Abstract
Hemoparasites of the apicomplexan family Plasmodiidae include the etiological agents of malaria, as well as a suite of non-human primate parasites from which the human malaria agents evolved. Despite the significance of these parasites for global health, little information is available about their ecology in multi-host communities. Primates were investigated in Kibale National Park, Uganda, where ecological relationships among host species are well characterized. Blood samples were examined for parasites of the genera Plasmodium and Hepatocystis using microscopy and PCR targeting the parasite mitochondrial cytochrome b gene, followed by Sanger sequencing. To assess co-infection, “deep sequencing” of a variable region within cytochrome b was performed. Out of nine black-and-white colobus (Colobus guereza), one blue guenon (Cercopithecus mitis), five grey-cheeked mangabeys (Lophocebus albigena), 23 olive baboons (Papio anubis), 52 red colobus (Procolobus rufomitratus) and 12 red-tailed guenons (Cercopithecus ascanius), 79 infections (77.5%) were found, all of which were Hepatocystis spp. Sanger sequencing revealed 25 different parasite haplotypes that sorted phylogenetically into six species-specific but morphologically similar lineages. “Deep sequencing” revealed mixed-lineage co-infections in baboons and red colobus (41.7% and 64.7% of individuals, respectively) but not in other host species. One lineage infecting red colobus also infected baboons, but always as the minor variant, suggesting directional cross-species transmission. Hepatocystis parasites in this primate community are a diverse assemblage of cryptic lineages, some of which co-infect hosts and at least one of which can cross primate species barriers.
Keywords: Plasmodiidae, Plasmodium, Hepatocystis, Malaria, Non-human primates
1. Introduction
Hemoparasites of the apicomplexan family Plasmodiidae include some of the most globally important human pathogens. Over 20 species of Plasmodium, which cause malaria, are known to infect non-human primates and at least four species, Plasmodium inui, Plasmodium simium, Plasmodium cynomolgi and Plasmodium knowlesi, are known to be transmissible to humans (Coatney et al., 2003; Cox-Singh et al., 2008). Human malaria parasites, including the particularly virulent Plasmodium falciparum, have zoonotic origins in non-human primates (Escalante et al., 1998; Rich et al., 2009; Duval et al., 2010; Krief et al., 2010; Liu et al., 2010; Prugnolle et al., 2011). However, the specific primate species from which each human malaria agent originated remains a matter of debate. Although the closest relative of P. falciparum was previously considered to be Plasmodium reichenowi, found in chimpanzees (Pan troglodytes), recent studies have discovered more closely related Plasmodium variants in other African apes and monkeys (Rich et al., 2009; Duval et al., 2010; Krief et al., 2010; Liu et al., 2010; Prugnolle et al., 2011).
Studies of Plasmodium and its relatives in non-human primates have been highly informative but influenced by the geography of sampling. Samples from captive animals close to humans may bias results towards zoonotic and reverse zoonotic transmission (Liu et al., 2010; Prugnolle et al., 2011). Broad surveys of primate hosts have yielded valuable data on geographic patterns of infection, but inferences about local transmission are difficult to make from such studies (Liu et al., 2010; Ayouba et al., 2012). Focused investigations of particular host species are useful for measuring local prevalence and parasite diversity but less so for examining transmission among species (Krief et al., 2010; Liu et al., 2010). Evaluating the propensity for Plasmodium and its relatives to cross species barriers in nature would require studying well-defined ecological communities where multiple host species are potentially exposed to the same vector populations (Prugnolle et al., 2011).
This study report results from the well-characterized primate community of Kibale National Park, Uganda (Kibale, hereafter) and the infection of its constituent host species with parasites of the genera Plasmodium and Hepatocystis. Like Plasmodium, Hepatocystis is a hemosporidian parasite within the family Plasmodiidae that infects primates (Desser and Bennett, 1993). Unlike Plasmodium, Hepatocystis is transmitted by biting midges of the genus Culicoides, rather than anopheline mosquitoes (Garnham, 1966). At least four species of Hepatocystis are known to infect African monkeys: Hepatocystis kochi, Hepatocystis simiae, Hepatocystis bouillezi and Hepatocystis cercopitheci (Seethamchai et al., 2008). Hepatocystis can cause parasitemia for up to 15 months without causing clinical signs (Vickerman, 2005), although early studies documented anemia and visible merocyst production, followed by scarring of the liver (Garnham, 1966). Primates can be infected repeatedly and prevalence increases with age (Vickerman, 2005). Hepatocystis is not known to infect humans.
Microscopy and molecular methods were used to assess infection in six species of monkeys in Kibale: black-and-white colobus (Colobus guereza), blue guenons (Cercopithecus mitis), grey-cheeked mangabeys (Lophocebus albigena), olive baboons (Papio anubis), red colobus (Procolobus rufomitratus), and red-tailed guenons (Cercopithecus ascanius). “Deep sequencing” was then used to assess co-infection of individual hosts with multiple parasite variants. To our knowledge, this is the first study of cross-species transmission of malaria-like parasites in a well-defined community of wild primates where host species overlap spatially and temporally and would thus encounter the same vector population.
