mechanisms that absorb external Pi for maintenance of Pi homeostasis. These mechanisms include change in the root systemic architecture (RSA), enhancement of Pi uptake activities, exudation of organic acids and phosphatases in order to solubilize organically and inorganically bound Pi.2–5
Recently, identification and characterization of major systemic regulators in Pi-starvation signaling and Pi homeostasis have been accomplished. Among the regulatory mechanisms are transcript levels of several miRNAs, among them miR156, miR158, miR163, miR319, miR399, miR447, miR778, miR827, miR866 and miR2111 that are induced by Pi limitation.6–9 However, molecular mechanisms of miRNAs are unknown with the exception of miR399, miR827 and miR2111. miR827 regulates anthocyanin synthesis in some cross-talk between Pi-homeostasis and homeostasis of other nutrients.7 miR2111 is involved in a regulatory role for plant survival under Pi limitation by the regulation of expression of a target, At3g27150, which encodes a kelch domain-containing F-box protein.8 Importantly, miR399 is a regulator of long-distance regulation of shoot-to-root communication under Pi limitating conditions. One target of miR399 is the transcript of PHO2/UBC24 encoding the ubiquitin-conjugating E2 enzyme in Arabidopsis. The PHO2/UBC24 gene is downregulated in Pi-deficiency responses, thereby activating the expression of root Pi-uptake transporters (e.g., PHT1;8 and PHT1;9).10 Thus, transgenic Arabidopsis overexpressing miR399 were impaired in Pi remobilization and enhanced Pi accumulation in the shoots.11
The miR399 family in Arabidopsis consists of six genes (a to f). All of them are strongly expressed by Pi starvation, and repress the transcript of the UBC24 target gene. To characterize the regulatory mechanism controlling miR399f expression, we performed in silico analysis of the presumptive miR399f promoter region using the PlantCARE database. This region included various cis-acting regulatory elements related to biotic- and abiotic-stress responses. We identified two putative MYB-binding sites (MBS, 5′-TAACTG-3′) on the hypothetical miR399f promoter. The more distal of these MBS specifically bound to the Arabidopsis MYB2 transcription factor. AtMYB2 binding in the identified miR399f promoter led to transcriptional activation of miR399f expression. In addition, transcriptional expression of AtMYB2, like miR399f expression, was induced by Pi-limitation in shoot and root. Significantly, AtMYB2 and miR399f were expressed in the same vascular tissues of cotyledons, leaves and roots, and their expressions were more strongly enhanced in those tissues under Pi starvation.
Several studies have provided evidence for cross-regulation between Pi-starvation responses (PSR), sugar signaling, and hormone signaling in plants.12–17 Exogenous cytokinin induced Pi uptake via the regulation of Pi transporters including PHT1;8, PHT1;9, PHT1;4, PHT1;5 and PHT3;1. CRE1/AHK4 plays an important role in suppressing the expression of phosphate starvation induced (PSI) genes, suggesting cross-talk between Pi and cytokinin signaling.14 Auxin and ethylene also play important roles in modulating the developmental adaptations of roots under Pi limitation.12,15 Application of gibberellic acid (GA) in Arabidopsis showed the Pi-limited phenomena including anthocyanin accumulation, root hair growth, and root systemic architecture and regulated the expression of PSI genes including AtPT1, AtPT2, At4 and AtIPS1 in Pi-starvation responses (PSR).16 Interestingly, ABA mediated the Pi-starvation responses (PSR) by the repression of transcriptional expression of At4 genes encoding low-Pi-induced ribo-regulators.17 These results strongly imply a connection between the Pi-starvation response and phytohormones. In addition, several transcription factors containing PHR1, WRKY75, MYB62, ZAT6 and BHLH32 have been identified as regulators in Pi-starvation responses. These transcription factors are also involved in the crosstalk between Pi-starvation signaling and signaling of phytohormones to regulate responses to Pi-limitation.13
AtMYB2 has initially been identified as a transcriptional activator of the dehydration-responsive gene, RD22 in ABA- and salt-stress signaling mechanisms. AtMYB2 is also involved in the cross-talk between apical dominance mechanism and auxin signaling, and it regulates cytokinin biosynthesis.18 The overexpression of AtMYB2 enhanced the expression of miR399f, thereby reducing the expression of UBC24, which serves as a target gene for miR399f, for Pi accumulation in the root and induced chlorosis symptoms in the leaves. In addition, AtMYB2-overexpressing plants showed a reduction of primary root growth under Pi-deficiency conditions and increased root hair density under Pi accumulation. Thus, these results indicate that phenotypes observed in the AtMYB2-overexpressing plants exhibit the same phenotypes as miRNA399-overexpressing plants, suggesting that AtMYB2 is a part of a Pi signaling pathway for maintaining Pi homeostasis within plants.
