Abstract
Hepatitis C virus interacts extensively with host factors not only to establish productive infection but also to trigger unique pathological processes. Our recent genome-wide siRNA screen demonstrated that IKKα is a critical host factor for HCV. Here we describe a novel NF-κB-independent and kinase-mediated nuclear function of IKKα in HCV assembly. HCV infection, through its 3’-untranslated region, interacts with DDX3X to activate IKKα, which translocates to the nucleus and induces a CBP/p300-mediated transcriptional program involving SREBPs. This novel innate pathway induces lipogenic genes and enhances core-associated lipid droplet formation to facilitate viral assembly. Chemical inhibitors of IKKα suppress HCV infection and IKKα-induced lipogenesis, offering a proof-of-concept approach for novel HCV therapeutic development. Our results show that HCV commands a novel mechanism to its advantage by exploiting intrinsic innate response and hijacking lipid metabolism, which likely contributes to a high chronicity rate and the pathological hallmark of steatosis in HCV infection.
Hepatitis C virus (HCV) infection is a leading cause of chronic liver disease associated with significant morbidity and mortality in the world1. To date a protective vaccine is not available, and current therapeutic regimen is suboptimal2. The virus has a unique propensity to cause persistent infection and induce progressive liver damage3. HCV exploits extensively host factors such as cellular lipid metabolic pathways for efficient propagation4,5. HCV has been shown to alter lipid metabolism of infected hepatocytes6, conferring a unique pathological feature of HCV infection – hepatic steatosis. Activation of sterol regulatory element-binding proteins (SREBPs), critical transcriptional regulators of cholesterol and fatty acid metabolism7, has been shown in HCV-infected hepatocytes8,9. The mechanistic basis of these functional effects remains unclear.
HCV activates host innate immunity that functions to limit viral infection. Recognition of viral pathogen-associated molecular patterns (PAMPs) by pattern recognition receptor (PRR) like RIG-I-like receptors (RLRs) and activation of various signaling pathways including IRF3 and NF-κB represent early steps of intrinsic innate immune response, with subsequent induction of interferons10. The NF-κB pathway is tightly regulated by the IκB kinase (IKK) complex, which consists of two catalytic subunits, IKKα and IKKβ, and a regulatory subunit NEMO (IKKγ)11. Activation of IKKβ and NEMO and subsequent IκB degradation are critical steps in the activation of the canonical NF-κB pathway12. IKKα preferentially phosphorylates NF-κB2 rather than IκB and leads to the activation of p52-RelB heterodimers that regulate a distinct subset of NF-κB target genes12. This alternative action is referred to as the non-canonical pathway13. IKKα is a remarkably versatile molecule involved in diverse and multiple signaling pathways to regulate gene expression; many of its actions are independent of NF-κB12,14. Unlike IKKβ, IKKα can shuttle between the cytoplasm and nucleus15,16. In the nucleus, IKKα interacts with CREB binding protein (CBP) and contributes to NF-κB-mediated gene expression through phosphorylation of histone H315,17,18. The transcription targets of IKKα, however, remain largely unknown.
We recently performed a genome-wide RNA interference (RNAi) screen to discover HCV host dependencies. One of the highly host-dependent factors identified is IKKα (under the name CHUK)19. Here we show that IKKα is requisite for productive HCV infection and upregulates lipogenesis of host cells for efficient viral assembly via transcriptional induction of SREBPs. HCV infection, through the action of 3’-untranslated region (3’UTR), interacts with DDX3X to activate IKKα and hence induces LD biogenesis. Our study provides a direct functional link between HCV infection, inflammation, innate immunity and hepatic lipid metabolism.
RESULTS
Identification of IKKα as a Novel Host Factor Required for HCV Infection
We applied a two-part viral infection protocol to characterize host dependencies associated with both early (part-one) and late (part-two) stages of HCV life cycle19. The effect of IKKα silencing was more pronounced in part-two (>85% inhibition) than part-one (~60% inhibition), implicating that IKKα acts more on the late stage of viral infection (Fig. 1a and Supplementary Fig. 1a). The effect of IKKα depletion was confirmed by testing four individual siRNAs of the pool (Fig. 1b and Supplementary Fig. 1b). Expression of a siRNA-resistant IKKα mutant (HA-IKKα MUT) restored HCV infection in IKKα siRNA-treated cells (Supplementary Fig. 1c), further validating the phenotype-specific role of IKKα in HCV infection.
