Abstract
Proliferation and apoptosis pathways are tightly regulated in a cell by the ubiquitin–proteasome system (UPS) and alterations in the UPS may result in cellular transformation or other pathological conditions. Indeed, the proteasome is often found to be overactive in cancer cells. It has also been found that cancer cells are more sensitive to proteasome inhibition than normal cells, and therefore proteasome inhibitors are pursued as antitumor drugs. The use of the proteasome inhibitor Bortezomib for treatment of multiple myeloma and mantle cell lymphoma has proved this principle. Recent studies have suggested that copper complexes can inhibit proteasome activity and induce apoptosis in some human cancer cells. However, the involved molecular mechanism is unknown. In this study, we investigated the biological activities of four amino acid Schiff base–copper(II) complexes by using human breast (MDA-MB-231 and MCF-7) and prostate (PC-3) cancer cells. The complexes C1 and C3, but not their counterparts C2 and C4, inhibit the chymotrypsin-like activity of purified 20S proteasome and human cancer cellular 26S proteasome, cause accumulation of proteasome target proteins Bax and IκB-α, and induce growth inhibition and apoptosis in concentration- and time-dependent manners. Docking analysis shows that C1, but not C2 has hydrophobic, pi–pi, pi–cation and hydrogen bond interactions with the proteasomal chymotrypsin-like pocket and could stably fit into the S3 region, leading to specific inhibition. Our study has identified the mechanism of action of these copper complexes on inhibiting tumor cell proteasome and suggested their great potential as novel anticancer agents.
Keywords: Anticancer, Drug discovery, Molecular modeling, Proteasome inhibitors, Apoptosis, Copper complexes
1. Introduction
Apoptosis or programmed cell death with distinct morphological characteristics occurs in all multicellular organisms. Apoptosis is a vital regulatory process responsible for the elimination of unwanted cells. In addition, it plays an essential role in human development, tissue homeostasis, and defense against mutations and viral infections [1–4]. Tumor cells are effective at evading apoptosis. The induction of apoptosis as an anti-cancer therapy has been actively pursued because tumor cells are more sensitive to apoptosis-inducing stimuli than normal cells [5–7].
The ubiquitin–proteasome system (UPS) plays an important role in a multitude of cellular processes including: cell cycle progression, DNA damage and repair, endocytosis, apoptosis, angiogenesis, drug resistance and differentiation [8,9]. The eukaryotic 26S proteasome is made up of two 19S regulatory particles and a catalytic 20S core. The 20S core consists of two identical non-catalytic α rings flanking two identical catalytic β rings. At least three distinct catalytic activities have been associated with the β-subunits of the 20S core: chymotrypsin-like (cleavage after hydrophobic residues by the β5 subunit), trypsin-like (cleavage after basic residues by the β2 subunit), and caspase-like or peptidyl-glutamyl peptidehydrolyzing-like (cleavage after acidic residues by the β1 subunit) [10–12]. It has been shown that inhibition of the proteasomal chymotrypsin-like, but not trypsin-like activity, is associated with induction of apoptosis in cancer cells [13–15]. Notably, the proteasomal subunits β4, β5, and β6 contribute to the full chymotrypsin-like active site in terms of the substrate recognition. However, catalysis occurs in β5 pocket by hydrolysis of a peptide bond at the C-terminus of hydrophobic substrates (site S1) by the nucleophilic OH group of the N-terminal threonine. The interface at β5/β6 plays a major role in conferring selectivity toward apolar peptide substrates (positions S2 and S3) [16].
Metal-containing drugs have existed for decades and cisplatin, a platinum containing compound, is known as one of the most effective antitumor drugs [17–20]. Since the approval of cisplatin by the U.S. Food and Drug Administration (FDA) in 1978, many researchers have focused their attention on this drug [21–23]. However, cisplatin-based chemotherapy leads to severe side effects (e.g., nephrotoxicity, ototoxicity, electrolyte disturbance and drug resistance) that severely limit its clinical use [24–26]. Therefore, many laboratories have been designing, synthesizing, and characterizing, from the biological point of view, new potential metal-based anticancer drugs to reduce toxicity and improve clinical effectiveness [27–29].
The Schiff base is a compound containing a carbon–nitrogen double bond (>C=N − R with R = aryl or alkyl group) as a functional group formed by condensation of an aldehyde or ketone with a primary amine. Schiff bases are able to stabilize many metals in various oxidation states coordinating them through the lone pair of the nitrogen atom of the >C=N − R moiety and additional functional groups [30]. It has been shown that the complexation of a metal with a Schiff base ligand improves the anticancer efficacy of the ligand [31,32]. Our previous work has focused on the biological activity of Schiff base–copper complexes and we have shown that several of these complexes have significant antitumor activity, associated with proteasome inhibition [33–36]. However, the detailed molecular mechanism responsible for proteasome inhibition by a Schiff base–copper complex remains unknown.
In the current study, we report the profile of cancer cell growth-inhibitory activity of four amino acid Schiff base–copper(II) complexes (Fig. 1) and their structure–activity relationships. We have found that the complexes C1 and C3, which contain Phen as the second ligand, inhibit proliferation and induce apoptosis in human breast and prostate cancer cells through inhibition of the ubiquitin–proteasome pathway. Furthermore, a computational docking study has suggested a novel mechanism of action for these active copper complexes to interact with and inhibit the tumor proteasome.
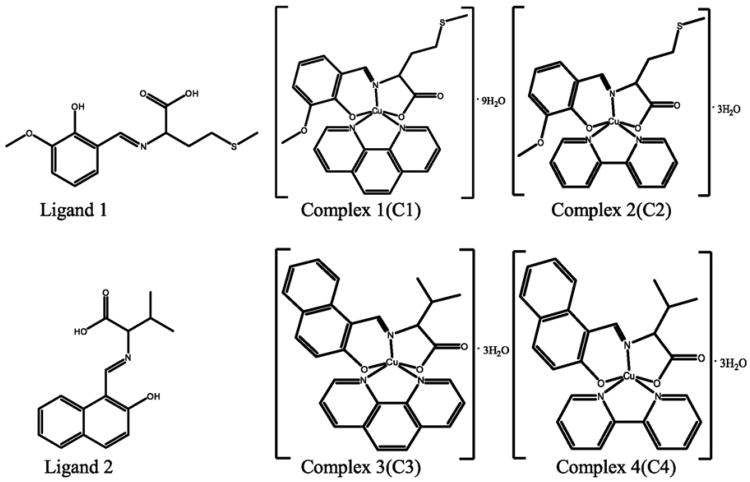
Fig. 1.
The chemical structures of H2L1, H2L2, C1, C2, C3 and C4. C1 andC2 are derived from H2L1 while C3 and C4 are from H2L2. Phen and Bpy present in complexes C1/C3 and C2/C4, respectively, are to chelate the copper atom.
