Abstract
A growing number of reports indicate the frequent presence of DNA sequences and gene products of human cytomegalovirus in various tumors as compared to adjacent normal tissues, the brain tumors being studied most intensely. The mechanisms underlying the tropism of human cytomegalovirus to the tumor cells or to the cells of tumor origin, as well as the role of the host’s genetic background in virus-associated oncogenesis are not well understood. It is also not clear why cytomegalovirus can be detected in many but not in all tumor specimens. Our in silico prediction results indicate that microRNA-34a may be involved in replication of some human DNA viruses by targeting and downregulating the genes encoding a diverse group of proteins, such as platelet-derived growth factor receptor-alpha, complement component receptor 2, herpes simplex virus entry mediators A, B, and C, and CD46. Notably, while their functions vary, these surface molecules have one feature in common: they serve as cellular entry receptors for human DNA viruses (cytomegalovirus, Epstein-Barr virus, human herpes virus 6, herpes simplex viruses 1 and 2, and adenoviruses) that are either proven or suspected to be linked with malignancies. MicroRNA-34a is strictly dependent on its transcriptional activator tumor suppressor protein p53, and both p53 and microRNA-34a are frequently mutated or downregulated in various cancers. We hypothesize that p53 – microRNA-34a axis may alter susceptibility of cells to infection with some viruses that are detected in tumors and either proven or suspected to be associated with tumor initiation and progression.
INTRODUCTION
Herpesviruses are a large family of DNA viruses that can cause latent or lytic infections in animals and humans, notably – often in immunocompromised patients. Epstein-Barr virus (EBV) and Kaposi’s sarcoma-associated herpes virus (KSHV) are proven to play an important role in the development of various cancers (1, 2). Though the direct role of other human herpesviruses in oncogenesis has not been proven yet, their proteins and/or DNA sequences are frequently detected in tumors but not in adjacent normal tissues. Human cytomegalovirus (HCMV) has been found in tumor cells of patients with malignant brain tumors (gliomas), breast cancer, colon cancer, cervical cancer, prostate carcinoma, and EBV-negative Hodgkin’s lymphoma (3). The finding of HSV-1 and-2 DNA in thyroid tumors (4) may be linked to the antiapoptotic activity of herpes simplex viruses (HSV) 1 and 2 and their ability to downregulate human telomerase reverse transcriptase (5–7). Human herpes virus 6 (HHV-6) has been connected with hematological malignancies (8, 9). It had been suggested that herpesviruses could initiate subtle changes, the so-called oncomodulation (10), which was defined as the ability of viral proteins and non-coding RNAs to promote oncogenic processes without direct oncotransformation but through disturbances in various intracellular signaling pathways. However, despite growing experimental evidence (11), the roles of HCMV, HHV-6 and HSV-1 and -2 as active participants in the tumorigenic processes continue to be debated. Additionally, it is not clear why the above-mentioned viruses can be detected in many but not in all tumor specimens.
Here we present a hypothesis that highlights a potential link between the major tumor suppressor p53, its transcriptional target microRNA-34a, and susceptibility of cells to infection with viruses that are either proven or actively studied for their possible role in the initiation or progression of cancers.
BACKGROUND
HCMV and human malignancies
HCMV is currently one of the most actively studied prooncogenic/oncomodulatory infectious agents (12). Its DNA sequences and proteins are detected in more than 90% of human gliomas, brain tumors with high morbidity and mortality. Many HCMV activities contribute to established hallmarks of cancer such as proliferative signaling, evasion of growth suppressors and resistance to cell death, genomic instability, activation of invasion, metastasis, angiogenesis, avoidance of immune destruction, and inflammation. However, HCMV is not characterized by the sustained expression of oncogenes or genomic integration, the features that are attributed to known oncogenic viruses (13). Hence, some additional factors, for example, genetic polymorphisms that render susceptibility to the oncomodulatory effects of HCMV, are proposed to play a role in the possible contribution of HCMV to oncogenesis. It has been hypothesized that, analogously to HSV-1, some haplotypes of HCMV-encoded Fc-like receptors may vary in their ability to bind to antibodies due to steric variations. The low-affinity binding interferes with the effector functions of antibodies, thus allowing HCMV-infected cells to avoid or to mitigate antibody-dependent cellular cytotoxicity, complement-dependent neutralization, and phagocytosis (14).
Another hypothesis is based on the fact that PDGFRA is an obligatory cellular entry receptor for HCMV. Viral attachment to PDGFRA elicits a potent cellular interferon-like response, which, in turn, activates downstream growth-factor-like receptor tyrosine kinase and integrin signaling pathways (13). Phosphorylation of PDGFRA upon its binding with HCMV glycoprotein B, and HCMV-mediated activation of human epidermal growth factor receptor trigger downstream signaling molecules PI3K/Akt and focal adhesion kinase, which are the components of the pro-oncogenic signaling network. PDGFRA deletion or blocking by antibodies, or targeted inhibition of its kinase activity abrogates HCMV internalization and gene expression, as well as the above-mentioned signaling cascade (13, 15). PDGFRA is essential for neural development, self-renewal of neural stem/progenitor cells and gliomagenesis (16–19). The extent of PDGFRA signaling apparently corresponds to tumor malignancy: PDGFRA amplification is detected in about 13% of brain tumors (20), and PDGFRA overexpression can be observed in all phenotypes of glioblastoma (21, 22), though it is rather associated with secondary than with primary glioblastomas (23). The PDGFRA promoter region contains a total of 10 polymorphic sites that give rise to five distinct haplotypes. As these haplotypes are characterized by varying levels of PDGFRA promoter activity, the second hypothesis suggests that PDGFRA haplotype differences confer differential susceptibility to HCMV infection as well as predisposition to gliomagenesis or progression of gliomas in humans (13, 18).
