Abstract
Significance: The discovery of the oxidoreductase disulfide bond protein A (DsbA) in 1991 opened the way to the unraveling of the pathways of disulfide bond formation in the periplasm of Escherichia coli and other Gram-negative bacteria. Correct oxidative protein folding in the E. coli envelope depends on both the DsbA/DsbB pathway, which catalyzes disulfide bond formation, and the DsbC/DsbD pathway, which catalyzes disulfide bond isomerization. Recent Advances: Recent data have revealed an unsuspected link between the oxidative protein-folding pathways and the defense mechanisms against oxidative stress. Moreover, bacterial disulfide-bond-forming systems that differ from those at play in E. coli have been discovered. Critical Issues: In this review, we discuss fundamental questions that remain unsolved, such as what is the mechanism employed by DsbD to catalyze the transfer of reducing equivalents across the membrane and how do the oxidative protein-folding catalysts DsbA and DsbC cooperate with the periplasmic chaperones in the folding of secreted proteins. Future Directions: Understanding the mechanism of DsbD will require solving the structure of the membranous domain of this protein. Another challenge of the coming years will be to put the knowledge of the disulfide formation machineries into the global cellular context to unravel the interplay between protein-folding catalysts and chaperones. Also, a thorough characterization of the disulfide bond formation machineries at work in pathogenic bacteria is necessary to design antimicrobial drugs targeting the folding pathway of virulence factors stabilized by disulfide bonds. Antioxid. Redox Signal. 19, 63–71.
Introduction
The oxidation reaction between two cysteine thiol groups leads to the formation of a disulfide bond and the simultaneous release of two electrons and two protons (i.e., two hydrogens). This process is critical for the correct folding and structural stability of many secreted and membrane proteins.
Together with peptidyl prolyl cis/trans isomerization (18), disulfide bond formation is often a rate-limiting step of the folding process of a protein. Therefore, cells contain dedicated pathways that catalyze disulfide bond formation to ensure fast and correct folding in vivo. The study of oxidative protein folding was initiated by Anfinsen and his colleagues in the early sixties, using ribonuclease A as a model. In a series of experiments that are now presented in some biochemistry textbooks, they discovered protein disulfide isomerase (PDI), the primary catalyst of disulfide bond formation in the endoplasmic reticulum (17). In this compartment, PDI catalyzes both disulfide bond formation and disulfide bond isomerization, ensuring the correct pairing of cysteine residues [reviewed in Ref. (19)].
The first bacterial catalyst of disulfide bond formation, disulfide bond protein A (DsbA), was identified in the periplasm of Escherichia coli by the group of Jon Beckwith (3) three decades after the discovery of PDI. In contrast to PDI, DsbA only functions as a disulfide-introducing protein and exhibits only very weak isomerase activity. In E. coli, the isomerization of non-native disulfides is ensured by another protein, DsbC.
The pathways of disulfide bond formation and isomerization in bacteria have been reviewed recently in several excellent articles, including some published in this journal (21, 24). We refer the reader to these reviews for in depth coverage of the initial experiments that led to the unraveling of the bacterial disulfide bond machineries. Here, we first briefly summarize the major achievements of the past 20 years and describe the most important properties of the oxidoreductases involved in protein disulfide formation in the E. coli periplasm. Then, we focus on the challenges ahead and on two fascinating mysteries that remain unsolved. We also highlight a new link that has been established recently between oxidative protein folding and the defense mechanisms against oxidative stress. In the last part of this review, we discuss the diversity of bacterial oxidative folding pathways.
Oxidative Protein Folding in E. coli
The proteins involved
Disulfide bond formation by the DsbA-DsbB machinery
Disulfide bonds are introduced into cell envelope proteins by the DsbA (3) (Fig. 1). In E. coli, about 300 proteins (∼40% of the envelope proteome) are known or predicted substrates of DsbA (15, 47). DsbA, like PDI and many other oxidoreductases, belongs to the thioredoxin superfamily. Proteins from that family have a canonical CXXC catalytic motif located in a highly conserved fold, which consists of four β-strands surrounded by three α-helices (37). The catalytic CXXC motif of DsbA is maintained in an oxidized state in vivo (22), which enables DsbA to react with proteins entering the periplasm to oxidize them. The oxidation reaction occurs in two successive steps. First, there is a nucleophilic attack by an active cysteine residue of the substrate on the first cysteine of the oxidized CXXC motif of DsbA. This results in the formation of a mixed-disulfide complex between DsbA and the target protein. Then, the mixed disulfide is attacked by another cysteine of the substrate, which results in the oxidation of the substrate and the reduction of DsbA (3). The increased stability of the reduced form of DsbA compared to its oxidized form endows DsbA with a strongly oxidizing redox potential compared to most structural disulfides in substrate proteins, so that DsbA readily oxidizes thiols to disulfide bonds in its substrates.
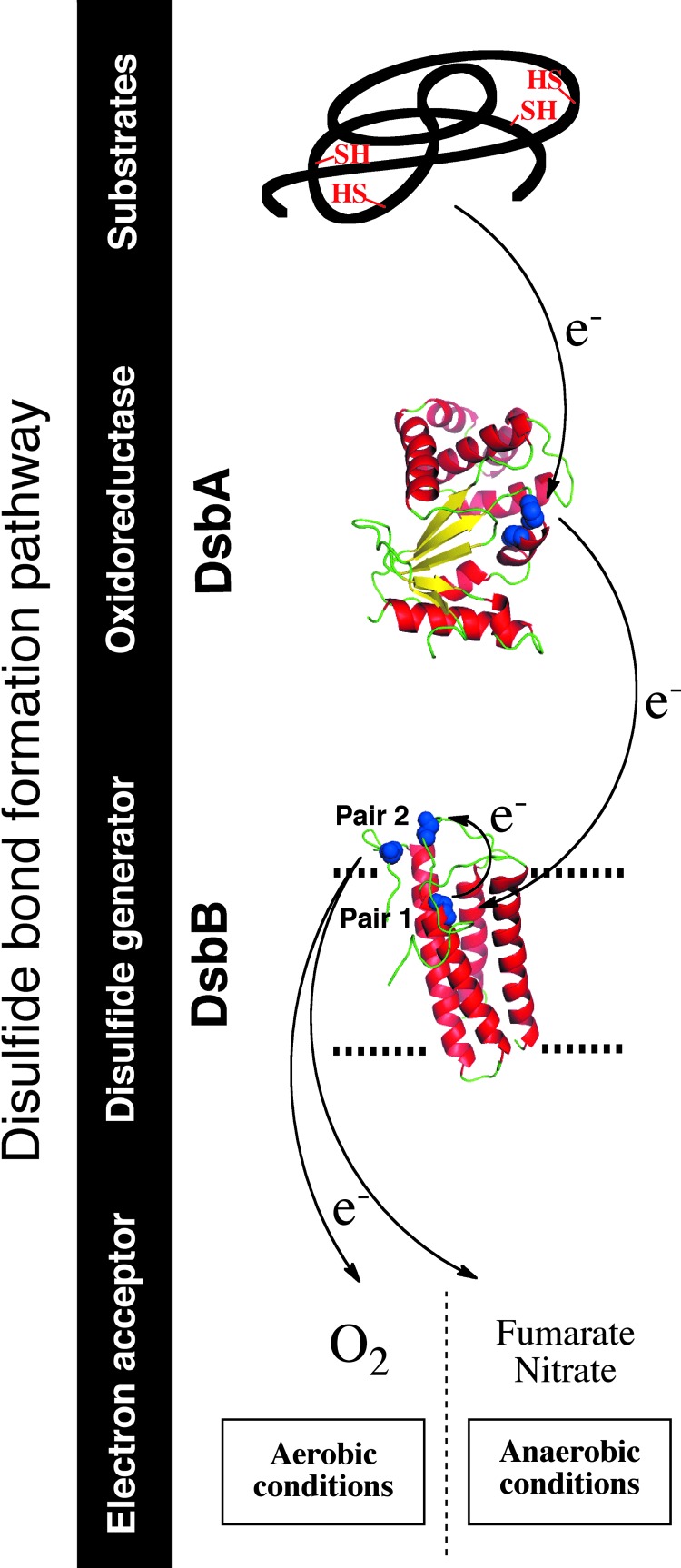
FIG. 1.