2. Results
2.1. Crystal structures of C1 and C2 complexes
We have previously shown that some copper complexes are able to inhibit proliferation and induce apoptosis in human cancer cells [33–36]. In the current study, we synthesized and characterized four amino acid Schiff base–copper(II) complexes, named complex (C) 1, 2, 3 and 4 (Fig. 1) to discover novel copper-based anticancer drugs and understand their mechanism of action.
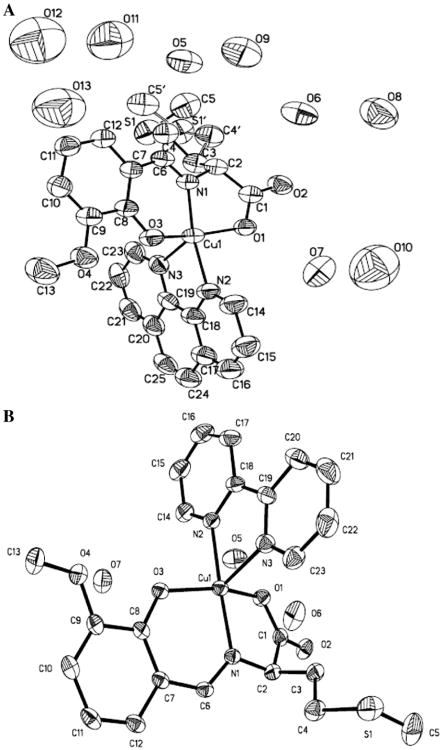
The crystal structures of C1 and C2 are shown in Fig. 2A and B, respectively. C1 crystallizes in the monoclinic system and the space group is C2/c. The positional disorder in the structure of C1 is realized by the presence of two positions for CH2–CH2–CH2–S–CH3 group. For this disordered group, S1/C4/C5/H3A/H3B/H4A/H4B/H5A/H5B/H5C, the occupation rate is 0.712 while for S1′/C4′/C5′/H3′A/H3′B/H4′1/H4′2/H5′1/H5′2/H5′3, the occupation rate is 0.288. The Cu center adopts a distorted square-pyramidal geometry by coordinating one carboxylate oxygen atom, one phenolate oxygen atom, one imine nitrogen atom of the l-methionine-o-vanillin Schiff base ligand (H2L1), and one nitrogen atom of Phen. The charge of Cu(II) is instead balanced by the hydroxyl and carboxylic groups. C2 has a similar coordination environment except that the space group is P21/c.
Fig. 2.
Thermal ellipsoid plot of C1 (A) and C2 (B), drawn at the 30% probability level. Selected bond distances (Å) and angles (deg) for C1: Cu–N1 1.918(6), Cu–N2 2.002(7), Cu–N3 2.261(7), Cu–O1 2.001(6), Cu–O3 1.936(6), N1–Cu–N2 171.0(3), N2–Cu–N3 76.6(3), and O1–Cu–O3 160.4(2), and for C2: Cu–N1 1.922(2), Cu–N2 1.996(2), Cu–N3 2.265(2), Cu–O1 2.0242 (18), Cu–O3 1.9327(18), N1–Cu–N2 171.92(9), N2–Cu–N3 76.93(9), and O1–Cu–O3 153.69(8).
2.2. Inhibition of proliferation of human breast cancer MDA-MB-231 cells by C1 but not C2
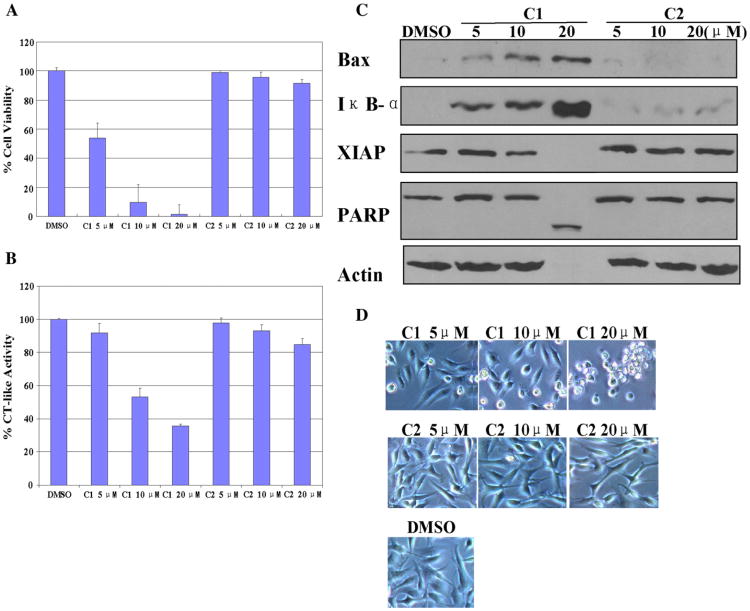
We first investigated and compared the effects of C1 and C2 on cancer cell growth. Human breast cancer MDA-MB-231 cells were treat-ed with C1 or C2 at different concentrations (5, 10 and 20 μM) for 24 h, followed by MTT assay. Cells treated with the solvent DMSO were used as control (Fig. 3A). We found that C1 had growth-inhibitory potency in a dose-dependent manner: at 5 μM it inhibited cell proliferation by 46%, and at 10 and 20 μM, it inhibited about 90 and 99% of breast cancer cell growth, respectively (Fig. 3A). As a sharp comparison, C2 did not have any inhibitory effect at even 20 μM (Fig. 3A).
Fig. 3.
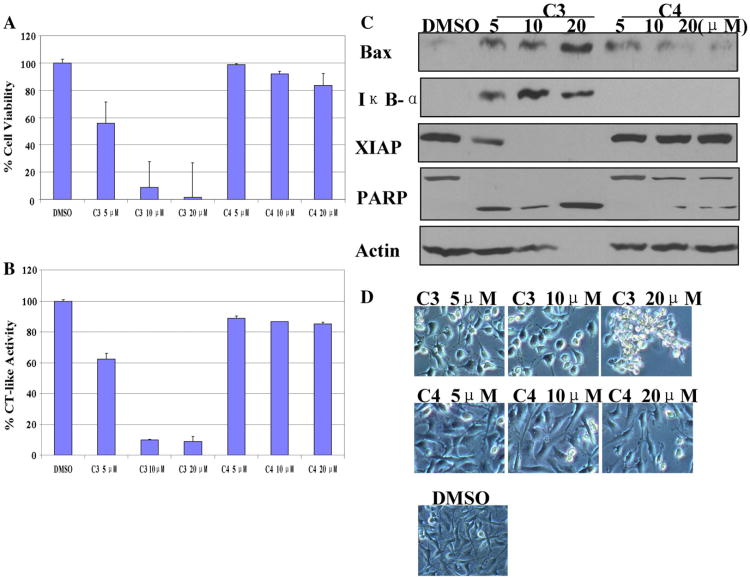
Dose response experiment using C1 and C2 in human breast cancer MDA-MB-231 cells. MDA-MB-231 cells were treated with either solvent DMSO or C1 and C2 at 5, 10, 20 μM for 24 h. (A) Anti-proliferative effects of C1 and C2. MDA-MB-231 cells were treated with C1 and C2 for 24 h, then medium was removed and cells were treated with MTT solution. (B) The inhibition of CT-like activity in MDA-MB-231 cells treated with C1 and C2. (C) Western blot analysis using antibodies to proteasome target proteins (Bax and IκB-α), XIAP, and PARP. (D) Cellular morphological changes visualized by phase-contrast imaging.