Tumor suppressor protein p53 and viral replication
p53, a key tumor suppressor protein and a master transcriptional regulator, influences the expression of a variety of genes that are involved in cell cycle progression, cell growth, differentiation and death, cell motility and migration, cellular senescence, DNA repair, energy metabolism, cell–cell communications, angiogenesis and immune response (24, 25), in particular, innate antiviral immunity (26, 27)
During the process of evolution, viruses adopted different strategies to manipulate the host cells to ensure that all the steps in the viral life cycle are complete (28). As cellular stress responses and apoptosis are mediated in large part by p53, viruses utilize a variety of mechanisms aimed to inactivate p53 in order to prevent cell death and abortion of viral replication. Among them are interactions between p53 and viral proteins, p53 phosphorylation, ubiquitination of p53 by viral E3-ubiquitin ligases, prevention of p53 acetylation, downregulation of p53 by interferons, interaction with p53 regulatory proteins, inhibition of p53 dependent transcription, and activation of Hdm2 (29–33). p53 is frequently mutated in many cancers in the so-called hotspots of its DNA-binding domain. These gain-of-function mutants act oppositely to their wild-type counterpart not only by failing to transactivate its usual target genes but also by de-repressing or transactivating a plethora of oncogenes (29, 34). Human immunodeficiency virus-1 replication (35–37), and the host response to hepatitis C virus (38, 39), respiratory syncytial virus (40), influenza (41), vesicular stomatitis virus (42), and Rift Valley fever virus (43) were reported to vary depending on whether the cells harbored wild-type or mutant p53. Integration of adeno-associated viral vectors was higher in p53-negative cells as compared to normal ones (44). Loss of p53 confers enhanced susceptibility to reoviral and myxoma infectivity and replication (45). The role of p53 status appears to be even more essential for the replication of viruses that are implicated in tumorigenesis: human papillomaviruses (46–48), hepatitis B virus (49–52), gallid herpesvirus 2, which causes T-cell lymphomas in chickens (53), human adenoviruses (54, 55), and HSV-1 (56).
However, little is known about the mechanisms by which p53 abnormalities (either mutant or nonfunctional “negative” p53) influence the susceptibility of cells to viral infection.
Though some HCMV effects are reported to be independent of p53 status (57), it has been observed that the pre-existing genetic lesions, p53 mutations in particular, could explain the differential response of glioma cell lines to HCMV infection or overexpression of HCMV proteins (58–61). p53 is involved in regulation of HCMV replication (62, 63), and specifically, the onset of a lytic cycle (64). It influences expression of 22 HCMV genes (65), inhibits cell division and DNA synthesis upon overexpression of HCMV IE86 protein (61, 66) and regulates HCMV UL94 gene, which is activated during productive HCMV infection (67). It has been suggested that p53 mutations might represent one of the mechanisms by which HCMV contributes to the transformation of primary baby rat kidney cells in cooperation with the adenovirus E1A protein (3, 68). Wild-type p53 has been shown to inhibit HCMV major immediate-early promoter-enhancer as well as several other viral promoters, including the HSV-1 UL9 promoter, and the long terminal repeat promoters of Rous sarcoma virus, human immunodeficiency virus-1, and human T-cell lymphotropic virus type 1. Conversely, mutations at any of the five “hotspot” amino acid positions 143, 175, 248, 273, and 281 release the repression of viral promoters to a variable extent (69–71). Altogether, p53 appears to provide wide-ranging impacts on the virus-host interaction from regulating expression of viral genes to maintaining the balance between latency and lytic infection.
MicroRNA-34a
MicroRNAs are short noncoding RNAs that regulate gene expression by targeting the 3’-untranslated region of mRNAs and inducing mRNA degradation or inhibiting its translation (72). microRNA-34a is a part of the p53-network (73), and a potent tumor suppressor, which is frequently downregulated in various cancers (74–78). microRNA-34a has previously been shown to control the same groups of genes that are regulated by p53 (79). It is encoded within the chromosome region 1p36, whose loss is shown to be associated with gliomas, neuroblastomas, pancreatic cancer, and chronic myeloid leukemia (80). According to the database TargetScan, microRNA-34a has a total of 512 conserved sites. In turn, p53 is believed to regulate between 1500 and 3000 genes (25). Either the loss of microRNA-34a or mutations within the p53-DNA binding domain that render p53 unable to transactivate its usual targets (including microRNA-34a) may result in significant changes of expression profiles across the whole genome.
