Abstract
Transmembrane exchange of materials is a fundamental process in biology. Molecular dynamics provides a powerful method to investigate in great detail various aspects of the phenomenon, particularly the permeation of small uncharged molecules, which continues to pose a challenge to experimental studies. We will discuss some of the recent simulation studies investigating the role of lipid-mediated and protein-mediated mechanisms in permeation of water and gas molecules across the membrane.
Introduction
The primary function of cellular membranes is to separate the interior of a living cell from its immediate surroundings. At the same time, the cell relies heavily on continuous exchange of materials and information with its environment, in order to sustain its activity and growth, and to communicate and coordinate with other cells within a cellular community, e.g., within a tissue, or in a whole organism. Transmembrane trafficking of materials, therefore, is one the most fundamental and highly regulated processes in the biology of all living organisms. As a matter of fact, a cell dedicates a large fraction of its energy and its genetic material to devising and implementing diverse mechanisms facilitating the translocation of materials across the membrane, most prominently through specialized proteins that mediate selective translocation of various molecular species between the two sides of the membrane, namely membrane channels and transporters.
The cellular membrane is primarily composed of lipids, which owing to their hydrophobic nature, create an energetic barrier against the diffusion of polar and charged species. Excluding exocytosis and endocytosis, which are entirely non-selective mechanisms accompanied by large-scale reorganization of the membrane structure, transmembrane exchange of molecules mainly occurs through one of the three mechanisms: (i) passive diffusion through the lipid bilayer; (ii) passive diffusion through membrane channels; and (iii) active (energy dependent) translocation mediated by membrane transporters. These three mechanisms along with some examples of the substrates and proteins involved in these processes are graphically presented in Fig. 1. Translocation of most molecular species, including ions, amino acids and other nutrients, neurotransmitters, as well as larger molecules such as drugs, peptides, and even proteins, across the membrane relies on the function of membrane channels and transporters. In contrast, the transport of very small molecules, e.g., O2, NO, and water, was long deemed to be mediated primarily through non-selective “pores” within the lipid molecules, and, thus, independent of membrane proteins. The discovery of aquaporins (AQPs) and the demonstration of their role in homeostasis of water in various cells and organs suggested that lipid-mediated transport of very small molecules, while providing an important means, might not be sufficient for the desired level of transport that is required, e.g., for physiologically optimal activity of certain cells (10, 77).
Figure 1.
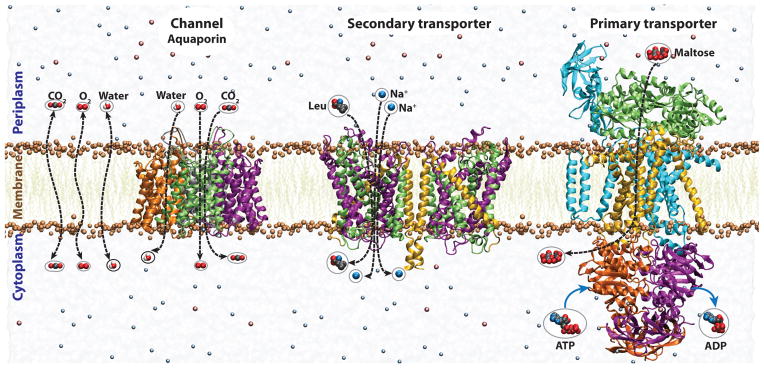
Various modes of transport across the cellular membrane. Three membrane proteins, a channel, mammalian aquaporin-1 (AQP1), a secondary transporter, leucine transporter (LeuT), and a primary transporter, maltose ABC transporter, are shown. The proteins are embedded in a lipid bilayer and surrounded by water (gray) and ions (Na+, blue, Cl−, red). Water and gas molecules can diffuse (downhill along the concentration gradient) either through the lipid bilayer or through AQP channels. In transporters, the transport is usually against the electrochemical gradient of the substrate, and therefore needs to be charged by another source of the cellular energy, namely ionic gradients in the case of secondary transporters, and ATP in primary transporters.
A detailed characterization of structural and dynamical properties of lipid bilayers and membrane proteins is necessary for studying transport processes in biological membranes at high resolution. Recent advances in experimental structural biology complemented by biophysical measurements have provided invaluable information on structural and dynamical elements involved in the function of membrane proteins, and in characterizing membrane processes. However, a significant number of functional aspects remain unresolved. The challenge is to achieve simultaneously the high temporal and spatial resolutions required for a detailed description of the molecular events and processes involved in membrane transport. Processes such as the mechanism of permeation and selectivity of membrane channels, co-transport of ions along with a neurotransmitter by a secondary transporter, and passive diffusion of hydrophobic molecules through the space (gaps) between the lipid molecules in a membrane can only be “visualized” by methods that offer a dynamical view of the system at hand, at an atomic resolution. Molecular dynamics (MD) simulations offer an unparalleled combination of these resolutions, and, thus, provide information that cannot be obtained currently by experimental methodologies.
In this article, we will review the application of MD to the study of transport of very small molecules across the membrane. Both lipid-mediated and protein-mediated modes of transport of these molecular species have been studied with MD and will be discussed here. Permeation of water or O2 through membrane channels represent fast phenomena (on the order of nanoseconds), and, thus, are ideal applications for MD simulations (21, 102, 113, 114, 116, 117). Given the simplicity of their primary function, i.e., water transport, and the wealth of structural information (36), AQPs have been extensively studied with MD simulations (21, 102, 113, 114, 116, 117). These studies have significantly contributed to our understanding of the mechanisms of permeation, substrate selectivity, and gating in these channels. AQP research has particularly benefited from close collaborative studies between experiment and computation where either new experiments were designed based on the results of the simulations or hypotheses developed by the simulations were corroborated by experiments (24, 26, 32, 75, 105, 121). Keeping the focus of the review on the application of MD to study permeation processes, we have refrained from a comprehensive discussion on various functional aspects of AQPs. Rather, we use AQPs as an example to demonstrate the application of MD and to showcase the various types of information, both qualitative and quantitative, that can be derived from the method.
Molecular Dynamics Simulations
Basic concepts of MD
Molecular dynamics (MD) and its application to molecular systems are based on basic concepts of physics and chemistry, in particular classical mechanics and statistical mechanics. From an algorithmic perspective, the method has evolved along with the developments in mathematics and computer science. MD is a numerical method that integrates the Newtonian equations of motion, e.g., for individual atoms in a given molecular system (3, 34). During a “classical” MD simulation, the interactions between the atoms are calculated using a pre-determined set of parameters that define a potential energy function, U, also known as a force field. The potential energy functions of various force fields available for biomolecular simulations could be different, but they all generally include terms that describe major bonded (bonds, angles, and dihedral angles), and non-bonded (van der Waals and electrostatic) modes of interaction:
The parameters used for these energy terms are derived from a combination of experimental data and quantum mechanical calculations (41) and are tuned to optimally reproduce the structure and vibrational modes of the molecular systems of interest, as well as their thermodynamic properties. At each time step during an MD simulation, the total force on each atom is calculated as the negative derivative of the potential energy with respect to its coordinates. Using these forces, the Newtonian equations of motion are then integrated to produce the trajectory of individual atoms. The result of an MD simulation is, therefore, a trajectory (collection of snapshots or configurations) of the system over a certain period of time, usually tens to hundreds of nanoseconds . These snapshots can be used to describe the dynamics of the system and to calculate macroscopic properties using principles of statistical mechanics.
Fig. 2 shows a typical membrane protein simulation system, featuring the water channel protein aquaporin-1 (AQP1) embedded in a lipid bilayer composed of 187 POPE lipids (~80,000 atoms). Explicit representation of lipid and water in these simulations provides a natural environment for the protein, and also allows one to perform the simulations under various pressures, when desired. In order to mimic the experimental conditions, MD simulations are often carried out under constant-temperature and constant-pressure conditions.
Figure 2.
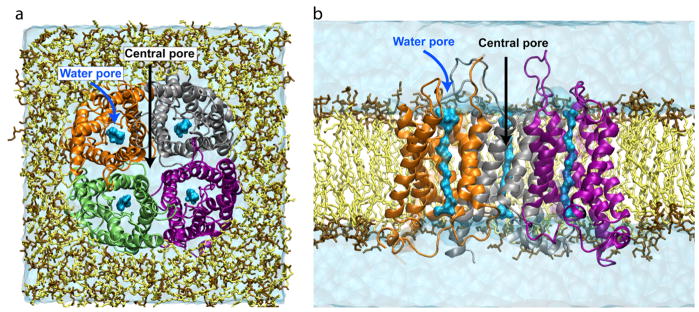
Membrane protein simulation system. Top (a) and side view (b) of the simulation system of a mammalian AQP1 tetramer embedded in a pure POPE bilayer. In the side view, the front monomer is reviewed for clarity. Water molecules permeating water pores within individual AQP1 monomers are represented by a blue space-filling representation, while bulk water is shown in a light blue transparent box. The locations of the water pores and the central pore are indicated by arrows.