Disulfide bond formation in the Escherichia coli periplasm. Disulfide bonds are introduced by disulfide bond protein A (DsbA), which is then recycled by the inner membrane (IM) protein DsbB. DsbB generates disulfides de novo from quinone reduction. The electrons are then transferred to the respiratory chain. The final electron acceptor is molecular oxygen (O2) under aerobic conditions and nitrate or fumarate under anaerobic conditions. Three-dimensional structures of E. coli DsbA (protein database [PDB] entry code 1A2L) and E. coli DsbB (PDB entry code 2ZUQ) are drawn in ribbon form. Cysteine residues are drawn in space-filling form and colored in blue. Black arrows indicate the flow of electrons. The figures were generated using MacPyMol (Delano Scientific LLC 2006). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars.
In E. coli, DsbA is recycled back to the oxidized form by the inner membrane protein DsbB (2) (Fig. 1). DsbB has four transmembrane segments and two periplasmic loops, each containing one pair of conserved cysteine residues that are maintained in an oxidized state (pairs 1 and 2). DsbB uses those cysteine residues to channel the electrons away from DsbA and to deliver them to bound quinone molecules. Thus, DsbB generates disulfides de novo with concomitant quinone reduction.
The reaction between DsbA and DsbB is initiated by the nucleophilic attack of the first cysteine residue of DsbA's CXXC motif on the oxidized cysteines of the second periplasmic loop of DsbB (pair 2), which leads to the formation of a DsbA-DsbB mixed-disulfide complex. The structure of this complex, which was solved by Inaba, Ito, and their collaborators (20), provided insightful information that significantly advanced our understanding of the mechanism used by DsbB to generate disulfides. The mixed disulfide is then transferred to DsbA, which releases the cysteine residues of pair 2 of DsbB in the reduced form. Reoxidation of these cysteines occurs by electron transfer to the cysteines of pair 1, which are finally recycled back to the oxidized state by transferring the electrons to an ubiquinone molecule. The electrons are then transferred to the electron transport chain, molecular oxygen being the terminal electron acceptor (1). The DsbA-DsbB system is also able to function anaerobically. Under these conditions, DsbB transfers the electrons to menaquinone and then to other terminal electron acceptors such as fumarate and nitrate (1) (Fig. 1).
Disulfide bond isomerization by the DsbC-DsbD system
DsbA is a powerful oxidant that preferentially introduces disulfides in a vectorial manner into proteins entering the periplasm (23), that is, between cysteine residues that are consecutive in the primary amino acid sequence. Therefore, proteins that require formation of nonconsecutive disulfides to attain their native structure are often incorrectly oxidized by DsbA. An isomerization system is present in the E. coli periplasm to catalyze the rearrangement of non-native disulfide bonds (39) (Fig. 2).
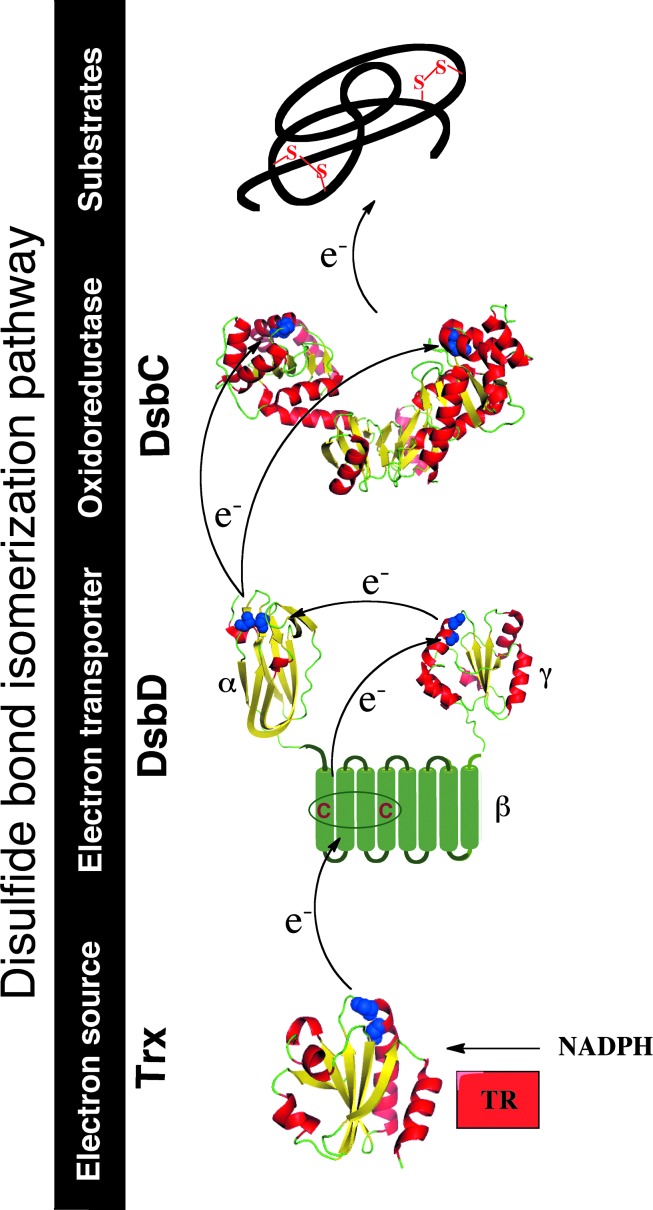
FIG. 2.
