Abstract
Structural characterization of protein–protein interactions across the broad spectrum of scales is key to our understanding of life at the molecular level. Low-resolution approach to protein interactions is needed for modeling large interaction networks, given the significant level of uncertainties in large biomolecular systems and the high-throughput nature of the task. Since only a fraction of protein structures in interactome are determined experimentally, protein docking approaches are increasingly focusing on modeled proteins. Current rapid advancement of template-based modeling of protein–protein complexes is following a long standing trend in structure prediction of individual proteins. Protein–protein templates are already available for almost all interactions of structurally characterized proteins, and about one third of such templates are likely correct.
There is nothing worse than a sharp image of a fuzzy concept. (Ansel Adam, US nature photographer, 1902–1984)
Introduction
The essence of the low-resolution approach to modeling of proteins and their interactions is in the epigraph to this paper. Indeed, when the high-resolution details of protein structure are highly unreliable, it is better not to include them in the picture. Structural characterization of protein–protein interactions (PPI) across the broad spectrum of scales is key to our understanding of life at the molecular level. Recently, the low-resolution/coarse-grained modeling approaches have been increasingly gaining popularity. Still, many biomolecular scientists will likely tell you that only the high resolution protein structures have an appreciable value, and the low-resolution ones add little to biology. Coincidentally, one could conclude that because computational modeling of large and heterogeneous macromolecular systems has limited accuracy, it is thus rarely useful. Such a notion, of course, is incompatible with the rapid development of the multiscale approaches to systems biology and, simply put, studies of the physical phenomena in general that inherently involve approximation.
Approximation is in the heart of physics, numerical analysis, and other branches of ‘exact sciences.’ Ironically, the most ‘inexact’ of them — molecular life science — is still not quite at home with this most basic concept. In fact, numerous studies involving low-resolution structural information have added a great amount of knowledge on the fundamental mechanisms of soluble and membrane proteins. Simply put, any level of physical characterization of a protein, as opposed to its absence, is valuable. The level of structure resolution is biologically relevant if it captures the functional elements of the structure. If such elements are large, then even ultralow resolution can provide important insights. When it comes to modeling of PPI, the high-resolution protein–protein docking is not necessary for a number of important biological questions where docking can be useful, such as the design of PPI inhibitors and other experimental and computational studies that take over from protein docking once it predicts the protein–protein interface.
Low resolution does not negate high-resolution. To the contrary, it is a prerequisite for our ability to obtain the high-resolution accuracy in modeling, through refinement of the low-resolution predictions. The whole paradigm of ‘refinement’ comes from the notion of an approximate model, which reflect the reality at a lower precision, and which is subject to the improvement of the precision. The low-resolution modeling of PPI is especially important in the efforts to model large PPI networks, up to the level of interactome, in the context of modeling the entire cell. The low-resolution approach is the only currently available tool for such modeling, given the high-throughput requirements of low computational cost per interaction, and the significant level of uncertainties inevitable in very large heterogeneous systems. The first credible model of a cell will be low resolution.
Low-resolution protein recognition factors
The geometric complementarity between interacting protein structures, the cornerstone of protein–protein docking methodology since its inception, is an essential predictor of interacting modes at low resolution [1]. A fundamental question concerning protein association is whether it is determined by local structural elements or whether there are also large-scale structural motifs that facilitate the formation of the complex. The local physicochemical and steric factors are responsible for the final ‘lock’ of the molecules when their binding sites are already in close proximity. At the same time, there are structural factors that contribute to bringing the binding sites to such proximity. An important insight into the basic rules of protein recognition is provided by the studies of large-scale structural recognition factors, such as recognition of proteins deprived of atom-size structural features [1,2], backbone complementarity in protein recognition [3], macromolecular assemblies [4], and binding- related anisotropy of protein shape [5,6]. The practical importance of the large-scale recognition factors for docking is that they often allow one to ignore local structural inaccuracies (e.g. those caused by conformational changes).
Intermolecular energy funnel is the ultimate low-resolution concept. The large-scale structural recognition factors in protein association have to do with the funnel-like intermolecular energy landscape [7]. It has been shown that simple energy functions, including coarse-grained (low-resolution) models, reveal major landscape characteristics, such as the number and distribution of the funnel-like energy basins, transition between low and high resolution, and funnel size [8]. The intermolecular energy landscapes are further characterized by conformational properties of interacting proteins [9–11].
Coarse-grained flexibility in protein interactions
The unbound/bound difference in protein backbone is often insignificant [12] and the formation of a complex can be described by the side-chain conformational changes [13–16]. However, the analysis of large-scale structural flexibility is important for understanding protein–protein association and our ability to model them [17, 18]. The coarse-graining of protein structures allows exploration of structural dynamics of large macromolecular systems at long time scales [19•,20]. It also allows comparison with low-resolution experimental data, which often are the only available structural information on the system [21]. Coarse-grained elastic networks modeling of structure fluctuations showed that, on average, the interface is more rigid than the rest of the protein surface [22,23], and the interface mobility is correlated with the interface type, size and obligate nature of the complex [23]. In structural modeling of protein–protein complexes, the coarse-graining approaches are used to model structural flexibility in protein assembly [10,19•, 24, 25]. Low-resolution allows implicit accounting for local conformational flexibility without sampling the internal degrees of freedom, and thus is useful in docking [1,26]. The residue frequencies in co-crystallized protein–protein complexes provide an opportunity to develop residue-residue statistical potentials for docking and scoring of PPI [27,28]. Such potentials provide a coarse-grained alternative to atomic-resolution statistical potentials, allowing greater tolerance to conformational changes.