2.3. Concentration-dependent proteasome inhibition and apoptosis induction by C1 but not C2 in MDA-MB-231 cells
To investigate whether the growth-inhibitory activity of C1 is associated with its ability to inhibit the proteasome activity and induce apoptotic cell death, MDA-MB-231 cells were treated with various concentrations (5, 10, and 20 μM) of C1 or C2 for 24 h, followed by measurement of the proteasome inhibition and apoptosis by multiple assays.
We found that C1 was able to inhibit the proteasomal chymotrypsin-like activity in a dose-dependent manner (Fig. 3B). Consistently, C1 treatment caused a dose-dependent accumulation of proteasome target proteins Bax and IκB-α (Fig. 3C). These results supported the conclusion that C1 inhibited cellular proteasome activity in intact MDA-MB-231 cells.
Additionally, when these cells were treated with C1, levels of XIAP, an endogenous caspase inhibitor protein, decreased at 10 μM and disappeared at 20 μM (Fig. 3C). In the same experiment, both cell death-associated PARP cleavage (Fig. 3C) and cellular morphological changes (shrunken cells and characteristic apoptotic blebbing) (Fig. 3D) were detected in MDA-MB-231 cells treated with C1. The PARP cleavage fragment was detected and 100% PARP protein was cleaved in cells when 20 μM of C1 was used (Fig. 3C). Morphological changes were also observed after 24 h treatment of C1 at 10 μM, which increased to 100% at 20 μM (Fig. 3D).
In contrast, C2 had no or little inhibition on the tumor cellular proteasome activity, as judged by high levels of the CT activity (Fig. 3B) and lack of accumulation of proteasome target proteins (Bax, IκB-α) (Fig. 3C). Consistent with its failure to inhibit proteasome activity, C2 induced no or little cell death in these breast cancer cells, as demonstrated by intact PARP protein (Fig. 3C) and small population of cells with apoptotic morphology (Fig. 3D).
2.4. Time-dependent proteasome inhibition and cell death induction by C1 but not C2 in MDA-MB-231 cells
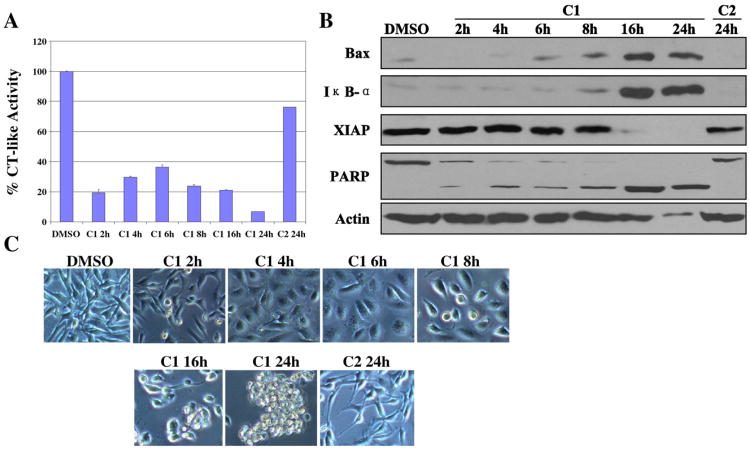
To detect the time-dependent effect of C1 on proteasome and apoptosis, MDA-MB-231 cells were treated with 20 μM of C1 from 2 to 24 h. Treatment with C2 for 24 h served as a control. We found that after only 2 h treatment with C1, the proteasome chymotrypsin-like activity was inhibited by 80% in these breast cancer cells (Fig. 4A). The chymotrypsin-like activity was recovered a little between 4 h and 6 h, but decreased again after 6 h (Fig. 4A).
Fig. 4.
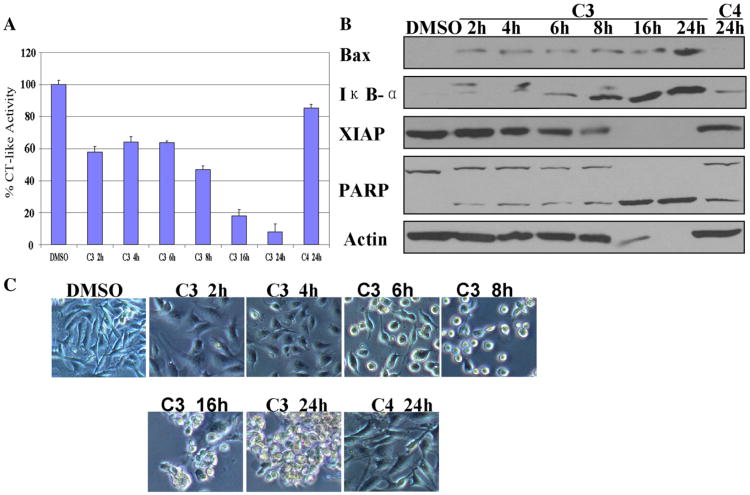
Time response experiment using C1 and C2 in human breast cancer MDA-MB-231 cells. MDA-MB-231 cells were treated with 20 μM of C1 and C2 for the indicated hours. (A) Kinetic studies on the inhibition of CT-like activity. (B) Western blottinganalysis using antibodies to proteasome target proteins (Bax and IκB-α), XIAP, and PARP. (C) Cellular morphological changes visualized by phase-contrast imaging.
Western blot analysis showed the levels of Bax and IκB-α proteins increased the highest in the cells treated with C1 at 16 h (Fig. 4B). We also found that the XIAP band disappeared after 16 and 24 h treatment (Fig. 4B). The PARP cleavage fragment appeared after 2 h treatment, and then the cleavage band increased to 100% after 16 and 24 h, and extensive apoptotic morphological changes were also observed after 8 h of treatment with C1 and increased to 100% after 24 h (Fig. 4B, C). In a sharp contrast, when the cells were treated with C2 for 24 h, although partial CT-like activity inhibition was observed, neither accumulation of proteasome target proteins (Bax and IκB-α) nor apoptosis was found (Fig. 4A–C).
2.5. C3 but not C4 has proteasome-inhibitory and apoptosis-inducing activities when tested in MDA-MB-231 cells
To further confirm our findings, we synthesized another pair of complexes, C3 and C4 (Fig. 1) and compared their biological activities in breast cancer MDA-MB-231 cells.