Disulfide bond isomerization in the E. coli periplasm. Non-native disulfides are repaired by the soluble homodimeric protein DsbC. DsbC is kept reduced in the periplasm by the membrane protein DsbD. DsbD receives electrons from cytoplasmic thioredoxin (Trx), which is kept reduced by thioredoxin reductase (TR), at the expense of reduced nicotine adenine dinucleotide phosphate (NADPH). The three-dimensional structures of E. coli DsbC (PDB entry code 1EEJ), DsbDα (PDB entry code 1JPE), and DsbDγ (PDB entry code 2FWF) are drawn in ribbon form. Cysteine residues are drawn in space-filling form and colored in blue. Black arrows indicate the flow of electrons. The figures were generated using MacPyMol (Delano Scientific LLC 2006). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars.
The main player of the E. coli isomerization pathway is the PDI DsbC. DsbC is a soluble homodimeric protein characterized by a V-shaped structure (36). Each monomer of DsbC presents an N-terminal dimerization domain and a C-terminal thioredoxin-like domain with a CXXC active site that is found predominantly in the reduced state in vivo (22). The dimerization of DsbC allows the formation of an uncharged cleft, which plays an important role in substrate binding. This is nicely illustrated by the fact that fusion of the N-terminal dimerization domain of DsbC to either DsbA or thioredoxin allows those proteins to function as disulfide isomerases in the periplasm [reviewed in Ref. (14)].
Upon binding of the substrate into DsbC's uncharged cleft, the N-terminal cysteine of the CXXC motif of DsbC performs a nucleophilic attack on an incorrect disulfide present in the substrate. This results in the formation of an unstable mixed-disulfide complex between DsbC and the substrate protein. This mixed disulfide is then attacked by another cysteine residue of the misfolded protein, creating a more stable disulfide in the target protein and releasing DsbC in the reduced state (here DsbC functions as an isomerase). Alternatively, the second cysteine of the CXXC motif of DsbC attacks the mixed disulfide, which results in the reduction of the substrate and the oxidation of DsbC (in that case, DsbC functions as a reductase).
The protein that recycles DsbC back to the reduced state is the inner membrane protein DsbD (39). DsbD is a 59-kDa monomeric protein composed of three distinct structural domains: an N-terminal periplasmic domain (DsbDα) with an immunoglobulin-like fold, a membrane-embedded domain with eight transmembrane segments (DsbDβ), and a C-terminal periplasmic thioredoxin-like domain (DsbDγ) (Fig. 2). Each of these domains contains a pair of conserved cysteine residues that are redox active and essential for the function of DsbD (see below) (26).
DsbD functions as an electron hub that dispatches reducing equivalents to various periplasmic oxidoreductases
The function of DsbD is to transfer reducing equivalents from the cytoplasmic thioredoxin system to the periplasm (39), by a mechanism that is still not fully understood (see below) (Fig. 2). The thioredoxin system, which involves thioredoxin and thioredoxin reductase, maintains cytoplasmic proteins reduced at the expense of nicotinamide adenine dinucleotide phosphate (NADPH) (10).
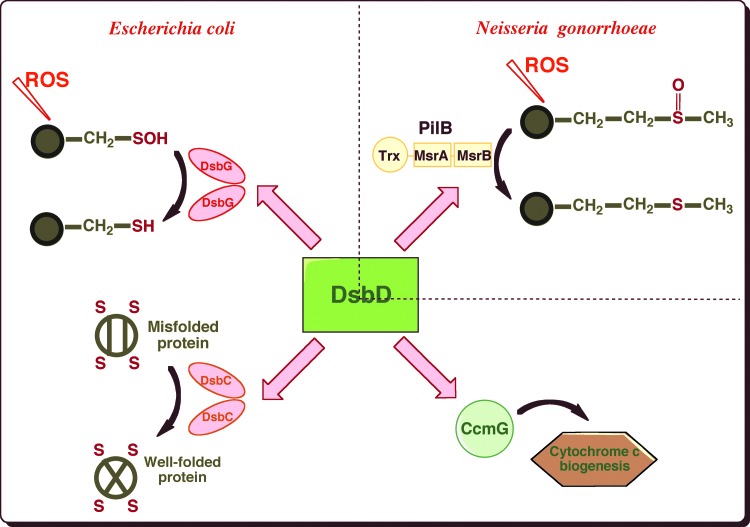
DsbC is not the only substrate of DsbD. In fact, this protein functions as an electron hub that dispatches reducing equivalents to various redox pathways present in the cell envelope (Fig. 3). For instance, DsbD provides reductants to CcmG, an oxidoreductase involved in the maturation of cytochrome c (46). Moreover, we recently found that DsbG, a homodimeric protein that is homologous to DsbC, uses electrons received from DsbD to control the global sulfenic acid content of the periplasm (13) (Fig. 3). Sulfenic acids are highly unstable modifications that form on cysteine residues exposed to reactive oxygen species (ROS). Further oxidation of sulfenic acids leads to sulfinic (−SO2H) and sulfonic (−SO3H) acids, which irreversibly modify proteins. The role of DsbG is to protect envelope proteins containing a single cysteine residue from oxidation to sulfenic acid to prevent their inactivation (13).
FIG. 3.
DsbD is an electron hub that transfers reducing equivalents to several oxidoreductases present in the cell envelope. In E. coli, DsbD provides reducing equivalents to DsbC, the isomerase that corrects non-native disulfides, to CcmG, an oxidoreductase involved in the maturation of cytochrome c and to DsbG, which controls the global sulfenic acid content of the periplasm. In Neisseria gonorrhoeae, DsbD shuttles electrons to the N-terminal domain of PilB, a multidomain protein that exhibits methionine sulfoxide reductase (Msr) activity (To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars.)
Another example of the role played by DsbD in the defense mechanisms against oxidative stress was reported in Neisseria gonorrhoeae. In this bacterium, DsbD provides electrons to the N-terminal domain of PilB (4), a multidomain protein that exhibits methionine sulfoxide reductase (Msr) activity (Fig. 3). Msrs are antioxidant enzymes that catalyze the reduction of methionine sulfoxides (MetO) back to the reduced state. MetO are formed upon exposure of methionine residues to ROS, which leads, if unrepaired, to alterations in protein conformation and loss of biological activity.