Knowledge about the functions of miRNAs in plants has increased in the past few years. miRNA399 has recently emerged as an important post-transcriptional regulator in controlling plant adaptive responses to Pi starvation. Pi starvation-induced miR399f promotes Pi uptake by cleavage of UBC24 mRNA, and mediates plant growth development and stress responses (Fig. 1). However, little is known about the upstream regulatory mechanisms. The observation of AtMYB2 to function as a transcriptional activator of miR399f expression is a new discovery in miRNA biogenesis for Pi homeostasis in plants. The finding suggests that AtMYB2 may function as a linker between the Pi homeostasis and abiotic stresses, such as NaCl and ABA or developmental regulation (Fig. 1). In this context we point to a GNATATNC sequence that is located 84 bp upstream on the miR399f promoter. The GNATATNC element is known as a putative binding site of PHR1, a MYB transcription factor in Pi starvation responses (PSR).19 It raises the possibility that AtMYB2 functions as a transcriptional activator together with PHR1 for miR399f expression. Also, the miR399f promoter region as defined by our study includes putative cis-acting regulatory elements related to defense-, hormone-, light- and water stress based on in silico analysis. Therefore, further investigations into the regulation of miRNAs in plants will be necessary, especially with respect to the characterization of cis-regulatory elements and their transcription factors. This will provide a better understanding on the roles of miRNAs in the cross-talks between nutrient acquisition, phytohormones, and abiotic stress signaling.
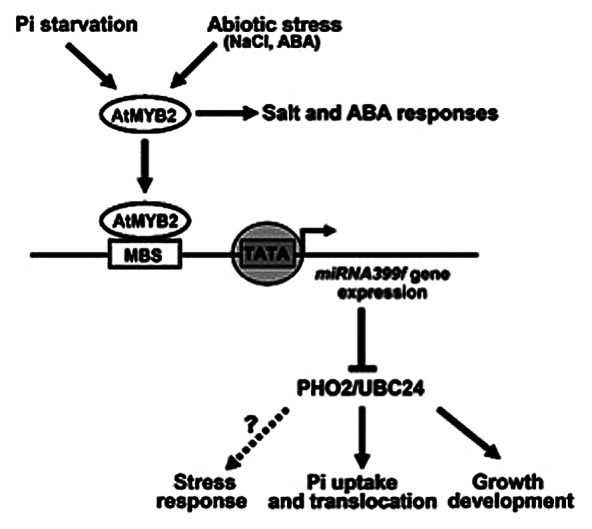
Figure 1. Model proposed for AtMYB2-regulated miR399f expression under Pi starvation and abiotic stresses. AtMYB2 directly binds in the promoter of the miR399f precursor and activates miR399f expression. It is expected that AtMYB2 may function as a linker between the Pi homeostasis and abiotic stresses, such as NaCl and ABA or developmental regulation. MBS: MYB binding site.