In silico analysis of microRNA-34a target sites
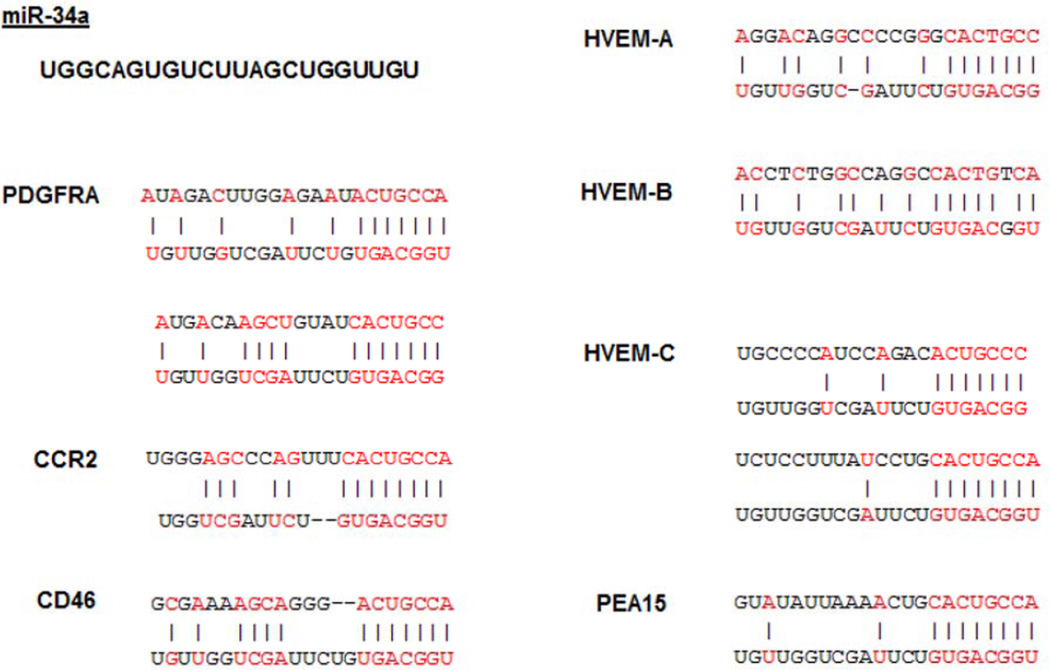
By analyzing the databases TargetScan and MicroCosm, we have found that microRNA-34a targets a group of genes with one common trait: they encode cellular entry receptors for human herpesviruses that are actively studied for their possible role in the initiation or progression of cancers. The target sites for microRNA-34a have been identified on the following genes: platelet-derived growth factor receptor-alpha (PDGFRA) – HCMV cellular entry receptor; complement component receptor 2 (CCR2) – EBV cellular entry receptor; herpes simplex virus entry mediators A, B, and C (HVEM-A, -B, -C); and CD46 protein, which is a cellular entry receptor for HHV-6 (Figure 1). The matching positions for microRNA-34a within 3’-UTR and the corresponding NCBI Reference Sequence numbers of the targeted mRNAs are shown in Table 1. It is noteworthy that analogously to herpesviruses human adenoviruses are also investigated for their possible contribution to oncogenic processes through oncomodulation. According to TargetScan and MicroCosm databases CXADR mRNA has no predicted target sites for microRNA-34a. However, microRNA-34a has a highly conserved target site on phosphoprotein enriched in astrocytes-15 (PEA15), which has been reported to upregulate the expression of coxsackievirus adenovirus receptor (CXADR) (81), the cellular entry receptor for all other serological groups of human adenoviruses. Of interest, is also the presence of a conserved miR-34a target site on CXADR pseudogene 2 (http://www.microrna.org).
Figure 1. Predicted microRNA-34a seed matches to viral entry receptors mRNAs.
Results of in silico analysis suggesting the presence of microRNA-34a (miR-34a) target sites on the genes encoding cellular entry receptors for the following viruses: platelet-derived growth factor receptor-alpha (PDGFRA) – cellular entry receptor for human cytomegalovirus; complement component receptor 2 (CCR2) – for Epstein-Barr virus; CD46 – for human herpes virus 6 and human adenoviruses group B; and herpes simplex virus entry mediators A, B, and C (HVEM-A, -B, -C). Phosphoprotein enriched in astrocytes-15 (PEA15) is a positive regulator for coxsackievirus adenovirus receptor, the cellular entry receptor for all other than group B serological groups of human adenoviruses.
Table 1.
MicroRNA-34a matching positions and NCBI reference numbers of the targeted mRNAs.
| Virus | Cellular receptor | Reference number |
Matching positions |
|---|---|---|---|
| HCMV | Platelet-derived growth factor receptor-alpha | NM_006206.4 | 6269–6278; 6292–6306 |
| EBV | Complement component receptor 2 | M26004.1 | 3364–3370 |
| HSV-1 and-2 | Herpes simplex virus entry mediator A | NM_003820.2 | 1531–1537 |
| HSV-1 and-2 | Herpes simplex virus entry mediator B | NG_029149.1 | 47320–47341 |
| HSV-1 and-2 | Herpes simplex virus entry mediator C | NM_002855 | 2585–2591; 3900–3910 |
| HHV-6, Human adenoviruses group B | CD46 | NM_172361.2 | 2463–2469 |
| Human adenoviruses | Coxsackievirus adenovirus receptor, upregulated by phosphoprotein enriched in astrocytes-15 (81) | NM_003768.3 | 1729–1736 |
HYPOTHESIS
We propose that tumor suppressor protein p53 regulates cellular entry receptors for HCMV as well as EBV, HSV-1 and -2, HHV-6 and adenoviruses. The effect of p53 on cellular entry receptors is mediated by its transcriptional target microRNA-34a. Thus, the status of the p53 – microRNA-34a axis may be considered as a pre-existing host condition that influences cell susceptibility to viral infections and accounts for the variable presence of HCMV and other herpesviruses in human tumors.