MTT assay shows that C3 but not C4 inhibited the tumor cell growth in a dose-dependent manner (Fig. 5A). C3 treatment also caused proteasome inhibition, evidenced by decreased levels of CT-like activity (Fig. 5B) and increased levels of Bax and IκB-α (Fig. 5C). Exposure of the cells to C3 also resulted in decreased levels of XIAP, associated with PARP cleavage (Fig. 5C) and apoptotic morphology changes (Fig. 5D). In contrast, C4 had little activity to inhibit the tumor cellular proteasome activity, as evidenced by low levels of CT-like activity inhibition and PARP cleavage. No accumulation of proteasome target proteins and little cell death were observed in cells treated with C4 (Fig. 5B–D).
Fig. 5.
Dose effects of C3 and C4 in human breast cancer MDA-MB-231 cells. MDA-MB-231 cells were treated with either solvent DMSO or C3 and C4 at indicated concentrations for 24 h, followed by the measurement of (A) anti-proliferative effects, (B) the proteasomal CT-like activity, and (C) levels of proteasome target proteins (Bax and IκB-α), XIAP, and PARP. (D) Cellular morphological changes.
Experiments using MDA-MB-231 cells treated with C3 showed time-dependent growth inhibition from 4 to 24 h (Fig. 6A), associated with increased levels of proteasome target proteins Bax and IκB-α, decreased expression of XIAP, and increased levels of cell death, all of which were time-dependent (Fig. 6B, C). As a sharp comparison, C4 treatment for 24 h failed to generate any of the above effects except a modest decrease in CT-like activity and a low level of PARP cleavage (Fig. 6).
Fig. 6.
Time effects of C3 and C4 in human breast cancer MDA-MB-231 cells. MDA-MB-231 cells were treated with 20 μM of C3 and C4 for the indicated hours, followed by the measurement of (A) the proteasomal CT-like activity, and (B) levels of proteasome target proteins (Bax and IκB-α), XIAP, and PARP. (C) Cellular morphological changes.
These results demonstrate that C1 and C3 inhibit proteasome activity and induce apoptosis in human breast MDA-MB-231 cells.
2.6. Effect of C1 and C3 in human breast cancer MCF-7 cells and prostate cancer PC-3 cells
To test the effect of these four copper complexes on other human cancer cells, human breast cancer MCF-7 cells and prostate cancer PC-3 cells were treated with each complex at various concentrations (5, 10 and 20 μM), followed by MTT (3-[4,5-dimethyltiazol-2-yl]-2,5-diphenyl-tetrazolium bromide) assay. Again we found that C1 and C3, but not C2 nor C4, inhibited the growth of these breast and prostate cancer cells in a dose-dependent manner (see Supplementary Information). Furthermore, apoptosis-associated morphological changes were observed in these cancer cells when treated with C1 and C3 at as low as 5 μM. C2 and C4 had no or little effect on both cancer cell lines (see Supplementary Information). These results strongly suggest that C1 and C3 inhibit proliferation and induce apoptosis in multiple human cancer cell lines.
2.7. Inhibition of the chymotrypsin-like activity of purified 20S proteasome by C1 and C3
On the basis of the cellular results showing proteasome inhibition, we then investigated whether these copper complexes could inhibit the purified 20S proteasome. We incubated each complex at increasing concentrations (5, 10, 20 and 50 M) with purified 20S proteasome for 2 h in the presence of a specific proteasome fluorogenic substrate for the chymotrypsin-like (CT-like) activity. Data shows that C1 and C3 are more potent inhibitors of the purified proteasomal CT-like activity with IC50 values of 13.3 and 11.3 μM, respectively, than their counterparts C2 and C4 with the IC50 values of 46.5 and 42.5 μM.
2.8. Docking studies of copper complex C1 as a proteasome inhibitor
In order to investigate the involved molecular mechanism for C1- or C3-mediated proteasome inhibition, we utilized molecular modeling to study the binding mode of these copper complexes in the CT-like active site. We investigated the docking of C1 and C2 (chosen as model compounds) and Bortezomib (Velcade) as a control (Fig. 7A). The computational model was built to correctly bind Velcade into the CT-like active site of the β5 subunit of the proteasome. The drug docks to the S1 site such that the boronic acid is proximal to Thr1 in agreement with crystal data (Fig. 7A). However, in the crystal structure solved by Groll and coworkers, a real covalent bond forms between the boron atom of Velcade and the −OH group coming from the side chain of Thr1 [37]. Among the 11 hydrogen bonds known, three are predicted with Ala49, Gly47, and Thr21. A predicted hydrogen bond between Asp-114 and the pyrizine ring of Velcade is not reproduced in the dock due to the rotation of the ring.
Fig. 7.
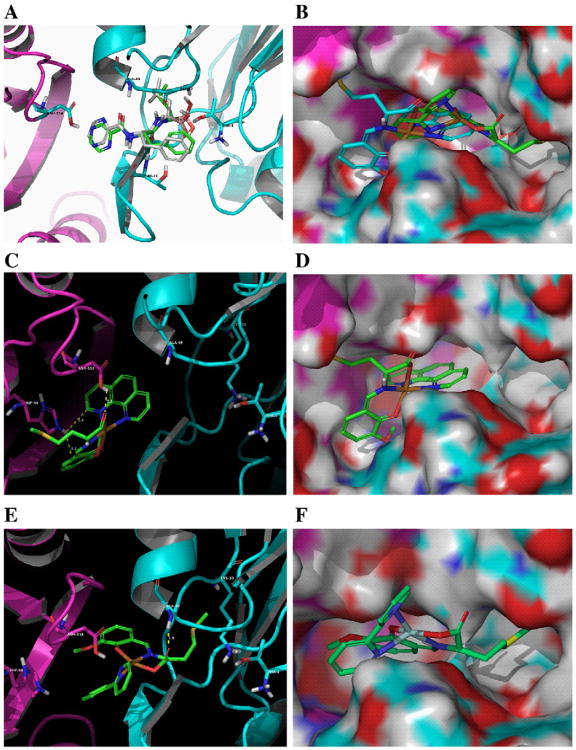
Docking poses obtained by means of software GLIDE. (A) Bortezomib (white carbons) compared to its crystal pose (green carbons) (PDB:2F16). (B) Overlap of C1 (blue carbons) and C2 (green carbons) by molecular surface visualization. Copper in orange, β5 in cyan, and β6 in purple. (C and D) C1 using a ribbon backbone representation and a molecular surface representation by space filling, respectively. (E and F) C2 using a ribbon backbone representation and a molecular surface representation by space filling, respectively. Distances of intermolecular interactions are in yellow.