Strengths and limitations of MD
In most all-atom MD simulations, the integration has to be performed every 1–2 femtoseconds (10−15 s) in order to describe properly the motion of atoms and to avoid numerical instability. As a result, current MD simulations of macromolecular systems are limited to the nanosecond-microsecond time scales, although consistent development in software and advances in computer hardware continue to tackle this limitation, as evidenced by very recent studies reporting simulations on the order of hundreds of microseconds (94). A second shortcoming associated with “classical” MD simulations is their inadequate treatment of electronic polarization effects (74) and simplified potential energy functions, precluding a complete description of inter-atomic interactions and accurate dynamics of the system. With regard to specific application of the method to membrane systems, such shortcomings have resulted, e.g., in underestimation of area per lipid (30, 31), and, thus, inaccuracies in the density and even the phase of the lipid bilayer under investigation. Moreover, the slow diffusion of lipid molecules continues to keep us from satisfactory sampling of mixed lipid bilayers.
Despite these limitations, MD has been successfully employed in studying a wide range of biomolecular systems and phenomena (66). In particular, and in regard to the subject of this review, membranes and membrane proteins have been widely studied by MD simulations (68, 73, 93, 118).
MD has the advantage of providing dynamical information of a system at very high spatial and temporal resolutions. Starting from an experimentally determined structure, e.g., those solved by x-ray crystallography or NMR measurements, MD allows the researcher to “watch” the natural motion of the molecular system and to monitor its dynamical behavior in real time. Therefore, MD can be considered as the in silico version of a conventional microscope, with the advantage of providing a live view of the molecule at an atomic resolution. Time-averaged properties computed from an MD trajectory can be compared to macroscopic quantities that are measured experimentally (5, 96, 125). Furthermore, MD offers researchers the opportunity to explore their systems under an unlimited number of artificial conditions that are often inaccessible experimentally. For instance, not only can one modify a residue to mimic the effect of an experimentally studied point mutation, but also can selectively screen, or even completely neutralize, the electrostatic forces generated by any given group of atoms (11, 102).
Enhanced sampling techniques
In the past few decades, MD has been used in the study of a wide range of biological phenomena, e.g., protein folding (25), ion conduction (90), and muscle elasticity (97). The rapid increase of the computational power and recent developments in simulation software have enabled MD studies of significantly larger systems, e.g., the ribosome in complex with a protein-conducting channel (2.7 million atoms) (40), and of much longer processes (hundreds of microseconds) (94). In spite of the striking progress, a substantial gap remains between the time scales currently accessible to MD simulations (on the order of nanosecond-microsecond) and the time scale of “slow” biological processes, e.g., those that involve large-scale domain motions of macromolecular systems (microseconds to milliseconds or even longer). In order to bridge this gap, special simulation and sampling techniques have been developed that allow an enhanced sampling of the configuration space, e.g., by enforcing certain molecular events in the system (5, 14, 38). Here, we briefly describe a number of enhanced sampling techniques that are used by studies reviewed in this article. For a more comprehensive discussion, we refer the reader to a number of excellent reviews on the subject (5, 14, 38).
In steered molecular dynamics (SMD) (57), external forces are applied to one or a group of atoms, in order to induce certain molecular events during the simulation, or to drive the system from one state to another. Some of the first applications of the method to biomolecular processes include mechanical characterization of the muscle protein titin in order to interpret the results of AFM experiments (37), and describing the unbinding pathway of a ligand from its receptor (59). More relevant examples to this review include SMD simulation of permeation of glycerol through AQPs (62), as well as induction of hydrostatic pressure difference for either water (103, 124–126) or gas molecules (113, 114, 116, 117), across the membrane by adding forces that drive these molecules toward the membrane on one side, and away from it on the other side, thus, generating a chemical potential gradient across the membrane. Apart from probing the system qualitatively, SMD trajectories have also been used to calculate the free energy difference (3Δ) between the states of the system (62, 85).
Other enhanced sampling techniques that are developed particularly with the purpose of calculating 3Δ include free energy perturbation (71, 127), thermodynamic integration (70, 98), and umbrella sampling (89), as well as the more recently developed methods of adaptive biasing force (ABF) (18, 47) and implicit ligand sampling (ILS) (15). The ILS method, which calculates a 3D free energy map from simulations without explicit presence of the ligand (e.g., gas molecules), has proven particularly useful in characterizing gas migration pathways within proteins (15) and lipid bilayers (113, 117) that might not be sampled during a standard MD simulation.
Simulation Studies of Water Conduction by Aquaporins
AQPs are a family of membrane proteins best characterized as water channels, since they increase the membrane's permeability to water and other small hydrophilic substrates. They form tetramers in the membrane, with each monomer acting as an independent water pore (1, 7, 65). An AQP monomer consists of six transmembrane helices and two half-membrane spanning repeats related by a quasi-two-fold symmetry. These two repeats, which line a large portion of the water pore, contain each a highly conserved asparagine-proline-alanine (NPA) motif (51) and meet approximately at the center of the pore (Fig. 3a). In the periplasmic half of the water pore, a region lined by aromatic residues and a conserved arginine forms the narrowest part of the pore, often referred to as the selectivity filter (SF) (91). To date, over 14 structures have been determined for different members of the AQP family, including E. coli GlpF (35, 102) and AqpZ (91), human AQP1 (79), AQP2 (92), AQP4 (53), and AQP5 (54), bovine AQP1 (99) and AQP0 (39), rat AQP4 (52), sheep AQP0 (44), spinach SoPIP2;1 (105), archaeal AqpM (72), yeast AQY1 (32), as well as PfAQP from the malarial parasite Plasmodium falciparum (83). As a result, AQPs have become the richest family of membrane channels with regard to the abundance of atomic-resolution structures.
Figure 3.
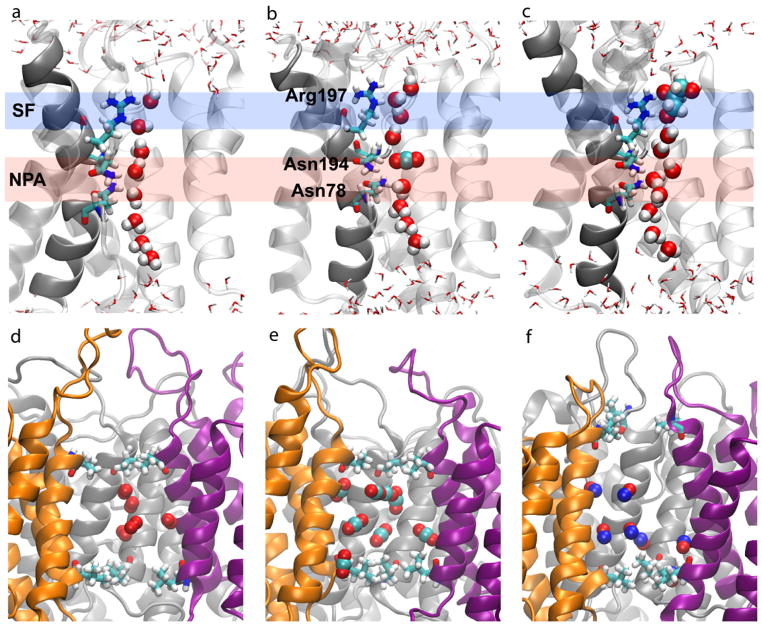
Conduction pathways for small molecules in AQPs. (a-c) Conduction of water (a), CO2 (b), and glycerol (c) by the water pores of AQPs (64, 113). (d-f) Spontaneous entrance and accumulation of O2 (d), CO2 (e), and NO (f) in the central pore captured during equilibrium MD simulations (113, 117). The bipolar orientation of water can be seen in (a), where water molecules in both halves of the channel point their oxygen towards the center of the pore. Two half-membrane spanning helices (dark gray) and the asparagine residues (Asn) from the NPA motifs, along with the conserved arginine (Arg) in the selectivity filter (SF) are shown in (a-c). The outermost hydrophobic residues defining the narrowest regions in the central pore are explicitly shown in (d-f).
The first set of MD simulations of AQPs (19, 102) were prompted by the high-resolution structures of GlpF (35) and AQP1 (79) and aimed at understanding the dynamics and selectivity of water permeation. These studies, which employed equilibrium simulation conditions, i.e., in the absence of any biasing force, found that water molecules form a continuous single file and move in a highly correlated manner within the water pores of individual AQP monomers (19, 64, 102). Every water molecule forms on the average two hydrogen bonds with neighboring water molecules in addition to a hydrogen bond with the protein (Fig. 3a). This fine-tuned hydrogen bond network, where water-water interaction is elegantly supplemented by protein-water interaction, allows AQPs to achieve a high efficiency in water conduction. The importance of the natural motions of pore-lining residues in water conduction by AQPs was also clearly demonstrated by simulations in which artificially confining the motion of these residues resulted in a complete block of water conduction (95, 115).
Apart from elucidating the dynamics of water permeation, MD simulations have made significant contribution to our understanding of the selectivity of AQPs. While conducting water at a rate close to its self-diffusion limit, AQPs strictly exclude protons, a property which is critical to the maintenance of the electrochemical potential across the membrane (122). However, given the high mobility of protons in bulk water, the mechanism of proton exclusion by AQPs remained a long-standing puzzle. A significant number of simulation studies (9, 11, 13, 20, 21, 67, 102) have provided a detailed insight into the fine-tuned electrostatic forces that form the basis for this selectivity mechanism. These studies have shown that water molecules within each water pore form a characteristic “bipolar” orientation (Fig. 3a) (102). This unique orientation of water reflects a positive electrostatic potential at the center of protein, which is mainly generated by the dipole moments of the two half-membrane spanning helices (11, 20, 102). Together with the desolvation effect (9, 67), the positive electrostatic potential effectively blocks the passage of protons, while leaving the rapid water conduction unaffected (22).