EVALUATION AND IMPLICATIONS
It is interesting to mention that HCMV immediate early 1 protein has been detected in nearly all glioblastomas and 82% of low-grade (less malignant) gliomas, while HCMV DNA could be identified in about 94% of clinical glioma specimens (13). These numbers are close to the frequency of altered p53 signaling in brain tumors – about 87% (19). As recently has been reported, repression of microRNA-34a upregulates PDGFRA (82). Overexpression of PDGFRA due to p53 or microRNA-34a abnormalities may increase the probability of HCMV infection of the tumor cells, as well as the potential cells of tumor origin. Furthermore, p53 inactivation or p53 mutations may significantly enhance HCMV replication due to de-repression of its promoter and thus create favorable conditions for HCMV to unfold its pro-oncogenic potential, which may further aggravate the already formed malignant glioma phenotype (11).
The microRNA-34a promoter has a bona fide binding site for wild-type p53, and microRNA-34a is recognized as a p53 downstream effector (73, 79, 83–85). In some cases, however, microRNA-34a levels can be elevated without functional p53. Inactivation of p53 by siRNAs was shown to lower basal levels of microRNA-34a transcript, but did not block microRNA-34a upregulation in response to oncogene-induced senescence (86). In p53-null K562 cells phorbol-esters could transactivate an alternative microRNA-34a promoter, which was located about 20 kb upstream of the classical microRNA-34a transcription start site, and produced a longer pri-microRNA-34a transcript (80). Recently it has been reported that CCAAT enhancer binding protein alpha (C/EBPα) and nuclear factor-kappa B (NF-κB) also bind to their appropriate sites and activate microRNA-34a, the NF-κB-mediated effect on microRNA-34a being p53-dependent (87, 88). Still, the overwhelming number of reports evidences microRNA-34a as a part of p53 network, thus allowing us to consider these two important tumor suppressor factors as the “p53-microRNA-34a axis.
Despite the in silico prediction data, the abilities of microRNAs to downregulate their transcriptional targets may vary significantly between tissues and depend on many factors including the accessibility of target sites, which is influenced by the complexity of RNA secondary structure and protein binding, on the levels of transcripts and other factors. (89). Contrary to the case with PDGFRA, the only route for HCMV, other herpesviruses may utilize alternative receptors that are not identified yet and are not controlled by microRNA-34a. Finally, expression of viral cellular entry receptors may vary in infected and non-infected cells, and even be downregulated upon their binding with viral proteins (90), probably in order to prevent superinfection followed by cell death. Notwithstanding the complexity of the issue, it should be acknowledged that the relatively slow evolution of DNA viruses and their long-term co-divergence with human hosts resulted in the development of mechanisms for controlling viral infectivity, replication and latency in host cells (91). It is recognized that though the known oncogenic viruses or other infectious agents are widely present in humans, only a small fraction of infected individuals develop cancer, apparently because of yet unidentified additional risk factors.
The ability of the viruses to enter non-infected cells in the first place is defined by the expression levels of their cellular entry receptors, which, in turn, may depend on the variations of the host’s genetic backgrounds. We suggest that p53 and microRNA-34a are important biomarkers that reflect genetic predisposition to HCMV infection and influence replication of HCMV and probably other human herpesviruses, such as EBV, HHV-6, HSV-1 and -2, and adenoviruses. Remarkably, all these viruses have been reported to have pro-oncogenic properties, although to a variable extent. Our speculations bring in another possible explanation of the inconstant detection of human herpesviruses, and specifically HCMV, in various tumors. Impaired p53 status, deletion of microRNA-34a, C/EBPα mutations, mutations within p53-binding sites, or aberrant CpG methylation in the microRNA-34a promoter (87, 88, 92) could probably expedite viral entry via upregulation of the appropriate cellular entry receptors. Consequently, herpesviruses, which cause lytic or latent infections, may further contribute on a circumstantial basis to oncotransformation, formation of malignant phenotypes and tumor progression. The relevance of our hypothesis needs to be validated on the appropriate cell models and clinical tumor samples.
In the case of HCMV and its debatable role in the development of malignant brain tumors it has been stressed that studies addressing possible environmental and/or genetic factors, which increase the risk and elucidate the mechanisms underlying the input of HCMV into glioma pathology, need to be conducted (13). The present hypothesis may stimulate further studies in order to address the questions of whether the p53-microRNA-34a status may account for the development of glioma in only a small percentage of the population with latent HCMV, and if matching between miR-34a and several viral receptors simply reflects an evolutionary juxtaposition of herpesviruses or substantially diminishes their pro-oncogenic potential.
Acknowledgments
Funding
This work was supported by the National Institutes of Health (R01 NS045209 and R01 CA134843 awarded to Roger Abounader, and R01 AI041644 669 awarded to Jay C. Brown).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTERESTS
None of the authors of the above manuscript has declared any conflict of interest within the last three years which may arise from being named as an author on the manuscript.