Docking of models
Direct experimental approaches to structure determination (primarily, X-ray crystallography and NMR) are capable of determining only a fraction of all protein structures. Thus the structures of most proteins in genomes have to be modeled by high throughput computational techniques. The major difference between the experimental structure and a model, in general, is a lower accuracy of the latter. The accuracy of the protein models may vary significantly, based on the availability of modeling templates and their similarity to the target, from ~1 Å RMSD (high-sequence similarity to templates) to >6Å RMSD (low-sequence similarity to templates, or no templates). Thus, in addition to computational efficiency (e.g. high-throughput, in case of large-scale modeling) the docking procedure has to be capable of tolerating significant structural inaccuracies. Docking cannot yield greater precision than the precision of the interacting proteins. However, even in the extreme case of low precision (~10 Å relative shift of the proteins) the results provide meaningful structural information on the interface location on one or both proteins and the general shape of the complex.
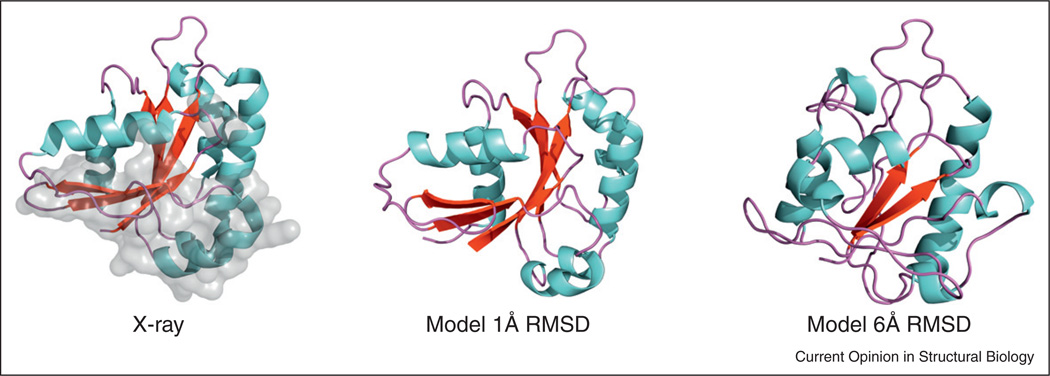
Computationally inexpensive methodology is required for structural modeling of the interactome. For systematic evaluation of expected accuracy in high-throughput modeling of binding sites, the analysis of target/template sequence alignments was performed on a representative protein–protein set [29•]. For most of the complexes, the alignments containing all interface residues were found, even in cases of poor overall alignments, inadequate for modeling of the whole proteins. The alignment of the interfaces significant enough to produce the binding site structure suitable for docking was found in about half of the complexes. An early study [30], systematically simulated structural inaccuracies of modeled proteins, starting from a representative set of co-crystallized proteins, and generating an array of distorted structures for each protein, with inaccuracies from 1 to 10Å. The models were docked at low-resolution and the results correlated with the accuracy of the models. The data showed that docking of even highly inaccurate protein models (~6 Å RMSD from the X-ray structure) still yields structurally meaningful results, accurate enough to predict binding interfaces and to serve as starting points for further structural analysis. The utility of the modeled proteins in protein–protein docking was further demonstrated by other systematic studies, involving docking approaches based on computational geometry [31,32] validated on benchmark protein–protein sets [31] and the nuclear pore complex [32], and Rosetta-based docking of antibody–antigen homology models [33]. The template-based docking approaches increasingly focus on the modeled structures as part of the docking protocol [34,35•] or the subject of structural alignment [36••,37••]. •]. Modeled proteins attract increasing attention as drug targets. Studies of binding pockets on modeled protein receptors, and docking of ligands to modeled receptors showed significant tolerance to the structural inaccuracies and the general utility of the modeled receptors [38–43]. In our most recent study, a new large benchmark suite of models with controlled distortions for 320 protein complexes was built using combination of homology modeling, low energy trajectories, and simulated annealing. For each X-ray monomer in the dataset, six models were generated with the pre-defined values of Cα RMSD between the native and the model structures (examples in Figure 1).
Figure 1.
Examples of simulated models. The X-ray structure is shown for comparison, with the binding site in gray.
The rise of the template-based docking
The physical principles of protein binding and folding are the same, thus their modeling shares many aspects. Prediction of individual protein structures has evolved from the ‘first principles’ approaches to the currently dominating template-based modeling, largely because of the difficulty the template-free methods face in delivering reliable solutions, and the explosive growth of the number of experimentally determined protein structures. Protein docking is significantly younger than the individual protein structure prediction, and much less advanced in its transition to the template-based approaches. The two factors that contributed to the evolution of the individual protein prediction determine the current lesser role of the template-based methodologies in protein docking. First, the template-free techniques have been relatively more successful in the prediction of protein complexes than in the prediction of individual proteins. The reason is that the docking first approximation (rigid-body docking), applicable in many cases, has to cope with only six degrees of freedom, which is incomparable with the number of degrees of freedom in the prediction of individual proteins at any meaningful level of approximation. Second, the number of experimentally determined structures of protein–protein complexes is far less than the number of such structures for individual proteins. However, with the advances in the experimental determination of protein complexes the situation is rapidly changing.
