Abstract
Purpose
Adolescent Idiopathic Scoliosis (AIS) is considered a complex genetic disease, in which malfunctioning or dysregulation of one or more genes has been proposed to be responsible for the expressed phenotype. However, to date, no disease causing genes has been identified and the pathogenesis of AIS remains unknown. The aim of this study is, therefore, to identify specific molecules with differing expression patterns in AIS compared to healthy individuals.
Methods
Microarray analysis and quantitative RT-PCR have examined differences in the gene transcription profile between primary osteoblasts derived from spinal vertebrae of AIS patients and those of healthy individuals.
Results
There are 145 genes differentially expressed in AIS osteoblasts. A drastic and significant change has been noted particularly in the expression levels of Homeobox genes (HOXB8, HOXB7, HOXA13, HOXA10), ZIC2, FAM101A, COMP and PITX1 in AIS compared to controls. Clustering analysis revealed the interaction of these genes in biological pathways crucial for bone development, in particular in the differentiation of skeletal elements and structural integrity of the vertebrae.
Conclusions
This study reports on the expression of molecules that have not been described previously in AIS. We also provide for the first time gene interaction pathways in AIS pathogenesis. These genes are involved in various bone regulatory and developmental pathways and many of them can be grouped into clusters to participate in a particular biological pathway. Further studies can be built on our findings to further elucidate the association between different biological pathways and the pathogenesis of AIS.
Keywords: Adolescent idiopathic scoliosis, Gene expression, Microarray, Bone development
Introduction
Idiopathic Scoliosis (IS) is characterized by a three-dimensional deformity of the spine and its incidence in the general population ranges from 0.15 to 4 % [1]. Adolescent IS (AIS) accounts for 80 % of IS [2]. The origin and cause of AIS remains unknown to date but there are several proposed etiological hypotheses [3] including melatonin deficiency [4] or signaling defect [5], connective tissue abnormalities [6], asymmetries in the central nervous system [7]. abnormal distribution and interaction between melatonin and calmodulin [5, 8] hormonal variation [9], diet, and posture. In addition, there is strong evidence of genetic predisposition to AIS. For instance, familial occurrences of AIS have been reported by many research groups, and concordance for this condition in twins’ studies further strengthens the genetic influence on the etiology of AIS. However, controversy exists as to whether the mode of inheritance is multifactorial trait an autosomal dominant trait, or even an X-linked dominant trait. AIS is considered as a complex genetic disease, in which one or more genes may be responsible for the expressed phenotype, and in which several modifying effects, such as age, sex, and environment, may play specific roles in the phenotypic variation between affected individuals; it is most likely premature to assign responsibility to a single gene.
Recently, chromosomal regions on 6, 10 and 18q [10], 17p11.2, 19p13.3, Xq23–26.1, 8q11, 9q31.2–q34.2, 17q25.3–qtel, 12p13.31 and recently 3q12.1 and 5q13.3 [11] have been associated with AIS. Even genome-wide association studies (GWAS) have recently been used to study genetic predisposition for AIS and although polymorphisms associated with AIS have been described in SNTG1 on 8q11.22, ESR1 on 6q25.1, MATN1 on 1p35, CHD7 on 8q12.1, MTNR1B on 11q21–q22 and CHL1 [12], no specific genes or proteins have been identified as players in the development of scoliosis. Therefore to gain an insight into the pathogenesis of AIS, we used a microarray approach to study specific alterations in the genetic expression profile of AIS osteoblasts. Our microarray results show that specific subsets of genes are differentially expressed in AIS, which was confirmed for the most part by reverse-transcription-quantitative PCR (RT-qPCR). Furthermore, we observe that the differentially regulated genes could be grouped and assigned to various functional categories, indicating that many regulatory pathways could be involved in AIS pathogenesis.
Materials and methods
Patients
Six unrelated individuals with AIS, and six controls (non-AIS individuals), all French-Canadian females from Quebec were studied. They were examined by the Adam’s test and by a standing upright radiograph of the spine. Two independent blinded orthopedic surgeons read the X-rays (clinical features of patients, Table 1). All AIS patients were selected by the same criteria, namely the spinal deformity was a right-thoracic progressive curve requiring corrective spinal surgery. The Cobb angle ranged between 30° and 84°. For the control patients, spinal deformity was excluded by X-ray and clinical examination: they were subjected to spine surgery for traumatic injury. For the experimental design, we choose individuals with the same features for each group to get homogeneous populations. Bone fragments excised during surgery were used to isolate osteoblasts as described below. Each participating subject or, in the case of minors, their legal guardian, gave informed consent. The research protocol was approved by the Research Ethics Committee of Sainte-Justine Hospital.
Table 1.
Clinical Features of AIS patients
| Patient | Sex | Age at presentation (years) | Location of primary curve | Cobb’s angle at diagnosis | Spinal surgery | Bracing | AIS Family history/ethnicity/origin |
|---|---|---|---|---|---|---|---|
| 1 | Female | 14.97 | Right thoracic | 30° | Yes | No | Yes/Caucasian/French Canadian |
| 2 | Female | 12.72 | Right thoracic | 63° | Yes | Yes | Yes/Caucasian/French Canadian |
| 3 | Female | 14.74 | Right thoracic | 32° | Yes | No | No/Caucasian/French Canadian |
| 4 | Female | 14.46 | Right thoracic | 78° | Yes | No | Yes/Caucasian/French Canadian |
| 5 | Female | 13.26 | Right-left thoracic | 58°–49° | Yes | No | Yes/Caucasian/French Canadian |
| 6 | Female | 17.78 | Right thoracic | 84° | Yes | Yes | Yes/Caucasian/French Canadian |
Six females with AIS were selected based upon described criteria. The table presents patient characteristics
Primary human osteoblast culture and RNA extraction
Briefly, bone fragments were cultivated in alpha-MEM (supplemented with 10 % (v/v) fetal bovine serum (FBS, Wisent) and 2 mM glutamine, with 100 U/mL penicillin and 100 μg/mL streptomycin (Invitrogen, Burlington, ON, Canada) as antibiotics) at 37 °C. 5 % CO2 for a period of 28 days, after which the osteoblasts derived from the bone pieces were separated from the remaining bone fragments by trypsinization. To confirm the osteoblasts phenotype, cells were stained for alkaline phosphatase, osteocalcin, osteopontin and collagen type I as we previously described in Letellier et al. [13]. RNA was extracted from osteoblasts using TRIzol Reagent (Invitrogen), according to the manufacturer’s instructions, and verification of RNA integrity and concentration were carried out with the Agilent Bioanalyzer 2100 in concert with the Agilent RNA 6,000 nano or pico kit (Agilent) (RNA Quality Testing Services, McGill University and Génome Québec Innovation Centre Montréal Canada).