REFERENCES
- 1.Michelow P, Wright C, Pantanowitz L. A review of the cytomorphology of Epstein-Barr virus-associated malignancies. Acta cytologica. 2012;56:1–14. doi: 10.1159/000334235. [DOI] [PubMed] [Google Scholar]
- 2.Sakakibara S, Tosato G. Regulation of angiogenesis in malignancies associated with Epstein-Barr virus and Kaposi's sarcoma-associated herpes virus. Future Microbiol. 2009;4:903–917. doi: 10.2217/fmb.09.49. [DOI] [PubMed] [Google Scholar]
- 3.Michaelis M, Doerr HW, Cinatl J. The story of human cytomegalovirus and cancer: increasing evidence and open questions. Neoplasia. 2009;11:1–9. doi: 10.1593/neo.81178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jensen K, Patel A, Larin A, et al. Human herpes simplex viruses in benign and malignant thyroid tumours. The Journal of pathology. 2010;221:193–200. doi: 10.1002/path.2701. [DOI] [PubMed] [Google Scholar]
- 5.Perng GC, Jones C, Ciacci-Zanella J, et al. Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript. Science. 2000;287:1500–1503. doi: 10.1126/science.287.5457.1500. [DOI] [PubMed] [Google Scholar]
- 6.Thompson RL, Sawtell NM. HSV latency-associated transcript and neuronal apoptosis. Science. 2000;289:1651. [PubMed] [Google Scholar]
- 7.Yang CT, Song J, Bu X, et al. Herpes simplex virus type-1 infection upregulates cellular promoters and telomerase activity in both tumor and nontumor human cells. Gene Ther. 2003;10:1494–1502. doi: 10.1038/sj.gt.3302005. [DOI] [PubMed] [Google Scholar]
- 8.Ablashi DV, Devin CL, Yoshikawa T, et al. Review Part 3: Human herpesvirus-6 in multiple non-neurological diseases. J Med Virol. 2010;82:1903–1910. doi: 10.1002/jmv.21860. [DOI] [PubMed] [Google Scholar]
- 9.Doniger J, Muralidhar S, Rosenthal LJ. Human cytomegalovirus and human herpesvirus 6 genes that transform and transactivate. Clin Microbiol Rev. 1999;12:367–382. doi: 10.1128/cmr.12.3.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cinatl J, Jr, Cinatl J, Vogel JU, Rabenau H, Kornhuber B, Doerr HW. Modulatory effects of human cytomegalovirus infection on malignant properties of cancer cells. Intervirology. 1996;39:259–269. doi: 10.1159/000150527. [DOI] [PubMed] [Google Scholar]
- 11.Kofman A, Marcinkiewicz L, Dupart E, et al. The roles of viruses in brain tumor initiation and oncomodulation. Journal of neuro-oncology. 2011;105:451–466. doi: 10.1007/s11060-011-0658-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soroceanu L, Cobbs CS. Is HCMV a tumor promoter? Virus Res. 2011;157:193–203. doi: 10.1016/j.virusres.2010.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dziurzynski K, Chang SM, Heimberger AB, et al. Consensus on the role of human cytomegalovirus in glioblastoma. Neuro-oncology. 2012;14:246–255. doi: 10.1093/neuonc/nor227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pandey JP. Genetic and viral etiology of glioblastoma--a unifying hypothesis. Cancer Epidemiol Biomarkers Prev. 2011;20:1061–1063. doi: 10.1158/1055-9965.EPI-11-0247. [DOI] [PubMed] [Google Scholar]
- 15.Soroceanu L, Akhavan A, Cobbs CS. Platelet-derived growth factor-alpha receptor activation is required for human cytomegalovirus infection. Nature. 2008;455:391–395. doi: 10.1038/nature07209. [DOI] [PubMed] [Google Scholar]
- 16.Assanah M, Lochhead R, Ogden A, Bruce J, Goldman J, Canoll P. Glial progenitors in adult white matter are driven to form malignant gliomas by platelet-derived growth factor-expressing retroviruses. J Neurosci. 2006;26:6781–6790. doi: 10.1523/JNEUROSCI.0514-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erlandsson A, Brannvall K, Gustafsdottir S, Westermark B, Forsberg-Nilsson K. Autocrine/paracrine platelet-derived growth factor regulates proliferation of neural progenitor cells. Cancer Res. 2006;66:8042–8048. doi: 10.1158/0008-5472.CAN-06-0900. [DOI] [PubMed] [Google Scholar]
- 18.Toepoel M, Joosten PH, Knobbe CB, et al. Haplotype-specific expression of the human PDGFRA gene correlates with the risk of glioblastomas. Int J Cancer. 2008;123:322–329. doi: 10.1002/ijc.23432. [DOI] [PubMed] [Google Scholar]
- 19.Thorarinsdottir HK, Santi M, McCarter R, et al. Protein expression of platelet-derived growth factor receptor correlates with malignant histology and PTEN with survival in childhood gliomas. Clin Cancer Res. 2008;14:3386–3394. doi: 10.1158/1078-0432.CCR-07-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanai N. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma. World Neurosurg. 2010;74:4–5. doi: 10.1016/j.wneu.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 22.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu Y, Parada LF. The molecular and genetic basis of neurological tumours. Nat Rev Cancer. 2002;2:616–626. doi: 10.1038/nrc866. [DOI] [PubMed] [Google Scholar]
- 24.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 25.Menendez D, Inga A, Resnick MA. The expanding universe of p53 targets. Nat Rev Cancer. 2009;9:724–737. doi: 10.1038/nrc2730. [DOI] [PubMed] [Google Scholar]