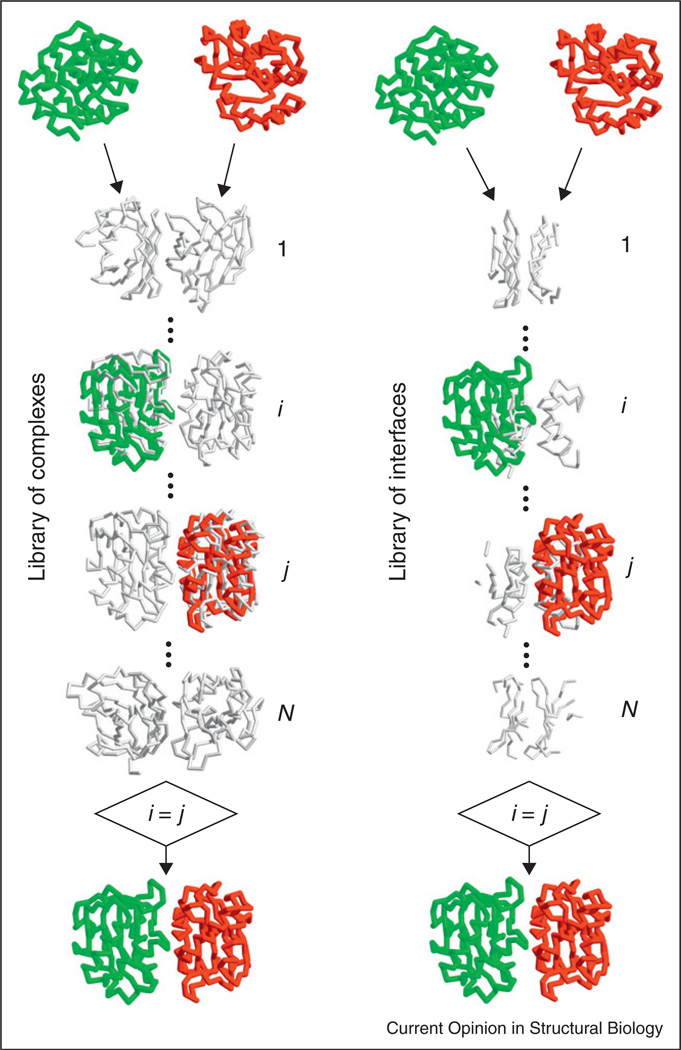
The template of the complex may be detected based on the sequence of the target proteins [34,35•]. However, since the docking problem assumes the knowledge of the components structure, a growing number of approaches take advantage of structural alignment techniques, for full and/or interface structure alignment [36••,44–53]. The template-based structure-comparison approaches (Figure 2) align backbones, secondary structure, and/or other coarse-grained elements of the structure. This reflects the low-resolution nature of the macrostructural recognition factors, fundamentally based on the backbone recognition, dating back to the early studies [3].
Figure 2.
Template-based docking by structural alignment.
To assess the predictive value of the template-based approach, it was benchmarked on protein–protein structures in PDB released in 2009–2011, utilizing template structures released before 2009. The templates were found for almost all new complexes, and more than a third of the new complexes were predicted correctly, with interface RMSD < 5 Å [37••]. The template-based docking, in general, performed in the community-wide assessment of docking techniques (Critical Assessment of PRediction of Interactions — CAPRI) with limited success [54], in sharp contrast with the significantly higher than the free docking success rates on the docking benchmark sets [53]. The reason is that CAPRI targets have high representation of novel structures, reflecting the effort of the crystallographers providing the targets to avoid ‘trivial’ complexes that are similar to the ones already in PDB. However, in typical ‘real case’ modeling of protein–protein complexes of biological interest, the novelty of the structure usually is not a consideration and the existence of homologous co-crystallized complexes is welcome. Thus, the docking benchmarks, which follow the increasing availability of co-crystallized homologous complexes, are representative of the biological community needs.
Since the experimentally determined structure of protein–protein complexes is generally more difficult to obtain than the structure of individual proteins, the availability of templates for protein–protein docking is a key issue. Comparative studies of protein–protein interfaces determined that the library of protein interfaces is close to complete [55••], and that it is generally possible to find representatives of the possible binding modes of a given protein [36••,56•]. Still, there are many structurally common binding regions among proteins that are not related to fold classification [57]. The direct way to assess the availability of templates for protein–protein complexes in PDB is to have the structural similarity metric that is correlated with the experimentally determined binding mode. Such a metric can be used in PPI datasets to see what percentage of PPI corresponds to the metric’s values for good templates. Recent results obtained in an all-to-all pairwise comparison of 989 co-crystallized complexes [37••] show a strikingly distinct phase transition to the same binding mode at minimal TM-score (the lowest of the two component proteins TM-scores [58]) of 0.4. Thus the values of the minimal TM-score > 0.4 can be used in detecting good templates of the complex. Remarkably, such structural templates were found for nearly all complexes in a database of known PPI, where the structure of the individual components of the interaction is determined by X-ray or can be built by homology [37••].
Proteome-scale modeling
To adequately model large systems of PPI, it is important to understand and simulate the environment in which the proteins interact in vivo. This environment is densely populated, which strongly affects protein diffusion, binding and conformational transitions. The investigations of the ‘crowding’ effects in such environment range from studies of protein stability and other conformational properties [59–61], and detection of binding regions [62], to the role of hydrodynamic interactions in cells [63•] and physical limits of cells and proteomes [64].