Microarray gene expression profiling
RNA samples were analyzed using 12 individual Illumina Human HT-12 v3 BeadChip microarrays, which contain probes for 48,804 unique gene expression sequences (from NCBI RefSeq build 38). with 99.99 % coverage specification. Preparatory cDNA synthesis and labeling. microarray hybridization reactions, and data collection were performed according to established protocols at the McGill University and Génome Québec Innovation Centre. Microarray expression data were subsequently analyzed using the FlexArray software (version 1.6.1) a front-end to R and Bioconductor. Probe intensity data were normalized across replicate arrays by robust multi-array average (RMA) and differential gene expression was calculated by empirical Bayes analyses of microarrays (EBAM) with Benjamini-Hochberg false-discovery rate (FDR) correction. Gene expression profiles from primary osteoblasts derived from spinal vertebrae of AIS patients (All AIS with right thoracic curve; n = 6) were compared with profiles from the same cells collected from age and sex-matched healthy individuals (n = 6). Microarray analysis was conducted on six AIS-Control sample pairs and the data were normalized. To determine those genes that were differentially expressed between AIS cells and control cells, fold-changes between AIS and control cells were calculated. All values were expressed as positive or negative fold changes. Genes that were differentially expressed >1.5-fold, relative to control patient levels, were considered as differentially regulated. Significance analyses of microarrays (SAM) algorithm were then used to calculate FDR-adjusted q-values according to the method of Storey; q-values <0.15 were considered statistically significant.
Functional classification clustering
To compare similarities in gene function in our list of differentially regulated genes, we used the Database for Annotation, Visualization, and Integrated Discovery (DAVID) functional gene clustering algorithm (version 6.7). 145 differentially regulated genes were selected based on our criteria as set forth, and this list was uploaded to the DAVID functional gene clustering web interface. The software compares the uploaded gene list to a gene–gene similarity matrix of over 75,000 functional annotation terms, and generates a cluster map of functionally similar genes using fuzzy heuristic partitioning.
Hierarchical clustering
To reveal potential gene–gene associations in our expression data, we performed hierarchical clustering analysis using Cluster 3.0 software. Briefly, we loaded our list of 145 differentially regulated genes, with the corresponding difference in fold change (as determined above), into the software. We then performed hierarchical clustering calculations using Euclidean distance as a similarity metric, with average linkage as the clustering method. The resulting dendrogram was visualized using Java TreeView (version 1.1.5r2).
Reverse-transcription quantitative PCR
To provide confirmation for our microarray results, the expression levels of a subset of up- and down-regulated genes (in AIS osteoblasts. compared to controls) were evaluated by RT-qPCR analysis. Quantitative PCR was performed for the following genes, selected from the list of genes with the highest fold-changes and those which seem interesting from clustering and functional analysis results. Table 2 displays the primers sequences. Total RNA was prepared from osteoblasts from three AIS patients and from three controls, as described above. Reverse transcription, using poly-deoxythymidine oligos (Invitrogen) as transcription primers, was then performed on 500 ng of RNA that had been treated with ribonuclease-free deoxyribonuclease I (Invitrogen). Quantitative PCR was performed, using SYBR GREEN chemistry as a marker for DNA amplification, on an ABI Prism 7900HT fast real-time PCR system, with 40 cycles of a stepwise amplification (once for 2 min at 50 °C, once for 10 min at 95 °C, 40 times for 15 s at 95 °C, followed by measurement for 1 min at 60 °C). Dissociation curve analysis was performed to ensure product specificity. The fold change of expression was calculated in relation to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an internal reference gene, and the expression level was then determined relative to control osteoblasts. Amplification plots, dissociation curves, and threshold cycle (Ct) values were generated by ABI Sequence Detection System software (version 2.4) after data collection, and expression fold-changes were calculated for each gene by the delta–delta Ct method. Individual genes were compared in between AIS patients and controls using Student’s t test.
Table 2.
RT-qPCR primers sequences for validated targets
| Gene name | Ref_Seq mRNA | Forward primer (5′–3′) | Reverse primer (3′–5′) |
|---|---|---|---|
| HOXB8 | NM_024016 | GTC CGT GCG CGC CAA TTA TTA | GCC CGT GGT AGA ACT CGT G |
| HOXB7 | NM_004502 | CCA GCC TCA AGT TCG GTT TTC | CGC GAA CGC GCT CCA TAG |
| HOXB5 | NM_002147 | AAC TCC TTC TCG GGG CGT TAT | CAT CGCATT GTA ATT GTA GCC GT |
| ZIC2 | NM_007129 | CAC AAC CAG TAC GGC CGCATG AA | AGC TCC TGC TTG ATG CAC TGC TG |