Calculation of the permeability coefficient pf from MD trajectories
The motion of individual water molecules in MD simulations can be closely monitored. Therefore, all the required information to calculate various kinetic properties of the process of water permeation through AQPs is fully accessible from an MD trajectory. Experimentally, the rate of water conduction is characterized by the osmotic permeability coefficient (pf ), a quantity that is measured in the presence of an osmotic (or hydrostatic) pressure gradient across the membrane. Several MD studies reported the calculation of pf from the simulated trajectories (22, 23, 45, 46, 61, 124, 126). Initially, a pressure gradient across the membrane was generated by applying small forces to individual water molecules in the simulation system, in order to bias their translocation, and hence, establish a net flow through an AQP channel (124, 126). Later studies (125) developed theories that related pf to the diffusive behavior of water inside the channel during equilibrium simulations, thereby, allowing for the calculation of pf from equilibrium MD simulations. The calculated value of pf for AQP1 from MD simulations is 7.1–7.5×10–14 cm3s–1 (22, 126), which is in good agreement with the experimental value of 5.43×10−14 cm−s−1 (112). Experimentally, pf values on the order of 10−14 cm3s−1 have been reported for different members of the AQP family (28, 120). A notable exception with this regard is AQP0, in which the water flow appears to be about an order of magnitude slower than in other AQPs (12, 120). Recent MD studies have suggested that the “phenolic barriers” formed by two luminal tyrosines are responsible for the reduced rate of water transport by AQP0 (42, 60).
Energetics of substrate permeation through the water pores
Given adequate sampling, the energetics associated with the molecular phenomenon at hand can be readily calculated from MD simulations. Since water permeation through the water pores of AQPs happens on a nanosecond time scale, all reported equilibrium MD simulations (21, 45, 63, 84, 101, 102, 105, 113, 117, 123) have been able to collect ample sampling on the dynamics of water inside the water pores. The relatively fast translocation of water in the water pores has resulted in adequate sampling of all points along the pore axis in all reported equilibrium MD simulations (21, 45, 63, 84, 101, 102, 105, 113, 117, 123), from which a free energy profile for water permeation can be reconstructed based on the probability distribution of water along the pore. Calculations based on this method have shown that water permeation through the water pores of AQPs requires crossing barriers of ~3 kcal/mol (see Table 1) (55, 117).
Table 1.
Free energy barriers against permeation of water and small gas molecules in AQPs and lipid bilayers. The values are collected from MD simulations performed on human AQP1 (55), bovine AQP1 (113), rat AQP4 (117), E. coli GlpF (55) and two model lipid bilayers, POPE (55, 117) and POPC (55).
| Water pore | Central pore | Lipid bilayer | ||||||
|---|---|---|---|---|---|---|---|---|
| Molecule | hAQP1 | bAQP1 | AQP4 | GlpF | bAQP1 | AQP4 | POPE | POPC |
| O2 | 6.5* | 5.7‡ | 4.9 | 3.0* | 3.6‡ | 2.9 | 0.4 | 1.0† |
| CO2 | 5.3* | – | – | 3.2* | 3.6‡ | – | 1.0† | 0.4† |
| NO | – | – | 5.8 | – | – | 2.9 | 0.3 | – |
| NH3 | 4.3* | – | – | 3.0* | – | – | 4.5† | 3.6† |
| H2O | 3.4 | – | 3.0 | 3.2 | – | – | 7.4† | 6.4† |
All values are in kcal/mol. *Statistical errors for these results are ≤0.6 kcal/mol (55).
Statistical errors for these values are ≤0.5 kcal/mol (55).
Statistical errors for these results are obtained using a special formula (15). The upper bound error is ≤ +0.25 kcal/mol, and the lower bound errors are ≤−1.7 kcal/mol and ≤−0.6 kcal/mol for the water pore and the centeral pore, respectively (113). No statistical errors were given for the remaining free energy results.
In addition to water, some AQPs are also permeable to other hydrophilic compounds. In particular, a subfamily of AQPs known as aquaglyceroporins are able to conduct glycerol and other small linear sugar molecules. The energetics associated with glycerol permeation through the E. coli aquaglyceroporin GlpF has also been calculated from MD simulations (48, 62). In this case, however, the significantly slower diffusion of glycerol along the pore did not allow the entire process, i.e., translocation of a glycerol molecule from one end of the channel to the other, to be captured in equilibrium MD simulations (63). As such, enhanced sampling methods had to be adopted to ensure glycerol occupancy of all points along the pore axis, in order to reconstruct the energetics associated with its permeation (48, 62). Jensen et al. performed SMD simulations to induce the permeation of glycerol with an external force (62). The free energy profile calculated from these simulations revealed a barrier of 7.3 kcal/mol (62), which is rougly comparable with the 9.6±1.5 kcal/mol Arrhenius activation energy estimated from experiments (8). Using the ABF method (18, 47), Henin et al. calculated a comparable energy barrier of 8.7 kcal/mol (48). We note that a notably smaller energy barrier (3.2 kcal/mol) was reported in a recent umbrella sampling study (55). Both Jensen et al. and Henin et al. also examined the stereoselectivity of the channel, and showed that the gauche-gauche conformation of glycerol is favored at the SF region, with the non-polar backbone of glycerol facing the hydrophobic side of the channel, and its three hydroxyl groups forming hydrogen bonds with the polar residues (Fig. 3c). Accordingly, it was suggested that the search for an optimal conformation at the SF region constitutes the rate-limiting step of glycerol permeation through GlpF (48).
Gating of the water pores
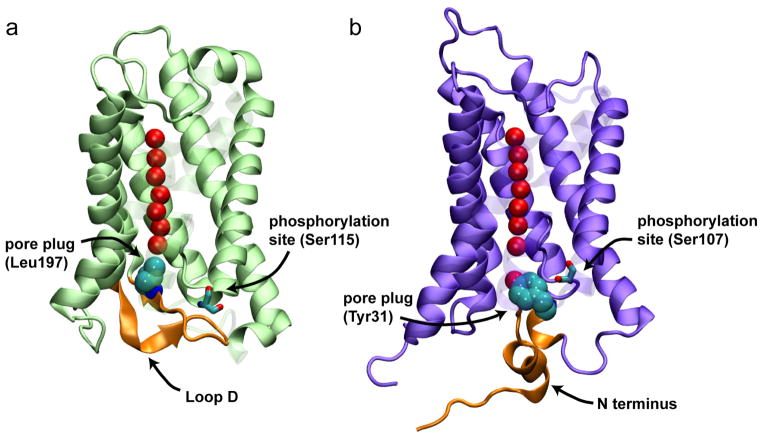
While largely known as always-open water channels, gating of the water pores has been reported for a number of AQPs, most prominently for plant AQPs (39, 75, 82, 106). Gating of AQPs is believed to provide the cell with a mechanism to modulate their permeability under conditions such as drought and flooding. It has been shown, for instance, that plant AQPs can be regulated by phosphorylation of highly conserved serine residues, or by variations of the intracellular pH (39, 82, 106). A recent combined experimental and MD study on yeast AQY1 suggests that AQPs might also be gated by changes in membrane curvature or surface tension, a mechanism reminiscent of the gating of the mechanosensitive ion channels (32). Regardless of the gating signal, a rather consistent gating mechanism has emerged from the structural and MD studies of these AQPs (Fig. 4). In spinach AQP, SoPIP2;1, the phosphorylation-mediated gating has been linked to the coupled movement of a pore plug (a conserved leucine residue that lines the pore in the cytoplasmic half) and one of the cytoplasmic loops that detects the phosphorylation signal or the intracellular pH change (69, 84, 105). In the yeast AQY1, a conserved tyrosine acts as a plug, while the extended N terminus has been suggested to act as the sensor of the membrane tension (32). As discussed in recent structural and MD studies (33, 69, 84, 109), gating of AQPs may be mediated by a diverse set of mechanisms which couple the motion of different signal-detecting domains (sensors) to the pore plug.
Figure 4.
Proposed gating mechanisms for a spinach aquaporin (a) and for yeast AQY1 (b). Upon phosphorylation of conserved serine residues the two AQP channels switch to an open state through the coupling of a plug residue (Leu197 in SoPIP2;1 and Tyr31 in AQY1) and a cytoplasmic region of the protein (loop D in SoPIP2;1 and the N-terminus in AQY1; shown in orange). The N terminus of AQY1 is also suggested to act as a sensor to the membrane tension (32).