- 26.Rivas C, Aaronson SA, Munoz-Fontela C. Dual Role of p53 in Innate Antiviral Immunity. Viruses. 2010;2:298–313. doi: 10.3390/v2010298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sato Y, Tsurumi T. Genome guardian p53 and viral infections. Rev Med Virol. 2012 doi: 10.1002/rmv.1738. [DOI] [PubMed] [Google Scholar]
- 28.Lazo PA, Santos CR. Interference with p53 functions in human viral infections, a target for novel antiviral strategies? Rev Med Virol. 2011 doi: 10.1002/rmv.696. [DOI] [PubMed] [Google Scholar]
- 29.Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009;9:701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- 30.McCormick F, Wood DA. The role of p53 in virally associated tumors. Trends Microbiol. 1997;5:181–182. doi: 10.1016/S0966-842X(97)85013-5. [DOI] [PubMed] [Google Scholar]
- 31.Muralidhar S, Doniger J, Mendelson E, et al. Human cytomegalovirus mtrII oncoprotein binds to p53 and down-regulates p53-activated transcription. J Virol. 1996;70:8691–8700. doi: 10.1128/jvi.70.12.8691-8700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neil JC, Cameron ER, Baxter EW. p53 and tumour viruses: catching the guardian off-guard. Trends Microbiol. 1997;5:115–120. doi: 10.1016/S0966-842X(96)10083-4. [DOI] [PubMed] [Google Scholar]
- 33.Wong M, Gruber J. Viral interactions with the p53 gene in human cancer: NCI workshop. J Natl Cancer Inst. 1994;86:177–182. doi: 10.1093/jnci/86.3.177. [DOI] [PubMed] [Google Scholar]
- 34.Weisz L, Oren M, Rotter V. Transcription regulation by mutant p53. Oncogene. 2007;26:2202–2211. doi: 10.1038/sj.onc.1210294. [DOI] [PubMed] [Google Scholar]
- 35.Sawaya BE, Khalili K, Mercer WE, Denisova L, Amini S. Cooperative actions of HIV-1 Vpr and p53 modulate viral gene transcription. J Biol Chem. 1998;273:20052–20057. doi: 10.1074/jbc.273.32.20052. [DOI] [PubMed] [Google Scholar]
- 36.Duan L, Ozaki I, Oakes JW, Taylor JP, Khalili K, Pomerantz RJ. The tumor suppressor protein p53 strongly alters human immunodeficiency virus type 1 replication. J Virol. 1994;68:4302–4313. doi: 10.1128/jvi.68.7.4302-4313.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mukerjee R, Claudio PP, Chang JR, Del Valle L, Sawaya BE. Transcriptional regulation of HIV-1 gene expression by p53. Cell Cycle. 2010;9:4569–4578. doi: 10.4161/cc.9.22.13836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamdi N, El-Akel W, El-Serafy M, Esmat G, Sarrazin C, Abdelaziz AI. Transcriptional response of MxA, PKR and SOCS3 to interferon-based therapy in HCV genotype 4-infected patients and contribution of p53 to host antiviral response. Intervirology. 2012;55:210–218. doi: 10.1159/000327783. [DOI] [PubMed] [Google Scholar]
- 39.Dharel N, Kato N, Muroyama R, et al. Potential contribution of tumor suppressor p53 in the host defense against hepatitis C virus. Hepatology. 2008;47:1136–1149. doi: 10.1002/hep.22176. [DOI] [PubMed] [Google Scholar]
- 40.Bian T, Gibbs JD, Orvell C, Imani F. Respiratory Syncytial Virus Matrix Protein Induces Lung Epithelial Cell Cycle Arrest through a p53 Dependent Pathway. PLoS One. 2012;7:e38052. doi: 10.1371/journal.pone.0038052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munoz-Fontela C, Pazos M, Delgado I, et al. p53 serves as a host antiviral factor that enhances innate and adaptive immune responses to influenza A virus. J Immunol. 2011;187:6428–6436. doi: 10.4049/jimmunol.1101459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munoz-Fontela C, Garcia MA, Garcia-Cao I, et al. Resistance to viral infection of super p53 mice. Oncogene. 2005;24:3059–3062. doi: 10.1038/sj.onc.1208477. [DOI] [PubMed] [Google Scholar]
- 43.Austin D, Baer A, Lundberg L, et al. p53 Activation following Rift Valley Fever Virus Infection Contributes to Cell Death and Viral Production. PLoS One. 2012;7:e36327. doi: 10.1371/journal.pone.0036327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zacharias J, Romanova LG, Menk J, Philpott NJ. p53 inhibits adeno-associated viral vector integration. Hum Gene Ther. 2011;22:1445–1451. doi: 10.1089/hum.2011.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim M, Williamson CT, Prudhomme J, et al. The viral tropism of two distinct oncolytic viruses, reovirus and myxoma virus, is modulated by cellular tumor suppressor gene status. Oncogene. 2010;29:3990–3996. doi: 10.1038/onc.2010.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown C, Kowalczyk AM, Taylor ER, Morgan IM, Gaston K. P53 represses human papillomavirus type 16 DNA replication via the viral E2 protein. Virol J. 2008;5:5. doi: 10.1186/1743-422X-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nguyen ML, Kraft RM, Aubert M, Goodwin E, DiMaio D, Blaho JA. p53 and hTERT determine sensitivity to viral apoptosis. J Virol. 2007;81:12985–12995. doi: 10.1128/JVI.01485-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Padlewska K, Ramoz N, Cassonnet P, et al. Mutation and abnormal expression of the p53 gene in the viral skin carcinogenesis of epidermodysplasia verruciformis. J Invest Dermatol. 2001;117:935–942. doi: 10.1046/j.0022-202x.2001.01515.x. [DOI] [PubMed] [Google Scholar]