| CXCL1 | NM_001511 | AGG GAA TTC ACC CCA AGA AC | ACT ATG GGG GA T GCA GGA TT |
| HOXB2 | NM_002145 | CGT TCC CGA CGT CAA cn CTT | CTC TTC CTC GGA AM AGG GAC |
| GDF15 | NM_004864 | CGC GGGACC CTC AGA GTT | CCG CAG CGT GGT TAG CA |
| DDIT4 | NM_019058 | AGG AAG CTC ATT GAG TTG TG | GGT ACA TGC TAC ACA CAC AT |
| SLC7A5 | NM_003486 | AGA AGG AAG AGG CGC GGG AGA AGA T | AAG ATG CGCGAG CCG ATA ATG GTC |
| TRIB3 | NM_021158 | GCC CTG CAC TGC CGTACA G | GGT ACC AGC CAG GAC CTC AGT |
| CBS | NM_000071 | ACA TGC TCT CGT CGC TGC TT | GTG AGG CGG ATC TGT TTG AAC T |
| PDGFRL | NM_006207 | TTG GGT GGA GCT ACC CTG CGT ATC | ACT GGC CGT AGC GCT CAT TCT G |
| TBX15 | NM_152380 | ATT CTG GAG ACC TCC TGT GCG C | CCA CAT TGA AAG TGT TGG GGG CC |
| HCLS1 | NM_005335 | GAC GGA GAA ACA CGA GTC CCA GAG | TGG TCG GGG CGT CCA TTT CAT TG |
| PITX1 | NM_002653 | MG TGG CGTAAG CGC GAG CGT AA | GAC AGC GGG CTC ATG GAG TTG AAG |
| COMP | NM_000095 | TAT CGT TGG TTC CTG CAG CAC CG | GCA TGG TTG TGT CCA AGA CCA CGT |
| BEX1 | NM_018476 | CAC TCG TGT CTC GCT ACC AG | CTG CTC GTT TCT CTT TGG ACT C |
| PCDH10 | NM_020815 | CAC AAA GTC GAC CAA CAA AA | ATG ATG ACT CCA TCC GAA AT |
| TGM2 | NM_004613 | GCC ACT TCA TTT TGC TCT TCA A | TCC TCT TCC GAG TCC AGG TAC A |
| MAB21L2 | NM_011839 | CAG CCG CTC AAC AAC TAC CA | CTC GTC CCA GTC CGT TTC TC |
| BST2 | NM_004335 | GAT GCA GAG AAG GCC CAA GGA CAA A | ACT TCT TGT CCG CGA TTC TCA CGC |
| ERAP2 | NM_001130140 | TGG ATG GGA CCA ACT CAT TAC A | TGC ACC AAC TAG CT AAA CAC |
| HOXA13 | NM_000522 | AGC GCG TGC CTT ATA CCA AG | GCC GCT CAG AGA GAT TCG T |
| HOXA10 | NM_018951 | AGC CTC GCC GGA GAA GGA TT | CCA GTG TCT GGT GCT TCG TGT AG |
Results
Microarray gene expression
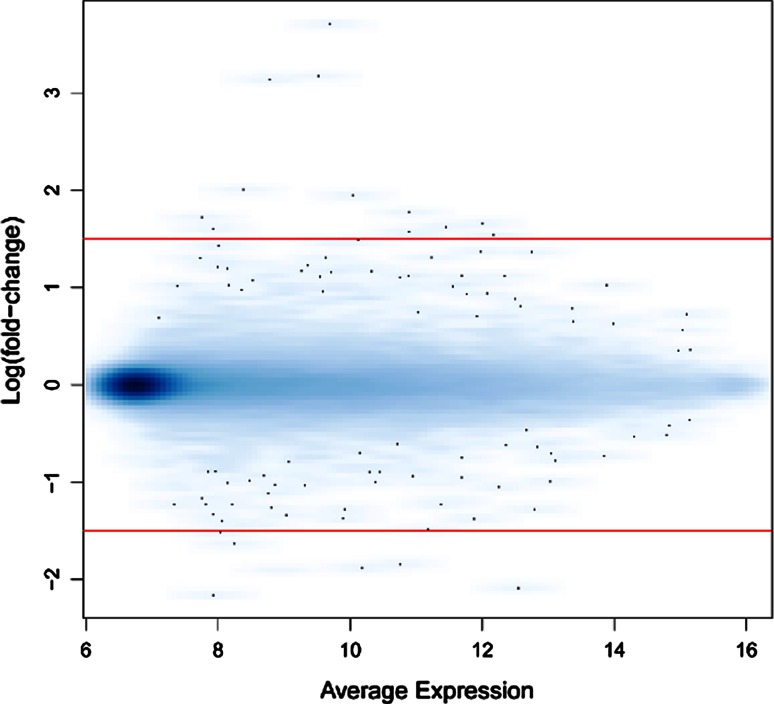
To screen for candidate genes that may contribute to the pathogenesis of AIS, we comparatively analyzed the gene expression patterns of AIS osteoblasts and healthy osteoblasts by microarray analysis. A scatter plot of the microarray data revealed significant gene expression changes in 145 genes in AIS osteoblasts, as compared to controls (Fig. 1; n = 6, empirical Bayes, p < 0.05). Among these 145 genes, 86 were up-regulated >1.5-fold, such as HOXB8, HOXB7, HOXB5, FLJ30375, ZIC2 and ZIC4 and 59 were down-regulated >1.5-fold, such as HOXA10, HOXA13, HOXA11, FAM101A, TINAGL1, ERAP2, COMP and PITX1. A complete list of up- and down-regulated genes is provided in Table 3.
Fig. 1.
Scatter plot of microarray gene expression data. Average signal intensity was plotted against the log2 (fold-change) for all 48.804 probes for the data set of AIS (n = 6) minus control (n = 6). Red lines indicate our threshold of 1.5-fold-change in expression
Table 3.
Differentially expressed genes
| Up-regulated genes | Down-regulated genes | ||||||
|---|---|---|---|---|---|---|---|
| TargetID | Entrez_gene_ID | log2 (Fold change) | P value | TargetID | Entrez_gene_ID | log2 (Fold-change) | P value |
| HOXB8 | 3218 | 6.35 | ≤0.001 | HOXA10 | 3206 | −5.06 | ≤0.001 |
| HOXB7 | 3217 | 6.25 | ≤0.001 | HOXA13 | 3209 | −3.24 | ≤0.001 |
| HOXB5 | 3215 | 5.32 | ≤0.001 | HOXA11 | 3207 | −2.88 | ≤0.001 |
| FLJ30375 | 440982 | 4.83 | ≤0.001 | FAM101A | 144347 | −2.87 | ≤0.001 |
| ZIC2 | 7546 | 4.31 | ≤0.001 | TINAGL1 | 64129 | −2.86 | ≤0.001 |
| ZIC4 | 84107 | 4.26 | ≤0.001 | ERAP2 | 64167 | −2.82 | ≤0.001 |
| HS.347185 | 3.29 | ≤0.001 | EPYC | 1833 | −2.82 | ≤0.001 | |
| RERG | 85004 | 2.97 | ≤0.001 | BST2 | 684 | −2.80 | ≤0.001 |
| HS.539440 | 2.89 | ≤0.001 | MAB21L2 | 10586 | −2.67 | 0.01 | |
| LOC404266 | 404266 | 2.85 | ≤0.001 | LOC130576 | 130576 | −2.48 | 0.04 |
| HOXB3 | 3213 | 2.76 | ≤0.001 | TGM2 | 7052 | −2.33 | 0.01 |
| HLA-A29.1 | 649853 | 2.59 | 0.03 | LRRN3 | 54674 | −2.29 | 0.01 |
| SMOC2 | 64094 | 2.53 | 0.01 | PCDH10 | 57575 | −2.26 | 0.03 |
| LOC644396 | 644396 | 2.51 | ≤0.001 | FMO3 | 2328 | −2.24 | 0.04 |
| CXCL1 | 2919 | 2.51 | 0.01 | BEX1 | 55859 | −2.24 | 0.02 |
| CHAC1 | 79094 | 2.48 | 0.01 | SHOX | 6473 | −2.23 | ≤0.001 |
| HOXB2 | 3212 | 2.47 | ≤0.001 | GPR116 | 221395 | −2.22 | 0.01 |
| ZNF608 | 57507 | 2.41 | ≤0.001 | COMP | 1311 | −2.19 | 0.01 |
| CNTNAP3B | 389734 | 2.38 | 0.01 | FMO3 | 2328 | −2.15 | 0.06 |
| CXCL2 | 2920 | 2.36 | 0.01 | HS.562127 | −2.13 | 0.02 | |
| FAM134B | 54463 | 2.30 | ≤0.001 | LYPD6 | 130574 | −2.10 | 0.01 |
| ADH1A | 124 | 2.28 | 0.08 | TSPAN13 | 27075 | −2.09 | 0.02 |
| EVI2A | 2123 | 2.18 | 0.01 | RELN | 5649 | −2.06 | ≤0.001 |
| G0S2 | 50486 | 2.16 | 0.01 | TM4SF20 | 79853 | −2.04 | 0.01 |
| HOXB6 | 3216 | 2.16 | ≤0.001 | EMX2 | 2018 | −2.03 | 0.01 |
| RASIP1 | 54922 | 2.14 | ≤0.001 | PCDH10 | 57575 | −2.01 | 0.06 |
| NGEF | 25791 | 2.09 | 0.03 | PGF | 5228 | −1.97 | ≤0.001 |
| MYLC2PL | 93408 | 2.08 | ≤0.001 | LRRN3 | 54674 | −1.96 | 0.02 |
| HOXA2 | 3199 | 2.06 | 0.01 | FLG | 2312 | −1.96 | 0.07 |
| FAM134B | 54463 | 2.03 | 0.01 | LOC284757 | 284757 | −1.93 | ≤0.001 |
| NOPE | 57722 | 2.02 | ≤0.001 | MYL4 | 4635 | −1.90 | ≤0.001 |
| HOXA2 | 3199 | 2.01 | ≤0.001 | HS.556994 | −1.85 | ≤0.001 | |
| HOXD4 | 3233 | 1.96 | ≤0.001 | PCDH7 | 5099 | −1.83 | 0.08 |
| HS.569104 | 1.96 | 0.01 | F3 | 2152 | −1.82 | 0.05 | |