Lipid- and Protein-Mediated Transmembrane Gas Conduction
Since the first report on the CO2 permeability of human AQP1 (86), the involvement of AQPs in gas conduction across cellular membranes has attracted marked attention from both experimentalists (6, 16, 17, 27, 29, 43, 49, 50, 58, 78, 80, 81, 104, 107, 108, 110, 119) and theoreticians (55, 56, 113, 117). Several studies have suggested that AQPs can conduct gas molecules across biological membranes, and that this conduction can be of physiological relevance. For instance, the effect of tobacco AQP, NtAQP1, on photosynthesis and stomatal opening has been attributed to its role in the facilitating transmembrane CO2 permeation (107). More recently, it was demonstrated that human AQP1 may conduct NO when reconstituted in liposomes (50). Later, through the measurement of acetylcholine-induced vasorelaxation in AQP1 knockout mice, the direct involvement of AQP1 in NO-dependent vasorelaxation was demonstrated in vivo (49). Apart from participating in gas permeation, relative CO2 and NH3 permeability measurements have suggested that AQPs might even exhibit selectivity for different gas molecules (80). However, contradicting bodies of evidence also exist (29, 78, 110, 119). For example, it has been shown that AQP1 deletion has no effect on CO2 transport in erythrocytes, lungs, or in kidneys (29, 119).
At the heart of the debate over the role of AQPs in gas conduction are two questions: first, whether AQPs can conduct gas molecules, and second, whether AQP-mediated gas conduction is of physiological relevance. These questions turn out to be very difficult to answer experimentally, primarily due to the fact that gas permeation through AQPs is often masked by the high gas permeability of the surrounding artificial lipid bilayer used to reconstruct the protein. While the answer to the second question (physiological relevance) remains to be determined experimentally, most likely in in vivo settings, MD simulations provide a powerful technique to address the first question, namely the availability of gas conduction pathways in AQPs, or in other membrane proteins for that matter. A number of reported MD studies (55, 56, 113, 117) have investigated this problem, and it is now well established that AQPs indeed are permeable to gas molecules. Interestingly, these studies have shown that the central pore of AQPs (the pore formed between the four monomers, see Fig. 2) provides a more favorable pathway for hydrophobic gas species than the water pores. The calculated energy barrier against O2 permeation through the water pores is found to be 5–6 kcal/mol, whereas the barrier for the same process through the central pore is only 3–4 kcal/mol. A summary of the energy barriers against gas permeation through AQPs calculated by MD simulations is given in Table 1. Below, after summarizing the results of the MD simulations of gas permeation through lipid bilayers and AQPs, we will discuss their implications with regard to the physiological relevance of AQP-mediated gas conduction.
Gas permeability of the water pores
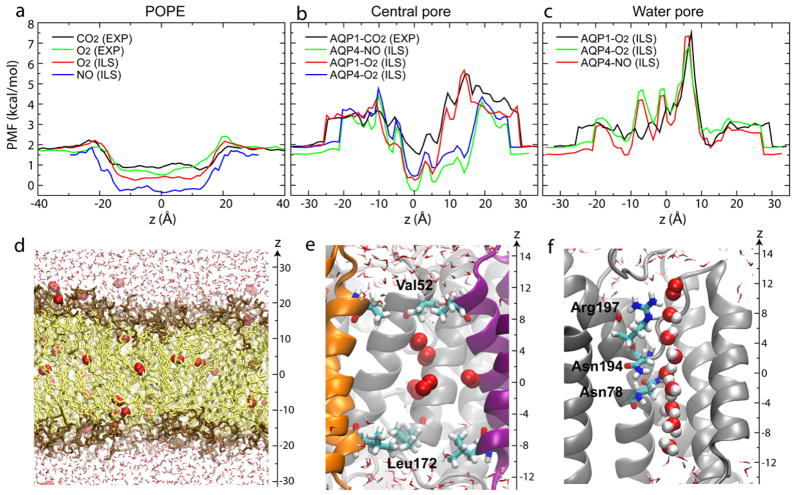
Using umbrella sampling, Hub and de Groot (56) showed that the highest energy barrier against CO2 permeation through the water pores of AQP1 is 5.5 kcal/mol and is located at the SF region (see Fig. 3). Using a combination of complementary sampling techniques, i.e., explicit ligand sampling, implicit ligand sampling (113), and umbrella sampling (117), comparable energy barriers (~5 kcal/mol) were calculated for the permeation of O2 and NO permeation through the water pores of AQP1 (113) and AQP4 (117) (Fig. 5c). Comparison of the energy barriers against gas permeation through the water pores calculated by various MD simulations (Table 1) shows that despite the difference in methodology, these studies have yielded a rather consistent picture, namely, an energy barrier of 5–6 kcal/mol for the permeation of hydrophobic species (CO2, O2 and NO) through the water pores of AQPs, and a lower barrier (4 kcal/mol) for the more hydrophilic NH3. These energy barriers are all located at the SF region (Fig. 5c) and appear to arise from the loss of favorable water-water and water-protein interactions in the SF (55) upon the insertion of the gas molecule into the single file of water within the water pores (Fig. 5f). The more hydrophilic NH3 partially compensates for this loss by NH3-protein interactions, and, hence, faces a smaller energy barrier during its permeation (55).
Figure 5.
Energetics of gas permeation through AQPs and pure lipid bilayers. (a-c) Free energy profiles of gas permeation (113, 117) through a pure POPE bilayer (a), the central pore (b) and the water pores of AQPs (c). (d-f) Snapshots of equilibrium simulations with oxygen molecule(s) inside the POPE bilayer (d), the central pore (e), and the water pore (f) of AQP1 (113). The reference point of all free energy profiles is the ideal gas phase (vacuum) (113). The scale used in (d) is different from that in (e) and (f).
It is noteworthy that the water pore of the aquaglyceroporin GlpF is found to have a significantly lower energy barrier against gas permeation (55). As shown in Table 1, a barrier of only ~3 kcal/mol is obtained for the permeation of different gas species through the water pores in GlpF (55). This low and non-discriminant barrier is explained by the wider and more hydrophobic SF region of GlpF when compared with pure water channels such as AQP1 and AQP4. Indeed, it has been shown that introduction of more hydrophobic residues in the SF of AQP1 can dramatically reduce the energy barrier against gas permeation (55). In human, AQP3, AQP7, and AQP9 are aquaglyceroporins (2). Although no atomic structures are currently available for these AQPs, they share a high level of sequence identity with GlpF, especially with regard to the SF region. Therefore, comparably high gas conduction properties can be expected for these human AQPs.
Gas permeability of the central pore
Besides the four monomeric pores, a fifth pore, named the central pore, is formed in the middle of the four monomers in an AQP tetramer (Fig. 2). While the function of the water pores has been well characterized, no functional implication has been experimentally established for the central pore. The central pore is largely lined by hydrophobic residues, which along with the small radius of the pore, result in an effective exclusion of water molecules. During the MD simulations, the central pore stays completely dry, effectively blocking the entrance and permeation of water molecules. Therefore, it is unlikely for the central pore to contribute to the experimentally measured overall water conduction rate of AQPs. The MD simulations, however, have revealed a novel property of the central pore, namely its high affinity for gas molecules; the pore acts as an ideal gas reservoir that is rapidly filled with gas molecules during the simulations. Furthermore, it serves as the most favorable pathway for permeation of hydrophobic gas molecules through the protein, specifically when compared to the water pores (56, 113). This property has been demonstrated by equilibrium MD simulations of AQP1 and AQP4, where the gas molecules initially placed in solution were shown to rapidly accumulate in the central pore (Fig. 3d-f) during the simulations (113, 117). Furthermore, free energy calculations have revealed an energy barrier of only 3–4 kcal/mol (56, 113, 117) against the permeation of CO2, O2, and NO through the central pore, supporting the view that the pore indeed serves as a favorable gas conduction pathway.
Physiological relevance of AQP-mediated gas conduction
The physiological significance of gas permeation through AQPs will depend on their relative gas permeability in comparison to the surrounding lipid bilayers. Recent experimental studies offer different views with this regard. Endeward et al. suggested that AQP1 is a major pathway for CO2 transport across the human erythrocyte membrane (27), while Missner et al. reported that the resistance to transmembrane CO2 permeation is mainly caused by the unstirred layer, instead of the membrane itself, thereby, suggesting that facilitation of CO2 transport by AQPs is not physiologically important (78). MD simulations performed on artificial lipid bilayers containing, e.g., 100% POPE or POPC, show that these pure phospholipid bilyers produce a very small energy barrier against the permeation of hydrophobic gas molecules such as O2 and CO2 (55, 56, 113, 117). For instance, the energy barrier of a pure POPE bilayer to O2 permeation is only 0.4 kcal/mol (117), which is essentially negligible compared to the barrier of O2 permeation through AQPs. The only exception with this regard seems to be the hydrophilic NH3, in which case a barrier of 4.5 kcal/mol for its permeation through a pure lipid bilayer has been calculated (55). These results support the notion that AQPs are unlikely to be of significance in the overall gas exchange across a “model” membrane. Nevertheless, “real” cellular membranes can differ dramatically, both in lipid composition and structure, from the model membranes used in the simulation and experimental studies. Therefore, the physiological relevance of AQP-mediated gas conduction will remain unknown until the composition of the lipid bilayers embedding AQPs in a living cell have been determined and used accordingly in experimental or computational studies.