2. Materials and methods
2.1. Study site and sampling
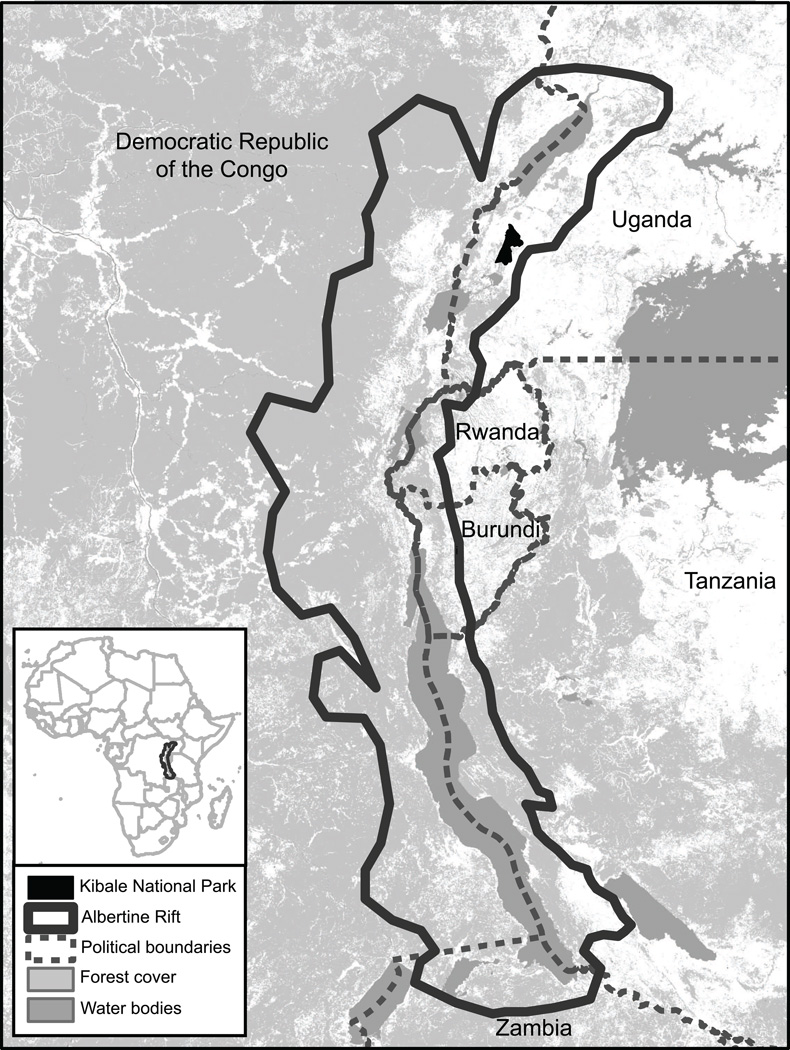
Kibale is a 795 km2 semi-deciduous, mid-altitude forest in western Uganda (0°13’−0°41’ N, 30°19’−30°32’ E; Chapman and Lambert, 2000) within the Albertine Rift, an area of exceptional biological diversity and conservation value (Plumptre et al., 2007; Fig. 1). Kibale’s primate community has been studied for over 40 years, such that a wealth of information exists on the ecology of its 13 constituent species (Chapman et al., 2005). Plasmodium infection has previously been documented in Kibale’s chimpanzees (Krief et al., 2010).
Fig. 1.
Map of Kibale National Park, Uganda, within the Albertine Rift, a region of exceptional biodiversity and conservation value shared by Uganda, Rwanda, Burundi, Tanzania, Zambia and the Democratic Republic of the Congo.
As part of a larger study of primate ecology, conservation and health (Goldberg et al., 2012), 34 animals in 2006 and 68 animals in 2010 were chemically immobilized as previously described (Goldberg et al., 2009; Lauck et al., 2011). Blood was drawn from the femoral vein, placed into an evacuated plasma collection tube (Becton, Dickinson and Company, Inc, Franklin Lakes, NJ, USA) and kept cool until processing. At the time of sampling, two thin blood smears were prepared, which were fixed and stained with Wright-Giesma stains within 5 h of collection. Blood was separated by centrifugation in a field laboratory and frozen immediately in liquid nitrogen for storage and transport. All animal procedures were approved by the Uganda National Council for Science and Technology, the Uganda Wildlife Authority, and the animal use committees of University of Wisconsin-Madison, USA and McGill University, Canada. Samples were shipped following International Air Transport Association (IATA) regulations under Ugandan Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) permit #002290.
2.2. Microscopy
Thin blood smears were viewed under 1000× oil immersion using a Nikon Eclipse e600 light microscope. For each slide, a field containing a representative monolayer of red blood cells (RBCs) was identified and all RBCs in that field were counted. This process was repeated five times, to produce an average number of RBCs per field. One hundred fields, or the maximum number of viewable fields on the slide, were then scanned for intra-erythrocytic parasites morphologically consistent with Plasmodium or Hepatocystis. Intensity of infection was calculated as: [total number of infected RBCs / (average number of RBCs per field × number of fields scanned)] × 100 (Warhurst and Williams, 1996).
2.3. PCR and sequencing
DNA was extracted from blood using a Zymo Quick-gDNA MiniPrep kit (Zymo Research, Irvine, CA, USA) according to the manufacturer’s instructions. PCR targeting the mitochondrial cytochrome b gene of Plasmodium and Hepatocystis was conducted using newly designed “universal” primers CytB3384F (5’-GTAATGCCTAGACGTATTCCTG-3’) and CytB3706R (5’ - GTTTGCTTGGGAGCTGTAATC -3’), which generate amplicons of predicted sizes 1,391 bp for Hepatocystis spp. and 1,254 bp for Plasmodium spp. PCRs were performed in 25 µl volumes using the FailSafe system (EpiCenter Biotechnologies, Madison, WI, USA), with reactions containing 1× FailSafe PCR PreMix with Buffer E, 1 Unit of FailSafe Enzyme Mix, 2.5 picomoles of each primer and 1 µl of template. Reactions were cycled in a Bio-Rad CFX96 platform (Bio-Rad Laboratories, Hercules, CA, USA) with the following temperature profile: 94°C for 1 min; 45 cycles of 94°C for 15 s, 50°C for 30 s, 72°C for 90 s; and a final extension at 72°C for 10 min. Amplicons were electrophoresed on agarose gels stained with ethidium bromide and purified from gels using the Zymoclean Ge l DNA Recovery Kit (Zymo Research, Irvine, CA, USA) according to the manufacturer’s instructions.
Products were Sanger sequenced in both directions using primers CytB3384F and CytB3706R, as well as internal sequencing primers CytB4081F (5’-TTACATTTACATGGTAGCAC-3’) and Cytb4062R (5’-GTGCTACCATGTAAATGTAA-3’) on ABI 3730xl DNA Analyzers (Applied Biosystems, Carlsbad CA, USA) at the University of Wisconsin Biotechnology Center DNA Sequence Facility. Sequences were hand-edited using Sequencher v4.9 (Gene Codes Corporation, Ann Arbor MI, USA) and all ambiguous bases were resolved by repeat PCR and re-sequencing, as described above. All new sequences were deposited in GenBank, under Accession Numbers KC262794–KC262867.