Unsolved mysteries
The work performed over the past 20 years has led to the identification and detailed characterization of most of the proteins that catalyze disulfide bond formation and isomerization in the E. coli periplasm. The oxidation and isomerization pathways have been reconstituted in vitro; the kinetic constants of the various electron transfer reactions have been determined, and the structures of most of the players involved have been solved [reviewed in Refs. (14, 24)]. However, despite these tremendous achievements, fundamental questions remain unanswered. Here, we focus on two major unsolved mysteries that require further investigation.
How does DsbD transfer reducing equivalents across the membrane?
A fascinating remaining mystery is the mechanism by which DsbD transfers reducing equivalents from the cytoplasmic thioredoxin system to periplasmic oxidoreductases such as DsbC and DsbG without using a cofactor. Elegant work by Beckwith and collaborators has revealed that the activity of DsbD depends on a cascade of thiol–disulfide exchange reactions involving the 3 pairs of cysteine residues of DsbD (26). First, thioredoxin reduces the disulfide bond involving the two cysteine residues of DsbDβ. Then, the electrons are transferred from those membrane-embedded cysteines to those of DsbDγ, DsbDα, and finally to the catalytic site of the periplasmic oxidoreductases (Fig. 2).
The reactions taking place in the periplasm, that is, the reduction of DsbDα by DsbDγ and the reaction between DsbDα and DsbC, have been thoroughly characterized [for instance, see Refs. (11, 40)]. We would like to highlight recent data reported by Redfield and coworkers, who characterized in detail the thiol–disulfide exchange reaction between the two soluble γ and α domains of DsbD (35). In particular, they demonstrated that the strength of the interaction between the two soluble subunits of DsbD depends on their respective redox states. They showed that complex formation is facilitated by a relatively high affinity (Kd of 86 μM) of the oxidized DsbDα for the reduced DsbDγ, whereas reduced DsbDα has a lowered affinity (Kd of 2.4–3.5 mM) for the oxidized DsbDγ. These oxidation state-dependent interactions between both partners avoid the formation of nonefficient complexes that would prevent the transfer of reductants across the membrane.
In contrast to the abundance of details regarding the reactions taking place in the periplasm, we have a much poorer understanding of the mechanism used by DsbDβ to transfer reducing equivalents across the inner membrane. This is clearly one of the most intriguing questions that remains to be solved regarding the bacterial oxidative folding pathways. It is likely that the solution to this problem will come from solving the structure of the membranous domain of DsbD, which is still unknown. The current working model for the structure of DsbDβ is a low-resolution structure based on the work of Cho et al. (6, 7, 9). Using small thiol-modifying reagents, these authors determined the accessibility of the catalytic cysteine residues of DsbDβ as well as that of membrane-embedded residues that they substituted by cysteines. Their data suggest that DsbDβ adopts an hourglass structure in which the catalytic cysteine residues are located at the juncture of the two cavities where they are exposed to both thioredoxin in the cytoplasm and DsbDγ in the periplasm. The redox states of these cysteines do not seem to affect the conformation of the membranous domain.
How do oxidative protein-folding catalysts cooperate with periplasmic chaperones?
A second fascinating mystery is how the protein-folding catalysts DsbA and DsbC coordinate their action with the periplasmic chaperones that assist the assembly of outer membrane (OM) proteins in Gram-negative bacteria. There are two major groups of proteins present in the OM: (i) lipoproteins that are present in the periplasm, but are anchored by a lipid moiety to the inner leaflet of the OM, and (ii) β-barrel proteins that are integral membrane proteins [reviewed in Ref. (31)]. After secretion to the periplasm, OM proteins are transported across this hydrophilic compartment by dedicated chaperones that deliver them to specific OM insertion machineries: lipoproteins are escorted by LolA, whereas SurA and Skp assist the assembly of ®-barrel proteins (44) (Fig. 4). Two OM proteins, the ®-barrel LptD and the lipoprotein RcsF, have recently been shown to require the formation of nonconsecutive disulfides for activity, which raises interesting questions regarding how and where disulfides are introduced into proteins destined for the OM.
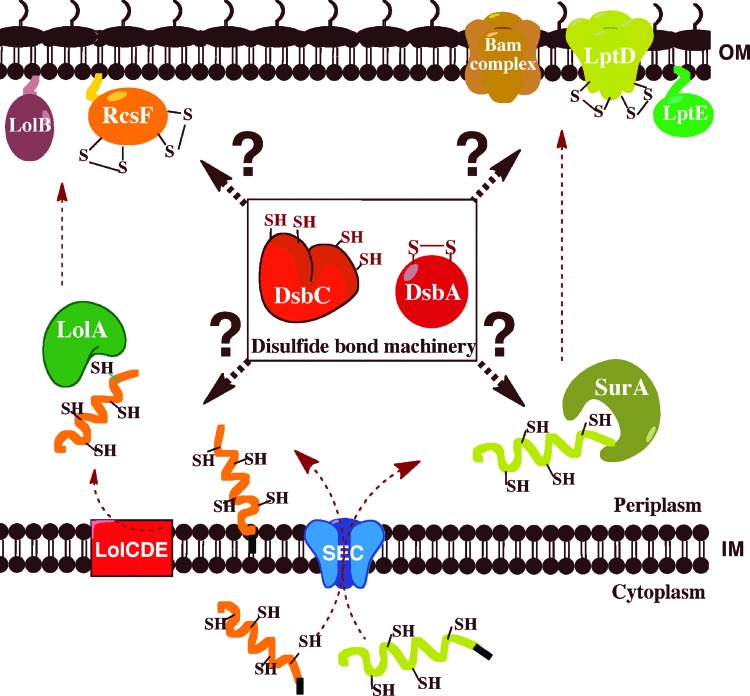
FIG. 4.
Protein-folding catalysts cooperate with periplasmic chaperones in the assembly of outer membrane (OM) proteins. The β-barrel protein LptD and the lipoprotein RcsF are synthesized in the cytoplasm. They are transported unfolded across the IM by the SEC translocon. RcsF is first addressed to the IM complex LolCDE and then transferred to the chaperone LolA. LolA transports RcsF across the periplasm and delivers it to LolB, which incorporates lipoproteins into the OM. After translocation by SEC, LptD is escorted across the periplasm by the chaperone SurA. SurA delivers β-barrel proteins to the Bam machinery, which inserts them in the OM. The assembly of LptD requires the formation of the LptD/LptE complex. The oxidative folding catalysts DsbA and DsbC are involved in the formation and isomerization of the nonconsecutive disulfides of RcsF and LptD. How they cooperate with LolA and SurA is not clearly established (To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars.)