| FGFBP2 | 83888 | 1.91 | 0.01 | PITX1 | 5307 | −1.80 | 0.05 |
| HS.122310 | 1.88 | 0.01 | TSPAN13 | 27075 | −1.80 | 0.02 | |
| DDX43 | 55510 | 1.87 | ≤0.001 | ACTC1 | 70 | −1.80 | 0.04 |
| WFDC3 | 140686 | 1.87 | ≤0.001 | FAM162B | 221303 | −1.79 | 0.08 |
| CX3CL1 | 6376 | 1.86 | 0.02 | LOC124220 | 124220 | −1.79 | 0.01 |
| DENND2A | 27147 | 1.85 | 0.01 | SAMD11 | 148398 | −1.78 | ≤0.001 |
| XG | 7499 | 1.82 | 0.03 | ECHDC3 | 79746 | −1.77 | 0.05 |
| AFAP1L2 | 84632 | 1.82 | ≤0.001 | S100P | 6286 | −1.75 | 0.02 |
| PDGFRL | 5157 | 1.81 | 0.07 | MYPN | 84665 | −1.74 | 0.02 |
| PAX9 | 5083 | 1.80 | ≤0.001 | TGM2 | 7052 | −1.73 | 0.03 |
| GDF15 | 9518 | 1.77 | 0.03 | LMNB1 | 4001 | −1.71 | 0.06 |
| DDIT4 | 54541 | 1.76 | 0.03 | DLX1 | 1745 | −1.70 | ≤0.001 |
| MEGF10 | 84466 | 1.76 | 0.02 | SPINK5L3 | 153218 | −1.70 | 0.09 |
| SLC7A5 | 8140 | 1.75 | 0.01 | HCLS1 | 3059 | −1.69 | 0.10 |
| FLJ10916 | 55258 | 1.74 | 0.09 | CALB2 | 794 | −1.69 | ≤0.001 |
| TRIB3 | 57761 | 1.73 | 0.03 | BARX1 | 56033 | −1.66 | 0.15 |
| WFDC3 | 140686 | 1.70 | 0.01 | TBX15 | 6913 | −1.66 | 0.01 |
| ZCCHC5 | 203430 | 1.70 | ≤0.001 | CDH6 | 1004 | −1.65 | 0.02 |
| CBS | 875 | 1.68 | 0.02 | MYPN | 84665 | −1.61 | 0.01 |
| DCLK1 | 9201 | 1.67 | ≤0.001 | TAF13 | 6884 | −1.56 | ≤0.001 |
| RIMS3 | 9783 | 1.67 | 0.05 | EFNB2 | 1948 | −1.54 | 0.03 |
| EPB41L3 | 23136 | 1.66 | 0.10 | ANGPTL7 | 10218 | −1.53 | 0.03 |
| RPL22L1 | 200916 | 1.66 | 0.03 | C2ORF40 | 84417 | −1.52 | 0.17 |
| NOPE | 57722 | 1.66 | 0.01 | LRRN1 | 57633 | −1.52 | 0.04 |
| ULBP1 | 80329 | 1.66 | 0.02 | NPAS1 | 4861 | −1.51 | ≤0.001 |
| CH25H | 9023 | 1.65 | 0.07 | ||||
| ZBTB46 | 140685 | 1.65 | ≤0.001 | ||||
| HMCN1 | 83872 | 1.65 | 0.01 | ||||
| HEY2 | 23493 | 1.65 | 0.09 | ||||
| F2RL2 | 2151 | 1.65 | 0.02 | ||||
| SHISA2 | 387914 | 1.65 | 0.01 | ||||
| SLC7A11 | 23657 | 1.64 | 0.05 | ||||
| PPL | 5493 | 1.63 | 0.04 | ||||
| HOXD1 | 3231 | 1.63 | 0.02 | ||||
| LSP1 | 4046 | 1.62 | 0.02 | ||||
| HOXB4 | 3214 | 1.61 | ≤0.001 | ||||
| REM1 | 28954 | 1.61 | 0.05 | ||||
| AGTR1 | 185 | 1.60 | 0.02 | ||||
| KCNG1 | 3755 | 1.60 | 0.01 | ||||
| PSAT1 | 29968 | 1.59 | 0.02 | ||||
| LOC285216 | 285216 | 1.58 | 0.01 | ||||
| IGFBP1 | 3484 | 1.58 | 0.02 | ||||
| PRPH2 | 5961 | 1.54 | 0.04 | ||||
| SFRP4 | 6424 | 1.54 | 0.03 | ||||
| GUCA1B | 2979 | 1.54 | 0.01 | ||||
| MAFB | 9935 | 1.53 | 0.05 | ||||
| IL18R1 | 8809 | 1.53 | 0.04 | ||||
| ALG11 | 440138 | 1.53 | 0.01 | ||||
| SPATA22 | 84690 | 1.53 | 0.15 | ||||
| MEOX2 | 4223 | 1.52 | 0.01 | ||||
| CNTN1 | 1272 | 1.51 | 0.10 | ||||
| AGTR1 | 185 | 1.50 | 0.01 | ||||
Genes with a fold-change >1.5 are listed with official NCBI gene symbols, Entrez gene ID numbers, the calculated fold-change, and the associated p value calculated as described. A positive log2 (fold-change) means that the gene was up-regulated in AIS osteoblasts, and a negative one means that the gene was down-regulated
Functional classification of up-regulated and down-regulated AIS genes
To identify biological pathways common to the differentially expressed genes in AIS osteoblasts, we performed a hierarchical clustering followed by a gene ontology (GO) analysis with DAVID (Database for Annotation, Visualization, and Integrated Discovery) on the 145 differentially expressed genes with a fold change values ≥1.5. Interestingly, hierarchical analysis revealed a very close genetic interaction between the down-regulated genes HOXA13 and HOXA10 genes, and between MAB21L2, BST2, EPYC, ERAP2, TINAGL1, FAM101A and HOXA11 genes (Fig. 2). In a second cluster, EVI2A, G0S2, HOXB6, RASIP1, NGEF. MYLC2PL and HOXA2 genes were found to interact together. Among the up-regulated genes, hierarchical analysis identified one interesting cluster of a closely related genetic interaction between HOXB8 and HOXB7, HOXB5 and FLJ30375 and between ZIC2 and ZIC4 genes (Fig. 2).
Fig. 2.

Hierarchical clustering of gene expression data. The dendrogram provides a measure of relatedness of between the 145 differentially expressed AIS genes. The figure depicts signal strengths for a representative gene. Colour indicates relative signal levels, with red indicating the highest (up regulated) and green indicating the lowest (down regulated) expression
GO analysis of the differentially expressed genes revealed five distinct groups based on similar molecular function and biological process (Group 1–5) (Table 4). Group 1 consists of 20 genes that are all transcription factors involved in organ development and morphogenesis, as well as in processes of segmentation and anterior/posterior pattern specification. Group 2 is composed of four genes involved in immune system development. Group 3 comprises four genes that are involved in cytokine signaling and secretion. Group 4 contains 17 genes. seven of which are involved in cellular signaling processes related to cell–cell adhesion and calcium ion binding. The remaining ten genes in cluster group four possess homology domains implicated in cell adhesion and membrane transport. Cluster group 5 is composed of four genes, all zinc finger proteins involved in signal transduction interactions and in ion binding.
Table 4.