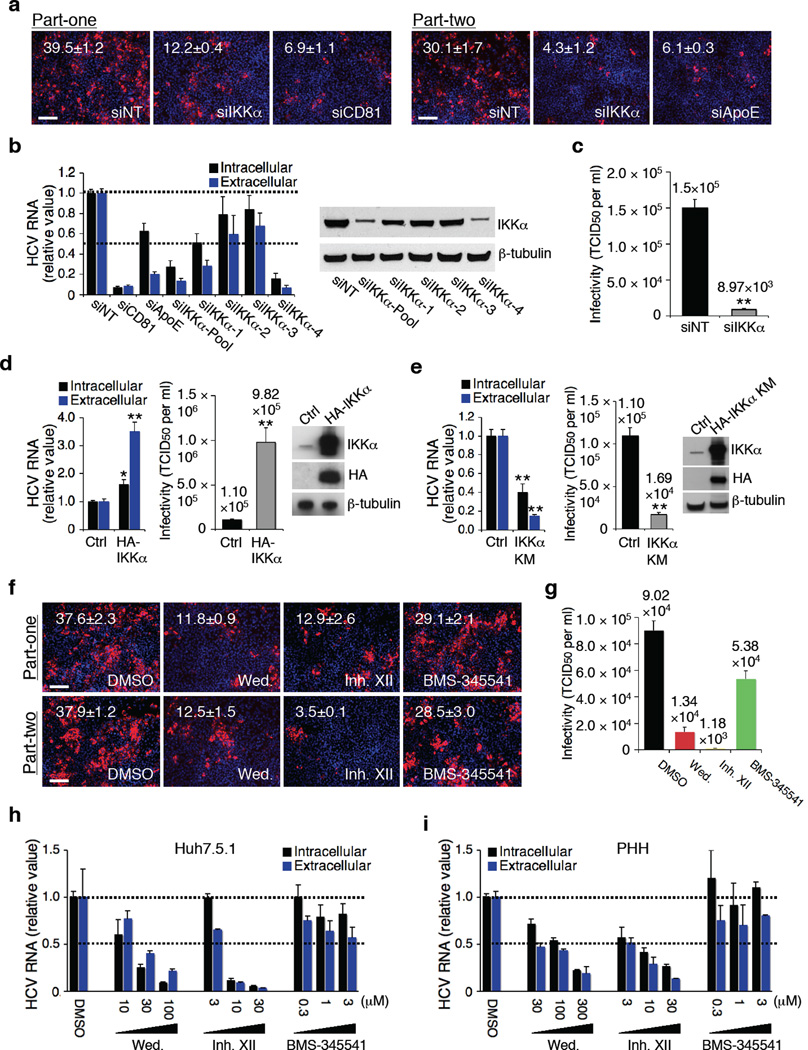
Figure 1.
Role IKKα in HCV infection. (a) Image illustration and quantitative analyses of HCV core staining part-one and part-two. Red: HCV core, blue: cell nuclei. Magnification 20 ×. (b) Efficacies of various IKKα siRNAs in silencing IKKα and restraining HCV RNA production. Values were normalized as relative to nontargeting siRNA (siNT) control. (c) Effect of IKKα depletion on infectious HCV production and secretion, assessed by limiting dilution assay. (d) Effect of over-expression of IKKα on HCV infection. (e) Effect of over-expression of the kinase-defective HA-IKKα KM on HCV infection. (f,g) Effects of wedelolactone (30 µM) and IKK inhibitor XII (10 µM) on HCV production (f) and viral infectivity (g). (h,i) Dose-response effects of wedelolactone and IKK inhibitor XII on HCV RNA production and secretion in Huh7.5.1 cells (h) and PHHs (i). Error bars represent ± s.d. of triplicate experiments. (a,f) Scale bars represent 100 µm.
IKKα siRNA significantly impaired production and secretion of infectious HCV by more than ten folds (Fig. 1c). Over-expression of IKKα by transfecting HA-IKKα plasmid substantially increased infectious HCV production (Fig. 1d). IKKα’s function requires its kinase activity and IKKα kinase-defective mutant (IKKα KM) behaves like a dominant negative mutant20. Transfection of IKKα KM blocked HCV propagation similar to the effect of IKKα siRNA (Fig. 1e).
To further investigate the role of IKKα in HCV infection, various IKK inhibitors were tested in HCV-infected Huh7.5.1 cells and primary human hepatocytes (PHHs) (Fig. 1f–i and Supplementary Fig. 2). Treatment of Huh7.5.1 cells with wedelolactone and IKK inhibitor XII, chemical inhibitors of both IKKα and β, drastically reduced HCV core protein staining and infectious viral particle production (Fig. 1f, g). Increasing concentrations of both compounds led to a dose-dependent decline in HCV RNA production and secretion in both Huh7.5.1 cells and PHHs that could not be accounted by cytotoxicity at high concentrations (Fig. 1h, i, and Supplementary Fig. 2b, c). BMS-345541, an IKKβ-specific inhibitor, had little effect on HCV production (Fig. 1f–i and Supplementary Fig. 2b, c).
IKKα’s Role in HCV Assembly and Lipid Droplet Formation
To investigate the step of HCV life cycle where IKKα is required, we applied multiple virologic assays. IKKα silencing preferentially affected extracellular HCV RNA levels in HCVcc infection system (Fig. 1b). We therefore specifically examined single-cycle replication by transfecting genomic HCV RNA into CD81-deficient Huh7 cells (Huh7.25)21 and showed that HCV replication was not affected by IKKα silencing (Fig. 2a). IKKα silencing had no effect on assays targeting individual steps of HCV life cycle including entry (HCV pseudovirus assay), translation (HCV IRES-driven reporter), and replication (subgenomic replicon) (Supplementary Fig. 3 and Fig. 2b), consistent with a predominant role of IKKα in the late stage of viral life cycle.
Figure 2.