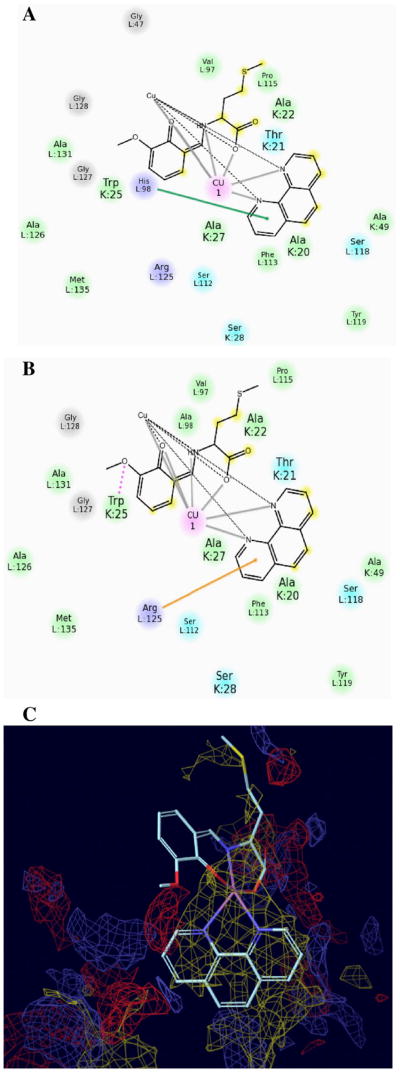
Unlike Velcade, C1 does not dock into the S1 region of the CT-like pocket but in the S2 and S3 regions between the subunits β5 and β6. There are four primary interactions that drive C1 binding in the S3 pocket of the proteasome. The most important feature is a hydrophobic interaction near the bottom of the pocket of the S3 binding site with the 1,10-phenanthroline ring system (Fig. 8C). Also coordinating the binding of C1 are three residues which are on the β6 subunit and encircle the S3 binding site: His98 (pi–pi), Arg125 (pi–cation), and Trp25 (hydrogen bond) (Figs. 7C, D and 8A, B). Mutations at H98A and R125A result in some binding mode degradation for C1. However, mutations at all three sites result in a complete disruption of the binding mode. This demonstrates that C1 binding is governed at least in part by four specific interactions. The hydrophobic character of the S3 pocket is the primary impetus for binding but the other three residues help stabilize the binding mode into S3 and show some redundancy. If all three residues are missing then a new binding mode appears that dramatically shifts C1 away from the S3 pocket. The results for C1's poses in mutant structures compared to wild-type structures are shown in Table 1.
Fig. 8.
The binding modes of C1 inside wild-type residue, mutant residue and S3 hydrophobic pocket. (A and B) C1 using two binding modes with the residues: His98 from the native H98 pose (pi–pi interaction), Arg125 from the mutant H98A pose (pi–cation interaction). The green line and orange line denote pi–pi and pi–cation interactions, respectively. (C) The phenanthroline ring of C1 has hydrophobic interaction with the bottom of the S3 binding region.
Table 1.
RMSD of calculated C1 binding modes in mutant structures compared to WT structures.
| H98A | R125A | W25A | RMSD |
|---|---|---|---|
| Y | – | – | 2.31 |
| Y | Y | – | 2.16 |
| Y | Y | Y | 10.47 |
With the mutation H98A, C1 replaces the lost pi–pi interaction with a pi–cation with R125. With the mutations H98A and R125A, C1 replaces the lost pi–cation interaction with a hydrogen bond with Trp25. When all three residues are mutated to alanine, the EModel scoring method finds a completely new binding mode for C1. This binding mode moves C1 away from the S3 pocket and into the S2 and S1 pockets. In this mode, C1 forms hydrogen bonds with Thr21 and the backbone oxygen of Gly47. Though C1 has a disordered side-chain, this disorder does not impact the final binding mode of C1 in the CT-active site. Molecular mechanics calculations and SP docking results show that C1′ (with the disordered side-chain located at lower occupation rate position) would give the same docked position even though the side-chain's initial position varies (data not shown). This is consistent with the behavior of GLIDE to test all possible configurations of flexible groups during docking.
Interestingly, the binding mode observed for C2 is significantly different (Fig. 7E, F). The pi–pi and pi–cation interactions identified for C1 are indeed absent in C2 docking. In addition, the H-bond between the carboxylic group of C2 and Trp25, or Thr21, or Gly47 has moved to Ala49 involving a backbone atom in lieu of the −OH group arising from the side chain of Asp.
3. Discussion
The toxicity of many anticancer agents is partially due to the inability to distinguish between normal and tumor cells. In order to eliminate toxicity, it is necessary to identify some specific properties of cancer cells different from normal cells [38]. Proteasome inhibition and anti-angiogenesis may be effective approaches in anticancer therapy due to the fact that the cancer cells are much more dependent on these processes than normal cells [5–7]. It has been reported that some copper complexes are capable of triggering proteasome inhibition and apoptosis induction in cancer cells [33–36]. In order to study the involved mechanisms of action of copper complexes, we synthesized four amino acid Schiff base–copper complexes, tested their biological activity, and investigated the structure–activity relationships among them.
First, we measured the antiproliferation activity of these complexes by the MTT assay, and found that C1 and C3 suppressed the proliferation of human breast and prostate cancer cells in a concentration-dependent manner. In addition, C1 and C3 at 20 μM inhibited proliferation of MDA-MB-231 cells by 99% and 98%, respectively, after 24 h treatment. We also found that C1 and C3 inhibited the proteasomal CT-like activity in intact MDA-MB-231 cells. Proteasome inhibition was confirmed by the increased levels of proteasome target proteins Bax and IκB-α.
We have reported that copper-based proteasome inhibitors potently induce apoptosis [33–36], however, the mechanism is unclear. Here we investigated whether the novel copper complexes C1 and C3 have a similar effect. The apoptosis-associated PARP cleavage and morphologic changes were found in the MDA-MB-231 cells treated by C1 and C3. Proteasome inhibition and apoptosis induction were also time-dependent. In contrast, C2 and C4 had no or little proteasome-inhibitory and cell death-inducing activities in MDA-MB-231 cells. We also found that C1 and C3, but not C2 and C4, had similar biological effects in other cancer cells, such as human breast cancer MCF-7 cells and prostate cancer PC-3 cells. Our data strongly suggests that the tumor proteasome is a target of the amino acid Schiff base–copper complexes C1 and C3, and that these complexes could inhibit proteasome activity and induce apoptosis in human cancer cells.
To investigate the mechanism of action, we focused our attention on the chemical structures of the copper complexes. It should be noted that only the complexes containing Phen as the second ligand (C1 and C3) triggered proteasome inhibition and cell death whereas the compounds C2 and C4, which involve Bpy as the second ligand, showed no or little anticancer activity, thus pointing out that the molecule chosen as the second ligand plays a major role in determining the cytotoxic activity of the corresponding copper-based compound. Furthermore, on the basis of structure-based comparisons, C1 and C3 are more hydrophobic than their counterparts C2 and C4. It is worth highlighting that CT-like activity is responsible for protein cleavage after hydrophobic amino acid residues, and therefore the presence of Phen as the second ligand in lieu of Bpy may account for the higher activity of C1 and C3.