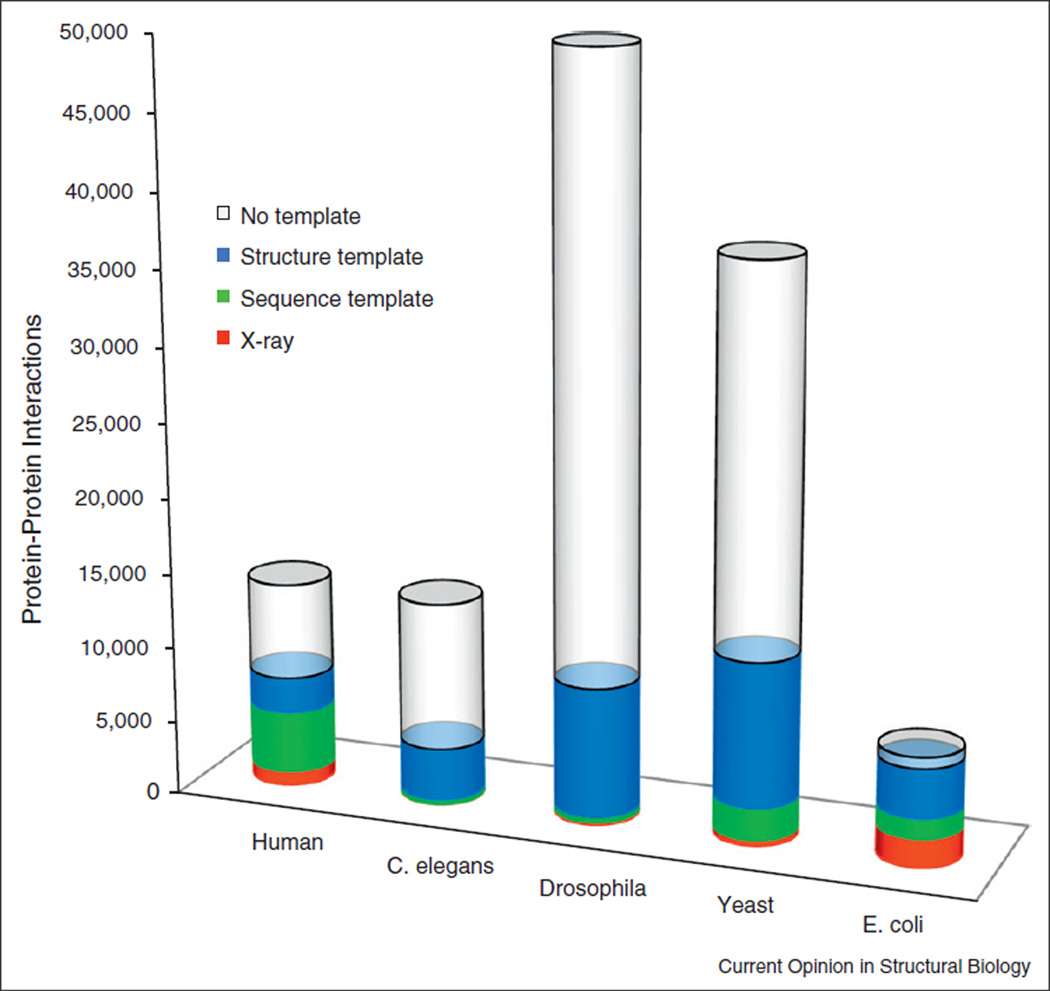
Structural characterization is essential for the proteome-scale modeling of PPI networks (Figure 3) [65,66•, 67•,68]. Modeling templates are available for a significant part of soluble proteins in genomes [69], including those in known PPI [37••]. The approaches to genome-wide structural modeling of PPI are either ‘traditional’ template-free docking [70,71] or the template-based docking [36••, 37••, 55••, 56•, 72–74]. The latter, while potentially providing much greater success rate [53], critically depends on the availability of the templates [36••, 37•• ,55••, 56•]. In a recent study [37••], the X-ray structures of the proteins were complemented by homology models and the templates for their complexes were detected in PDB. Figure 4 shows the results for five genomes with the largest number of known PPI. Structural alignments yielded a dramatic increase in the structural coverage of complexes, from the coverage provided by the sequence alignment. The structural templates were found for nearly all (33 537 out of 33 840, or 99%) complexes in which both components could be built. ‘No template’ in Figure 4 indicates no template for individual proteins, not for the complex. Thus, contrary to the common perception of rarity of the templates for complexes, as opposed to the structure prediction of individual proteins where the template-based modeling has long been the default approach, the limiting factor in interactome modeling is actually the availability of the templates for the individual proteins (more protein–protein templates are still needed for greater accuracy of modeling). The structural coverage of interactome should increase with more structures of individual proteins experimentally determined, and with more sophisticated modeling of individual proteins at lower levels of target/template similarity. The ability to detect templates for almost all complexes is a consequence of the proteins modeling by sequence similarity, followed by protein–protein modeling by structure similarity (which is significantly broader in scope than the sequence similarity, since structure is more conserved than sequence).
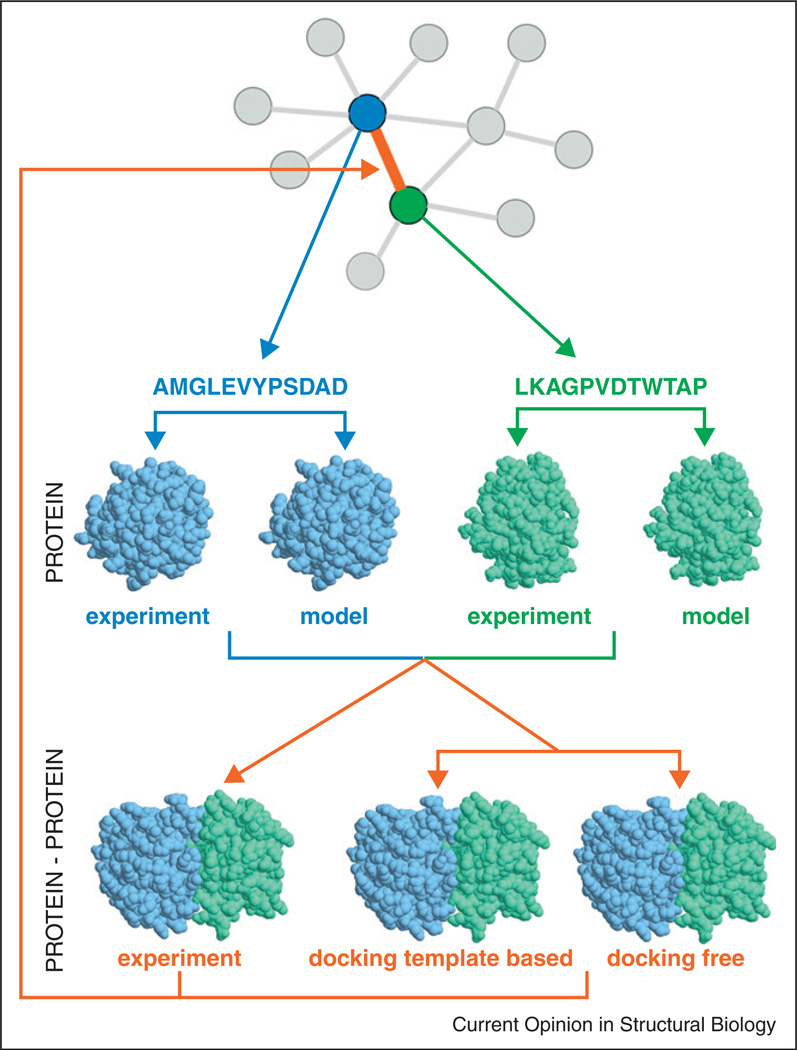
Figure 3.