It should be also noted that despite a lower permeability of AQPs for gas molecules in comparison to pure lipid bilayers, AQPs might occupy a significant portion of the membrane surface. Under these conditions, the AQP-mediated gas permeation might become physiologically significant. AQP4 presents a case in which, this scenario might apply. AQP4 is the predominant water channel in the central nervous system (CNS) (76, 111), where it is most abundantly present in astroglial cells that surround capillaries and form glia limitans, as well as in ependymal cells lining ventricles (4, 52, 100). Freeze fracture and immunogold labeling experiments have shown that AQP4 forms extensive 2D square arrays in the membrane of these cells (87, 88), where large sets of tightly packed AQP4 proteins occupy a significant portion of the membrane. The high density of AQP4 in these cells suggests that a non-trivial fraction of gas exchange in certain regions in the CNS might take place through pathways provided by the protein. Interestingly, recent experimental studies report that AQP1 can mediate NO permeation both in vitro and in vivo (49, 50). Consistent with the intriguing notion that AQP4 might play a role in the conduction of this important signaling molecule in the CNS, MD simulations have found a small energy barrier against NO permeation through AQP4 (117).
Concluding Remarks
In this article, we have reviewed some of the recent molecular dynamics (MD) studies of membranes and membrane proteins, specifically seeking to showcase the power of the method in describing molecular events involved in the translocation of small molecules across the cellular membrane. As exemplified, a wide range of phenomena can be captured at a high resolution by MD simulations. The calculated trajectories have been used to reveal, e.g., a novel and unique configuration of water molecules in AQPs, and the basis of substrate selectivity and proton exclusion. MD simulations continue to provide a powerful platform to develop and test new hypotheses regarding the relevant functional aspects of the system at hand, for instance, the involvement of electrostatic forces in the selectivity of AQPs. On the more quantitative side, MD simulations have been used to calculate key thermodynamical and kinetic properties, e.g., the permeability coefficients of different AQPs, and the free energy profiles associated with the permeation of various substrates along different conduction pathways.
With regard to the exchange of very small molecules (water, CO2, O2 and NO) across the membrane, there is currently no evidence suggesting the involvement of active (energy-dependent) transporters in the process. Therefore, the exchange of these molecules across the cellular membrane appears to be merely mediated by their passive diffusion through transient or permanent pathways in the membrane. Such pathways can be in principle provided by either the lipid bilayer itself, or by proteins that are embedded in the membrane. For charged molecules, as well as for the majority of polar molecules, the hydrophilicity of the pathway is critical for their efficient permeation, an aspect that explains why the conduction of such species is exclusively through membrane proteins (channels). Very small molecules, in particular hydrophobic gas molecules, on the other hand, seem to favor hydrophobic translocation pathways, e.g., transient pores that constantly form between the lipid molecules due to their structural fluctuation, or hydrophobic pores and pathways within membrane proteins which are expected to be more permanent in nature due to the relatively stable structure of proteins when compared to lipids. Extensive MD simulations of AQPs have provided strong evidence that AQPs, and likely other membrane proteins, can indeed conduct gas molecules, although the physiological significance of the phenomenon remains to be established. It is interesting that the central (tetrameric) pore of AQPs, for which no function has been demonstrated experimentally, turns out to be the most favorable conduction pathway for gas molecules. The formation of an additional pore that is involved in the permeation of substrates other than the main substrate of the channel (water) might be even speculated to be one of the reasons for oligomerization of AQPs, and maybe for other oligomeric membrane proteins.
Acknowledgments
The authors wish to acknowledge support from the NIH (grants R01-GM086749 and R01-GM067887), as well as computational resources provided by TeraGrid (grant number MCA06N060).
References
- 1.Agre P. The aquaporin water channels. Proc Am Thorac Soc. 2006;3:5–13. doi: 10.1513/pats.200510-109JH. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agre P, Bonhivers M, Borgnia MJ. The aquaporins, blueprints for cellular plumbing systems. J Biol Chem. 1998;273:14659–14662. doi: 10.1074/jbc.273.24.14659. [DOI] [PubMed] [Google Scholar]
- 3.Allen MP, Tildesley DJ. Computer Simulation of Liquids. New York: Oxford University Press; 1987. [Google Scholar]
- 4.Amiry-Moghaddam M, Otsuka T, Hurn P, Traystman R, Haug F, Froehner S, Adams M, Neely J, Agre P, Ottersen O, Bhardwaj A. An alpha-syntrophin-dependent pool of AQP4 in astroglial end-feet confers bidirectional water flow between blood and brain. Proc Natl Acad Sci USA. 2003;100:2106–2111. doi: 10.1073/pnas.0437946100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beveridge DL, DiCapua FM. Free energy via molecular simulation: Applications to chemical and biological systems. Annu Rev Biophys Biophys Chem. 1989;18:431–492. doi: 10.1146/annurev.bb.18.060189.002243. [DOI] [PubMed] [Google Scholar]
- 6.Blank M, Ehmke H. Aquaporin-1 and HCO3−-Cl− transporter-mediated transport of CO2 across the human erythrocyte membrane. J Physiol. 2003;550:419–29. doi: 10.1113/jphysiol.2003.040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borgnia M, Nielsen S, Engel A, Agre P. Cellular and molecular biology of the aquaporin water channels. Annu Rev Biochem. 1999;68:425–458. doi: 10.1146/annurev.biochem.68.1.425. [DOI] [PubMed] [Google Scholar]
- 8.Borgnia MJ, Agre P. Reconstitution and functional comparison of purified GlpF and AqpZ, the glycerol and water channels from Escherichia coli. Proc Natl Acad Sci USA. 2001;98:2888–2893. doi: 10.1073/pnas.051628098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burykin A, Warshel A. On the origin of the electrostatic barrier for proton transport in aquaporin. FEBS Lett. 2004;570:41–46. doi: 10.1016/j.febslet.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 10.Carbrey JM, Agre P. Discovery of the aquaporins and development of the field. Handb Exp Pharmacol. 2009;190:3–28. doi: 10.1007/978-3-540-79885-9_1. [DOI] [PubMed] [Google Scholar]
- 11.Chakrabarti N, Tajkhorshid E, Roux B, Pomès R. Molecular basis of proton blockage in aquaporins. Structure. 2004;12:65–74. doi: 10.1016/j.str.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 12.Chandy G, Zampighi G, Kreman M, Hall J. Comparison of the water transporting properties of MIP and AQP1. J Membr Biol. 1997;159:29–39. doi: 10.1007/s002329900266. [DOI] [PubMed] [Google Scholar]
- 13.Chen H, Wu Y, Voth GA. Origins of proton transport behavior from selectivity domain mutations of the aquaporin-1 channel. Biophys J. 2006;90:L73–5. doi: 10.1529/biophysj.106.084061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christen M, van Gunsteren W. On searching in, sampling of, and dynamically moving through conformational space of biomolecular systems: A review. J Comp Chem. 2008;29:157–66. doi: 10.1002/jcc.20725. [DOI] [PubMed] [Google Scholar]
- 15.Cohen J, Arkhipov A, Braun R, Schulten K. Imaging the migration pathways for O2, CO, NO, and Xe inside myoglobin. Biophys J. 2006;91:1844–1857. doi: 10.1529/biophysj.106.085746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooper G, Boron W. Effect of PCMBS on CO2 permeability of Xenopus Oocytes expressing aquaporin 1 or its C189S mutant. Am J Physiol. 1998;275:C1481–C1486. doi: 10.1152/ajpcell.1998.275.6.C1481. [DOI] [PubMed] [Google Scholar]
- 17.Cooper G, Zhou Y, Bouyer P, Grichtchenko I, Boron W. Transport of volatile solutes through AQP1. J Physiol. 2002;542:17–29. doi: 10.1113/jphysiol.2002.023218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darve E, Pohorille A. Calculating free energies using average force. J Chem Phys. 2001;115:9169–9183. [Google Scholar]