2.4. Pyrosequencing
To assess co-infection, 40 samples were chosen for multiplexed pyrosequencing based on phylogenetic analyses of Sanger sequences, such that all combinations of lineages and host species were represented. The primers 454cytbF (5’-3’) GAGAATTATGGAGTGGATGGTGTT and 454cytbR (5’-3’) ATGCTGTATCATACCCTAAAGGATT were designed to amplify 420 bp of a variable and phylogenetically informative region of cytochrome b within Hepatocystis sequences from Kibale, based on analysis of Sanger sequences. To allow multiplexing, unique identification tag sequences (10 bp) and adaptor sequences (25 bp) were designed for each of the 40 individuals. PCRs were conducted with the Phusion High-Fidelity PCR Kit (Finnzymes, Espoo, Finland) in 20 µl volumes containing 1× Phusion HF buffer, 10 mmol of dNTPs, 1 unit of Phusion DNA Polymerase, 5.0 pmol of each primer and 1 µl of original DNA extract as template. Reactions were cycled in a Bio-Rad CFX96 platform (Bio-Rad Laboratories, Hercules, CA, USA) with the following temperature profile: 98°C for 30 s; 30 cycles of 98°C for 10 s, 57°C for 15 s, 72°C for 150 s; and a final extension at 72°C for 5 min. Amplicons were purified as described above [section 2.3] and further cleaned using the AMPure XP PCR purification system (Beckman Coulter, Brea CA, USA). DNA was quantitated using the Quant-iT dsDNA HS Assay kit (Life Technologies, Madison, WI, USA), concentrations were normalized to the least concentrated sample, and samples were pooled at 2×106 molecules/µl. Pooled samples were then subjected to 454 emulsion PCR, and products were pyrosequenced on a GS Junior instrument (Roche 454 Life Sciences, Branford, CT, USA).
2.5. Statistical analyses
Frequencies of infection and co-infection were calculated as simple ratios, with 95% confidence limits estimated using the modified Wald method (Agresti and Coull, 1998). Mean and median intensities of infection were calculated and 95% confidence limits were generated using the computer program Quantitative Parasitology 3.0 (Rózsa et al., 2000), which was also used to perform Mood’s median tests (Mood, 1954) to compare intensities of infection among groups. Frequencies of infection among groups were compared using Fisher’s exact tests (Fisher, 1925) and other statistical comparisons were performed using the computer program SPSS Statistics 17.0 (SPSS, Inc., Chicago, IL, USA).
Sanger sequences were aligned using the computer program ClustalX (Larkin et al., 2007) with minor manual adjustment. To construct phylogenetic trees, the neighbor joining method was applied (Saitou and Nei, 1987) using the computer program MEGA5 (Tamura et al., 2011), with a maximum composite likelihood distance correction and 1,000 bootstrap replicates of the data to assess robustness of phylogenetic groupings. Phylogenetic trees were constructed both from full-length alignments of newly generated Hepatocystis sequences (1,178 positions) and from alignments trimmed to 726 bp to allow inclusion of shorter published reference sequences (Ayouba et al., 2012).
To assess co-infection of individual primates with multiple Hepatocystis lineages using pyrosequencing data, individual sequences from each animal were mapped to newly generated and published Hepatocystis cytochrome b sequences using CLC Genomics Workbench 5.1 (CLC bio, Aarhus, Denmark) with stringent settings, including minimum 95% similarity, 50% length fraction, and insertion and deletion costs of three. Sequences were mapped to each animal’s Sanger consensus sequence, to all other Hepatocystis variants identified in Kibale primates as described above, and to Hepatocystis reference sequences available in GenBank. An individual was considered to be co-infected when at least 5% of sequences within that individual that mapped to a Hepatocystis lineage other than the lineage defined by that animal’s Sanger consensus sequence. To guard against false positive results, a filtering step was applied to exclude low quality and truncated sequences (less than 300 bases).
3. Results
3.1. Microscopy and PCR
Microscopy identified intraerythrocytic parasites in 75% of blood smears across six primate species in Kibale (Table 1). Parasites displayed trophozoite and gametocyte forms that were morphologically similar in all host species (Fig. 2). Prevalence ranged from 22.2% in black-and-white colobus to 91.3% in baboons (Table 1). Mean intensity (percentage of infected red blood cells) across all species was 0.058%, ranging from 0.052% in baboons to 0.090% in black-and-white colobus. PCR yielded amplicons of the predicted size in 77.5% of blood DNA extracts across all host species, with species-specific pr evalence agreeing closely with microscopy (Table 1). Based on both microscopy and PCR, prevalence varied significantly across the four host species in which infection was detected (Table 1; Fisher’s exact P < 0.001 in both cases). However, intensity was only marginally different across the same four host species (Table 1; Mood’s exact P = 0.059).
Table 1.
Prevalence and intensity of Hepatocystis infection in six primate hosts from Kibale National Park, Uganda.
| Prevalence (%)a | Intensity (%)b | ||||
|---|---|---|---|---|---|
| Host | nc | Microscopy | PCR | Mean | Median |
| Black-and-white colobus (Colobus guereza) | 9 | 22.2 (5.3 – 55.7) | 22.2 (5.3 – 55.7) | 0.090 (0.083 – 0.090) | 0.090 (undefined) |
| Blue guenon (Cercopithecus mitis) | 1 | 0 (0.0 – 83.3) | 0 (0.0 –83.3) | -- | -- |
| Grey-cheeked mangabey (Lophocebus albigena) | 5 | 0 (0.0 – 48.9) | 0 (0.0 – 48.9) | -- | -- |
| Olive baboon (Papio anubis) | 23 | 91.3 (72.0 –98.8) | 95.7 (77.3 – 99.9) | 0.052 (0.030 – 0.088) | 0.025 (95.7%:0.010 – 0.032) |
| Red colobus (Procolobus rufomitratus) | 46 | 89.1 (76.5 – 95.7) | 88.5 (76.7 – 95.0) | 0.058 (0.043 – 0.100) | 0.038 (95.6%:0.028 –0.064) |
| Red-tailed guenon (Cercopithecus ascanius) | 12 | 66.7 (38.8 – 86.5) | 75.0 (46.2 – 91.7) | 0.069 (0.035 – 0.121) | 0.045 (96.1%:0.013 – 0.158) |
| All | 102 | 75.0 (65.4 – 82.6) | 77.5 (68.4 – 84.5) | 0.058 (0.045 – 0.078) | 0.032 (95.6%:0.024 – 0.041) |
Numbers in parentheses are 95% confidence limits calculated using the modified Wald method (Agresti and Coull, 1998).
Intensity was measured as the percentage of infected red blood cells in positive animals, based on microscopic examination. Numbers in parentheses are bootstrap 95% confidence limits (mean) and “exact” confidence intervals (median), with the precise level of confidence around each median indicated (the confidence interval for black-and-white colobus was undefined because only two individuals were infected).
Numbers indicate sample sizes of thin smears examined microscopically and blood DNA extracts used for PCR. Thin smears from six red colobus were not readable and were thus excluded.