LptD has received much attention lately. It is an essential β-barrel protein that inserts lipopolysaccharides (LPS) into the OM [reviewed in Ref. (42)] and that has recently been shown to be the cellular target of a new class of peptidomimetic antibiotics (45). Although the mechanism used by LptD to insert LPS in the OM has not been elucidated yet, some key results have been obtained regarding the assembly pathway of this protein (Fig. 4). First, LptD has been shown to interact preferentially with the chaperone SurA (48): the chaperone is thought to bind to unfolded LptD upon entry into the periplasm and to deliver the protein to the Bam assembly complex for insertion in the OM (27, 50). Second, LptD forms a very stable complex with the lipoprotein LptE in the OM (5), which is required for LptD assembly. Third, LptD contains two pairs of cysteine residues that have recently been shown to form two nonconsecutive disulfides (connecting C1 to C3 and C2 to C4) bridging the N-terminal periplasmic soluble domain of the protein to the C-terminal domain that likely adopts a membrane-embedded β-barrel conformation (41). Interestingly, disulfide bond formation does not appear to be required for LptD folding and stability. However, formation of at least one of the nonconsecutive disulfides is required for function (41). Both DsbA and DsbC have been trapped in complex with LptD (12, 25), indicating that these oxidoreductases play a role in the assembly pathway of the protein. However, it is not known when oxidation of LptD takes place. Does DsbA introduce the disulfides into LptD upon entry of the protein in the periplasm, as would be suggested by the current model for disulfide bond formation? Or, does SurA interact with LptD first, the disulfides being introduced upon or after assembly of LptD in the OM, and what is the role of DsbC? The fact that the formation of the LptD/LptE complex in the OM is required for proper oxidation of LptD (5) suggests that the oxidation of this protein is only completed after LptD reaches the OM. Clearly, further investigation is required to firmly establish the oxidative folding pathway of this protein and to determine how DsbA and DsbC assist the assembly of ®-barrel proteins.
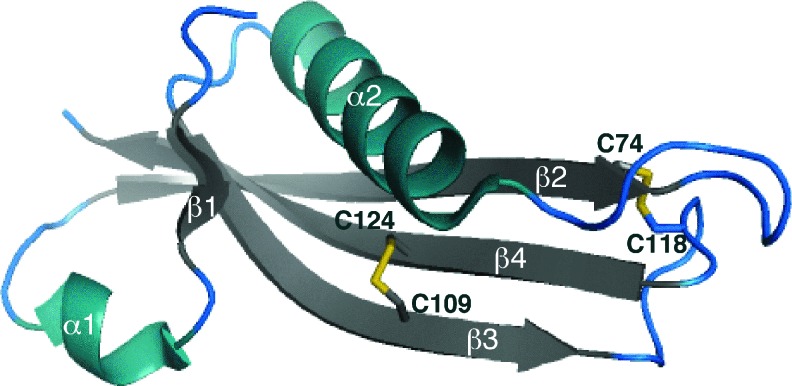
Similar questions can be raised regarding the small OM lipoprotein RcsF (14 kDa). RcsF functions as a sensor that activates a signaling cascade controlling the transcription of genes involved in cell division, motility, and biofilm formation in response to various envelope stresses [reviewed in Ref. (34)]. We recently solved the structure of RcsF, which revealed that the protein has two nonconsecutive disulfide bonds (C1–C3 and C2–C4) essential for folding and activity (Fig. 5) (30). The formation of these disulfides depends on DsbA and DsbC (25, 30). Interestingly, one of the nonconsecutive disulfides bridges, two cysteine residues that are on two adjacent β-strands, forms a so-called cross-strand disulfide (CSD) (49). CSD usually plays a functional role, suggesting that the activity of RcsF might be redox regulated. Here also, further work is required to elucidate how DsbA, DsbC, and LolA, the chaperone dedicated to lipoproteins, cooperate in the assembly process of RcsF (Fig. 4).
FIG. 5.
Three-dimensional structure of the RcsF protein. Two nonconsecutive disulfides are present in the three-dimensional structure of RcsF, which is drawn in ribbon form (PDB entry code 2Y1B). The first disulfide bond involves Cys74 (C1) and Cys118 (C3) and links an amino acid in the β2-strand to a residue in the β3-β4 loop. The second disulfide between Cys109 (C2) and Cys124 (C4) is a cross-strand disulfide (CSD) that connects the β3 and β4 strands. Cysteine residues are indicated on the structure. The figures were generated using MacPyMol (Delano Scientific LLC 2006). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars.
A Variety of Disulfide Bond-Forming Systems Among Gram-Negative Bacteria
The E. coli Dsb system is generally considered as the paradigm of oxidative protein-folding pathways in bacteria. However, bioinformatic analysis of whole-genome sequencing data, completed by molecular studies of disulfide bond machineries in bacteria other than E. coli, has revealed that there is a significant diversity of bacterial disulfide bond-forming systems [reviewed in Ref. (43)].
First, many bacterial genomes encode a repertoire of thiol–disulfide oxidoreductases that is significantly expanded compared to the E. coli K12 system. For instance, the genome of Neisseria meningitidis, a causative agent of epidemic meningitis, encodes three DsbA proteins, two of which are anchored in the inner membrane by a lipid moiety, while the third one is soluble like E. coli DsbA. Neisserial DsbAs differ in terms of redox properties, surface characteristics, and substrate specificity (29). In other bacteria, additional redox pairs that are responsible for the oxidation of specific substrates are present in addition to the classical DsbA-DsbB and DsbC-DsbD pathways. For instance, in Salmonella enterica serovar Typhimurium, DsbL and DsbI, which are homologous to DsbA and DsbB, respectively, specifically oxidize a periplasmic aryl sulfate sulfotransferase, an enzyme that detoxifies phenolic substances and antibiotics (33). Alternatively, other disulfide bond systems diverge from the E. coli machinery by the fact that they lack an isomerization pathway, such as in Wolbachia pipientis, an insect parasite (28).