- 19.de Groot BL, Engel A, Grubmüller H. A refined structure of human aquaporin-1. FEBS Lett. 2001;504:206–211. doi: 10.1016/s0014-5793(01)02743-0. [DOI] [PubMed] [Google Scholar]
- 20.de Groot BL, Frigato T, Helms V, Grubmüller H. The mechanism of proton exclusion in the aquaporin-1 water channel. J Mol Biol. 2003;333:279–293. doi: 10.1016/j.jmb.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 21.de Groot BL, Grubmüller H. Water permeation across biological membranes: Mechanism and dynamics of aquaporin-1 and GlpF. Science. 2001;294:2353–2357. doi: 10.1126/science.1066115. [DOI] [PubMed] [Google Scholar]
- 22.de Groot BL, Grubmüller H. The dynamics and energetics of water permeation and proton exclusion in aquaporins. Curr Opin Struct Biol. 2005;15:176–183. doi: 10.1016/j.sbi.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 23.de Groot BL, Tieleman DP, Pohl P, Grubmüller H. Water permeation through gramicidin A: Desformylation and the double helix: A molecular dynamics study. Biophys J. 2002;82:2934–2942. doi: 10.1016/S0006-3495(02)75634-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Detmers FJM, de Groot BL, Mller EM, Hinton A, Konings IBM, Sze M, Flitsch SL, Grubmller H, Deen PMT. Quaternary Ammonium Compounds as Water Channel Blockers. Journal of Biological Chemistry. 2006;281:14207–14214. doi: 10.1074/jbc.M513072200. [DOI] [PubMed] [Google Scholar]
- 25.Duan Y, Kollman P. Pathways to a protein folding intermediate observed in a 1 microsecond simulation in aqueous solution. Science. 1998;282:740–744. doi: 10.1126/science.282.5389.740. [DOI] [PubMed] [Google Scholar]
- 26.Dynowski M, Mayer M, Moran O, Ludewig U. Molecular determinants of ammonia and urea conductance in plant aquaporin homologs. FEBS Letters. 2008;582:2458–2462. doi: 10.1016/j.febslet.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 27.Endeward V, Musa-Aziz R, Cooper G, Chen L, Pelletier M, Virkki L, Supuran C, King L, Boron W, Gros G. Evidence that Aquaporin 1 is a major pathway for CO2 transport across the human erythrocyte membrane. FASEB J. 2006;20:1974–1981. doi: 10.1096/fj.04-3300com. [DOI] [PubMed] [Google Scholar]
- 28.Engel A, Stahlberg H. Aquaglyceroporins: channel proteins with a conserved core, multiple functions, and variable surfaces. Int Rev Cytol. 2002;215:75–104. doi: 10.1016/s0074-7696(02)15006-6. [DOI] [PubMed] [Google Scholar]
- 29.Fang X, Yang B, Matthay M, Verkman A. Evidence against aquaporin-1-dependent CO2 permeability in lung and kidney. J Physiol. 2002;542:63–9. doi: 10.1113/jphysiol.2001.013813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feller SE. Molecular dynamics simulations of lipid bilayers. Curr Opin Coll & Interf Sci. 2000;5:217–223. [Google Scholar]
- 31.Feller SE, Pastor RW. Constant surface tension simulations of lipid bilayers: The sensitivity of surface areas and compressibilities. J Chem Phys. 1999;111:1281–1287. [Google Scholar]
- 32.Fischer G, Kosinska-Eriksson U, Aponte-Santamar C, Palmgren M, Geijer C, Hedfalk K, Hohmann S, de Groot BL, Neutze R, Lindkvist-Petersson K. Crystal structure of a yeast aquaporin at 1.15 Å reveals a novel gating mechanism. 3. PLoS Biol. 2009;7:e1000130. doi: 10.1371/journal.pbio.1000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fischer M, Kaldenhoff R. On the pH regulation of plant aquaporins. J Biol Chem. 2008;283:33889–33892. doi: 10.1074/jbc.M803865200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frenkel D, Smit B. Understanding Molecular Simulation From Algorithms to Applications. California: Academic Press; 2002. [Google Scholar]
- 35.Fu D, Libson A, Miercke LJW, Weitzman C, Nollert P, Krucinski J, Stroud RM. Structure of a glycerol conducting channel and the basis for its selectivity. Science. 2000;290:481–486. doi: 10.1126/science.290.5491.481. [DOI] [PubMed] [Google Scholar]
- 36.Fu D, Lu M. The structural basis of water permeation and proton exclusion in aquaporins. Mol Memb Biol. 2007;24:366–374. doi: 10.1080/09687680701446965. [DOI] [PubMed] [Google Scholar]
- 37.Gao M, Wilmanns M, Schulten K. Steered molecular dynamics studies of titin I1 domain unfolding. Biophys J. 2002;83:3435–3445. doi: 10.1016/S0006-3495(02)75343-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gilson M, Given J, Bush B, McCammon J. The statistical-thermodynamic basis for computation of binding affinities: A critical review. Biophys J. 1997;72:1047–1069. doi: 10.1016/S0006-3495(97)78756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gonen T, Sliz P, Kistler J, Cheng Y, Walz T. Aquaporin-0 membrane junctions reveal the structure of a closed water pore. Nature. 2004;429:193–197. doi: 10.1038/nature02503. [DOI] [PubMed] [Google Scholar]
- 40.Gumbart J, Trabuco LG, Schreiner E, Villa E, Schulten K. Regulation of the protein-conducting channel by a bound ribosome. Structure. 2009;17:1453–1464. doi: 10.1016/j.str.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guvench O, MacKerell AD. Comparison of protein force fields for molecular dynamics simulations. Methods Mol Biol. 2008;443:63–88. doi: 10.1007/978-1-59745-177-2_4. [DOI] [PubMed] [Google Scholar]
- 42.Han BG, Guliaev AB, Walian PJ, Jap BK. Water transport in AQP0 aquaporin: molecular dynamics studies. J Mol Biol. 2006;360:285–296. doi: 10.1016/j.jmb.2006.04.039. [DOI] [PubMed] [Google Scholar]
- 43.Hanba YT, Shibasaka M, Hayashi Y, Hayakawa T, Kasamo K, Terashima I, Katsuhara M. Overexpression of the barley aquaporin HvPIP2;1 increases internal CO2 conductance and CO2 assimilation in the leaves of transgenic rice plants. Plant Cell Physiol. 2004;45:521–529. doi: 10.1093/pcp/pch070. [DOI] [PubMed] [Google Scholar]
- 44.Harries WEC, Akhavan D, Miercke LJW, Khademi S, Stroud R. The channel architecture of aquaporin 0 at a 2.2-angstrom resolution. Proc Natl Acad Sci USA. 2004;101:14045–14050. doi: 10.1073/pnas.0405274101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hashido M, Ikeguchi M, Kidera A. Comparative simulations of aquaporin family: AQP1, AQPZ, AQP0 and GlpF. FEBS Lett. 2005;579:5549–5552. doi: 10.1016/j.febslet.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 46.Hashido M, Kidera A, Ikeguchi M. Water transport in aquaporins: osmotic permeability matrix analysis of molecular dynamics simulations. Biophys J. 2007;93:373–385. doi: 10.1529/biophysj.106.101170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hénin J, Chipot C. Overcoming free energy barriers using unconstrained molecular dynamics simulations. J Chem Phys. 2004;121:2904–2914. doi: 10.1063/1.1773132. [DOI] [PubMed] [Google Scholar]
- 48.Henin J, Tajkhorshid E, Schulten K, Chipot C. Diffusion of glycerol through Escherichia coli aquaglyceroporin GlpF. Biophys J. 2008;94:832–839. doi: 10.1529/biophysj.107.115105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Herrera M, Garvin J. Novel role of AQP-1 in NO-dependent vasorelaxation. Am J Physiol Renal Physiol. 2007;292:F1443–51. doi: 10.1152/ajprenal.00353.2006. [DOI] [PubMed] [Google Scholar]
- 50.Herrera M, Hong NJ, Garvin JL. Aquaporin-1 transports NO across cell membranes. Hypertension. 2006;48:157–164. doi: 10.1161/01.HYP.0000223652.29338.77. [DOI] [PubMed] [Google Scholar]
- 51.Heymann J, Engel A. Structural clues in the sequences of the aquaporins. JMB. 2000;295:1039–1053. doi: 10.1006/jmbi.1999.3413. [DOI] [PubMed] [Google Scholar]
- 52.Hiroaki Y, Tani K, Kamegawa A, Gyobu N, Nishikawa K, Suzuki H, Walz T, Sasaki S, Mitsuoka K, Kimura K, Mizoguchi A, Fujiyoshi Y. Implications of the Aquaporin-4 structure on array formation and cell adhesion. J Mol Biol. 2006;355:628–639. doi: 10.1016/j.jmb.2005.10.081. [DOI] [PubMed] [Google Scholar]
- 53.Ho JD, Yeh R, Sandstrom A, Chorny I, Harries WEC, Robbins RA, Miercke LJW, Stroud RM. Crystal structure of human aquaporin 4 at 1.8 Å and its mechanism of conductance. Proc Natl Acad Sci USA. 2009;106:7437–442. doi: 10.1073/pnas.0902725106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Horsefield R, Nordén K, Fellert M, Backmark Å, Törnroth-Horsefield S, van Scheltinga AT, Kvassman J, Kjellbom P, Johanson U, Neutze R. High-resolution X-ray structure of human aquaporin 5. Proc Natl Acad Sci USA. 2008;105:13327–32. doi: 10.1073/pnas.0801466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hub J, de Groot B. Mechanism of selectivity in aquaporins and aquaglyceroporins. Proc Natl Acad Sci USA. 2008;105:1198–203. doi: 10.1073/pnas.0707662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hub JS, de Groot BL. Does CO2 permeate through Aquaporin-1? Biophys J. 2006;91:842–848. doi: 10.1529/biophysj.106.081406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Isralewitz B, Gao M, Schulten K. Steered molecular dynamics and mechanical functions of proteins. Curr Opin Struct Biol. 2001;11:224–230. doi: 10.1016/s0959-440x(00)00194-9. [DOI] [PubMed] [Google Scholar]