Gene interaction networks and gene clusters
| official_gene_symbol | Gene name | Entrez_gene_ID | log2 (Fold-change) |
|---|---|---|---|
| Gene group 1 enrichment score: 8.84 | |||
| HOXB8 | Homeobox B8 | 3218 | 6.35 |
| HOXB7 | Homeobox B7 | 3217 | 6.25 |
| HOXB5 | Homeobox B5 | 3215 | 5.32 |
| HOXB3 | Homeobox B3 | 3213 | 2.76 |
| HOXB2 | Homeobox B2 | 3212 | 2.47 |
| HOXB6 | Homeobox B6 | 3216 | 2.16 |
| HOXA2 | Homeobox A2 | 3199 | 2.06 |
| HOXD4 | Homeobox D4 | 3233 | 1.96 |
| HOXD1 | Homeobox D1 | 3231 | 1.63 |
| HOXB4 | Homeobox B4 | 3214 | 1.61 |
| MEOX2 | Mesenchyme homeobox 2 | 4223 | 1.52 |
| TBX15 | T-box 15 | 6913 | −1.66 |
| BARX1 | BARX homeobox 1 | 56033 | −1.66 |
| DLX1 | Distal-less homeobox 1 | 1745 | −1.70 |
| PITX1 | Paired-like homeodomain 1 | 5307 | −1.80 |
| EMX2 | Empty spiracles homeobox 2 | 2018 | −2.03 |
| SHOX | Short stature homeobox | 6473 | −2.23 |
| HOXA11 | Homeobox A11 | 3207 | −2.88 |
| HOXA13 | Homeobox A13 | 3209 | −3.24 |
| HOXA10 | Homeobox A10 | 3206 | −5.06 |
| Gene group 2 enrichment score: 2.68 | |||
| WFDC3 | WAP four-disulfide core domain 3 | 140686 | 1.87 |
| PDGFRL | Platelet-derived growth factor receptor-like | 5157 | 1.81 |
| ANGPTL7 | Angiopoietin-like 7 | 10218 | −1.53 |
| LYPD6 | LY6/PLAUR domain containing 6 | 130574 | −2.10 |
| Gene group 3 enrichment score: 2.32 | |||
| FGFBP2 | Fibroblast growth factor binding protein 2 | 83888 | 1.91 |
| GDF15 | Growth differentiation factor 15 | 9518 | 1.77 |
| SFRP4 | Secreted frizzled-related protein 4 | 6424 | 1.54 |
| ANGPTL7 | Angiopoietin-like 7 | 10218 | −1.53 |
| Gene group 4 enrichment score: 1.20 | |||
| CNTNAP3B | Contactin associated protein-like 3B | 389734 | 2.38 |
| FAM134B | FAMILY WITH SEQUENCE SIMILARITY (134 MEMBER B) | 54463 | 2.30 |
| EVI2A | Ecotropic viral integration site 2A | 2123 | 2.18 |
| XG | Xg blood group | 7499 | 1.82 |
| F2RL2 | Coagulation factor II (thrombin) receptor-like 2 | 2151 | 1.65 |
| SHISA2 | Shisa homolog 2 (Xenopus laevis) | 387914 | 1.65 |
| SLC7A11 | Solute carrier family 7. member 11 | 23657 | 1.64 |
| IL18R1 | Interleukin 18 receptor 1 | 8809 | 1.53 |
| LRRN1 | leucine rich repeat neuronal 1 | 57633 | −1.52 |
| EFNB2 | Ephrin-B2 | 1948 | −1.54 |
| CDH6 | CADHERIN 6. TYPE 2. K-CADHERIN (FETAL KIDNEY) | 1004 | −1.65 |
| FAM162B | Family with sequence similarity (162 member B) | 221303 | −1.79 |
| TSPAN13 | TETRASPANIN 13 | 27075 | −1.80 |
| PCDH7 | Protocadherins (7) | 5099 | −1.83 |
| PCDH10 | Protocadherins (10) | 57575 | −2.01 |
| TM4SF20 | Transmembrane 4 L six family member 20 | 79853 | −2.04 |
| GPR116 | G protein-coupled receptor 116 | 221395 | −2.22 |
| GENE group 5 enrichment score: 0.39 | |||
| ZIC4 | Zinc finger protein. ZNF of the cerebellum 4 | 84107 | 4.26 |
| ZNF608 | Zinc finger protein. ZNF608 | 57507 | 2.41 |
| ZCCHC5 | Zinc finger protein with CCHC domain containing 5 | 203430 | 1.70 |
| ZBTB46 | Zinc finger protein with BTB domain containing 46 | 140685 | 1.65 |
The 46 genes that were clustered by the DAVID algorithm are listed in their five gene cluster groupings with gene enrichment scores, with associated fold-changes per gene. Gene cluster analysis of differentially regulated genes in primary human osteoblasts. Up-regulated genes are shown in red, Down-regulated genes are shown in green
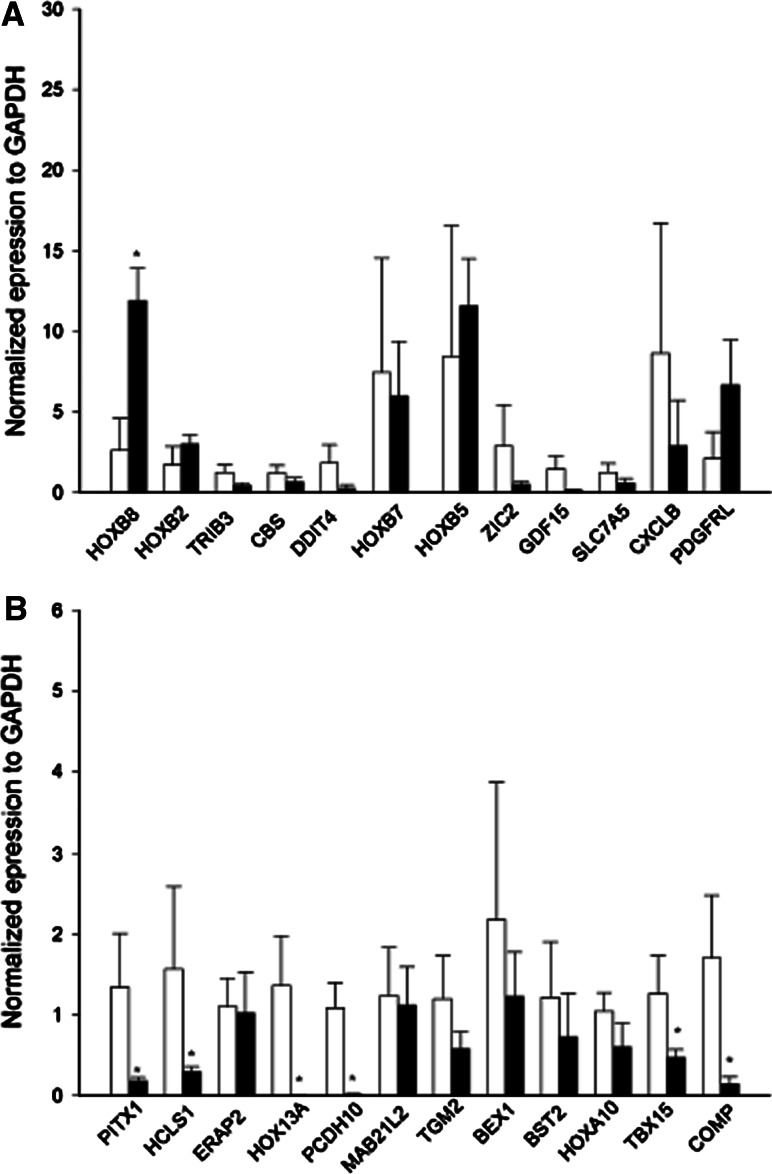
RT-qPCR validation of gene expression
To confirm the microarray results 24 genes were chosen for RT-qPCR validation. They were selected because they were the highest fold-changed and they appeared in either functional classification or clustering analyses. Similar patterns of gene expression differences were seen for all genes determined to be down-regulated by our microarray analyses (Fig. 3a, b), thereby confirming our conclusions. Some of the up-regulated by our microarray analyses were also confirmed by RT-qPCR experiments, however no significant up-regulation was observed for HOXB7, ZIC2, SLC7A5, DDIT4, TRIB3, CBS, GDF15 and CXCL1 (Fig. 3a).