One of the most interesting findings in this area was reported recently by Dutton et al. (15, 16). The observation that some bacteria, including cyanobacteria and certain actinobacteria, ɛ-, and ™-proteobacteria, possess a DsbA homolog, but lack a homolog of DsbB, has led them to the identification of a protein with a DsbB-like activity. This protein is the bacterial homolog of the eukaryotic vitamin K epoxide reductase (VKOR), an enzyme that catalyzes the reduction of vitamin K epoxide, a quinone derivative, to vitamin K. Vitamin K is required as an essential cofactor for enzyme-catalyzed carboxylation reactions responsible for blood coagulation (38). Interestingly, the VKOR homolog from Mycobacterium tuberculosis (Mtb VKOR) can efficiently restore disulfide bond formation in E. coli dsbB mutants in a DsbA-dependent manner (15). These results suggest that Mtb VKOR and E. coli DsbB function in an analogous way in vivo, and that VKOR can substitute for DsbB in bacteria that do not encode the dsbB gene. It is noteworthy that when expressed in E. coli, Mtb VKOR is inhibited by warfarin, an anticoagulant that blocks the human VKOR function, whereas DsbB activity is unaffected by this drug (16).
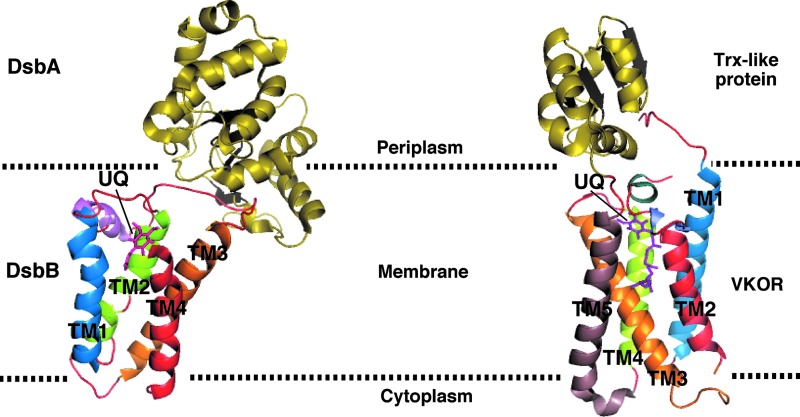
Recently, the structure of a natural fusion protein from Synechococcus sp. has been solved, in which a VKOR homolog and its DsbA-like redox partner are found in complex (32) (Fig. 6). The structure revealed that VKOR and DsbB share three-dimensional structure similarities, which was unexpected because of the absence of sequence homology between these two proteins. The structural similarities suggest that DsbB and VKOR generate disulfide bonds via a similar mechanism of action.
FIG. 6.
Architecture comparison between the E. coli DsbB-DsbA complex and Synechococcus vitamin K epoxide reductase (VKOR) fused to its Trx-like partner. Three-dimensional structures of the E. coli DsbB-DsbA complex (PDB entry code 2HI7) and Synechococcus VKOR fused to its Trx-like partner (PDB entry code 3KP9) are shown by ribbon diagram. E. coli DsbB is characterized by a four-helix bundle scaffold, whereas Synechococcus VKOR has five transmembrane α-helices. The electron-accepting cofactor, ubiquinone (UQ), is represented by magenta sticks. Dotted lines indicate membrane boundaries. The figures were generated using MacPyMol (Delano Scientific LLC 2006). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars.
We also would like to highlight a recent work that revealed the diversity of the proteins from the DsbD superfamily. Using bioinformatic programs to seek homologs of E. coli DsbD, Cho et al. detected a new class of reductive proteins whose prototype is Salmonella typhimurium ScsB (8). Although the ScsB class has domains comparable to those of DsbD, the N-terminal periplasmic domain of ScsB is quite different from DsbDα, suggesting that ScsB proteins act on a different array of substrate proteins from those already known for DsbD. Indeed, Caulobacter crescentus ScsB was found to transfer reducing equivalents to ScsC, a novel bacterial disulfide isomerase, and to TlpA, a thioredoxin-like protein that in turn reduces a peroxidase PprX. These results reveal that the range of electron transfer reactions involving DsbD-like proteins is much broader than has been understood. Furthermore, they suggest that the unraveling of the redox pathways at work in bacteria other than E. coli will likely lead to the identification of novel types of DsbD substrates.
Conclusions
We have a good knowledge of the pathways of disulfide bond formation in E. coli: the proteins involved have been identified, and their major characteristics have been determined. Understanding the details of the mechanism employed by DsbD to transport reducing equivalents across the membrane probably remains the most important problem to be solved to complete the molecular characterization of the E. coli Dsb proteins.
The unraveling of the E. coli disulfide bond formation machinery has allowed discovery of the basic principles of the bacterial oxidative folding pathways in general. It is now established that in bacteria, disulfides are generated by membrane proteins such as DsbB or VKOR that connect oxidative protein folding to the electron transport chain. The generated disulfides are then donated to folding proteins by oxidoreductases from the thioredoxin superfamily. In some bacteria, such as E. coli, an isomerization pathway exists next to the oxidation pathway. This reducing pathway uses electrons derived from the cytoplasmic pool of NADPH to correct wrongly formed disulfides.
One of the challenges of the coming years will be to put the knowledge of the disulfide formation machineries into the global cellular context. For instance, it will be interesting to determine how the oxidative protein folding catalysts cooperate with the other proteins involved in protein assembly. Another important challenge will be to characterize in details the oxidative folding pathways at work in bacteria other than E. coli, including pathogens. As many virulence factors are stabilized by disulfide bonds, this could lead to the design of new antibiotics.
Abbreviations Used
- CSD
cross-strand disulfide
- Dsb
disulfide bond
- e−
electron
- IM
inner membrane
- LPS
lipopolysaccharide
- Msr
methionine sulfoxide reductase
- NADPH
nicotinamide adenine dinucleotide phosphate
- O2
molecular oxygen
- OM
outer membrane
- PDB
protein database
- PDI
protein disulfide isomerase
- ROS
reactive oxygen species
- TR
thioredoxin reductase
- Trx
thioredoxin
- UQ
ubiquinone
- VKOR
vitamin K epoxide reductase
Acknowledgments
We would like to thank Lloyd Ruddock and Robert Freedman for the invitation. We thank Pauline Leverrier, Seung-Hyun Cho, Alexandra Gennaris, and Carole Linster for critical reading of the manuscript. JFC is Chercheur Qualifié of the FRS-FNRS. KD is a research fellow of the FRIA. This work was supported by the European Research Council (FP7/2007–2013) ERC independent researcher starting grant 282335–Sulfenic and by grants from the FRS-FNRS to JFC. We apologize to authors whose work was not cited directly due to reference limitations.