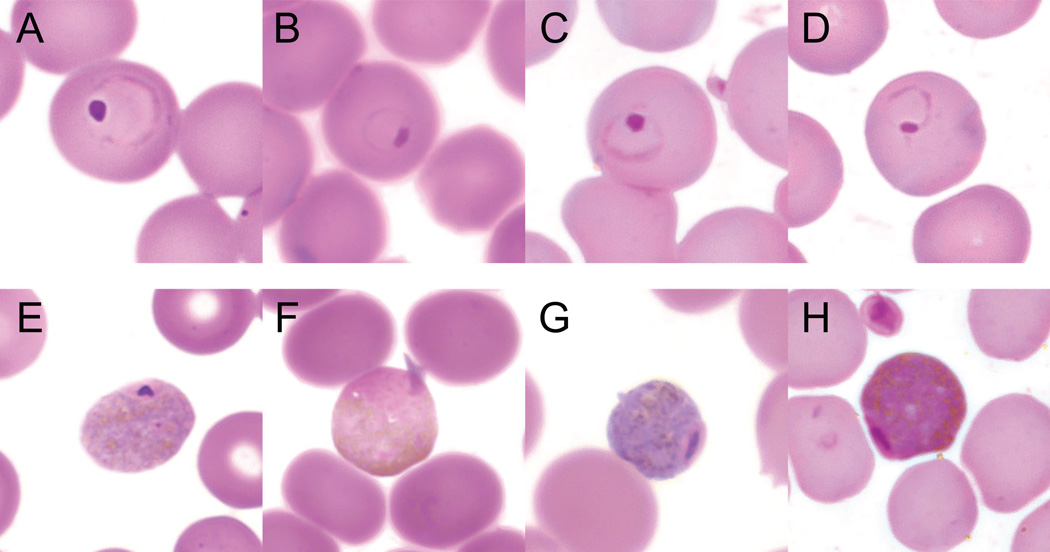
Fig. 2.
Trophozoite and gametocyte stages of Hepatocystis in four primate host species. Trophozoites (A–D) and gametocytes (E–H) were photographed at 1,000× magnification in representative thin blood smears stained with Wright-Giesma stain for each of the four Kibale primate species positive for Hepatocystis: olive baboon (A, E), black-and-white colobus (B, F), red colobus (C, G) and red-tailed guenon (D, H).
3.2. Sanger sequences
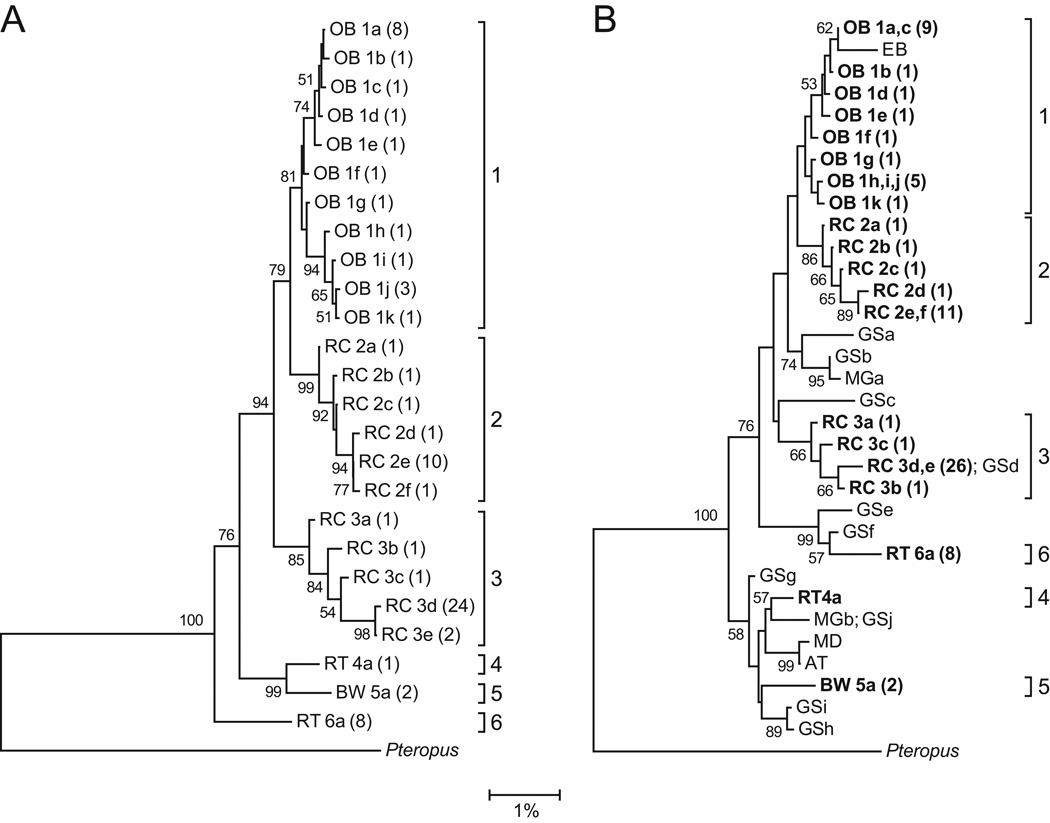
Analysis of Sanger sequences revealed 25 Hepatocystis haplotypes within the 79 PCR-positive Kibale primate samples. No primates were infected with Plasmodium. Phylogenetic analysis sorted these haplotypes into six host-specific lineages, arbitrarily numbered 1–6 (Fig. 3A). Lineage 1 infected baboons, lineages 2 and 3 infected red colobus, lineages 4 and 6 infected red-tailed guenons, and lineage 5 infected black-and-white colobus. Bootstrap values indicated strong to moderate support for this pattern of clustering. Intensity of infection (measured using microscopy; see above) was not statistically different across the six lineages (Table 1; Mood’s exact P = 0.100).
Fig. 3.