Fig. 3.
Validation of most up-regulated (a) and down-regulated (b) genes by RT-qPCR. The relative level of mRNA of regulated genes was analyzed by quantitative RT-PCR. Gene expression results are depicted as ∆Ct values, normalized to GAPDH. *p < 0.05, Student’s t test, AIS (■) versus control (□) expression levels
Profiled genes versus previously reported AIS candidate regions
Several genetic linkage and genome-wide association studies have identified chromosomal loci predisposing to AIS. But to date, no genes have been clearly identified as causative in AIS. We therefore sought to identify if there were any significantly differentially regulated genes in AIS osteoblasts within the reported loci. 32 genes were identified as corresponding with previously reported AIS candidate loci (Table 5). Within region 19p13.3 [14], we identified two genes to be up-regulated: MKNK2 and CD70, HCLS1 and COL8A1 genes were down-regulated and F2R, F2RL2 and BHMT2 genes were up-regulated in the chromosomal regions identified by Edery et al. [11]. Within locus 1p35 [15], TINAGL1 gene was found down-regulated. Finally, in the AIS candidate region 9q31.2–q34.2 [16], OLFML2A was up-regulated. However, none of these genes had a significant fold change ≥1.5 (Table 3).
Table 5.
Transcriptome profile of genes within reported AIS candidate loci
| Candidate region | References | Candidate gene | Ref_Seq | Chromosomal Localization | Fold-Change |
|---|---|---|---|---|---|
| 11q21–q22 | Qiu et al. [30] | MMP13 | NM_002427.2 | 11q22.2b | 0.62 |
| 1p35 | Montanaro et al. [15] | TINAGL1 | NM_022164.1 | 1p35.2a | −1.05 |
| IFI6 | NM_022872.2 | 1p35.3b | −0.84 | ||
| SESN2 | NM_031459.3 | 1p35.3b | 0.76 | ||
| 6q25.1 | Wu et al. [31] | ULBP1 | NM_025218.2 | 6q25.1b | 0.9 |
| 19p13.3 | Chan et al. [14] | CD70 | NM_001252.3 | 19p13.3a | 0.71 |
| Alden et al. [32] | MKNK2 | NM_017572.2 | 19p13.3 h | 0.93 | |
| 12p13.31 | Raggio et al. [33] | CD4 | NM_000616.3 | 12p13.31d | −0.32 |
| NTF3 | NM_002527.3 | 12p13.31e | 0.89 | ||
| 17q25.3–qtel | Ocaka et al. [16] | ARL16 | NM_001040025.1 | 17q25.3f | −0.27 |
| 9q31.2–q34.2 | Ocaka et al. [16] | PTGS1 | NM_080591.1 | 9q33.2b | −0.86 |
| OLFML2A | NM_182487.2 | 9q33.3a | 0.96 | ||
| Xq23–26.1 | Justice et al. [34] | GRIA3 | NM_000828.3 | Xq25b | −0.56 |
| GPC4 | NM_001448.2 | Xq26.2b | 0.62 | ||
| MGC16121 | XM_001128419.1 | Xq26.3a | 0.68 | ||
| 17p11.2 | Salehi et al. [35] | EPPB9 | NM_015681.2 | 17p11.2e | −0.48 |
| SPECC1 | NM_001033554.1 | 17p11.2d-p11.2c | −0.31 | ||
| 18q | Wise et al. [10] | LOC284293 | XM_209104.2 | 18q21.33b | −0.32 |
| BCL2 | NM_000633.2 | 18q21.33b | 0.48 | ||
| 10q | Wise et al. [10] | DDIT4 | NM_019058.2 | 10q22.1f | 1.62 |
| DKK1 | NM_012242.2 | 10q21.1a | −1.28 | ||
| EMX2 | NM_004098.2 | 10q26.11a | −0.89 | ||
| SVIL | NM_003174.3 | 10p11.23b | 1.01 | ||
| 6q | Wise et al. [10] | FAM162B | NM_001085480.1 | 6q22.2a | −0.9 |
| SMOC2 | NM_022138.1 | 6q27d–q27e | 1.03 | ||
| 3q13.3 | Edery et al. [11] | COL8A1 | NM_001850 | 3q12.1b–q12.1c | −0.97 |
| HCLS1 | NM_005335.3 | 3q13.33c | −1.42 | ||
| 5q13 | Edery et al. [11] | FOXD1 | NM_004472.2 | 5q13.2c | 0.57 |
| F2R | NM_001992.2 | 5q13.3d | 1.04 | ||
| F2RL2 | NM_004101.2 | 5q13.3d | 0.97 | ||
| BHMT2 | NM_017614.3 | 5q14.1c | 0.68 |
The table presents a list of genes within AIS reported loci and their corresponding fold-changes. As revealed by microarray analysis
Discussion
In the present work, we have used gene expression profiling to identify differentially expressed genes in AIS compared with non-AIS osteoblasts. Our study provides a previously unrecognized list of genes and related potential pathways that merit further investigation, such as identification of variants in these genes, as putative AIS causative genes. These genes were grouped in terms of their biological function, and clustered by gene–gene interactions. We identified at least four particular pathways that might be important in AIS: the developmental/growth-differentiation of skeletal elements (HOXB8, HOXA2, HOXB2, MEOX2 and PITX1); cellular signaling (HOXA11 and BARX1), connecting structural integrity of the extracellular matrix to the structural integrity of a bone or a muscle fiber (COMP, HOXA2 and HOXA11); and cellular signaling and cartilage damage (GDF15). Among the differentially expressed genes, some could act on processes directly related to the causes of AIS (associated with embryogenesis/morphogenesis), while others may play contributory roles (related to spinal deformity progression). Yet others may be condition-specific genes (differentially expressed genes as a consequence of disease).
Our results revealed that the most up- and down-regulated genes involved in the AIS pathology are members of the Homeobox (HOX) gene family. The HOX genes are, in general, implicated in the regulation of patterns of development (morphogenesis). We identified differential expression of HOXA group genes (10, 11 and 13), of HOXB group genes (2–8) and of HOXD (1–4). Interestingly, knockdown of Hoxd1 generates defects in hindbrain and neural crest derivatives [17]. The over-expression of Hoxd4 has resulted in severe cartilage defects in mice [18], while over-expression of Hoxb8 in transgenic mouse embryos has resulted in defects in the vertebrae [19]. HOXA10 plays a key role in regulating target genes for osteoblast differentiation and bone formation in the postnatal skeleton [20]. HOXA13 gene is involved particularly in segment identity specification along the limb axis in vertebrates [21]. These suggest that HOX genes are important in vertebral development and abnormal expression of these genes as we observed in AIS patients could play a role in curvation of the spine.