References
- 1.Bader M. Muse W. Ballou DP. Gassner C. Bardwell JC. Oxidative protein folding is driven by the electron transport system. Cell. 1999;98:217–227. doi: 10.1016/s0092-8674(00)81016-8. [DOI] [PubMed] [Google Scholar]
- 2.Bardwell JC. Lee JO. Jander G. Martin N. Belin D. Beckwith J. A pathway for disulfide bond formation in vivo. Proc Natl Acad Sci U S A. 1993;90:1038–1042. doi: 10.1073/pnas.90.3.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardwell JC. McGovern K. Beckwith J. Identification of a protein required for disulfide bond formation in vivo. Cell. 1991;67:581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- 4.Brot N. Collet JF. Johnson LC. Jonsson TJ. Weissbach H. Lowther WT. The thioredoxin domain of Neisseria gonorrhoeae PilB can use electrons from DsbD to reduce downstream methionine sulfoxide reductases. J Biol Chem. 2006;281:32668–32675. doi: 10.1074/jbc.M604971200. [DOI] [PubMed] [Google Scholar]
- 5.Chimalakonda G. Ruiz N. Chng SS. Garner RA. Kahne D. Silhavy TJ. Lipoprotein LptE is required for the assembly of LptD by the beta-barrel assembly machine in the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A. 2011;108:2492–2497. doi: 10.1073/pnas.1019089108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho SH. Beckwith J. Mutations of the membrane-bound disulfide reductase DsbD that block electron transfer steps from cytoplasm to periplasm in Escherichia coli. J Bacteriol. 2006;188:5066–5076. doi: 10.1128/JB.00368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho SH. Beckwith J. Two snapshots of electron transport across the membrane: insights into the structure and function of DsbD. J Biol Chem. 2009;284:11416–11424. doi: 10.1074/jbc.M900651200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho SH. Parsonage D. Thurston C. Dutton RJ. Poole LB. Collet JF. Beckwith J. A new family of membrane electron transporters and its substrates, including a new cell-envelope peroxiredoxin, reveal a broadened reductive capacity of the oxidative bacterial cell envelope. mBio. 2012;3 doi: 10.1128/mBio.00291-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho SH. Porat A. Ye J. Beckwith J. Redox-active cysteines of a membrane electron transporter DsbD show dual compartment accessibility. EMBO J. 2007;26:3509–3520. doi: 10.1038/sj.emboj.7601799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collet JF. Messens J. Structure, function, and mechanism of thioredoxin proteins. Antioxid Redox Signal. 2010;13:1205–1216. doi: 10.1089/ars.2010.3114. [DOI] [PubMed] [Google Scholar]
- 11.Collet JF. Riemer J. Bader MW. Bardwell JC. Reconstitution of a disulfide isomerization system. J Biol Chem. 2002;277:26886–26892. doi: 10.1074/jbc.M203028200. [DOI] [PubMed] [Google Scholar]
- 12.Denoncin K. Vertommen D. Paek E. Collet JF. The protein-disulfide isomerase DsbC cooperates with SurA and DsbA in the assembly of the essential beta-barrel protein LptD. J Biol Chem. 2010;285:29425–29433. doi: 10.1074/jbc.M110.119321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Depuydt M. Leonard SE. Vertommen D. Denoncin K. Morsomme P. Wahni K. Messens J. Carroll KS. Collet JF. A periplasmic reducing system protects single cysteine residues from oxidation. Science. 2009;326:1109–1111. doi: 10.1126/science.1179557. [DOI] [PubMed] [Google Scholar]
- 14.Depuydt M. Messens J. Collet JF. How proteins form disulfide bonds. Antioxid Redox Signal. 2011;15:49–66. doi: 10.1089/ars.2010.3575. [DOI] [PubMed] [Google Scholar]
- 15.Dutton RJ. Boyd D. Berkmen M. Beckwith J. Bacterial species exhibit diversity in their mechanisms and capacity for protein disulfide bond formation. Proc Natl Acad Sci U S A. 2008;105:11933–11938. doi: 10.1073/pnas.0804621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dutton RJ. Wayman A. Wei JR. Rubin EJ. Beckwith J. Boyd D. Inhibition of bacterial disulfide bond formation by the anticoagulant warfarin. Proc Natl Acad Sci U S A. 2010;107:297–301. doi: 10.1073/pnas.0912952107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldberger RF. Epstein CJ. Anfinsen CB. Acceleration of reactivation of reduced bovine pancreatic ribonuclease by a microsomal system from rat liver. J Biol Chem. 1963;238:628–635. [PubMed] [Google Scholar]
- 18.Gothel SF. Marahiel MA. Peptidyl-prolyl cis-trans isomerases, a superfamily of ubiquitous folding catalysts. Cell Mol Life Sci. 1999;55:423–436. doi: 10.1007/s000180050299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatahet F. Ruddock LW. Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxid Redox Signal. 2009;11:2807–2850. doi: 10.1089/ars.2009.2466. [DOI] [PubMed] [Google Scholar]
- 20.Inaba K. Murakami S. Suzuki M. Nakagawa A. Yamashita E. Okada K. Ito K. Crystal structure of the DsbB-DsbA complex reveals a mechanism of disulfide bond generation. Cell. 2006;127:789–801. doi: 10.1016/j.cell.2006.10.034. [DOI] [PubMed] [Google Scholar]
- 21.Ito K. Inaba K. The disulfide bond formation (Dsb) system. Curr Opin Struct Biol. 2008;18:450–458. doi: 10.1016/j.sbi.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 22.Joly JC. Swartz JR. In vitro and in vivo redox states of the Escherichia coli periplasmic oxidoreductases DsbA and DsbC. Biochemistry. 1997;36:10067–10072. doi: 10.1021/bi9707739. [DOI] [PubMed] [Google Scholar]
- 23.Kadokura H. Beckwith J. Detecting folding intermediates of a protein as it passes through the bacterial translocation channel. Cell. 2009;138:1164–1173. doi: 10.1016/j.cell.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kadokura H. Beckwith J. Mechanisms of oxidative protein folding in the bacterial cell envelope. Antioxid Redox Signal. 2010;13:1231–1246. doi: 10.1089/ars.2010.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kadokura H. Tian H. Zander T. Bardwell JC. Beckwith J. Snapshots of DsbA in action: detection of proteins in the process of oxidative folding. Science. 2004;303:534–537. doi: 10.1126/science.1091724. [DOI] [PubMed] [Google Scholar]
- 26.Katzen F. Beckwith J. Transmembrane electron transfer by the membrane protein DsbD occurs via a disulfide bond cascade. Cell. 2000;103:769–779. doi: 10.1016/s0092-8674(00)00180-x. [DOI] [PubMed] [Google Scholar]