Although Fig. 2 shows that copper centers in C1 and C2 are five-coordinated, such a transition metal can increase its coordination number to 6, leading to an octahedral geometry as what occurs for the Cu(II)-hexaquo complex, [Cu(H2O)6]2+ with Jahn–Teller distortion. It is possible that a bond could take place between the nucleophile amino acid residue of the proteasomal β5 subunit and the copper ion of the complex, occupying the active site of the enzymatic pocket, thus decreasing or completely blocking the proteolytic activity. In addition, if a bond forms between amino acid residue of the proteasomal β5 subunit and the copper ion, a greater stabilizing electronic delocalization effect could occur in C1 than C2, because the former contains Phen as the second ligand, which is a molecule (completely planar) with a higher conjugation extent and more rigid if compared to Bpy.
To further test our above hypothesis, we investigated the effect of each copper complex on the purified 20S proteasome. Similar to cellular data, C1 and C3 showed lower IC50 values than C2 and C4. Therefore, only compounds having Phen as a ligand have more potent proteasome inhibitory activity.
The docking analysis provided new insights on a possible mechanism of proteasome inhibition. The molecular modeling results may account for the data observed both in vitro and in the purified proteasome studies (e.g., C1 and C3 proved more potent than C2 and C4). It is worth highlighting that the binding score values predict that C1 should have greater inhibitory capability than C2 in a cell-free system (Fig. 7B). Due to intermolecular interactions, C1 exactly fits into the CT-like binding pocket, thus blocking the S3 region and leading to reversible inhibition of activity. In fact, the high affinity of C1 for the sites S2 and S3, important for the selectivity toward hydrophobic substrates, could hamper the protein substrate access into the pocket. On the contrary, C2 lacks important intermolecular interactions with the enzymatic pocket and is not stable in the proteasome. These differences between C1 and C2 suggest a binding mode that is less stable for C2, resulting in decreased inhibitory potential for C2 and that binding in S3 may be significant with respect to inhibitory activity. Although we observed two distinct binding modes for C1 and C2, the copper ion seems to play the same role in both cases. In fact, the metal center maintains the ligands in fixed positions, thus pointing them to specific amino acid residues. Such a structural role proved unexpected since metal centers are usually reactive. However, the formation of a covalent bond between the copper ion and a residue such as Thr1 or Asp114 cannot be excluded a priori. At a molecular level, it is likely that novel copper complexes bearing less bulky ligands show different mechanisms of inhibition, such as direct binding to Thr1 or occupation of the S1 site similar to Bortezomib. Notably, scaffold size and accessibility to the metal center play key roles to enable the binding to some amino acid residues.
4. Conclusions
We investigated the growth-inhibitory activity of four copper complexes along with their mechanism of action. Both cellular and computational analyses of the results show that C1 and C3 are potent proteasome inhibitors and apoptosis inducers in human cancer cells. Our study strongly suggests that the amino acid Schiff base–copper complexes which use 1,10-phenanthroline as the second ligand could be used as novel anticancer drugs in the future.
5. Experimental section
5.1. Synthesis of copper complexes 1–4
All chemicals used for synthesis of copper complexes were purchased from Acros.
[Cu(L1)(Phen)]·9H2O (1). O-vanillin (1.52 g, 10 mmol) was added to a methanol solution (50 mL) containing l-methionine (1.49 g, 10 mmol) and KOH (0.56 g, 10 mmol) at 70 °C. The mixture was stirred for 6 h before adding Cu(OAc)2·H2O (1.99 g, 10 mmol) and 1,10-phenanthroline (Phen, 1.80 g, 10 mmol). The resulting green solution was allowed to evaporate slowly at room temperature for two weeks, yielding dark green block crystals, which were collected by filtration, washed with Et2O and dried in vacuum (4.61 g. 67%). H2L1 is l-methionine-o-vanillin. IR (KBr, cm−1): 3446 (br, w), 3055 (w), 2911 (w), 2830 (w), 2358 (s), 1634 (vs), 1540 (m), 1437 (s), 1362 (m), 1217 (s), 1081 (m), 849 (m), 728 (s), 667 (w), 547 (w), 419 (m). Elemental analysis calculated (%) for C25H41CuN3O13S: C, 43.66; H, 6.01; N, 6.11. Found: C, 43.39; H, 6.19; N, 5.96.
[Cu(L1)(Bpy)]·3H2O (2). Complex 2 was prepared in the same way as for 1, except for the use of 2,2′-bipyridine (Bpy, 1.56 g, 10 mmol) instead of 1,10-phenanthroline. Dark green block crystals were obtained within 10 days (3.38 g, 61%). IR (KBr, cm−1): 3426 (br, m), 3061 (w), 2916 (w), 2833 (w), 2359 (s), 1633 (vs), 1539 (m), 1439 (s), 1367 (m), 1220 (s), 1086 (m), 1027 (m), 853 (m), 772 (s), 669 (w), 472 (w), 418 (m). Elemental analysis calculated (%) for C23H29CuN3O7S: C, 49.77; H, 5.27; N, 7.57. Found: C, 49.53; H, 5.46; N, 7.49.
[Cu(L2)(Phen)] ·3H2O (3). Complex 3 was prepared in the same way as for 1, except for the use of l-Valine (1.17 g, 10 mmol) instead of l-methionine, 2-hydroxy-1-naphthaldehyde (1.72 g, 10 mmol) instead of o-vanillin. Dark green precipitation was obtained (4.08 g, 72%). H2L2 is l-Valine-2-Hydroxy-1-Naphthaldehyde. IR (KBr, cm−1): 3388 (br, m), 2957 (w), 2356 (s), 1620 (vs), 1513 (m), 1426 (m), 1343 (m), 1185 (m), 840(m), 726 (s), 668 (m), 569 (w), 428 (w). Elemental analysis calculated (%) for C28H29CuN3O6: C, 59.31; H, 5.16; N, 7.41. Found: C, 59.06; H, 5.29; N, 7.26.
[Cu(L2)(Bpy)]·3H2O (4). Complex4 was prepared in the same way as for 3, except for the use of 2,2′-bipyridine (1.56 g, 10 mmol) instead of 1,10-phenanthroline. Dark green precipitation was obtained (4.29 g, 79%). IR (KBr, cm−1): 3383 (br, s), 2945 (w), 2361 (s), 1621 (vs), 1540 (w),1437 (s),1389(m), 1186 (m), 1031 (m), 830 (m), 754 (s), 667 (m), 617 (w), 454 (w). Elemental analysis calculated (%) for C26H29CuN3O6: C, 57.51; H, 5.38; N, 7.74. Found: C, 57.36; H, 5.49; N, 7.63.
5.2. Biological reagents and antibodies
Stock solutions of compounds were prepared by dissolution in DMSO (20 mM) and stored at −20 °C DMEM/F-12, RPMI-1640, penicillin, and streptomycin were purchased from Invitrogen (Carlsbad, CA). Fetal bovine serum was purchased from Tissue Culture Biologicals (Tulare, CA). MTT and other chemicals were purchased from Sigma Aldrich (St. Louis). Fluorogenic peptide substrate Suc-LLVY-AMC was purchased from Calbio-chem (San Diego, CA). Mouse monoclonal antibody against human poly (ADP-ribose) polymerase (PARP) was purchased from Biomol International LP (Plymouth Meeting, PA). Mouse monoclonal antibodies against Bax (B-9), IκB-α (H-4), and XIAP (A-7), and goat polyclonal antibody against actin (C-11) and all secondary antibodies were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). The concentrations of all antibodies were 200 μg/mL.