Phylogenetic trees of Hepatocystis spp. (A) The tree was constructed from a 1,178 bp alignment of the mitochondrial cytochrome b gene for all PCR-positive primates from Kibale National Park, Uganda. Numbers indicate bootstrap values (%) estimated from 1,000 resamplings of the sequence data; bootstrap values ≤ 50% are not shown. Taxon names indicate the host species from which each sequence was recovered (OB = olive baboon; RC = red colobus; RT = red-tailed guenon; BW = black-and-white colobus), followed by an arbitrary haplotype designation and the number of individuals infected with that haplotype in parentheses. Brackets indicate species-specific lineages, arbitrarily labeled 1–6. (B) The tree was constructed using identical methods, but with sequences trimmed to 726 bp to allow incorporation of shorter published reference sequences from an Ethiopian baboon (EB), 10 greater spot-nosed guenons (GSa-h), two mustached guenons (MGa,b), a mandrill (MD) and an Angolan talapoin (AT) (GenBank accession numbers AF069626, JF923757, JF923758, JF923759, JF923760, JQ070819, JQ070820, JQ070869, JQ070892, JQ070901, JQ070903, JQ070910, JQ070938, JQ070951 and JQ070953). Haplotypes sequenced in this study are shown in bold type. The trees are rooted with an outgroup Hepatocystis sequence from a small flying fox, Pteropus hypomelanus (GenBank accession number FJ168565). The scale bar indicates the percentage of nucleotide substitutions per site.
A second phylogenetic tree was generated using sequences trimmed to 726 bp to incorporate 16 unique published Hepatocystis reference sequences (Ayouba et al., 2012; Escalante et al., 1998; Prugnolle et al., 2011). All six species-specific lineages identified in Fig. 3A were retained in this second phylogeny (Fig. 3B). However, the position of lineage 6 shifted, due to the exclusion of a region at the 3’ end of the full-length alignment within which lineage 6 contained several point substitutions and a deletion. A reference sequence from an Ethiopian baboon (Papio sp.) clustered within lineage 1, which infects Kibale baboons, and a reference sequence from a greater spot-nosed guenon (Cercopithecus nictitans) was identical to a haplotype within lineage 3, which infects Kibale red c olobus. Other reference sequences from greater spot-nosed guenons, mustached guenons (Cercopithecus cephus), a mandrill (Mandrillus sphinx), and a talapoin (Miopithecus talapoin) clustered separately from the six Kibale lineages but fell within the overall diversity circumscribed by the Kibale lineages.
3.3. Pyrosequencing and co-infection
Multiplexed pyrosequencing of 40 Hepatocystis 420 bp amplicons yielded an average of 646 high-quality sequences per individual (S.D. = 232; range 278–1,198) of average length 398 bp (S.D. = 25; range 302 – 420 bp). Mapping individual sequences to Sanger sequences from each Kibale primate and published reference sequences identified mixed-lineage co-infections in 21 of 40 individuals (52.5%). All co-infections occurred in either baboons (5/12; 41.7%) or red colobus (11/17; 64.7%%). There was no evidence of co-infections in red-tailed guenons (n = 9) or black-and-white colobus (n = 2). Frequencies of mixed-lineage co-infection were statistically different across host species (Fisher’s exact P = 0.004). The number of sequences within singly infected individuals (mean = 701 ± 265) and co-infected individuals (mean = 564 ± 141) did not differ statistically (Mann-Whitney U = 153.5; z = 1.05; P = 0.294), demonstrating that assessment of co-infection was not biased by variation in sequencing “depth” among individuals.
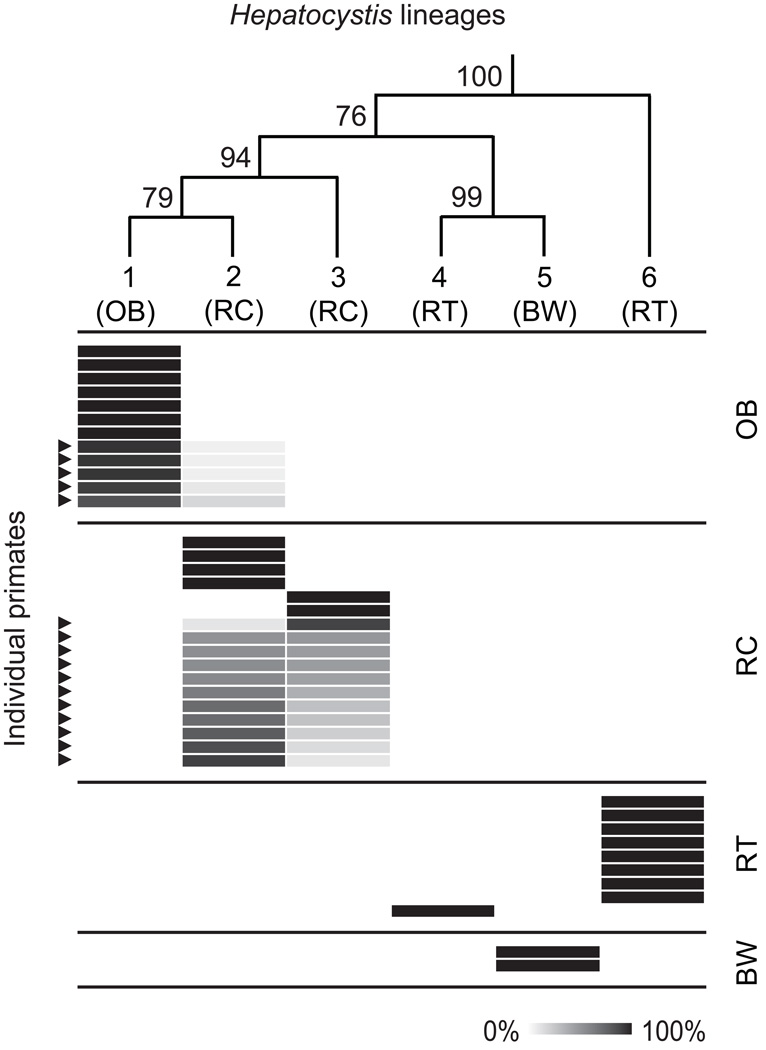
Fig. 4 shows the pattern of single lineage and mixed lineage infection and co-infection, respectively, for the 40 individuals subjected to pyrosequencing. Black-and-white colobus were singly infected only with lineage 5. Red-tailed guenons were singly infected with either lineage 4 (1/9; 11.1%) or lineage 6 (8/9; 88.9%). Red colobus were either singly infected with lineage 2 (4/17; 23.5%) or lineage 3 (2/17; 11.8%), or co-infected with a mixture of lineages 2 and 3 (11/17; 64.7%). The ratio of lineage 2: lineage 3 sequences within the 11 co-infected red colobus ranged from 1:8.5 to 7.7:1. Baboons were either singly infected with lineage 1 (7/12; 58.3%) or co-infected with a mixture of lineages 1 and 2 (5/12; 41.7%). In co-infected baboons, lineage 2 was always the minor variant; the ratio of lineage 1: lineage 2 sequences within the five co-infected baboons ranged from 14.4:1 to 4.6:1. There was no evidence of co-infection of any Kibale primate with any Hepatocystis lineage that was not detected in the primates of Kibale using Sanger sequencing.