IKKα function in HCV assembly and HCV-induced LD formation. (a) Effects of various siRNAs on HCV JFH-1/P7-Luc RNA replication in CD81-deficient Huh7.25 cells. (b) Effects of various siRNAs on HCV subgenomic replicon assay. (a,b) Values were normalized as relative units to siNT control, and error bars represent ± s.d. of quintuplicate experiments. (c) LD contents (BODIPY) and HCV core expression in Huh7.5.1 cells treated with siNT or siIKKα. LD number: mean of >150 cells ± s.d. Percent of LD-positive area: mean of >150 cells ± s.d. LD mean fluorescence intensity: mean of >300 cells ± s.d. (d) Huh7.5.1 cells were transfected with control, HA-IKKα WT or HA-IKKα KM plasmid and then stained for HA-tagged IKKα expression and LD contents. LD numbers, positive area and mean fluorescence intensity were quantified: mean of >30 cells per condition ± s.d. (e) Effect of IKKα silencing on HCV 3’UTR-mediated elevation of LD contents in Huh7.5.1 cells. For all microscopic images, scale bars represent 20 µm. **, P < 0.01. NS, not significant.
We thus hypothesized that IKKα is involved in the assembly or secretion of infectious viral particles. The lipid droplet has been shown to play a crucial role in HCV assembly22–24. As expected, HCV proteins, particularly core and NS5A, were observed in close proximity to LDs in HCV-infected cells (Fig. 2c and Supplementary Fig. 4a, b). Cells infected with HCV exhibited a marked increase of LD numbers, LD-positive area, LD fluorescence intensity and triglyceride contents, while IKKα silencing significantly blocked this increase (Fig. 2c and Supplementary Fig. 4b, c). In HCV-infected cells treated with IKKα siRNA, the core protein expression was modestly reduced, but more importantly, the association between core protein and the LD was diminished substantially (Fig. 2c and Supplementary Fig. 4b), suggesting that IKKα is important for the association of LD with HCV proteins during the assembly process. As expected, IKK inhibitors wedelolactone and XII but not BMS-345541, showed strong inhibition of LD formation and co-localization of core and LD (Supplementary Fig. 4d). IKKα over-expression also significantly increased the number, size and fluorescence intensity of LDs (Fig. 2d). Conversely, transfection of HA-IKKα KM mutant strongly reduced LD formation (Fig. 2d). Collectively, these data indicate that IKKα affects predominantly HCV-induced LD formation and viral assembly, although additional effect on secretion cannot be ruled out completely.
HCV 3’UTR Mediates Activation of IKKα and LD Formation
To define the mechanism of HCV-responsive activation of IKKα and induction of lipogenesis, we examined the role of 3’UTR of viral genome, which contains a previously identified HCV PAMP molecule, a poly-U/C sequence25. Transfection of HCV 3’UTR RNA demonstrated a strong induction of LD formation, and IKKα silencing by siRNA markedly diminished HCV 3’UTR-mediated increase of LD content (Fig. 2e and Supplementary Fig. 5a). Poly(I:C), a synthetic PAMP, also enhanced LD formation (Supplementary Fig. 5a). Transfection of HA-IKKα plasmid and HCV 3’UTR RNA together showed increased phosphorylated IKKα, whereas IKKα phosphorylation was not detected in cells transfected with HA-IKKα KM (Supplementary Fig. 5b). Therefore HCV infection induces LD biogenesis through viral RNA-triggered activation of an IKKα-dependent pathway.
IKKα’s Function in HCV infection is Independent of NF-κB