- 49.Kubicka S, Kuhnel F, Zender L, et al. p53 represses CAAT enhancer-binding protein (C/EBP)-dependent transcription of the albumin gene. A molecular mechanism involved in viral liver infection with implications for hepatocarcinogenesis. J Biol Chem. 1999;274:32137–32144. doi: 10.1074/jbc.274.45.32137. [DOI] [PubMed] [Google Scholar]
- 50.Lee H, Kim HT, Yun Y. Liver-specific enhancer II is the target for the p53-mediated inhibition of hepatitis B viral gene expression. J Biol Chem. 1998;273:19786–19791. doi: 10.1074/jbc.273.31.19786. [DOI] [PubMed] [Google Scholar]
- 51.Kirby GM, Batist G, Fotouhi-Ardakani N, et al. Allele-specific PCR analysis of p53 codon 249 AGT transversion in liver tissues from patients with viral hepatitis. Int J Cancer. 1996;68:21–25. doi: 10.1002/(SICI)1097-0215(19960927)68:1<21::AID-IJC4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 52.Hsu IC, Tokiwa T, Bennett W, et al. p53 gene mutation and integrated hepatitis B viral DNA sequences in human liver cancer cell lines. Carcinogenesis. 1993;14:987–992. doi: 10.1093/carcin/14.5.987. [DOI] [PubMed] [Google Scholar]
- 53.Stik G, Laurent S, Coupeau D, et al. A p53-dependent promoter associated with polymorphic tandem repeats controls the expression of a viral transcript encoding clustered microRNAs. Rna. 2010;16:2263–2276. doi: 10.1261/rna.2121210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang H, Zheng Z, Zhao LY, Li Q, Liao D. Downregulation of Mdm2 and Mdm4 enhances viral gene expression during adenovirus infection. Cell Cycle. 2012;11:582–593. doi: 10.4161/cc.11.3.19052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Royds JA, Hibma M, Dix BR, et al. p53 promotes adenoviral replication and increases late viral gene expression. Oncogene. 2006;25:1509–1520. doi: 10.1038/sj.onc.1209185. [DOI] [PubMed] [Google Scholar]
- 56.Wilcock D, Lane DP. Localization of p53, retinoblastoma and host replication proteins at sites of viral replication in herpes-infected cells. Nature. 1991;349:429–431. doi: 10.1038/349429a0. [DOI] [PubMed] [Google Scholar]
- 57.Cinatl J, Scholz M, Kotchetkov R, Vogel JU, Doerr HW. Molecular mechanisms of the modulatory effects of HCMV infection in tumor cell biology. Trends Mol Med. 2004;10:19–23. doi: 10.1016/j.molmed.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 58.Cobbs CS, Soroceanu L, Denham S, Zhang W, Kraus MH. Modulation of oncogenic phenotype in human glioma cells by cytomegalovirus IE1-mediated mitogenicity. Cancer Res. 2008;68:724–730. doi: 10.1158/0008-5472.CAN-07-2291. [DOI] [PubMed] [Google Scholar]
- 59.Kalejta RF, Bechtel JT, Shenk T. Human cytomegalovirus pp71 stimulates cell cycle progression by inducing the proteasome-dependent degradation of the retinoblastoma family of tumor suppressors. Molecular and cellular biology. 2003;23:1885–1895. doi: 10.1128/MCB.23.6.1885-1895.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanchez V, Spector DH. Subversion of cell cycle regulatory pathways. Curr Top Microbiol Immunol. 2008;325:243–262. doi: 10.1007/978-3-540-77349-8_14. [DOI] [PubMed] [Google Scholar]
- 61.Stinski MF, Petrik DT. Functional roles of the human cytomegalovirus essential IE86 protein. Curr Top Microbiol Immunol. 2008;325:133–152. doi: 10.1007/978-3-540-77349-8_8. [DOI] [PubMed] [Google Scholar]
- 62.Casavant NC, Luo MH, Rosenke K, Winegardner T, Zurawska A, Fortunato EA. Potential role for p53 in the permissive life cycle of human cytomegalovirus. J Virol. 2006;80:8390–8401. doi: 10.1128/JVI.00505-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luo MH, Fortunato EA. Long-term infection and shedding of human cytomegalovirus in T98G glioblastoma cells. J Virol. 2007;81:10424–10436. doi: 10.1128/JVI.00866-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zydek M, Hagemeier C, Wiebusch L. Cyclin-dependent kinase activity controls the onset of the HCMV lytic cycle. PLoS Pathog. 2010;6:e1001096. doi: 10.1371/journal.ppat.1001096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hannemann H, Rosenke K, O'Dowd JM, Fortunato EA. The presence of p53 influences the expression of multiple human cytomegalovirus genes at early times postinfection. J Virol. 2009;83:4316–4325. doi: 10.1128/JVI.02075-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Song YJ, Stinski MF. Inhibition of cell division by the human cytomegalovirus IE86 protein: role of the p53 pathway or cyclin-dependent kinase 1/cyclin B1. J Virol. 2005;79:2597–2603. doi: 10.1128/JVI.79.4.2597-2603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wing BA, Johnson RA, Huang ES. Identification of positive and negative regulatory regions involved in regulating expression of the human cytomegalovirus UL94 late promoter: role of IE2-86 and cellular p53 in mediating negative regulatory function. J Virol. 1998;72:1814–1825. doi: 10.1128/jvi.72.3.1814-1825.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shen Y, Zhu H, Shenk T. Human cytomagalovirus IE1 and IE2 proteins are mutagenic and mediate "hit-and-run" oncogenic transformation in cooperation with the adenovirus E1A proteins. Proc Natl Acad Sci U S A. 1997;94:3341–3345. doi: 10.1073/pnas.94.7.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Deb D, Lanyi A, Scian M, et al. Differential modulation of cellular and viral promoters by p73 and p53. Int J Oncol. 2001;18:401–409. [PubMed] [Google Scholar]