PITX1 (pituitary homeobox 1) gene encodes for a protein that is a member of RIEG/PITX homeobox family with transcriptional properties that have been defined for number of late downstream target genes in the pituitary gland [22]. As a member of this family, PITX1 gene is involved in limb and organ development and in left–right asymmetry [23]. The down-regulated expression of PITX1 in our study confirms that this protein plays a crucial role in bone development and probably in AIS. Furthermore, Cartilage oligomeric matrix protein (COMP) is a novel gene to consider in the context of AIS pathogenesis. This gene is essential for the normal development of cartilage and for its conversion to bone during growth. For instance, COMP also interacts with the transcription factor SOX-9, which plays an important role in normal skeletal development. Mutations in COMP produce clinical phenotypes of pseudoachondroplasia (PSACH) and multiple epiphyseal dysplasia (MED). These disorders are characterized by disproportionate short stature, brachydactyly, joint hyper-mobility, early-onset osteoarthritis, and scoliosis [24]. Consistent with our study, COMP was recently found to be down-regulated by fourfold in AIS compared to unaffected individuals and it was proposed as an important and novel biomarker in predicting scoliosis development [25]. Interestingly, COMP and HOXA10 interact closely in embryonic limb morphogenesis (GO: 0030326. http://amigo.geneontology.org) and with ERAP2; which is associated with familial ankylosing spondylitis and it affects joints and can cause eventual fusion of the spine [26]. Altogether, these data suggest that low expression of COMP and its molecular interactions are associated with AIS.
Other modulated genes in our experiment include BST2, HCLS1, TBX15, PCDH10 and GDF15. Although these genes are not directly involved in bone and cartilage development, they are involved in immune process and Wnt, tyrosine kinase signaling pathways that are important in the embryonic development and may be associated with AIS.
Our study screened candidate genes that may contribute to the pathogenesis of AIS, and provided a new list of genes that merit further investigation. such as the epigenetic interactions (that could modify the expression of specific genes) as well as the identification of variants in these genes, as possible AIS contributing genes. We found that the gene expression patterns of primary osteoblasts derived from spinal vertebrae of AIS patients were different from those of healthy individuals. Gene mutations can affect gene transcripts. In addition, the expression of specific genes by cells can be modified by various ways: enzymes methylate DNA to modulate transcription, histone modification resulting in inducing or repression of target sequences, non-coding small RNA which could attach to messengers RNA to modify gene expression of specific genes [27, 28]. Therefore, it is likely that these mechanisms might play an important role in the altered gene expression patterns in AIS osteoblasts. Genes with altered expression in AIS could be grouped into specific subsets based on their biological functions and gene–gene interactions, suggesting the possible involvement of various pathways in AIS pathogenesis. Interestingly, patients with AIS with similar gene profiles may have varying severity of spinal curve suggesting that genetic factors likely control disease susceptibility and course, but not disease pattern. Hormonal and environmental factors may also affect the clinical phenotype and the severity of curve [29] although patients may have the similar gene profiles but this area remains unexplored and beyond the scope of this study.
Taken together our study revealed gene expression changes in AIS osteoblasts. These findings help to gain further insights into potential genes and molecular pathways that could contribute to understanding the pathophysiology of idiopathic scoliosis. Furthermore, although bone contributes significantly in developing AIS, the intervertebral discs (IVD) and muscles also have significant mechanisms in the pathogenesis of AIS. Therefore, our approach can be helpful to study the gene expression profile of AIS IVD and muscles and to further add to our understanding of AIS etiology. Identification of the underlying mechanisms that leads to the observed clinical features of scoliosis remains the crucial next step to further advance understanding of AIS pathogenesis.
Acknowledgments
FM and PE were supported by Yves Cotrel Foundation and SAP was supported by CHU Sainte-Justine and Foundation of the Stars scholarship.
Conflict of interest
None.
References
- 1.Je L. Idiopathic Scoliosis. In: Winter RB, Ogilive JW, editors. Lonstein JE BD. Philadelphia: Moe’s textbook of scoliosis and other spinal deformaties. WB Saunders; 1995. pp. 219–256. [Google Scholar]
- 2.Cavallaro Goodman C, Fuller KS. Pathology: implications for the physical therapist. St. Louis: Saunders. WB/CO; 2009. [Google Scholar]
- 3.Winter RB. Posterior spinal fusion in scoliosis: indications technique and results. Orthop Clin N Am. 1979;10:787–800. [PubMed] [Google Scholar]
- 4.Dubousset JQP, Thillard M. Experimental scoliosis induced by pineal and diencephalic lesions in young chickens: its relation with clinical findings. Orthop Trans. 1983;7:7–12. [Google Scholar]
- 5.Machida M, Dubousset J, Imamura Y, Miyashita Y, Yamada T, Kimura J. Melatonin a possible role in pathogenesis of adolescent idiopathic scoliosis. Spine. 1996;21:1147–1152. doi: 10.1097/00007632-199605150-00005. [DOI] [PubMed] [Google Scholar]
- 6.Taylor TK, Ghosh P, Bushell GR. The contribution of the intervertebral disk to the scoliotic deformity. Clin Orthop Relat Res. 1981;156:79–90. [PubMed] [Google Scholar]
- 7.Sahlstrand T, Petruson B. A study of labyrinthine function in patients with adolescent idiopathic scoliosis I. An electro-nystagmographic study. Acta orthop Scand. 1979;50:759–769. doi: 10.3109/17453677908991307. [DOI] [PubMed] [Google Scholar]
- 8.Kindsfater K, Lowe T, Lawellin D, Weinstein D, Akmakjian J. Levels of platelet calmodulin for the prediction of progression and severity of adolescent idiopathic scoliosis. J Bone Jt Surg Am. 1994;76:1186–1192. doi: 10.2106/00004623-199408000-00009. [DOI] [PubMed] [Google Scholar]
- 9.Nissinen M, Heliovaara M, Seitsamo J, Poussa M. Trunk asymmetry. posture. growth. and risk of scoliosis. A three-year follow-up of Finnish prepubertal school children. Spine. 1993;18:8–13. doi: 10.1097/00007632-199301000-00002. [DOI] [PubMed] [Google Scholar]