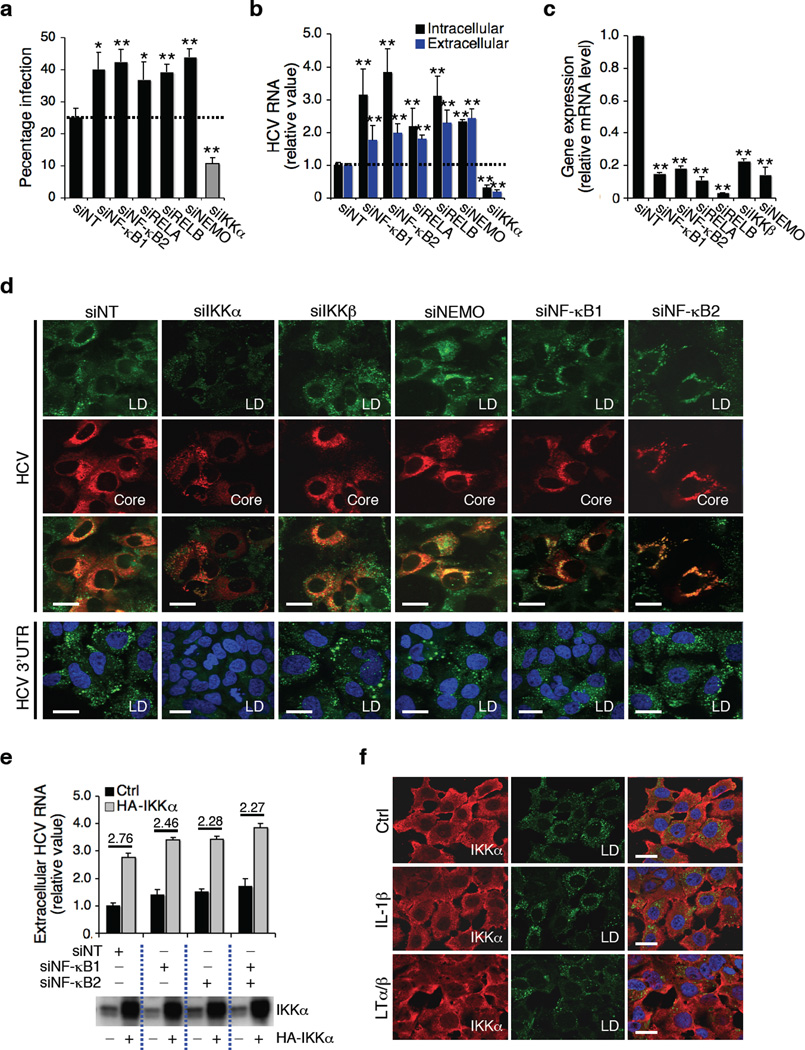
IKKα is a component of IKK controlling NF-κB activation but it has also been implicated in NF-κB-independent functions12. To dissect the function of the NF-κB pathway in connection to IKKα in HCV infection, we first silenced four major factors of the NF-κB family (NF-κB1, NF-κB2, RELA, and RELB), and two essential IKK catalytic units for NF-κB activation (IKKβ and NEMO)11 (Fig. 3a–c). We observed an increase of HCV infection and replication (Fig. 3a, b), which is opposite to the observed effect of IKKα silencing. Second, silencing of the above NF-κB genes did not affect IKKα-mediated induction of LD formation and enhancement of HCV propagation (Fig. 3d, e). Third, signals known to activate the NF-κB pathway, such as IL1-β and lymphotoxin (LT)α/β, did not induce LD formation (Fig. 3f and Supplementary Fig. 5c). SiRNA against LTβ (LTB) or its receptor (LTBR), known to be involved in non-canonical activation of NF-κB via IKKα12, had little or no effect on HCV replication or LD induction (Supplementary Fig. 5d–g).
Figure 3.
IKKα’s function and NF-κB pathway. (a–c) Effects of knockdown of NF-κB pathway on HCV infection. Values represent the means ± s.d., n = 3. **, P < 0.01. (d) Effects of NF-κB silencing on HCV core-associated LD contents and 3’UTR-induced LD formation. (e) Huh7.5.1 cells were treated with indicated siRNAs, and then transfected with control or HA-IKKα plasmid before infection with HCV. Extracellular HCV RNA levels were subsequently measured and normalized to samples treated with siNT and control plasmid. (f) Huh7.5.1 cells were incubated with IL-1β (0.1 µg ml−1) or LTα/β (0.1 µg ml−1) for 30 min, and stained for LD contents and IKKα. Scale bars represent 20 µm.
DDX3X’s Role in HCV Assembly and LD Formation
We next explored the signaling pathway that mediates the specific effect of HCV 3’UTR on IKKα activation. RIG-I has been shown to be the key PRR for HCV PAMP that resides in the 3’UTR25. Because RIG-I is nonfunctional in Huh7.5.1 cells25, we tested the roles of two other RLRs – MDA5 and LGP2, the RLR signaling-adaptor molecule MAVS, and a major downstream cytosolic kinase TBK1. Depletion of these factors by siRNA had little or no effect on HCV infection- or HCV 3’UTR-triggered LD formation (Supplementary Fig. 6a–c). The effects on HCV infection were somewhat variable, with siMDA5 causing an increase and siLGP2 a modest decrease (Supplementary Fig. 6d), which we attribute to a positive role of MDA5 and a negative role of LGP2 in RLR signaling26.
We then investigated whether DDX1, a putative PRR sensing dsRNA27, is involved in HCV 3’UTR-mediated effects and showed that siDDX1 had no effect (Supplementary Fig. 6a–c). DDX3X, another DExD/H helicase, has recently been implicated in innate immunity28,29. DDX3X has been shown to be important for HCV infection30,31. It was also identified as a strong proviral factor in our genome-wide siRNA screen but affected more the late stage of viral life cycle19. Silencing of DDX3X led to a more profound inhibition in extracellular than intracellular HCV RNA level and a significant impairment of viral infectivity (Fig. 4a), whereas over-expression of DDX3X increased extracellular level of HCV RNA more than the intracellular level (Fig. 4b). Depletion of DDX3X also abrogated the induction of LD biogenesis mediated by HCV 3’UTR and core-LD association in the context of HCV infection (Supplementary Fig. 6a–c).