- 70.Deb S, Jackson CT, Subler MA, Martin DW. Modulation of cellular and viral promoters by mutant human p53 proteins found in tumor cells. J Virol. 1992;66:6164–6170. doi: 10.1128/jvi.66.10.6164-6170.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Subler MA, Martin DW, Deb S. Inhibition of viral and cellular promoters by human wild-type p53. J Virol. 1992;66:4757–4762. doi: 10.1128/jvi.66.8.4757-4762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Godlewski J, Newton HB, Chiocca EA, Lawler SE. MicroRNAs and glioblastoma; the stem cell connection. Cell Death Differ. 2010;17:221–228. doi: 10.1038/cdd.2009.71. [DOI] [PubMed] [Google Scholar]
- 73.He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen HC, Chen GH, Chen YH, et al. MicroRNA deregulation and pathway alterations in nasopharyngeal carcinoma. Br J Cancer. 2009;100:1002–1011. doi: 10.1038/sj.bjc.6604948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Corney DC, Hwang CI, Matoso A, et al. Frequent downregulation of miR-34 family in human ovarian cancers. Clin Cancer Res. 2010;16:1119–1128. doi: 10.1158/1078-0432.CCR-09-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kumar B, Yadav A, Lang J, Teknos TN, Kumar P. Dysregulation of MicroRNA-34a Expression in Head and Neck Squamous Cell Carcinoma Promotes Tumor Growth and Tumor Angiogenesis. PLoS One. 2012;7:e37601. doi: 10.1371/journal.pone.0037601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guessous F, Zhang Y, Kofman A, et al. microRNA-34a is tumor suppressive in brain tumors and glioma stem cells. Cell Cycle. 2010;9 doi: 10.4161/cc.9.6.10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li Y, Guessous F, Zhang Y, et al. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 2009;69:7569–7576. doi: 10.1158/0008-5472.CAN-09-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chang TC, Wentzel EA, Kent OA, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Navarro F, Gutman D, Meire E, et al. miR-34a contributes to megakaryocytic differentiation of K562 cells independently of p53. Blood. 2009;114:2181–2192. doi: 10.1182/blood-2009-02-205062. [DOI] [PubMed] [Google Scholar]
- 81.Botta G, Perruolo G, Libertini S, et al. PED/PEA-15 modulates coxsackievirus-adenovirus receptor expression and adenoviral infectivity via ERK-mediated signals in glioma cells. Hum Gene Ther. 2010;21:1067–1076. doi: 10.1089/hum.2009.181. [DOI] [PubMed] [Google Scholar]
- 82.Silber J, Jacobsen A, Ozawa T, et al. miR-34a repression in proneural malignant gliomas upregulates expression of its target PDGFRA and promotes tumorigenesis. PLoS One. 2012;7:e33844. doi: 10.1371/journal.pone.0033844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bommer GT, Gerin I, Feng Y, et al. p53-Mediated Activation of miRNA34 Candidate Tumor-Suppressor Genes. Curr Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 84.Raver-Shapira N, Marciano E, Meiri E, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 85.Tarasov V, Jung P, Verdoodt B, et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 86.Christoffersen NR, Shalgi R, Frankel LB, et al. p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differ. 2010;17:236–245. doi: 10.1038/cdd.2009.109. [DOI] [PubMed] [Google Scholar]
- 87.Li J, Wang K, Chen X, et al. Transcriptional activation of microRNA-34a by NF-kappa B in human esophageal cancer cells. BMC Mol Biol. 2012;13:4. doi: 10.1186/1471-2199-13-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pulikkan JA, Peramangalam PS, Dengler V, et al. C/EBPalpha regulated microRNA-34a targets E2F3 during granulopoiesis and is down-regulated in AML with CEBPA mutations. Blood. 2010;116:5638–5649. doi: 10.1182/blood-2010-04-281600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kuhn DE, Martin MM, Feldman DS, Terry AV, Jr, Nuovo GJ, Elton TS. Experimental validation of miRNA targets. Methods. 2008;44:47–54. doi: 10.1016/j.ymeth.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gredmark S, Straat K, Homman-Loudiyi M, Kannisto K, Soderberg-Naucler C. Human cytomegalovirus downregulates expression of receptors for platelet-derived growth factor by smooth muscle cells. J Virol. 2007;81:5112–5120. doi: 10.1128/JVI.02197-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Holmes EC. Evolutionary history and phylogeography of human viruses. Annual review of microbiology. 2008;62:307–328. doi: 10.1146/annurev.micro.62.081307.162912. [DOI] [PubMed] [Google Scholar]
- 92.Feinberg-Gorenshtein G, Avigad S, Jeison M, et al. Reduced levels of miR-34a in neuroblastoma are not caused by mutations in the TP53 binding site. Genes Chromosomes Cancer. 2009;48:539–543. doi: 10.1002/gcc.20662. [DOI] [PubMed] [Google Scholar]