- 58.Ivanov II, Loktyushkin AV, Guskova RA, Vasilev NS, Fedorov GE, Rubin AB. Oxygen channels of erythrocyte membrane. Dokl Biochem Biophys. 2007;414:137–40. doi: 10.1134/s160767290703012x. [DOI] [PubMed] [Google Scholar]
- 59.Izrailev S, Stepaniants S, Balsera M, Oono Y, Schulten K. Molecular dynamics study of unbinding of the avidin-biotin complex. Biophys J. 1997;72:1568–1581. doi: 10.1016/S0006-3495(97)78804-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jensen M, Dror R, Xu H, Borhani D, Arkin I, Eastwood M, Shaw D. Dynamic control of slow water transport by aquaporin 0: implications for hydration and junction stability in the eye lens. Proc Natl Acad Sci USA. 2008;105:14430–5. doi: 10.1073/pnas.0802401105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jensen MØ, Mouritsen OG. Single-channel water permeabilities of Escherichia coli aquaporins AqpZ and GlpF. Biophys J. 2006;90:2270–2284. doi: 10.1529/biophysj.105.073965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jensen MØ, Park S, Tajkhorshid E, Schulten K. Energetics of glycerol conduction through aquaglyceroporin GlpF. Proc Natl Acad Sci USA. 2002;99:6731–6736. doi: 10.1073/pnas.102649299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jensen MØ, Tajkhorshid E, Schulten K. The mechanism of glycerol conduction in aquaglyceroporins. Structure. 2001;9:1083–1093. doi: 10.1016/s0969-2126(01)00668-2. [DOI] [PubMed] [Google Scholar]
- 64.Jensen MØ, Tajkhorshid E, Schulten K. Electrostatic tuning of permeation and selectivity in aquaporin water channels. Biophys J. 2003;85:2884–2899. doi: 10.1016/S0006-3495(03)74711-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jung JS, Preston GM, Smith BL, Guggino WB, Agre P. Molecular structure of the water channel through aquaporin CHIP – the hourglass model. J Biol Chem. 1994;269:14648– 14654. [PubMed] [Google Scholar]
- 66.Karplus M, McCammon JA. Molecular dynamics simulations of biomolecules. Nat Struct Biol. 2002;265:654–652. doi: 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
- 67.Kato M, Pisliakov AV, Warshel A. The barrier for proton transport in aquaporins as a challenge for electrostatic models: the role of protein relaxation in mutational calculations. Proteins: Struct, Func, Bioinf. 2007;93:373–385. doi: 10.1002/prot.21012. [DOI] [PubMed] [Google Scholar]
- 68.Khalili-Araghi F, Gumbart J, Wen PC, Sotomayor M, Tajkhorshid E, Schulten K. Molecular dynamics simulations of membrane channels and transporters. Curr Opin Struct Biol. 2009;19:128–137. doi: 10.1016/j.sbi.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khandelia H, Jensen MØ, Mouritsen OG. To gate or not to gate: using molecular dynamics simulations to morph gated plant aquaporins into constitutively open conformations. J Phys Chem B. 2009;113:5239–5244. doi: 10.1021/jp809152c. [DOI] [PubMed] [Google Scholar]
- 70.Kirkwood J. Statistical mechanics of fluid mixtures. J Chem Phys. 1935;3:300–313. [Google Scholar]
- 71.Kollman P. Free energy calculations: Applications to chemical and biochemical phenomena. Chem Rev. 1993;93:2395–2417. [Google Scholar]
- 72.Lee JK, Kozono D, Remis J, Kitagawa Y, Agre P, Stroud R. Structural basis for conductance by the archaeal aquaporin AqpM at 1.68 Å. Proc Natl Acad Sci USA. 2005;102:18932– 18937. doi: 10.1073/pnas.0509469102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lindahl E, Sansom MSP. Membrane proteins: molecular dynamics simulations. Curr Opin Struct Biol. 2008;18:425–431. doi: 10.1016/j.sbi.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 74.Lopes PEM, Roux B, MacKerell AD., Jr Molecular modeling and dynamics studies with explicit inclusion of electronic polarizability: theory and applications. Theoret Chim Acta. 2009;124:11–28. doi: 10.1007/s00214-009-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ludewig U, Dynowski M. Plant aquaporin selectivity: where transport assays, computer simulations and physiology meet. Cell Mol Life Sci. 2009;66:3161–3175. doi: 10.1007/s00018-009-0075-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Manley GT, Binder DK, Papadopoulos MC, Verkman AS. New insights into water transport and edema in the central nervous system from phenotype analysis of aquaporin-4 null mice. Neuroscience. 2004;129:983–991. doi: 10.1016/j.neuroscience.2004.06.088. [DOI] [PubMed] [Google Scholar]
- 77.Maurel C, Verdoucq L, Luu DT, Santoni V. Plant aquaporins: membrane channels with multiple integrated functions. Annu Rev Plant Biol. 2008;59:595–624. doi: 10.1146/annurev.arplant.59.032607.092734. [DOI] [PubMed] [Google Scholar]
- 78.Missner A, Küler P, Saparov S, Sommer K, Matthai J, Zeidel M, Pohl P. Carbon dioxide transport through membranes. J Biol Chem. 2008 doi: 10.1074/jbc.M800096200. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Murata K, Mitsuoka K, Hirai T, Walz T, Agre P, Heymann JB, Engel A, Fujiyoshi Y. Structural determinants of water permeation through aquaporin-1. Nature. 2000;407:599–605. doi: 10.1038/35036519. [DOI] [PubMed] [Google Scholar]
- 80.Musa-Aziz R, Chen LM, Pelletier MF, Boron WF. Relative CO2/NH3 selectivities of AQP1, AQP4, AQP5, AmtB, and RhAG. Proc Natl Acad Sci USA. 2009;106:5406–5411. doi: 10.1073/pnas.0813231106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nakhoul N, Davis B, Romero M, Boron W. Effect of expressing the water channel aquaporin-1 on the CO2 permeability of Xenopus oocytes. Am J Physiol. 1998;274:C543–548. doi: 10.1152/ajpcell.1998.274.2.C543. [DOI] [PubMed] [Google Scholar]
- 82.Nemeth-Cahalan K, Hall J. pH and calcium regulate the water permeability of aquaporin 0. J Biol Chem. 2000;275:6777–82. doi: 10.1074/jbc.275.10.6777. [DOI] [PubMed] [Google Scholar]
- 83.Newby ZER, Joseph O'Connell III, Robles-Colmenare Y, Khadem S, Mierck LJ, Stroud RM. Crystal structure of the aquaglyceroporin PfAQP from the malarial parasite Plasmodium falciparum. Nat Struct Mol Biol. 2008;15:619–625. doi: 10.1038/nsmb.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nyblom M, Frick A, Wang Y, Ekvall M, Hallgren K, Hedfalk K, Neutze R, Tajkhorshid E, Törnroth-Horsefield S. Structural and functional analysis of SoPIP2;1 mutants add insight into plant aquaporin gating. J Mol Biol. 2009;387:653–668. doi: 10.1016/j.jmb.2009.01.065. [DOI] [PubMed] [Google Scholar]
- 85.Park S, Khalili-Araghi F, Tajkhorshid E, Schulten K. Free energy calculation from steered molecular dynamics simulations using Jarzynski's equality. J Chem Phys. 2003;119:3559– 3566. [Google Scholar]
- 86.Prasad GVT, Coury L, Finn F, Zeidel M. Reconstituted aquaporin 1 water channels transport CO2 across membranes. J Biol Chem. 1998;273:33123–33126. doi: 10.1074/jbc.273.50.33123. [DOI] [PubMed] [Google Scholar]
- 87.Rash J, Davidson K, Yasumura T, Furman C. Freeze-fracture and immunogold analysis of aquaporin-4 (AQP4) square arrays, with models of AQP4 lattice assembly. Neuroscience. 2004;129:915–34. doi: 10.1016/j.neuroscience.2004.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rash J, Yasumura T, Hudson C, Agre P, Nielsen S. Direct immunogold labeling of aquaporin-4 in square arrays of astrocyte and ependymocyte plasma membranes in rat brain and spinal cord. Proc Natl Acad Sci USA. 1998;95:11981–6. doi: 10.1073/pnas.95.20.11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Roux B. The calculation of the potential of mean force using computer simulations. Computer Physics Communications. 1995;91:275–282. [Google Scholar]
- 90.Roux B. Ion conduction and selectivity in K+ channels. Annu Rev Biomol Struc Dyn. 2005;34:153–171. doi: 10.1146/annurev.biophys.34.040204.144655. [DOI] [PubMed] [Google Scholar]