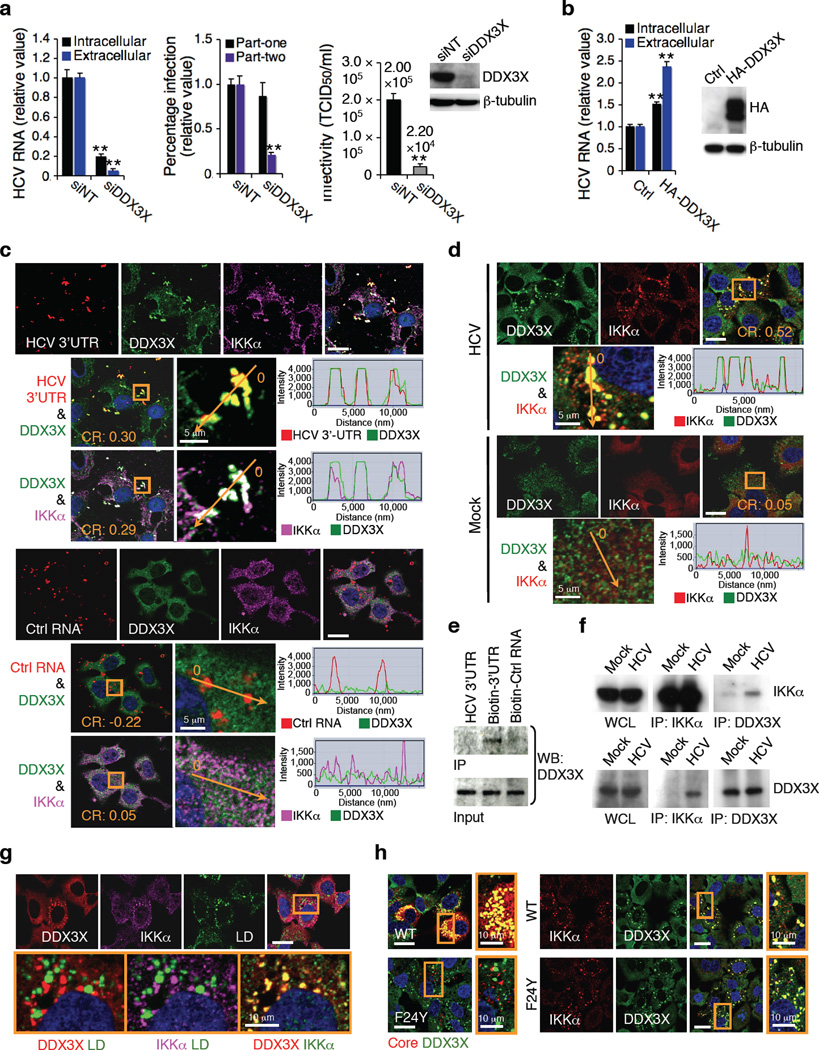
Figure 4.
Interaction of DDX3X with HCV 3’UTR and its role in HCV infection. (a) Effect of DDX3X siRNA on HCV RNA production, infectious HCV production as determined by part-two core staining and HCV infectivity (left, middle, and right panels, respectively). (b) Effect of over-expression of DDX3X on HCV RNA production. (a,b) Error bars represent ± s.d. of triplicate experiments. **, P < 0.01. (c) Cy3-labelled RNA was transfected into Huh7.5.1 cells for 4 h and examined by confocal microscopy for co-localization with DDX3X and IKKα. The control RNA is derived from a negative-strand transcript of an EGFP plasmid. (d) HCV infection and DDX3X and IKKα co-localization in Huh7.5.1 cells. (c,d) The orange arrow or box corresponds to the co-localization analysis of fluorescence intensities that were measured by ZNF2009 software, and shown next to each of the image. CR – Pearson's correlation coefficient (R). (e) Biotinylated RNA was bound to streptavidin beads, and incubated with lysate from Huh7.5.1 cells. Protein eluted from the beads was subjected to Western blot with α-DDX3X antibody. (f) Association of DDX3X and IKKα in HCV-infected cells, determined co-immunoprecipitation. (g) Huh7.5.1 cells were transfected with HCV 3’UTR for 24 h, and then stained for DDX3X, IKKα and LDs. (h) Huh7.5.1 cells were transfected with wild-type (WT) HCV RNA or F24Y HCV RNA for 48 h, and then stained for core, DDX3X and IKKα. Unless otherwise indicated, scale bars represent 20 µm.
DDX3X Interacts with HCV 3’UTR and Activates IKKα-mediated Lipogenesis
In HCV 3’UTR-treated or HCV-infected cells, DDX3X redistributed to form speckle-like cytoplasmic structures (Fig. 4c, d and Supplementary Fig. 7, 8a), albeit its overall expression remained unaltered (Supplementary Fig. 8b, c). DDX3X co-localized specifically with transfected Cy3-labelled HCV 3’UTR RNA but not with the control RNA (Fig. 4c and Supplementary Fig. 7). HCV 3’UTR RNA but not the control RNA formed a complex with DDX3X in cell lysate (Fig. 4e). HCV 3’UTR or infection induced the formation of IKKα aggregates, which co-localized extensively with the DDX3X-HCV 3’UTR complexes (Fig. 4c, d and Supplementary Fig. 7a, 8d). Poly(I:C) also induces DDX3X-IKKα association (Supplementary Fig. 8d, e).
In HCV-infected cells, increased interaction of DDX3X and IKKα was verified by co-immunoprecipitation (Fig. 4f). Silencing of DDX3X markedly reduced the co-localization of HCV 3’UTR and IKKα, whereas siIKKα had no effect on the complex formation between HCV 3’UTR and DDX3X (Supplementary Fig. 7a), suggesting that interaction of HCV 3’UTR with DDX3X is required for the subsequent recruitment of IKKα to the complex. Silencing of MAVS, which has been suggested to interact with DDX3X in activation of innate immunity28,32, had no effect on the interaction of HCV 3’UTR, DDX3X and IKKα (Supplementary Fig. 7a). These data indicate that in HCV-infected Huh7.5.1 cells, DDX3X signals predominantly through the IKKα-mediated lipogenic pathway to play a proviral role in HCV propagation.