5.3. X-ray structure determination for C1 and C2
Suitable single crystals of C1 and C2 were analyzed on a computer-controlled Bruker SMART-1000 CCD diffractometer equipped with graphite monochromated MoKa (λ=0.71073 Å) radiation at 298(2) K. The structures were solved by a direct method and refined with the full-matrix least-squares technique using the SHELXTL-97 program package. The non-hydrogen atoms were refined anisotropically by full-matrix least-squares calculations on F2. The hydrogen atoms were added theoretically, riding on the concerned atoms and not refined.
5.4. Cell cultures and whole-cell extract preparation
MDA-MB-231, MCF-7 and PC-3 lines were purchased from American Type Culture Collection (Manassas, VA). MDA-MB-231 human breast cancer cells were cultured in DMEM (Dulbecco's Modified Eagle Medium)/F-12 media. MCF-7 human breast cancer cells and PC-3 human prostate cells were cultured in RPMI-1640 media. All media were supplemented with 10% fetal calf serum, 100 U/mL of penicillin, and 100 mg/mL of streptomycin. All cells were maintained at 37 °C and 5% CO2. Whole cell extracts were prepared as described previously [39]. Briefly, cells were harvested, washed with PBS (Phosphate Buffered Saline) and homogenized in a lysis buffer (50 mM Tris–HCl, Ph=7.5, 150 mM NaCl, 0.5% NP-40, 0.5 mM phenyl-methylsulfonyl fluoride, and 0.5 mM dithiothreitol) for 30 min at 4 °C. Afterwards, the lysates were centrifuged at 12,000 g for 12 min at 4 °C and the supernatants collected as whole-cell extracts.
5.5. Cell proliferation assay
MDA-MB-231 cells were seeded in triplicate in a 96-well plate and grown until 70–80% confluence, followed by treatment with indicated agents for 24 h. Then, the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay was performed as described previously [40].
5.6. Inhibition of purified 20S proteasome activity
The chymotrypsin-like activity of purified 20S proteasome was measured as previously described. Purified rabbit 20S proteasome (35 ng) was incubated in 100 μL assay buffer (20 mmol/L Tris–HCl, pH=7.5) with different concentrations of complexes and 20 μM of the fluorogenic peptide substrate Suc-LLVY-AMC for the proteasomal chymotrypsin-like activity for 2 h at 37 °C. Equivalent volume of solvent DMSO was used as control. After incubation, production of hydrolyzed AMC groups was measured with a Wallac Victor 3 Multilabel Counter with an excitation filter of 365 nm and an emission filter of 460 nm.
5.7. Proteasomal chymotrypsin-like activity in cell extracts
Whole cells extracts (4 μg) were incubated for 2 h at 37 °C in 100 μL of assay buffer (50 mM Tris–HCl, pH=7.5), with 20 μmol/L fluorogenic peptide substrate Suc-LLVY-AMC for assessment of proteasomal chymotrypsin-like activity. After incubation, production of hydrolyzed AMC groups was measured with a Wallac Victor 3 Multilabel Counter with an excitation filter of 365 nm and emission filter of 460 nm. Changes in fluorescence were calculated against the DMSO treated control.
5.8. Cellular morphologic analysis
A Zeiss Axiovert-25 microscope was used for all microscopic imaging with phase contrast for cellular morphology. Cells that became round and detached were considered to be apoptotic.
5.9. Western blot analysis
Various breast and prostate cancer cell lines were treated as indicated in the figure legend. Equal amounts of cell lysate (40 μg) were separated by SDS-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane. Western blot analysis was done using specific antibodies against Bax, IκB-α, XIAP, PARP and actin, followed by visualization with the enhanced chemiluminescence reagent.
5.10. In silico docking analysis
The docking investigation was carried out by means of the software GLIDE from Schrödinger's discovery Suite (2011 release) and the Protein Data Bank file PDB:2F16 (yeast 20S proteasome with Bortezomib covalently bound). Bortezomib was used as a control to verify that the docking procedure was able to reproduce the crystal position of Bortezomib. Grids were constructed using the Bortezomib crystal position as the center. Grid size was set to maximum to allow un-biased exploration of the binding site. Water molecules in the crystal structure and the covalent bond between Thr1 and Velcade were removed.
Amino acid residues Asp, His and Glu were set as protonated due to the noted hydrogen bond between Asp114 (subunit β6, site S2) and Velcade. The hydrogen bond network was optimized so that the proton on Asp114 was pointed to the pyrizine ring of Velcade. Hydroxyl groups on key residues (Thr1, Thr21, and Asp114) were set to freely rotate owing to the low crystal structure resolution (2.8 Å). The crystal structure underwent a constrained energy minimization in order to optimize the overall structure to a local energy minimum. The model proved able to correctly place Bortezomib. The pose with the lowest estimated free energy of binding yields the best binding mode for the molecule compared to the original crystal located position [Root-Mean-Square Deviation or RMSD of 1.21 for GScore −9.190 Kcal/mol].
To compare the mutations for changes in binding modes, two different scoring methods were used to determine the most favorable mode for the complex under each condition: the standard scoring method (GScore) and a refined scoring method (EModel; more heavily weights hydrophobic interactions). The results for Bortezomib's EModel poses in comparison with crystal position are shown in Table 2.
Table 2.
RMSD of calculated Bortezomib binding mode compared to crystal position.
| H98A | R125A | W25A | RMSD |
|---|---|---|---|
| Y | – | – | 1.13 |
| Y | Y | – | 11.20 |
| Y | Y | Y | 6.43 |
This shows that Velcade is more sensitive to mutations at Arg125 and other positions than with the His98. Additionally, its binding mode degrades substantially in the mutated proteasome. This control provides confidence that any binding mode changes due to mutations can be detected by the docking software.
Supplementary Material
Acknowledgments
This research was supported partially by the National Natural Science Foundation of China (grant nos. 20971115 to Yuhua Fan and 21071134 to Caifeng Bi), NIH/NCI (1R01CA120009, 3R01CA120009-04S1 and 5R01CA127258 to Q. Ping Dou), and a scholarship from the Chinese Scholarship Council (to Jian Zuo).
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jinorgbio.2012.10.006.
Contributor Information
Yuhua Fan, Email: fanyuhua301@163.com.