Structural modeling of interactome. The flowchart shows modeling of two proteins (blue and green) interaction (orange) from the PPI network, through determination of their individual structures (X-ray structure from PDB or modeling), and the structure of their complex (X-ray structure from PDB or modeling by template-based or free docking).
Figure 4.
Structural coverage of PPI in five genomes with the largest number of known protein interactions. Complexes with the X-ray structure are in red, and complexes with a sequence template are in green. Complexes for which the structure of the monomers is known or can be built by homology are in blue — structural templates are available for 99% of such complexes
Future of PPI modeling
The quasi-complete low-resolution description of interactome is likely not that far down the road. Templates are already available for almost all interactions of structurally characterized proteins, and about one third of such templates are likely correct. The limiting factor is the availability of templates for individual proteins. With more experimentally determined protein structures becoming available, more accurate genome-wide maps of PPIs, and the growing computational resources allowing application of more sophisticated template detection approaches, our ability to structurally model PPI at the level of interactome should rapidly develop.
With the advance of the template-based docking, the free docking will not fade away — there are many protein encounters in the crowded cell environment, which are not likely to correspond to energetically stable co-crystallized templates. And the high-resolution modeling of PPI will be there too, as the next step in our ability to reveal the full picture, in all its clarity.
Acknowledgements
This study was supported by grant R01GM074255 from the NIH. The author thanks Petras Kundrotas and Ivan Anishchenko for their help in the preparation of the manuscript.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
• • of outstanding interest
- 1.Vakser IA, Matar OG, Lam CF. A systematic study of low-resolution recognition in protein–protein complexes. Proc Natl Acad Sci U S A. 1999;96:8477–8482. doi: 10.1073/pnas.96.15.8477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Q, Sanner M, Olson AJ. Shape complementarity of protein–protein complexes at multiple resolutions. Proteins. 2009;75:453–467. doi: 10.1002/prot.22256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vakser IA. Main-chain complementarity in protein–protein recognition. Protein Eng. 1996;9:741–744. doi: 10.1093/protein/9.9.741. [DOI] [PubMed] [Google Scholar]
- 4.Lasker K, Sali A, Wolfson HJ. Determining macromolecular assembly structures by molecular docking and fitting into an electron density map. Proteins. 2010;78:3205–3211. doi: 10.1002/prot.22845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vacha R, Frenkel D. Relation between molecular shape and the morphology of self-assembling aggregates: a simulation study. Biophys J. 2011;100:1432–1439. doi: 10.1016/j.bpj.2011.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicola G, Vakser IA. A simple shape characteristic of protein– protein recognition. Bioinformatics. 2007;23:789–792. doi: 10.1093/bioinformatics/btm018. [DOI] [PubMed] [Google Scholar]
- 7.Tovchigrechko A, Vakser IA. How common is the funnel-like energy landscape in protein–protein interactions? Protein Sci. 2001;10:1572–1583. doi: 10.1110/ps.8701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vakser IA. Low-resolution recognition factors determine major characteristics of the energy landscape in protein–protein interaction. In: Schreiber G, Nussinov R, editors. In Computational Protein–Protein Interactions. Taylor and Francis, CRC Press; 2009. pp. 21–42. [Google Scholar]
- 9.Trizac E, Levy Y, Wolynes PG. Capillarity theory for the fly-casting mechanism. Proc Natl Acad Sci U S A. 2010;107:2746–2750. doi: 10.1073/pnas.0914727107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ravikumar KM, Huang W, Yang S. Coarse-grained simulations of protein–protein association: an energy landscape perspective. Biophys J. 2012;103:837–845. doi: 10.1016/j.bpj.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, Faeder JR, Camacho CJ. Toward a quantitative theory of intrinsically disordered proteins and their function. Proc Natl Acad Sci U S A. 2009;106:19819–19823. doi: 10.1073/pnas.0907710106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao Y, Douguet D, Tovchigrechko A, Vakser IA. DOCKGROUND system of databases for protein recognition studies: unbound structures for docking. Proteins. 2007;69:845–851. doi: 10.1002/prot.21714. [DOI] [PubMed] [Google Scholar]
- 13.Ruvinsky AM, Kirys T, Tuzikov AV, Vakser IA. Side-chain conformational changes upon protein–protein association. J Mol Biol. 2011;408:356–365. doi: 10.1016/j.jmb.2011.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirys T, Ruvinsky A, Tuzikov AV, Vakser IA. Rotamer libraries and probabilities of transition between rotamers for the side chains in protein–protein binding. Proteins. 2012;80:2089–2098. doi: 10.1002/prot.24103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirys T, Ruvinsky AM, Tuzikov AV, Vakser IA. Correlation analysis of the side-chains conformational distribution in bound and unbound proteins. BMC Bioinformatics. 2012;13:236. doi: 10.1186/1471-2105-13-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beglov D, Hall D, Brenke R, Shapovalov MV, Dunbrack RL, Kozakov D, Vajda S. Minimal ensembles of side chain conformers for modeling protein–protein interactions. Proteins. 2011;80:591–601. doi: 10.1002/prot.23222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Csermely P, Palotai R, Nussinov R. Induced fit, conformational selection and independent dynamic segments: an extended view of binding events. Trends Biochem Sci. 2010;35:539–546. doi: 10.1016/j.tibs.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abyzov A, Bjornson R, Felipe M, Gerstein M. RigidFinder: a fast and sensitive method to detect rigid blocks in large macromolecular complexes. Proteins. 2010;78:309–324. doi: 10.1002/prot.22544. [DOI] [PubMed] [Google Scholar]
- 19. Saunders MG, Voth GA. Coarse-graining of multiprotein assemblies. Curr Opin Struct Biol. 2012;22:144–150. doi: 10.1016/j.sbi.2012.01.003. The review describes recent advances in coarse-graining methods for multiprotein assemblies. The methods involve mapping, which uses information from one scale of representation to parameterize a lower resolution model, and bridging, which connect different scales during simulation. The paper discusses a large number of approaches to information transfer between scales.