Fig. 4.
Heatmap showing infection and co-infection of individual primates with multiple Hepatocystis lineages. Rows represent 40 individual primates subjected to pyrosequencing of a 420 bp amplicon of the parasite cytochrome b gene. The cladogram on the top of the figure shows the phylogeny of the six Kibale Hepatocystis lineages, with numbers representing bootstrap values (from Fig. 3A). Cells are shaded in proportion to the percentage of Hepatocystis sequences within an indivdual primate host mapping to each lineage. Host species: OB, olive baboon; RC, red colobus; RT, red-tailed guenon; BW, black-and-white colobus; and lineage numbers (1–6) follow Fig. 3. Arrows indicate primates with mixed-lineage co-infections, as evidenced by sequences mapping to more than one lineage.
4. Discussion
This study examined a community of wild primates in Uganda where ecological relationships among host species are well defined and where spatio-temporal overlap would make cross-species transmission of vector-borne parasites possible. Microscopy and PCR indicated rates of infection with Hepatocystis ranging from approximately 20% to 90% across host species. This finding is consistent with published data from wild greater spot-nosed guenons in Cameroon, which documented a prevalence of Hepatocystis infection of 49% (Ayouba et al., 2012). Even though Hepatocystis infection was common, there was no evidence of Plasmodium infection, despite the fact that Plasmodium infects chimpanzees from the same location (Krief et al., 2010). These results suggest that primate hosts vary in their susceptibility to parasites of the family Plasmodiidae, even when those hosts live in the same environment.
Despite morphological similarity among parasites, phylogenetic analyses of cytochrome b sequences yielded strong evidence that multiple, host-specific Hepatocystis lineages infect Kibale primates (Fig. 3A). These results are in contrast to the aforementioned study of Cameroonian greater spot-nosed guenons, which showed a lack of phylogenetic clustering of Hepatocystis haplotypes infecting this species (Ayouba et al., 2012). However, the results of this study are consistent with findings of host specificity in parasites in the subgenus Laverania infecting wild chimpanzees and lowland gorillas (Gorilla gorilla) also from eastern and central Africa (Liu et al., 2010). The current findings may have resulted from the longer cytochrome b sequences generated in this study, relative to previously published methods (Liu et al., 2010). However, sequences were trimmed to allow comparison with shorter published reference sequences (Fig. 2B), the general pattern from the analysis of full-length sequences persisted.
These results suggest that the evolution of the Hepatocystis genus is shaped principally by host factors, rather than by geography. Kibale, a forest park of only 795 km2, contains as much Hepatocystis cytochrome b phylogenetic diversity as has previously been documented in primates across sub-Saharan Africa (Prugnolle et al., 2011; Ayouba et al., 2012). The Albertine Rift wherein Kibale lies is a reservoir of biological diversity (Plumptre et al., 2007); perhaps the diversity of Hepatocystis lineages within Kibale reflects the unique biogeography of this region. It is also noteworthy that one published reference sequence from an Ethiopian baboon clusters within Kibale lineage 1, which infects olive baboons, a related species (Fig. 2B), again suggesting a strong role of host factors in shaping Hepatocystis evolution.
To our knowledge, this is the first study to use “deep sequencing” to investigate co-infection and cross-species transmission in a malaria-like parasite. Pyrosequencing of a portion of cytochrome b showed that mixed-lineage co-infection is common in Kibale red colobus and baboons. No mixed-lineage co-infections were detected in red-tailed guenons, despite roughly equal sampling intensity. The ability to detect mixed-lineage co-infections in black-and-white colobus was limited by the small number of infected individuals. These results complement a previous study of Asian crab-eating macaques (Macaca fasicularis), showing that one animal out of 99 was infected with two distinct Hepatocystis clones (Seethamchai et al., 2008).
In red colobus, individuals were singly and dually infected with two Hepatocystis lineages. Within co-infected red colobus, some individuals were co-infected with approximately equal proportions of both lineages. In contrast, there were a lower proportion of co-infected baboons, and all such baboons were co-infected with one of the red-colobus-associated lineages as the minor variant. Furthermore, no baboon was singly infected with this same red colobus associated lineage. Thus, despite co-infection of baboons with red colobus-associated Hepatocystis, no red colobus were co-infected with the corresponding baboon-associated lineage. These results imply that red colobus-associated Hepatocystis can cross primate species barriers to infect baboons, but that baboon-associated Hepatocystis does not show a similar pattern of transmission in the other direction.
Unfortunately, little information exists on the vectors that transmit these parasites in Kibale. Hepatocystis is thought to be transmitted by biting midges of the genus Culicoides (Garnham et al., 1961). The patterns of infection observed for Hepatocystis in Kibale primates could reflect the distribution and/or differential host feeding preferences of vectors. Clearly, investigations of midges and other potential insect vectors in Kibale are in order, perhaps coupled with blood meal analyses to identify patterns of host preference (Molaei and Andreadis, 2006; Lassen et al., 2011). Primates in Kibale often form mixed species groups, also known as polyspecific associations, perhaps to guard against predation (Chapman and Chapman, 1996). In such groups, red colobus associate preferentially with red-tailed guenons, and less so with black-and-white colobus (Chapman and Chapman, 2000). Baboons are largely terrestrial and do not form polyspecific associations with these three arboreal species. The patterns of cross-species transmission between red colobus and baboons described herein cannot therefore be explained by patterns of association among host species in Kibale.
It should be noted that a conservative analysis of pyrosequencing data may have underestimated rates of mixed-lineage co-infection. For example, small numbers of truncated, low quality sequences were found in additional baboons and red colobus consistent with the patterns shown in Fig. 4; these would have increased estimates of the frequency of co-infection had they been included. Haplotype-specific PCR followed by Sanger sequencing may have higher sensitivity than pyrosequencing for detecting mixed-lineage co-infection, although perhaps with higher error rates. With improvements in “next generation” sequencing technologies, it should be possible to identify very low levels of mixed lineage co-infection using methods similar to those described herein (Shokralla et al., 2012), and without the need for labor-intensive cloning (Seethamchai et al., 2008). It should also be noted that sequence-level mitochondrial heteroplasmy has been documented in certain parasite taxa (e.g. Bowles et al., 1995; van Herwerden et al., 2000; Curtis et al., 2001; Le et al., 2002; Messenger et al., 2012). However, the host-specific and asymmetrical patterns of infection documented for Hepatocystis in Kibale primates are inconsistent with this type of variation, as well as with the low levels of random genetic variation that would be expected from processes such as mutation or sequencing error.