- 91.Savage DF, Egea PF, Robles-Colmenares Y, O'Connell JD, III, Stroud RM. Architecture and selectivity in aquaporins: 2.5 Å X-ray structure of aquaporin Z. PLoS Biol. 2003;1:e72. doi: 10.1371/journal.pbio.0000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schenk A, Werten P, Scheuring S, de Groot B, Muller S, Stahlberg H, Philippsen A, Engel A. The 4.5 Å structure of human AQP2. J Mol Biol. 2005;350:278–89. doi: 10.1016/j.jmb.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 93.Shaikh SA, Wen PC, Enkavi G, Huang Z, Tajkhorshid E. Capturing Functional Motions of Membrane Channels and Transporters with Molecular Dynamics Simulation. J Comput Theor Nanosci. doi: 10.1166/jctn.2010.1636. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shaw DE, Dror RO, Salmon JK, Grossman J, Mackenzie KM, Bank JA, Young C, Deneroff MM, Batson B, Bowers KJ, Chow E, Eastwood MP, Ierardi DJ, Klepeis JL, Kuskin JS, Larson RH, Lindorff-Larsen K, Maragakis P, Moraes MA, Piana S, Shan Y, Towles B. Millisecond-scale molecular dynamics simulations on anton. Proceedings of the ACM/IEEE Conference on Supercomputing; 2009. [Google Scholar]
- 95.Smolin N, Li B, Beck DA, Daggett V. Side-chain dynamics are critical for water permeation through aquaporin-1. Biophys J. 2008;95:1089–1098. doi: 10.1529/biophysj.107.125187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sotomayor M, Corey DP, Schulten K. In search of the hair-cell gating spring: Elastic properties of ankyrin and cadherin repeats. Structure. 2005;13:669–682. doi: 10.1016/j.str.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 97.Sotomayor M, Schulten K. Single-molecule experiments in vitro and in silico. Science. 2007;316:1144–1148. doi: 10.1126/science.1137591. [DOI] [PubMed] [Google Scholar]
- 98.Straatsma T, McCammon J. Multiconfiguration thermodynamic integration. J Chem Phys. 1991;95:1175–1188. [Google Scholar]
- 99.Sui H, Han BG, Lee JK, Walian P, Jap BK. Structural basis of water-specific transport through the AQP1 water channel. Nature. 2001;414:872–878. doi: 10.1038/414872a. [DOI] [PubMed] [Google Scholar]
- 100.Sulyok E, Vajda Z, Dóczi T, Nielsen S. Aquaporins and the central nervous system. Acta Neurochir (Wien) 2004;146:955–60. doi: 10.1007/s00701-004-0319-z. [DOI] [PubMed] [Google Scholar]
- 101.Tajkhorshid E, Aksimentiev A, Balabin I, Gao M, Isralewitz B, Phillips JC, Zhu F, Schulten K. Large scale simulation of protein mechanics and function. In: Richards FM, Eisenberg DS, Kuriyan J, editors. Advances in Protein Chemistry. Vol. 66. New York: Elsevier Academic Press; 2003. pp. 195–247. [DOI] [PubMed] [Google Scholar]
- 102.Tajkhorshid E, Nollert P, Jensen MØ, Miercke LJW, O'Connell J, Stroud RM, Schulten K. Control of the selectivity of the aquaporin water channel family by global orientational tuning. Science. 2002;296:525–530. doi: 10.1126/science.1067778. [DOI] [PubMed] [Google Scholar]
- 103.Tajkhorshid E, Zhu F, Schulten K. Kinetic theory and simulation of single-channel water transport. In: Yip S, editor. Handbook of Materials Modeling, Vol. I: Methods and Models. Netherlands: Springer; 2005. pp. 1797–1822. [Google Scholar]
- 104.Terashima I, Ono K. Effects of HgCl2 on CO2 dependence of leaf photosynthesis: Evidence indicating involvement of aquaporins in CO2 diffusion across the plasma membrane. Plant Cell Physiol. 2002;43:70–78. doi: 10.1093/pcp/pcf001. [DOI] [PubMed] [Google Scholar]
- 105.Törnroth-Horsefield S, Wang Y, Hedfalk K, Johanson U, Karlsson M, Tajkhorshid E, Neutze R, Kjellbom P. Structural mechanism of plant aquaporin gating. Nature. 2006;439:688–694. doi: 10.1038/nature04316. [DOI] [PubMed] [Google Scholar]
- 106.Tournaire-Roux C, Sutka M, Javot H, Gout E, Gerbeau P, Luu D, Bligny R, Maurel C. Cytosolic pH regulates root water transport during anoxic stress through gating of aquaporins. Nature. 2003;425:393–397. doi: 10.1038/nature01853. [DOI] [PubMed] [Google Scholar]
- 107.Uehlein N, Lovisolo C, Siefritz F, Kaldenhoff R. The tobacco aquaporin NtAQP1 is a membrane CO2 pore with physiological functions. Nature. 2003;425:734–737. doi: 10.1038/nature02027. [DOI] [PubMed] [Google Scholar]
- 108.Uehlein N, Otto B, Hanson D, Fischer M, McDowell N, Kaldenhoff R. Function of nicotiana tabacum aquaporins as chloroplast gas pores challenges the concept of membrane CO2 permeability. Plant Cell. 2008;20:648–57. doi: 10.1105/tpc.107.054023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Verdoucq L, Grondin A, Maurel C. Structure-function analysis of plant aquaporin AtPIP2;1 gating by divalent cations and protons. Biochem J. 2008;415:409–416. doi: 10.1042/BJ20080275. [DOI] [PubMed] [Google Scholar]
- 110.Verkman A. Does aquaporin-1 pass gas? An opposing view. J Physiol. 2002;542:31. doi: 10.1113/jphysiol.2002.024398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Verkman A. More than just water channels: unexpected cellular roles of aquaporins. J Chem Soc. 2005;118:3225–32. doi: 10.1242/jcs.02519. [DOI] [PubMed] [Google Scholar]
- 112.Walz T, Smith BL, Zeidel ML, Engel A, Agre P. Biologically active two-dimensional crystals of aquaporin chip. J Biol Chem. 1994;269:1583–1586. [PubMed] [Google Scholar]
- 113.Wang Y, Cohen J, Boron WF, Schulten K, Tajkhorshid E. Exploring gas permeability of cellular membranes and membrane channels with molecular dynamics. J Struct Biol. 2007;157:534–544. doi: 10.1016/j.jsb.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 114.Wang Y, Ohkubo YZ, Tajkhorshid E. Gas conduction of lipid bilayers and membrane channels. In: Feller S, editor. Current Topics in Membranes: Computational Modeling of Membrane Bilayers. chapter 60. Elsevier; 2008. pp. 343–367. [Google Scholar]
- 115.Wang Y, Schulten K, Tajkhorshid E. What makes an aquaporin a glycerol channel: A comparative study of AqpZ and GlpF. Structure. 2005;13:1107–1118. doi: 10.1016/j.str.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 116.Wang Y, Tajkhorshid E. Molecular mechanisms of conduction and selectivity in aquaporin water channels. J Nutr. 2007;137:1509S–1515S. doi: 10.1093/jn/137.6.1509S. [DOI] [PubMed] [Google Scholar]
- 117.Wang Y, Tajkhorshid E. Nitric oxide conduction by the brain aquaporin AQP4. Proteins: Struct, Func, Bioinf. 2009 doi: 10.1002/prot.22595. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wen PC, Huang Z, Enkavi G, Wang Y, Gumbart J, Tajkhorshid E. Molecular mechanisms of active transport across the cellular membrane. In: Biggin P, Sansom M, editors. Molecular Simulations and Biomembranes: From Biophysics to Function. Royal Society of Chemistry; 2009. [Google Scholar]
- 119.Yang B, Fukuda N, van Hoek A, Matthay MA, Ma T, Verkman AS. Carbon dioxide permeability of aquaporin-1 measured in erythrocytes and lung of aquaporin-1 null mice and in reconstituted proteoliposomes. J Biol Chem. 2000;275:2682–92. doi: 10.1074/jbc.275.4.2686. [DOI] [PubMed] [Google Scholar]
- 120.Yang B, Verkman AS. Water and glycerol permeabilities of aquaporins 1–5 and MIP determined quantitatively by expression of epitope-tagged constructs in Xenopus oocytes. J Biol Chem. 1997;272:16140–16146. doi: 10.1074/jbc.272.26.16140. [DOI] [PubMed] [Google Scholar]
- 121.Yu J, Yool AJ, Schulten K, Tajkhorshid E. Mechanism of gating and ion conductivity of a possible tetrameric pore in Aquaporin-1. Structure. 2006;14:1411–1423. doi: 10.1016/j.str.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 122.Zeidel ML, Nielsen S, Smith BL, Ambudkar SV, Maunsbach AB, Agre P. Ultra-structure, pharmacological inhibition, and transport selectivity of aquaporin channel-forming integral protein in proteoliposomes. Biochemistry. 1994;33:1606–1615. doi: 10.1021/bi00172a042. [DOI] [PubMed] [Google Scholar]
- 123.Zhu F, Tajkhorshid E, Schulten K. Molecular dynamics study of aquaporin-1 water channel in a lipid bilayer. FEBS Lett. 2001;504:212–218. doi: 10.1016/s0014-5793(01)02749-1. [DOI] [PubMed] [Google Scholar]
- 124.Zhu F, Tajkhorshid E, Schulten K. Pressure-induced water transport in membrane channels studied by molecular dynamics. Biophys J. 2002;83:154–160. doi: 10.1016/S0006-3495(02)75157-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhu F, Tajkhorshid E, Schulten K. Collective diffusion model for water permeation through microscopic channels. Phys Rev Lett. 2004;93:224501. doi: 10.1103/PhysRevLett.93.224501. (4 pages) [DOI] [PubMed] [Google Scholar]
- 126.Zhu F, Tajkhorshid E, Schulten K. Theory and simulation of water permeation in aquaporin-1. Biophys J. 2004;86:50–57. doi: 10.1016/S0006-3495(04)74082-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zwanzig RW. High-temperature equation of state by a perturbation method. I. Nonpolar gases. J Chem Phys. 1954;22:1420–1426. [Google Scholar]