- 27.Knowles TJ. Scott-Tucker A. Overduin M. Henderson IR. Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nat Rev Microbiol. 2009;7:206–214. doi: 10.1038/nrmicro2069. [DOI] [PubMed] [Google Scholar]
- 28.Kurz M. Iturbe-Ormaetxe I. Jarrott R. Shouldice SR. Wouters MA. Frei P. Glockshuber R. O'Neill SL. Heras B. Martin JL. Structural and functional characterization of the oxidoreductase alpha-DsbA1 from Wolbachia pipientis. Antioxid Redox Signal. 2009;11:1485–1500. doi: 10.1089/ars.2008.2420. [DOI] [PubMed] [Google Scholar]
- 29.Lafaye C. Iwema T. Carpentier P. Jullian-Binard C. Kroll JS. Collet JF. Serre L. Biochemical and structural study of the homologues of the thiol-disulfide oxidoreductase DsbA in Neisseria meningitidis. J Mol Biol. 2009;392:952–966. doi: 10.1016/j.jmb.2009.07.056. [DOI] [PubMed] [Google Scholar]
- 30.Leverrier P. Declercq JP. Denoncin K. Vertommen D. Hiniker A. Cho SH. Collet JF. Crystal structure of the outer membrane protein RcsF, a new substrate for the periplasmic protein-disulfide isomerase DsbC. J Biol Chem. 2011;286:16734–16742. doi: 10.1074/jbc.M111.224865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leverrier P. Vertommen D. Collet JF. Contribution of proteomics toward solving the fascinating mysteries of the biogenesis of the envelope of Escherichia coli. Proteomics. 2010;10:771–784. doi: 10.1002/pmic.200900461. [DOI] [PubMed] [Google Scholar]
- 32.Li W. Schulman S. Dutton RJ. Boyd D. Beckwith J. Rapoport TA. Structure of a bacterial homologue of vitamin K epoxide reductase. Nature. 2010;463:507–512. doi: 10.1038/nature08720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin D. Kim B. Slauch JM. DsbL and DsbI contribute to periplasmic disulfide bond formation in Salmonella enterica serovar Typhimurium. Microbiology. 2009;155:4014–4024. doi: 10.1099/mic.0.032904-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Majdalani N. Gottesman S. The Rcs phosphorelay: a complex signal transduction system. Annu Rev Microbiol. 2005;59:379–405. doi: 10.1146/annurev.micro.59.050405.101230. [DOI] [PubMed] [Google Scholar]
- 35.Mavridou DA. Saridakis E. Kritsiligkou P. Goddard AD. Stevens JM. Ferguson SJ. Redfield C. Oxidation state-dependent protein-protein interactions in disulfide cascades. J Biol Chem. 2011;286:24943–24956. doi: 10.1074/jbc.M111.236141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCarthy AA. Haebel PW. Torronen A. Rybin V. Baker EN. Metcalf P. Crystal structure of the protein disulfide bond isomerase, DsbC, from Escherichia coli. Nat Struct Biol. 2000;7:196–199. doi: 10.1038/73295. [DOI] [PubMed] [Google Scholar]
- 37.Messens J. Collet JF. Pathways of disulfide bond formation in Escherichia coli. Int J Biochem Cell Biol. 2006;38:1050–1062. doi: 10.1016/j.biocel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 38.Oldenburg J. Bevans CG. Muller CR. Watzka M. Vitamin K epoxide reductase complex subunit 1 (VKORC1): the key protein of the vitamin K cycle. Antioxid Redox Signal. 2006;8:347–353. doi: 10.1089/ars.2006.8.347. [DOI] [PubMed] [Google Scholar]
- 39.Rietsch A. Bessette P. Georgiou G. Beckwith J. Reduction of the periplasmic disulfide bond isomerase, DsbC, ocurs by passage of electrons from cytoplasmic thioredoxin. J Bacteriol. 1997;179:6602–6608. doi: 10.1128/jb.179.21.6602-6608.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rozhkova A. Stirnimann CU. Frei P. Grauschopf U. Brunisholz R. Grutter MG. Capitani G. Glockshuber R. Structural basis and kinetics of inter- and intramolecular disulfide exchange in the redox catalyst DsbD. EMBO J. 2004;23:1709–1719. doi: 10.1038/sj.emboj.7600178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruiz N. Chng SS. Hiniker A. Kahne D. Silhavy TJ. Nonconsecutive disulfide bond formation in an essential integral outer membrane protein. Proc Natl Acad Sci U S A. 2010;107:12245–12250. doi: 10.1073/pnas.1007319107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruiz N. Kahne D. Silhavy TJ. Transport of lipopolysaccharide across the cell envelope: the long road of discovery. Nat Rev Microbiol. 2009;7:677–683. doi: 10.1038/nrmicro2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shouldice SR. Heras B. Walden PM. Totsika M. Schembri MA. Martin JL. Structure and function of DsbA, a key bacterial oxidative folding catalyst. Antioxid Redox Signal. 2011;14:1729–1760. doi: 10.1089/ars.2010.3344. [DOI] [PubMed] [Google Scholar]
- 44.Silhavy TJ. Kahne D. Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srinivas N. Jetter P. Ueberbacher BJ. Werneburg M. Zerbe K. Steinmann J. Van der Meijden B. Bernardini F. Lederer A. Dias RL. Misson PE. Henze H. Zumbrunn J. Gombert FO. Obrecht D. Hunziker P. Schauer S. Ziegler U. Kach A. Eberl L. Riedel K. DeMarco SJ. Robinson JA. Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science. 2010;327:1010–1013. doi: 10.1126/science.1182749. [DOI] [PubMed] [Google Scholar]
- 46.Stirnimann CU. Rozhkova A. Grauschopf U. Grutter MG. Glockshuber R. Capitani G. Structural basis and kinetics of DsbD-dependent cytochrome c maturation. Structure. 2005;13:985–993. doi: 10.1016/j.str.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 47.Vertommen D. Depuydt M. Pan J. Leverrier P. Knoops L. Szikora JP. Messens J. Bardwell JC. Collet JF. The disulphide isomerase DsbC cooperates with the oxidase DsbA in a DsbD-independent manner. Mol Microbiol. 2008;67:336–349. doi: 10.1111/j.1365-2958.2007.06030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vertommen D. Ruiz N. Leverrier P. Silhavy TJ. Collet JF. Characterization of the role of the Escherichia coli periplasmic chaperone SurA using differential proteomics. Proteomics. 2009;9:2432–2443. doi: 10.1002/pmic.200800794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wouters MA. Fan SW. Haworth NL. Disulfides as redox switches: from molecular mechanisms to functional significances. Antioxid Redox Signal. 2010;12:53–91. doi: 10.1089/ars.2009.2510. [DOI] [PubMed] [Google Scholar]
- 50.Wu T. Malinverni J. Ruiz N. Kim S. Silhavy TJ. Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]