Overall, the results of this study show that Hepatocystis exists in the Kibale primate community as a diverse assemblage of cryptic lineages, some of which are capable of co-infecting hosts and at least one of which can cross primate species barriers. These observations, together with the lack of Plasmodium infection in Kibale monkeys despite its presence in Kibale chimpanzees (Krief et al., 2010), demonstrate that the cross-species transmission potential of the Plasmodiidae varies among parasite lineages, even when hosts live in the same environment. Understanding why certain parasites within the Plasmodiidae cross species barriers in nature could help predict which new or poorly studied members of this critically important parasite family are most likely to emerge.
Highlights.
Malaria-like parasites of primates can evolve into significant human pathogens.
Primates in Kibale National Park, Uganda were screened for malaria-like parasites.
Multiple, species-specific lineages of the parasite Hepatocystis were discovered.
Two Hepatocystis lineages co-infect hosts and one moves between primate species.
Certain malaria-like parasites can cross primate species barriers in nature.
Acknowledgments
The authors gratefully acknowledge the Uganda Wildlife Authority and the Uganda National Council for Science and Technology for granting us permission to conduct this research. The authors also thank P. Omeja and L. Kilby for assistance with logistics; J. Byaruhanga, P. Katurama, A. Mbabazi, A. Nyamwija, E. Nyamwija, J. Rusoke and K. Cameron for assistance in the field; A. McCord for assistance in the laboratory; D. Mills for assistance with image preparation; J. Bleecker for assistance with map creation; A. Plumptre, the Wildlife Conservation Society and the ESA GlobCover 2009 Project for map data; and S. Perkins for helpful comments on the manuscript. This work was funded by National Institutes of Health (NIH), USA grant TW009237 as part of the joint NIH-NSF Ecology of Infectious Disease program and the UK Economic and Social Research Council, by the Wisconsin Partnership Program through the Wisconsin Center for Infectious Disease (WisCID), by the Morris Animal Foundation under Award No. MSN142182, and by the Merial-NIH Veterinary Summer Scholars program at the University of Wisconsin-Madison, USA. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
New sequences generated during this study were deposited in GenBank, under Accession 23 Numbers KC262794–KC262867.
References
- Agresti A, Coull BA. Approximate is better than "exact" for interval estimation of binomial proportions. Am. Stat. 1998;52:119–126. [Google Scholar]
- Ayouba A, Mouacha F, Learn GH, Mpoudi-Ngole E, Rayner JC, Sharp PM, Hahn BH, Delaporte E, Peeters M. Ubiquitous Hepatocystis infections, but no evidence of Plasmodium falciparum-like malaria parasites in wild greater spot-nosed monkeys (Cercopithecus nictitans) Int. J. Parasitol. 2012;42:709–713. doi: 10.1016/j.ijpara.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles J, Blair D, McManus DP. A molecular phylogeny of the genus Echinococcus. Parasitology. 1995;110(Pt 3):317–328. doi: 10.1017/s0031182000080902. [DOI] [PubMed] [Google Scholar]
- Chapman CA, Chapman LJ. Mixed-species primate groups in Kibale forest: ecological constraints on association. Int. J. Primatol. 1996;17:31–50. [Google Scholar]
- Chapman CA, Chapman LJ. Interdemic variation in mixed-species association patterns: common diurnal primates of Kibale National Park, Uganda. Behav. Ecol. Sociobiol. 2000;47:129–139. [Google Scholar]
- Chapman CA, Lambert JE. Habitat alteration and the conservation of African primates: case study of Kibale National Park, Uganda. Am. J. Primatol. 2000;50:169–185. doi: 10.1002/(SICI)1098-2345(200003)50:3<169::AID-AJP1>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Chapman CA, Struhsaker TT, Lambert JE. Thirty years of research in Kibale National Park, Uganda, reveals a complex picture for conservation. Int. J. Primatol. 2005;26:539–555. [Google Scholar]
- Coatney GR, Collins WEMW, Contacos PG. Centers for Disease Control and Prevention. Atlanta, GA: Division of Parasitic Disease; 2003. The primate malarias [CD-ROM; original book published 1971] [Google Scholar]
- Cox-Singh J, Davis TM, Lee KS, Shamsul SS, Matusop A, Ratnam S, Rahman HA, Conway DJ, Singh B. Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening. Clin. Infect. Dis. 2008;46:165–171. doi: 10.1086/524888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis J, Fraga LA, de Souza CP, Correa-Oliveira R, Minchella DJ. Widespread heteroplasmy in schistosomes makes an mtVNTR marker "nearsighted". J. Hered. 2001;92:248–253. doi: 10.1093/jhered/92.3.248. [DOI] [PubMed] [Google Scholar]
- Desser SS, Bennett GF. The genera Leucocytozoon, Haemoproteus, and Hepatocystis. In: Kreier JP, editor. Parasitic Protozoa. Volume 4. San Diego: Academic Press Inc.; 1993. pp. 273–307. [Google Scholar]
- Duval L, Fourment M, Nerrienet E, Rousset D, Sadeuh SA, Goodman SM, Andriaholinirina NV, Randrianarivelojosia M, Paul RE, Robert V, Ayala FJ, Ariey F. African apes as reservoirs of Plasmodium falciparum and the origin and diversification of the Laverania subgenus. Proc. Natl. Acad. Sci. USA. 2010;107:10561–10566. doi: 10.1073/pnas.1005435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante AA, Freeland DE, Collins WE, Lal AA. The evolution of primate malaria parasites based on the gene encoding cytochrome b from the linear mitochondrial genome. Proc. Natl. Acad. Sci. USA. 1998;95:8124–8129. doi: 10.1073/pnas.95.14.8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. Statistical Methods for Research Workers. Edinburgh: Oliver & Boyd; 1925. [Google Scholar]