Q. Ping Dou, Email: doup@karmanos.org.
References
- 1.Jacobson MD, Weil M, Raff MC. Cell. 1997;88:347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- 2.Thatte U, Dahanukar S. Drugs. 1997;54:511–532. doi: 10.2165/00003495-199754040-00002. [DOI] [PubMed] [Google Scholar]
- 3.Song Z, Steller H. Trends Cell Biol. 1999;9:M49–M52. [PubMed] [Google Scholar]
- 4.Dou QP, Smith DM, Daniel KG, Kazi A. Prog Cell Cycle Res. 2003;5:441–446. [PubMed] [Google Scholar]
- 5.Adams J. Drug Discov Today. 2003;8:307–315. doi: 10.1016/s1359-6446(03)02647-3. [DOI] [PubMed] [Google Scholar]
- 6.Dou QP, Li B. Drug Resist Updat. 1999;2:215–223. doi: 10.1054/drup.1999.0095. [DOI] [PubMed] [Google Scholar]
- 7.Almond JB, Cohen GM. Leukemia. 2002;16:433–443. doi: 10.1038/sj.leu.2402417. [DOI] [PubMed] [Google Scholar]
- 8.Orlowski RZ, Dees EC. Breast Cancer Res. 2003;5:1–7. doi: 10.1186/bcr460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landis-Piwowar KR, Milacic V, Chen D, Yang H, Zhao Y, Chan TH, Yan B, Dou QP. Drug Resist Updat. 2006;9:263–273. doi: 10.1016/j.drup.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Milacic V, Chen D, Giovagnini L, Diez A, Fregona D, Dou QP. Toxicol Appl Pharmacol. 2008;231:24–33. doi: 10.1016/j.taap.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voges D, Zwickl P, Baumeister W. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 12.Seemüller E, Lupas A, Stock D, Löwe J, Huber R, Baumeister W. Science. 1995;268:579–582. doi: 10.1126/science.7725107. [DOI] [PubMed] [Google Scholar]
- 13.An B, Goldfarb RH, Siman R, Dou QP. Cell Death Differ. 1998;5:1062–1075. doi: 10.1038/sj.cdd.4400436. [DOI] [PubMed] [Google Scholar]
- 14.Lopes UG, Erhardt P, Yao R, Copper GM. J Biol Chem. 1997;272:12893–12896. doi: 10.1074/jbc.272.20.12893. [DOI] [PubMed] [Google Scholar]
- 15.Daniel KG, Gupta P, Harbach RH, Guida WC, Dou QP. Biochem Pharmacol. 2004;67:1139–1151. doi: 10.1016/j.bcp.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 16.Tsvetkov L, Nanjundan M, Domino M, Daniel KG. Expert Opin Drug Discov. 2010;5:1221–1236. doi: 10.1517/17460441.2010.530654. [DOI] [PubMed] [Google Scholar]
- 17.Ronconi L, Sadler PJ. Coord Chem Rev. 2007;251:1633–1648. doi: 10.1016/j.ccr.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams DR. Chem Rev. 1972;72:203–213. doi: 10.1021/cr60277a001. [DOI] [PubMed] [Google Scholar]
- 19.Guo ZJ, Sadler PJ. Angew Chem Int Ed. 1999;38:1513–1531. [Google Scholar]
- 20.Kelland LR, Farell NP, Spinelli S. Uses of Inorganic Chemistry in Medicine. 1999:109–123. [Google Scholar]
- 21.Thompson KH, Orvig C. Science. 2003;300:936–939. doi: 10.1126/science.1083004. [DOI] [PubMed] [Google Scholar]
- 22.Rosenberg B, Vancamp L, Krigas T. Nature. 1965;205:698–699. doi: 10.1038/205698a0. [DOI] [PubMed] [Google Scholar]
- 23.Wong E, Giandomenico CM. Chem Rev. 1999;99:2451–2466. doi: 10.1021/cr980420v. [DOI] [PubMed] [Google Scholar]
- 24.Baird RD, Kaye SB. Eur J Cancer. 2003;39:2450–2461. doi: 10.1016/s0959-8049(03)00619-1. [DOI] [PubMed] [Google Scholar]
- 25.Wang D, Lippard SJ. Nat Rev Drug Discov. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 26.Hartmann JT, Lipp HP. Expert Opin Pharmacother. 2003;4:889–901. doi: 10.1517/14656566.4.6.889. [DOI] [PubMed] [Google Scholar]
- 27.Hartinger CG, Zorbas-Seifried S, Jakupec MA, Kynast B, Zorbas H, Keppler BK. J Inorg Biochem. 2006;100:891–904. doi: 10.1016/j.jinorgbio.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 28.Yan YK, Melchart M, Habtemariam A, Sadler PJ. Chem Commun. 2005:4764–4776. doi: 10.1039/b508531b. [DOI] [PubMed] [Google Scholar]
- 29.Ang WH, Dyson PJ. Eur J Inorg Chem. 2006;20:4003–4018. [Google Scholar]
- 30.Cozzi PG. Chem Soc Rev. 2004;33:410–421. doi: 10.1039/b307853c. [DOI] [PubMed] [Google Scholar]
- 31.Holla BS, Veerendra B, Shivananda MK, Poojary B. Eur J Med Chem. 2003;38:759–767. doi: 10.1016/s0223-5234(03)00128-4. [DOI] [PubMed] [Google Scholar]
- 32.Creaven BS, Duff B, Egan DA, Kavanagh K, Rosair G, Thangella VR, Walsh M. Inorg Chim Acta. 2010;363:4048–4058. [Google Scholar]
- 33.Zhang X, Bi CF, Fan YH, Cui QZ, Chen D, Xiao Y, Dou QP. Int J Mol Med. 2008;22:677–682. [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao Y, Bi CF, Fan YH, Cui C, Zhang X, Dou QP. Int J Oncol. 2008;33:1073–1079. [PubMed] [Google Scholar]
- 35.Adsule S, Barve V, Chen D, Ahmed F, Dou QP, Padhye S, Sarkar FH. J Med Chem. 2006;49:7242–7246. doi: 10.1021/jm060712l. [DOI] [PubMed] [Google Scholar]
- 36.Padhye S, Yang HJ, Jamadar A, Cui QZC, Chavan D, Dominiak K, McKinney J, Banerjee S, Dou QP, Sarkar FH. Pharm Res. 2009;26:1874–1880. doi: 10.1007/s11095-009-9900-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Groll M, Berkers CR, Ploegh HL, Ovaa H. Structure. 2006;14:451–456. doi: 10.1016/j.str.2005.11.019. [DOI] [PubMed] [Google Scholar]
- 38.Chen D, Peng FY, Cui QZC, Daniel KG, Orlu S, Liu JG, Dou QP. Front Biosci. 2005;10:2932–2939. doi: 10.2741/1749. [DOI] [PubMed] [Google Scholar]
- 39.An B, Dou QP. Cancer Res. 1996;56:438–442. [PubMed] [Google Scholar]
- 40.Daniel KG, Chen D, Orlu S, Cui QC, Miller FR, Dou QP. Breast Cancer Res. 2005;7:R897–R908. doi: 10.1186/bcr1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.