Previous studies showed that HCV core binds to and redistributes DDX3X to the viral assembly sites around LDs; however, this interaction appears to be dispensable for HCV replication33. In HCV-infected cells, redistributed DDX3X partially co-localized with core or LDs (Supplementary Fig. 8f). Treatment of Huh7.5.1 cells with HCV 3’UTR or poly(I:C) also induced DDX3X-IKKα association but the DDX3X-IKKα complexes did not localize to the LDs (Fig. 4g and Supplementary Fig. 8e). To confirm that core is not involved in this DDX3X-IKKα pathway, we studied a HCV mutant with a core amino acid substitution (F24Y) that abolishes its interaction with DDX3X33, and showed that in the absence of core-DDX3X binding, DDX3X-IKKα interaction is still induced by HCV infection without LD association (Fig. 4h and Supplementary Fig. 8g).
HCV Induces IKKα Phosphorylation and Nuclear Translocation
We next examined the consequence of DDX3X-IKKα interaction induced by HCV infection. Huh7.5.1 cells were infected with HCV and harvested at varying times (4 to 72 h) for determination of IKKα gene expression. There was no change in either mRNA or protein levels of IKKα in the presence of HCV infection (Supplementary Fig. 9a, b). However, a significant increase in phosphorylated IKKα representing the active form of IKKα was noted 12–24 h after HCV infection (Fig. 5a and Supplementary Fig. 9c). IKKα, when activated, shuttles from the cytoplasm to nucleus15,16. We examined the distribution of IKKα in nuclear and cytosolic fractions of HCV-infected cells. HCV infection by 12 h resulted in a marked nuclear accumulation of IKKα (Fig. 5b). As expected, treatment of cells with TNF-α, a bona fide activator of IKKα15, significantly enhanced IKKα’s phosphorylation and nuclear translocation, while the overall expression of IKKα remained unchanged (Fig. 5a, b and Supplementary Fig. 9b). Confocal microscopy confirmed nuclear accumulation of IKKα in HCV-infected or HCV 3’UTR-transfected cells (Fig. 5c and Supplementary Fig. 9d, e), and these cells displayed speckle-like association of IKKα and DDX3X (Fig. 5c), and elevated LD contents (Supplementary Fig. 9e, f), whereas the majority of IKKα resided in the cytoplasm of uninfected cells. Phosphorylation of IKKα is required for its nuclear translocation because HA-IKKα KM mutant did not show nuclear localization (Supplementary Fig. 9g).
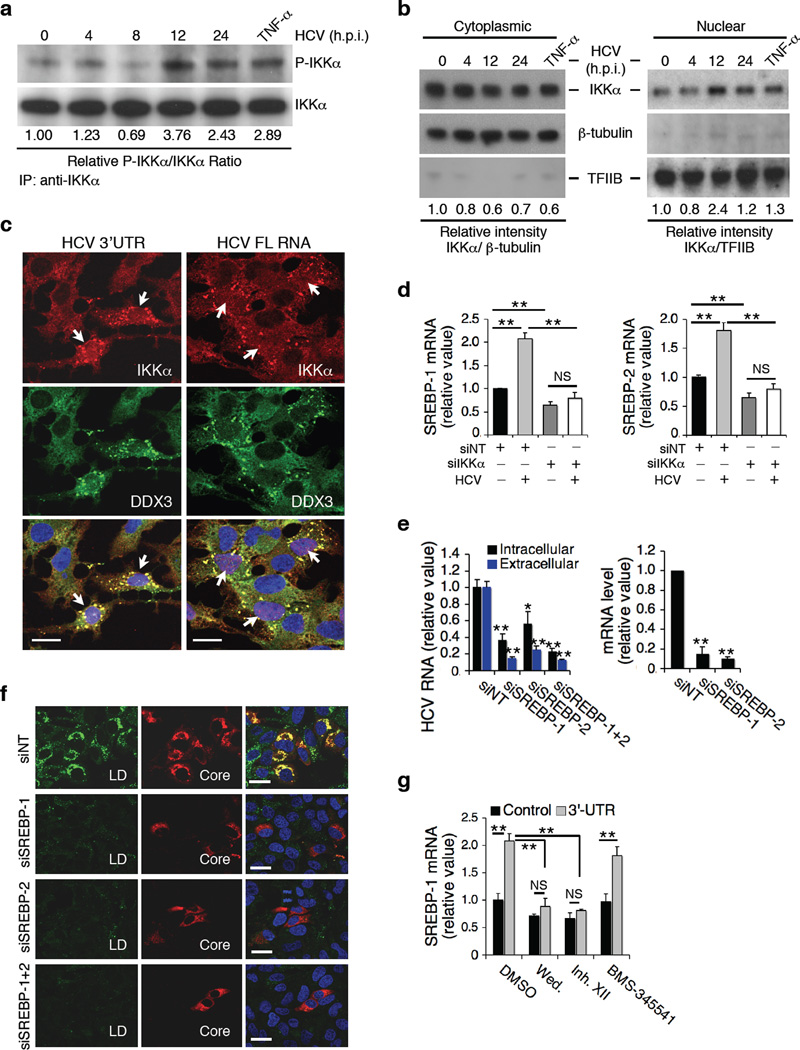
Figure 5.