- Garnham PC, Heisch RB, Minter DM. The vector of Hepatocystis (Plasmodium) kochi; the successful conclusion of observations in many parts of tropical Africa. Trans. R. Soc. Trop. Med. Hyg. 1961;55:497–502. doi: 10.1016/0035-9203(61)90071-2. [DOI] [PubMed] [Google Scholar]
- Garnham PCC. Malaria Parasites and Other Haemosporidia. Blackwell Scientific. United Kingdom: Oxford; 1966. [Google Scholar]
- Goldberg TL, Paige SB, Chapman CA. The Kibale EcoHealth Project: exploring connections among human health, animal health, and landscape dynamics in western Uganda. In: Aguirre AA, Daszak P, Ostfeld RS, editors. New Directions in Conservation Medicine: Applied Cases of Ecological Health. New York: Oxford University Press; 2012. pp. 452–465. [Google Scholar]
- Goldberg TL, Sintasath DM, Chapman CA, Cameron KM, Karesh WB, Tang S, Wolfe ND, Rwego IB, Ting N, Switzer WM. Coinfection of Ugandan red colobus (Procolobus [Piliocolobus] rufomitratus tephrosceles) with novel, divergent delta-, lenti-, and spumaretroviruses. J. Virol. 2009;83:11318–11329. doi: 10.1128/JVI.02616-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krief S, Escalante AA, Pacheco MA, Mugisha L, Andre C, Halbwax M, Fischer A, Krief JM, Kasenene JM, Crandfield M, Cornejo OE, Chavatte JM, Lin C, Letourneur F, Gruner AC, McCutchan TF, Renia L, Snounou G. On the diversity of malaria parasites in African apes and the origin of Plasmodium falciparum from Bonobos. PLoS Pathog. 2010;6:e1000765. doi: 10.1371/journal.ppat.1000765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lassen SB, Nielsen SA, Skovgard H, Kristensen M. Molecular identification of bloodmeals from biting midges (Diptera: Ceratopogonidae: Culicoides Latreille) in Denmark. Parasitol. Res. 2011;108:823–829. doi: 10.1007/s00436-010-2123-4. [DOI] [PubMed] [Google Scholar]
- Lauck M, Hyeroba D, Tumukunde A, Weny G, Lank SM, Chapman CA, O'Connor DH, Friedrich TC, Goldberg TL. Novel, divergent simian hemorrhagic fever viruses in a wild Ugandan red colobus monkey discovered using direct pyrosequencing. PLoS One. 2011;6:e19056. doi: 10.1371/journal.pone.0019056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TH, Blair D, McManus DP. Mitochondrial genomes of parasitic flatworms. Trends Parasitol. 2002;18:206–213. doi: 10.1016/s1471-4922(02)02252-3. [DOI] [PubMed] [Google Scholar]
- Liu W, Li Y, Learn GH, Rudicell RS, Robertson JD, Keele BF, Ndjango JB, Sanz CM, Morgan DB, Locatelli S, Gonder MK, Kranzusch PJ, Walsh PD, Delaporte E, Mpoudi-Ngole E, Georgiev AV, Muller MN, Shaw GM, Peeters M, Sharp PM, Rayner JC, Hahn BH. Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature. 2010;467:420–425. doi: 10.1038/nature09442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenger LA, Llewellyn MS, Bhattacharyya T, Franzen O, Lewis MD, Ramirez JD, Carrasco HJ, Andersson B, Miles MA. Multiple mitochondrial introgression events and heteroplasmy in Trypanosoma cruzi revealed by maxicircle MLST and next generation sequencing. PLoS Negl. Trop. Dis. 2012;6:e1584. doi: 10.1371/journal.pntd.0001584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molaei G, Andreadis TG. Identification of avian-and mammalian-derived bloodmeals in Aedes vexans and Culiseta melanura (Diptera : Culicidae) and its implication for West Nile virus transmission in Connecticut, USA. J. Med. Entomol. 2006;43:1088–1093. doi: 10.1603/0022-2585(2006)43[1088:IOAAMB]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Mood AM. On the asymptotic efficiency of certain non-parametric two-sample tests. Ann. Math. Stat. 1954;25:514–522. [Google Scholar]
- Plumptre AJ, Davenport TRB, Behangana M, Kityo R, Eilu G, Ssegawa P, Ewango C, Meirte D, Kahindo C, Herremans M, Peterhans JK, Pilgrimg JD, Wilson M, Languy M, Moyer D. The biodiversity of the Albertine Rift. Biol. Conserv. 2007;134:178–194. [Google Scholar]
- Prugnolle F, Ollomo B, Durand P, Yalcindag E, Arnathau C, Elguero E, Berry A, Pourrut X, Gonzalez JP, Nkoghe D, Akiana J, Verrier D, Leroy E, Ayala FJ, Renaud F. African monkeys are infected by Plasmodium falciparum nonhuman primate-specific strains. Proc. Natl. Acad. Sci. USA. 2011;108:11948–11953. doi: 10.1073/pnas.1109368108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich SM, Leendertz FH, Xu G, LeBreton M, Djoko CF, Aminake MN, Takang EE, Diffo JL, Pike BL, Rosenthal BM, Formenty P, Boesch C, Ayala FJ, Wolfe ND. The origin of malignant malaria. Proc. Natl. Acad. Sci. USA. 2009;106:14902–14907. doi: 10.1073/pnas.0907740106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rózsa L, Reiczigel J, Majoros G. Quantifying parasites in samples of hosts. J. Parasitol. 2000;86:228–232. doi: 10.1645/0022-3395(2000)086[0228:QPISOH]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Seethamchai S, Putaporntip C, Malaivijitnond S, Cui L, Jongwutiwes S. Malaria and Hepatocystis species in wild macaques, southern Thailand. Am. J. Trop. Med. Hyg. 2008;78:646–653. [PubMed] [Google Scholar]
- Shokralla S, Spall Jl, Gibson JF, Hajibabaei M. Next-generation sequencing technologies for environmental DNA research. Mol. Ecol. 2012;21:1794–1805. doi: 10.1111/j.1365-294X.2012.05538.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Herwerden L, Blair D, Agatsuma T. Multiple lineages of the mitochondrial gene NADH dehydrogenase subunit 1 (ND1) in parasitic helminths: implications for molecular evolutionary studies of facultatively anaerobic eukaryotes. J. Mol. Evol. 2000;51:339–352. doi: 10.1007/s002390010096. [DOI] [PubMed] [Google Scholar]
- Vickerman K. The lure of life cycles: C yril Garnham and the malaria parasites of primates. Protist. 2005;156:433–449. doi: 10.1016/j.protis.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Warhurst DC, Williams JE. Laboratory diagnosis of malaria. J. Clin. Pathol. 1996;49:533–538. doi: 10.1136/jcp.49.7.533. [DOI] [PMC free article] [PubMed] [Google Scholar]