- 20.Bahar I, Lezon TR, Yang LW, Eyal E. Global dynamics of proteins: bridging between structure and function. Ann Rev Biophys. 2010;39:23–42. doi: 10.1146/annurev.biophys.093008.131258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z, Voth GA. Coarse-grained representations of large biomolecular complexes from low-resolution structural data. J Chem Theory Comput. 2010;6:2990–3002. doi: 10.1021/ct100374a. [DOI] [PubMed] [Google Scholar]
- 22.Ruvinsky AM, Vakser IA. Sequence composition and environment effects on residue fluctuations in protein structures. J Chem Phys. 2010;133:155101. doi: 10.1063/1.3498743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zen A, Micheletti C, Keskin O, Nussinov R. Comparing interfacial dynamics in protein–protein complexes: an elastic network approach. BMC Struct Biol. 2010;10:26. doi: 10.1186/1472-6807-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karaca E, Bonvin AMJJ. Multidomain flexible docking approach to deal with large conformational changes in the modeling of biomolecular complexes. Structure. 2011;19:555–565. doi: 10.1016/j.str.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 25.Burton B, Zimmermann MT, Jernigan RL, Wang Y. A computational investigation on the connection between dynamics properties of ribosomal proteins and ribosome assembly. PLoS Comp Biol. 2012;8:e1002530. doi: 10.1371/journal.pcbi.1002530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gray JJ, Moughon S, Wang C, Schueler-Furman O, Kuhlman B, Rohl CA, Baker D. Protein–protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J Mol Biol. 2003;331:281–299. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 27.Liu S, Vakser IA. DECK: Distance and environment-dependent, coarse-grained, knowledge-based potentials for protein– protein docking. BMC Bioinformatics. 2011;12:280. doi: 10.1186/1471-2105-12-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vreven T, Hwang H, Pierce BG, Weng Z. Prediction of protein– protein binding free energies. Protein Sci. 2012;21:396–404. doi: 10.1002/pro.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kundrotas PJ, Vakser IA. Accuracy of protein–protein binding sites in high-throughput template-based modeling. PLoS Comp Biol. 2010;6:e1000727. doi: 10.1371/journal.pcbi.1000727. For systematic evaluation of expected accuracy in high-throughput modeling of binding sites, the analysis of target/template sequence alignments was performed on a representative protein–protein set. For most of the complexes, the alignments containing all interface residues were found, even in cases of poor overall alignments, inadequate for modeling of the whole proteins. The alignment of the interfaces significant enough to produce the binding site structure suitable for docking was found in about half of the complexes.
- 30.Tovchigrechko A, Wells CA, Vakser IA. Docking of protein models. Protein Sci. 2002;11:1888–1896. doi: 10.1110/ps.4730102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li B, Kihara D. Protein docking prediction using predicted protein–protein interface. BMC Bioinformatics. 2012;13:7. doi: 10.1186/1471-2105-13-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dreyfus T, Doye V, Cazals F. Assessing the reconstruction of macromolecular assemblies with toleranced models. Proteins. 2012;80:2125–2136. doi: 10.1002/prot.24092. [DOI] [PubMed] [Google Scholar]
- 33.Sircar A, Gray JJ. SnugDock: paratope structural optimization during antibody–antigen docking compensates for errors in antibody homology models. PLoS Comp Biol. 2010;6:e1000644. doi: 10.1371/journal.pcbi.1000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu L, Lu H, Skolnick J. MULTIPROSPECTOR: an algorithm for the prediction of protein–protein interactions by multimeric threading. Proteins. 2002;49:350–364. doi: 10.1002/prot.10222. [DOI] [PubMed] [Google Scholar]
- 35. Mukherjee S, Zhang Y. Protein–protein complex structure predictions by multimeric threading and template recombination. Structure. 2011;13:955–966. doi: 10.1016/j.str.2011.04.006. The paper presents a threading-based approach to model protein–protein complexes. The query sequences are aligned to complex templates using a modified dynamic programming algorithm. The monomer alignments are shifted to the multimeric template framework by structural alignments. The approach outperforms conventional homology modeling algorithms.