HCV infection, IKKα activation, SREBP induction and LD formation. (a) Effect of HCV infection or TNF-α treatment on phosphorylation of IKKα, as shown by immunoprecipitation followed by Western blot. Phospho-IKKα and IKKα bands were quantified using ImageJ software and the ratios of P-IKKα over IKKα are provided beneath the blots. (b) Nucleus/cytoplasm (N/C) ratio of IKKα determined by dividing the relative intensity of nuclear IKKα normalized to TFIIB over that of cytoplasmic IKKα normalized to β-tubulin. The highest N/C ratio was achieved at 12 h post-infection (3.98). Control TNF-α treatment led to an increased N/C ratio of 2.15. (c) Huh7.5.1 cells were treated with HCV 3’UTR or full-length (FL) HCV RNA for 4 h, immunostaining for DDX3X and IKKα was performed. White arrows indicate nuclear translocation of IKKα in viral RNA-treated cells showing co-localization of IKKα and DDX3X. (d) SREBP expression in IKKα-deficient cells. SREBP mRNA levels were measured at 72 h after siRNA treatment, 24 h after HCV infection or after both treatments, and were normalized as relative values to siNT-treated samples in the absence of HCV infection. (e,f) Effects of SREBP silencing in Huh7.5.1 cells on HCV infection (e) and LD formation (f). (g) Effects of various IKK inhibitors (wedelolactone, 30 µM; IKK Inhibitor XII, 10 µM; BMS-345541, 1µM) on HCV 3’UTR-mediated induction of SREBP-1. (d,e,g) Error bars represent ± s.d. of triplicate experiments. **, P < 0.01. *, P < 0.05. NS, not significant. (c,f) Scale bars represent 20 µm.
IKKα Activation Induces SREBP-Mediated Lipogenesis and LD Formation
We performed microarray gene expression profiling and identified cellular genes that are transcriptionally regulated by IKKα. Huh7.5.1 cells were treated with either non-targeting (NT) control or IKKα siRNA in the absence or presence of HCV infection (four conditions). The knockdown efficiency of IKKα and effect on HCV infection were confirmed by measuring IKKα mRNA and HCV RNA levels (Supplementary Fig. 10a, b). Lipogenic genes, particularly SREBPs and SREBP target genes involved in fatty acid and triglyceride synthesis were upregulated by HCV, and their inductions were abrogated in IKKα-silenced cells (Fig. 5d and Supplementary Fig. 10c–i). Over-expression of IKKα WT or KM mutant, upregulated and downregulated the expression of SREBPs, respectively (Supplementary Fig. 10j). Under our experimental conditions, the effects of HCV infection and transfection tend to be underestimated, because the efficiency of infection and transfection is only about 40–50% of cells under the best circumstance.
We compared our microarray data set with a recent publication, which identified all SREBP-1-regulated lipid metabolism and human hepatic genes34. We showed that the genes regulated by SREBP-1 are significantly affected by HCV infection or IKKα silencing (Supplementary Fig. 10k, l), implying that IKKα plays a major role in SREBP-1 regulation of hepatic lipid metabolism genes. We further demonstrated that SREBP knockdown has a similar effect as IKKα silencing on HCV infection. After depletion of SREBP-1 and -2, there was a significant impairment of HCV production (Fig. 5e), likely stemming from the apparent diminution of LD formation (Fig. 5f). IKKα silencing and IKK inhibitors (wedelolactone and XII) also abrogated HCV 3’UTR-induced SREBP induction (Fig. 5g and Supplementary Fig. 10i). In PHHs, IKK inhibitors significantly reduced the mRNA levels of SREBP-1 and -2 (Supplementary Fig. 10m). As expected, silencing of DDX3X, IKKα, SREBP-1 or -2 in PHHs led to a significant reduction of extracellular than intracellular HCV RNA levels (Supplementary Fig. 11a, b).