Acknowledgments
We thank Dr. Hans J. Bohnert for critical reading and insightful comments. This work was supported by grants from the World class University Program (R32-10148) funded by the Ministry of Education, Science and Technology and Next-Generation BioGreen21 Program (SSAC, grant#: PJ008025), Rural Development Administration, Republic of Korea.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/23488
References
- 1.Vance CP, Uhde-Stone C, Allan DL. Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol. 2003;157:423–47. doi: 10.1046/j.1469-8137.2003.00695.x. [DOI] [PubMed] [Google Scholar]
- 2.Raghothama KG. Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:665–93. doi: 10.1146/annurev.arplant.50.1.665. [DOI] [PubMed] [Google Scholar]
- 3.Poirier Y, Bucher M. Phosphate transport and homeostasis in Arabidopsis. In CR Somerville and EM Meyerowitz, eds, The Arabidopsis Book (Rockville, MD: American Society of Plant Biologist) 2002; doi/10.1199/tab.0009, http://www.aspb.org/publication/arabidopsis [DOI] [PMC free article] [PubMed]
- 4.Yuan H, Liu D. Signaling components involved in plant responses to phosphate starvation. J Integr Plant Biol. 2008;50:849–59. doi: 10.1111/j.1744-7909.2008.00709.x. [DOI] [PubMed] [Google Scholar]
- 5.Péret B, Clément M, Nussaume L, Desnos T. Root developmental adaptation to phosphate starvation: better safe than sorry. Trends Plant Sci. 2011;16:442–50. doi: 10.1016/j.tplants.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 6.Hammond JP, White PJ. Sugar signaling in root responses to low phosphorus availability. Plant Physiol. 2011;156:1033–40. doi: 10.1104/pp.111.175380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pant BD, Musialak-Lange M, Nuc P, May P, Buhtz A, Kehr J, et al. Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol. 2009;150:1541–55. doi: 10.1104/pp.109.139139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsieh LC, Lin SI, Shih AC, Chen JW, Lin WY, Tseng CY, et al. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009;151:2120–32. doi: 10.1104/pp.109.147280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK. A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol. 2005;15:2038–43. doi: 10.1016/j.cub.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 10.Pant BD, Buhtz A, Kehr J, Scheible WR. MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 2008;53:731–8. doi: 10.1111/j.1365-313X.2007.03363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF, Su CL. Regulation of phosphate homeostasis by MicroRNA in Arabidopsis. Plant Cell. 2006;18:412–21. doi: 10.1105/tpc.105.038943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rubio V, Bustos R, Irigoyen ML, Cardona-López X, Rojas-Triana M, Paz-Ares J. Plant hormones and nutrient signaling. Plant Mol Biol. 2009;69:361–73. doi: 10.1007/s11103-008-9380-y. [DOI] [PubMed] [Google Scholar]
- 13.Rouached H, Arpat AB, Poirier Y. Regulation of phosphate starvation responses in plants: signaling players and cross-talks. Mol Plant. 2010;3:288–99. doi: 10.1093/mp/ssp120. [DOI] [PubMed] [Google Scholar]
- 14.Franco-Zorrilla JM, Martín AC, Leyva A, Paz-Ares J. Interaction between phosphate-starvation, sugar, and cytokinin signaling in Arabidopsis and the roles of cytokinin receptors CRE1/AHK4 and AHK3. Plant Physiol. 2005;138:847–57. doi: 10.1104/pp.105.060517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Pérez-Torres A, Rampey RA, Bartel B, et al. An auxin transport independent pathway is involved in phosphate stress-induced root architectural alterations in Arabidopsis. Identification of BIG as a mediator of auxin in pericycle cell activation. Plant Physiol. 2005;137:681–91. doi: 10.1104/pp.104.049577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang C, Gao X, Liao L, Harberd NP, Fu X. Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol. 2007;145:1460–70. doi: 10.1104/pp.107.103788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shin H, Shin HS, Chen R, Harrison MJ. Loss of At4 function impacts phosphate distribution between the roots and the shoots during phosphate starvation. Plant J. 2006;45:712–26. doi: 10.1111/j.1365-313X.2005.02629.x. [DOI] [PubMed] [Google Scholar]
- 18.Guo Y, Gan S. AtMYB2 regulates whole plant senescence by inhibiting cytokinin-mediated branching at late stages of development in Arabidopsis. Plant Physiol. 2011;156:1612–9. doi: 10.1104/pp.111.177022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, et al. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001;15:2122–33. doi: 10.1101/gad.204401. [DOI] [PMC free article] [PubMed] [Google Scholar]