- 36. Zhang QC, Petrey D, Deng L, Qiang L, Shi Y, Thu CA, Bisikirska B, Lefebvre C, Accili D, Hunter T, et al. : Structure-based prediction of protein–protein interactions on a genome-wide scale. Nature. 2012;490:556–560. doi: 10.1038/nature11503. Structural information was used to predict protein–protein interactions with an accuracy and coverage that are superior to predictions based on non-structural evidence. An algorithm, which combines structural information with other functional clues, was shown to be comparable in accuracy to high-throughput experiments. The effectiveness of the structural information is attributed to the use of homology models and both close and remote geometric relationships between proteins.
- 37. Kundrotas PJ, Zhu Z, Janin J, Vakser IA. Templates are available to model nearly all complexes of structurally characterized proteins. Proc Natl Acad Sci U S A. 2012;109:9438–9441. doi: 10.1073/pnas.1200678109. The results of a large scale, systematic study show that, surprisingly, in spite of the limited number of crystallographically determined protein– protein complexes, docking templates can be found for complexes representing almost all known protein–protein interactions, provided the components themselves have a known structure or can be homology- built. About one-third of the templates are of good quality when compared to experimental structures in benchmark sets.
- 38.Brylinski M, Skolnick J. Q-DockLHM: low-resolution refinement for ligand comparative modeling. J Comput Chem. 2010;31:1093–1105. doi: 10.1002/jcc.21395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee HS, Zhang Y. BSP-SLIM: a blind low-resolution ligand– protein docking approach using predicted protein structures. Proteins. 2012;80:93–110. doi: 10.1002/prot.23165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao J, Dundas J, Kachalo S, Ouyang Z, Liang J. Accuracy of functional surfaces on comparatively modeled protein structures. J Struct Funct Genomics. 2011;12:97–107. doi: 10.1007/s10969-011-9109-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bordogna A, Pandini A, Bonati L. Predicting the accuracy of protein–ligand docking on homology models. J Comput Chem. 2011;32:81–98. doi: 10.1002/jcc.21601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Novoa EM, de Pouplana LR, Barril X, Orozco M. Ensemble docking from homology models. J Chem Theory Comput. 2010;6:2547–2557. doi: 10.1021/ct100246y. [DOI] [PubMed] [Google Scholar]
- 43.Vorobjev YN. Blind docking method combining search of low-resolution binding sites with ligand pose refinement by molecular dynamics-based global optimization. J Comput Chem. 2010;31:1080–1092. doi: 10.1002/jcc.21394. [DOI] [PubMed] [Google Scholar]
- 44.Gunther S, May P, Hoppe A, Frommel C, Preissner R. Docking without docking: ISEARCH — prediction of interactions using known interfaces. Proteins. 2007;69:839–844. doi: 10.1002/prot.21746. [DOI] [PubMed] [Google Scholar]
- 45.Gao M, Skolnick J. iAlign: a method for the structural comparison of protein–protein interfaces. Bioinformatics. 2010;26:2259–2265. doi: 10.1093/bioinformatics/btq404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghoorah AW, Devignes MD, Smaïl-Tabbone M, Ritchie DW. Spatial clustering of protein binding sites for template based protein docking. Bioinformatics. 2011;27:2820–2827. doi: 10.1093/bioinformatics/btr493. [DOI] [PubMed] [Google Scholar]
- 47.Jordan RA, EL-Manzalawy Y, Dobbs D, Honavar V. Predicting protein–protein interface residues using local surface structural similarity. BMC Bioinformatics. 2012;13:41. doi: 10.1186/1471-2105-13-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koike R, Ota M. SCPC: a method to structurally compare protein complexes. Bioinformatics. 2012;28:324–330. doi: 10.1093/bioinformatics/btr654. [DOI] [PubMed] [Google Scholar]
- 49.Konc J, Depolli M, Trobec R, Rozman K, Janezic D. Parallel-ProBiS: fast parallel algorithm for local structural comparison of protein structures and binding sites. J Comput Chem. 2012;33:2199–2203. doi: 10.1002/jcc.23048. [DOI] [PubMed] [Google Scholar]
- 50.Tuncbag N, Keskin O, Nussinov R, Gursoy A. Fast and accurate modeling of protein–protein interactions by combining template-interface-based docking with flexible refinement. Proteins. 2012;80:1239–1249. doi: 10.1002/prot.24022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pang B, Zhao N, Korkin D, Shyu CR. Fast protein binding site comparisons using visual words representation. Bioinformatics. 2012;28:1345–1352. doi: 10.1093/bioinformatics/bts138. [DOI] [PubMed] [Google Scholar]
- 52.Sinha R, Kundrotas PJ, Vakser IA. Protein docking by the interface structure similarity: how much structure is needed? PloS One. 2012;7:e31349. doi: 10.1371/journal.pone.0031349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sinha R, Kundrotas PJ, Vakser IA. Docking by structural similarity at protein–protein interfaces. Proteins. 2010;78:3235–3241. doi: 10.1002/prot.22812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lensink MF, Wodak SJ. Blind predictions of protein interfaces by docking calculations in CAPRI. Proteins. 2010;78:3085–3095. doi: 10.1002/prot.22850. [DOI] [PubMed] [Google Scholar]
- 55. Gao M, Skolnick J. Structural space of protein–protein interfaces is degenerate, close to complete, and highly connected. Proc Natl Acad Sci U S A. 2010;107:22517–22522. doi: 10.1073/pnas.1012820107. The paper describes development and application of an efficient structural alignment method, to study the structural similarity of representative protein– protein interfaces involving interactions between dimers. It finds that the degeneracy of interface space is largely due to the packing of compact, hydrogen-bonded secondary structure elements. The comparative study of artificial and native interfaces suggests that the library of protein interfaces is close to complete and consists of ~1000 distinct interface types