- 10.Wise CA, Barnes R, Gillum J, Herring JA, Bowcock AM, Lovett M. Localization of susceptibility to familial idiopathic scoliosis. Spine. 2000;25:2372–2380. doi: 10.1097/00007632-200009150-00017. [DOI] [PubMed] [Google Scholar]
- 11.Edery P, Margaritte-Jeannin P, Biot B, Labalme A, Bernard JC, Chastang J, Kassai B, Plais MH, Moldovan F, Clerget-Darpoux F. New disease gene location and high genetic heterogeneity in idiopathic scoliosis. Eur J Hum Genet. 2011;19:865–869. doi: 10.1038/ejhg.2011.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma S, Gao X, Londono D, Devroy SE, Mauldin KN, Frankel JT, Brandon JM, Zhang D, Li QZ, Dobbs MB, Gurnett CA, Grant SF, Hakonarson H, Dormans JP, Herring JA, Gordon D, Wise CA. Genome-wide association studies of adolescent idiopathic scoliosis suggest candidate susceptibility genes. Hum Mol Genet. 2011;20:1456–1466. doi: 10.1093/hmg/ddq571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Letellier K, Azeddine B, Parent S, Labelle H, Rompre PH, Moreau A, Moldovan F. Estrogen cross-talk with the melatonin signaling pathway in human osteoblasts derived from adolescent idiopathic scoliosis patients. J Pineal Res. 2008;45:383–393. doi: 10.1111/j.1600-079X.2008.00603.x. [DOI] [PubMed] [Google Scholar]
- 14.Chan V, Fong GC, Luk KD, Yip B, Lee MK, Wong MS, Lu DD, Chan TK. A genetic locus for adolescent idiopathic scoliosis linked to chromosome 19p13.3. Am J Hum Genet. 2002;71:401–406. doi: 10.1086/341607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montanaro L, Parisini P, Greggi T, Di Silvestre M, Campoccia D, Rizzi S, Arciola CR. Evidence of a linkage between matrilin-1 gene (MATN1) and idiopathic scoliosis. Scoliosis. 2006;1:21. doi: 10.1186/1748-7161-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ocaka L, Zhao C, Reed JA, Ebenezer ND, Brice G, Morley T, Mehta M, O’Dowd J, Weber JL, Hardcastle AJ, Child AH. Assignment of two loci for autosomal dominant adolescent idiopathic scoliosis to chromosomes 9q31.2-q34.2 and 17q25.3-qtel. J Med Genet. 2008;45:87–92. doi: 10.1136/jmg.2007.051896. [DOI] [PubMed] [Google Scholar]
- 17.McNulty CL, Peres JN, Bardine N, van den Akker WM, Durston AJ. Knockdown of the complete Hox paralogous group 1 leads to dramatic hindbrain and neural crest defects. Development. 2005;132:2861–2871. doi: 10.1242/dev.01872. [DOI] [PubMed] [Google Scholar]
- 18.Pollock RA, Sreenath T, Ngo L, Bieberich CJ. Gain of function mutations for paralogous Hox genes: implications for the evolution of Hox gene function. Proc Natl Acad Sci USA. 1995;92:4492–4496. doi: 10.1073/pnas.92.10.4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang X, Ji X, Shi X, Cao X. Smad1 domains interacting with Hoxc-8 induce osteoblast differentiation. J Biol Chem. 2000;275:1065–1072. doi: 10.1074/jbc.275.2.1065. [DOI] [PubMed] [Google Scholar]
- 20.Riggs BL, Khosla S, Melton LJ., 3rd Sex steroids and the construction and conservation of the adult skeleton. Endocr Rev. 2002;23:279–302. doi: 10.1210/er.23.3.279. [DOI] [PubMed] [Google Scholar]
- 21.Mortlock DP, Innis JW. Mutation of HOXA13 in hand-foot-genital syndrome. Nat Genet. 1997;15:179–180. doi: 10.1038/ng0297-179. [DOI] [PubMed] [Google Scholar]
- 22.Logan M, Tabin CJ. Role of Pitx1 upstream of Tbx4 in specification of hindlimb identity. Science. 1999;283:1736–1739. doi: 10.1126/science.283.5408.1736. [DOI] [PubMed] [Google Scholar]
- 23.Gurnett CA, Alaee F, Kruse LM, Desruisseau DM, Hecht JT, Wise CA, Bowcock AM, Dobbs MB. Asymmetric lower-limb malformations in individuals with homeobox PITX1 gene mutation. Am J Hum Genet. 2008;83:616–622. doi: 10.1016/j.ajhg.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Briggs MD, Chapman KL. Pseudoachondroplasia and multiple epiphyseal dysplasia.: mutation review molecular interactions, and genotype to phenotype correlations. Hum Mutat. 2002;19:465–478. doi: 10.1002/humu.10066. [DOI] [PubMed] [Google Scholar]
- 25.Fendri K, Moldovan F. Potential role of COMP as a biomarker for adolescent idiopathic scoliosis. Med Hypotheses. 2011;76:762–763. doi: 10.1016/j.mehy.2011.01.038. [DOI] [PubMed] [Google Scholar]
- 26.Tsui FW, Haroon N, Reveille JD, Rahman P, Chiu B, Tsui HW, Inman RD. Association of an ERAP1 ERAP2 haplotype with familial ankylosing spondylitis. Ann Rheum Dis. 2010;69:733–736. doi: 10.1136/ard.2008.103804. [DOI] [PubMed] [Google Scholar]
- 27.Hamilton JP. Epigenetics: principles and practice. Dig Dis. 2011;29:130–135. doi: 10.1159/000323874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silahtaroglu A, Stenvang J. MicroRNAs epigenetics and disease. Essays Biochem. 2010;48:165–185. doi: 10.1042/bse0480165. [DOI] [PubMed] [Google Scholar]
- 29.Burwell RG, Dangerfield PH, Moulton A, Grivas TB. Adolescent idiopathic scoliosis (AIS), environment. exposome and epigenetics: a molecular perspective of postnatal normal spinal growth and the etiopathogenesis of AIS with consideration of a network approach and possible implications for medical therapy. Scoliosis. 2011;6:26. doi: 10.1186/1748-7161-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiu XS, Tang NL, Yeung HY, Lee KM, Hung VW, Ng BK, Ma SL, Kwok RH, Qin L, Qiu Y, Cheng JC (2007) Melatonin receptor 1B (MTNR1B) gene polymorphism is associated with the occurrence of adolescent idiopathic scoliosis. Spine (Phila Pa 1976) 32(16):1748–1753 [DOI] [PubMed]
- 31.Wu J, Qiu Y, Zhang L, Sun Q, Qiu X, He Y (2006) Association of estrogen receptor gene polymorphisms with susceptibility to adolescent idiopathic scoliosis. Spine (Phila Pa 1976) 31(10):1131–1136 [DOI] [PubMed]
- 32.Alden KJ, Marosy B, Nzegwu N, Justice CM, Wilson AF, Miller NH (2006) Idiopathic scoliosis: identification of candidate regions on chromosome 19p13. Spine (Phila Pa 1976) 31(16):1815–1819 [DOI] [PubMed]
- 33.Raggio CL, Giampietro PF, Dobrin S, Zhao C, Dorshorst D, Ghebranious N, Weber JL, Blank RD (2009) A novel locus for adolescent idiopathic scoliosis on chromosome 12p. J Orthop Res 27:1366–1372 [DOI] [PMC free article] [PubMed]
- 34.Justice CM, Miller NH, Marosy B, Zhang J, Wilson AF (2003) Familial idiopathic scoliosis: evidence of an X-linked susceptibility locus. Spine 28:589–594 [DOI] [PubMed]
- 35.Salehi LB, Mangino M, De Serio S, De Cicco D, Capon F, Semprini S, Pizzuti A, Novelli G, Dallapiccola B (2002) Assignment of a locus for autosomal dominant idiopathic scoliosis (IS) to human chromosome 17p11. Hum Genet 111:401–404 [DOI] [PubMed]