- 56. Zhang QC, Petrey D, Norel R, Honig BH. Protein interface conservation across structure space. Proc Natl Acad Sci U S A. 2010;107:10896–10901. doi: 10.1073/pnas.1005894107. The paper explores the range of applicability of structural information to prediction of protein–protein interactions by analyzing the extent to which the location of binding sites on protein surfaces is conserved among structural neighbors. The study finds that interface conservation is most significant among proteins that have a clear evolutionary relationship, but that there is a significant level of conservation even among remote structural neighbors. A new procedure predicts binding sites on protein structures
- 57.Teyra J, Hawkins J, Zhu H, Pisabarro MT. Studies on the inference of protein binding regions across fold space based on structural similarities. Proteins. 2011;79:499–508. doi: 10.1002/prot.22897. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2303–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dong H, Qin S, Zhou HX. Effects of macromolecular crowding on protein conformational changes. PLoS Comp Biol. 2010;6:e1000833. doi: 10.1371/journal.pcbi.1000833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McGuffee SR, Elcock AH. Diffusion, crowding and protein stability in a dynamic molecular model of the bacterial cytoplasm. PLoS Comp Biol. 2010;6:e1000694. doi: 10.1371/journal.pcbi.1000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feig M, Sugita Y. Variable interactions between protein crowders and biomolecular solutes are important in understanding cellular crowding. J Phys Chem B. 2012;16:599–605. doi: 10.1021/jp209302e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li X, Moal IH, Bates PA. Detection and refinement of encounter complexes for protein–protein docking: taking account of macromolecular crowding. Proteins. 2010;78:3189–3196. doi: 10.1002/prot.22770. [DOI] [PubMed] [Google Scholar]
- 63. Ando T, Skolnick J. Crowding and hydrodynamic interactions likely dominate in vivo macromolecular motion. Proc Natl Acad Sci U S A. 2010;107:18457–18462. doi: 10.1073/pnas.1011354107. The paper explores the principles of intermolecular dynamics in the crowded environment of cells by examining possible mechanisms responsible for the reduction in diffusion constants of macromoleculesin vivofrom that at infinite dilution. The study considered hydrodynamic interactions and the effects of nonspecific attractive interactions. It determined qualitative differences that can differentiate the importance of hydrodynamic versus nonspecific attractive interactions in macromolecular motion in cells.
- 64.Dill KA, Ghosh K, Schmit JD. Physical limits of cells and proteomes. Proc Natl Acad Sci U S A. 2011;108:17876–17882. doi: 10.1073/pnas.1114477108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuzu G, Keskin O, Gursoy A, Nussinov R. Constructing structural networks of signaling pathways on the proteome scale. Curr Opin Struct Biol. 2012;22:367–377. doi: 10.1016/j.sbi.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 66. Stein A, Mosca R, Aloy P. Three-dimensional modeling of protein interactions and complexes is going ‘omics. Curr Opin Struct Biol. 2011;21:200–208. doi: 10.1016/j.sbi.2011.01.005. The review describes recent developments in the field of structural bioinformatics applied to modeling of protein interactions and complexes, from large macromolecular machines to domain–domain and peptide-mediated interactions, focusing on proteome-wide applications
- 67. Kar G, Keskin O, Nussinov R, Gursoy A. Human proteome-scale structural modeling of E2–E3 interactions exploiting interface motifs. J Proteome Res. 2012;11:1196–1207. doi: 10.1021/pr2009143. The paper describes modeling of the interactions of ubiquitin-conjugating and ubiquitin-ligating enzymes in a large, proteome-scale strategy based on interface structural motifs, which allows prediction of interacting partners, as well as their interaction modes, in the human ubiquitination pathway
- 68.Wass MN, David A, Sternberg MJE. Challenges for the prediction of macromolecular interactions. Curr Opin Struct Biol. 2011;21:382–390. doi: 10.1016/j.sbi.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 69.Levitt M. Nature of the protein universe. Proc Natl Acad Sci U S A. 2009;106:11079–11084. doi: 10.1073/pnas.0905029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mosca R, Pons C, Fernandez-Recio J, Aloy P. Pushing structural information into the yeast interactome by high-throughput protein docking experiments. PLoS Comp Biol. 2009;5:e1000490. doi: 10.1371/journal.pcbi.1000490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhu Z, Tovchigrechko A, Baronova T, Gao Y, Douguet D, O’Toole N, Vakser IA. Large-scale structural modeling of protein complexes at low resolution. J Bioinformatics Comp Biol. 2008;6:789–810. doi: 10.1142/s0219720008003679. [DOI] [PubMed] [Google Scholar]
- 72.Aloy P, Bottcher B, Ceulemans H, Leutwein C, Mellwig C, Fischer S, Gavin AC, Bork P, Superti-Furga G, Serrano L, et al. : Structure-based assembly of protein complexes in yeast. Science. 2004;303:2026–2029. doi: 10.1126/science.1092645. [DOI] [PubMed] [Google Scholar]
- 73.Kundrotas PJ, Zhu Z, Vakser IA. GWIDD: a comprehensive resource for genome-wide structural modeling of protein– protein interactions. Hum Genomics. 2012;6:7. doi: 10.1186/1479-7364-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kundrotas PJ, Zhu Z, Vakser IA. GWIDD: genome-wide protein docking database. Nucleic Acids Res. 2010;38:D513–D517. doi: 10.1093/nar/gkp944. [DOI] [PMC free article] [PubMed] [Google Scholar]