IKKα Induces Expression of SREBP through CBP/p300
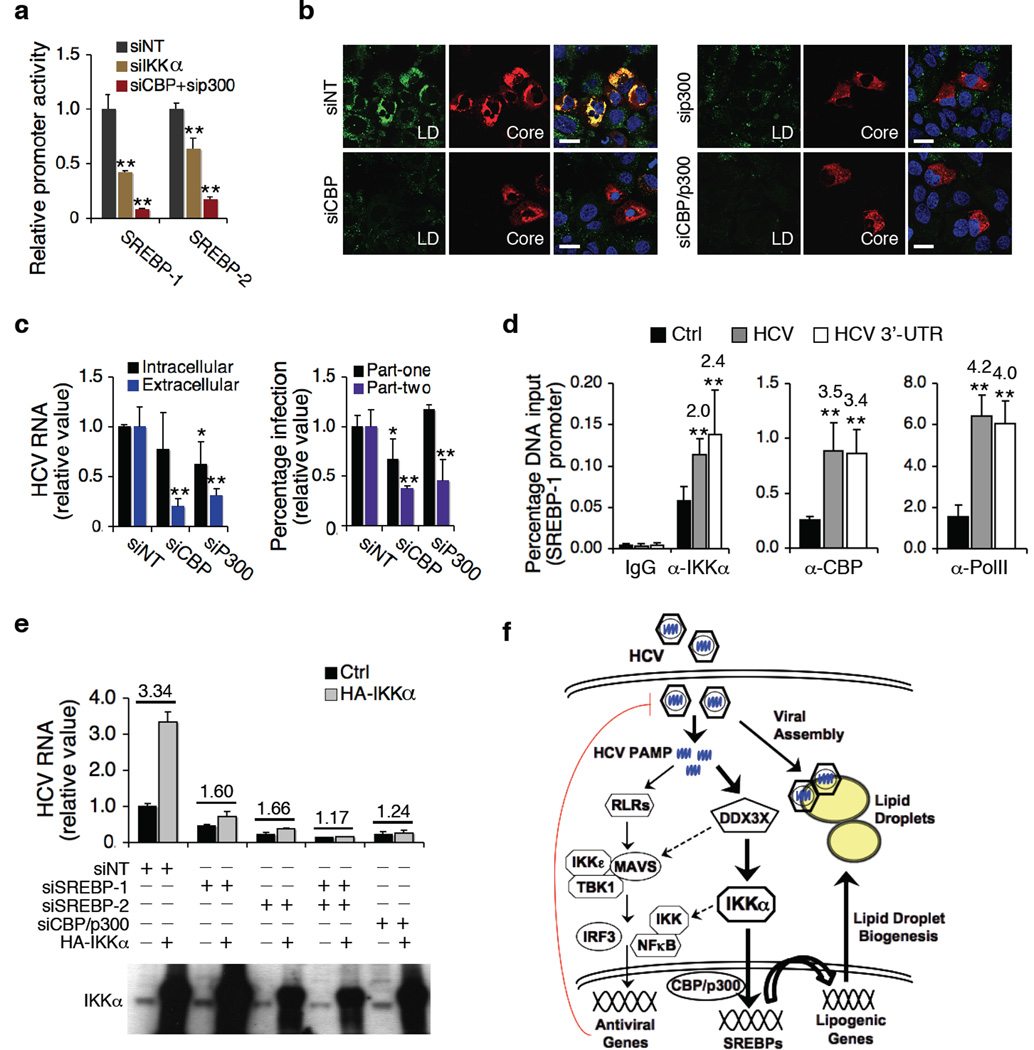
Activated IKKα has been shown to phosphorylate CBP and upregulates its activities in the nucleus18. CBP/p300 is also involved in SREBP transcriptional activity35,36. CBP/p300 over-expression dramatically upregulates the expression of both SREBP-1 and SREBP-237. We showed that both IKKα and CBP/p300 siRNAs significantly reduced SREBP promoter activities and mRNA levels (Fig. 5d, 6a and Supplementary Fig. 11c, d). We showed that IKKα exerts its effect on SREBP induction and lipogenesis through recruiting CBP/p300 complex. First, CBP/p300 depletion by siRNA significantly decreased cytosolic LD contents and HCV production (Fig. 6b, c). Second, siRNAs against CBP/p300 markedly diminished HCV 3’UTR-mediated SREBP-1 induction and LD formation (Supplementary Fig. 11d, e). Third, chromatin immunoprecipitation (ChIP) assay revealed that SREBP-1 promoter bindings of both IKKα and CBP were enhanced by HCV infection or HCV 3’UTR transfection (Fig. 6d and Supplementary Fig. 11f). In addition, enhanced viral production by IKKα over-expression was substantially compromised in cells treated with CBP/p300 or SREBP siRNA (Fig. 6e), indicating that CBP/p300 and SREBPs are intimately involved in the IKKα-mediated proviral effect. Finally, to show that the DDX3X-IKKα-SREBP pathway is indeed crucial for productive HCV infection, we silenced these genes in combination and demonstrated that HCV infection was markedly reduced (>100-fold) as we knocked out the entire pathway (Supplementary Fig. 11g).
Figure 6.
Signaling pathway involved in IKKα-mediated lipogenic induction of HCV infection. (a) Effects of IKKα or CBP/p300 siRNA on SREBP luciferase reporter activities. (b) LD contents and HCV core expression in CBP or p300 siRNA-treated Huh7.5.1 cells prior to HCV infection. Scale bars represent 20 µm. (c) Effects of CBP/p300 silencing on HCV infection. Left: intracellular and extracellular HCV RNA levels; right: HCV core quantification in part-one and part-two of HCV cc assay. Western blot of CBP and p300 protein levels is shown in Supplementary Fig. 12c. (d) Huh7.5.1 cells were untreated, infected with HCV or transfected with HCV 3’UTR RNA for 48 h, and chromatin immunoprecipitation (ChIP) assays were performed with the indicated antibodies. Only data for SREBP-1 promoter is shown here and data for IL-8 (positive control) and actin (negative control) promoters are shown in Supplementary Fig. 12d. Data are presented as means ± s.d., n = 4. (e) IKKα over-expression in HCV-infected cells deprived of SREBP-1, SREBP-2, or CBP/p300. (a,c,d,e) Error bars represent ± s.d. of triplicate experiments. **, P < 0.01, and *, P < 0.05, as compared to control. (f) A proposed model of innate antiviral response and HCV-induced lipogenesis and LD formation in HCV assembly. The thickness of the arrows represents the putative magnitude of the two pathways (proviral > antiviral) in Huh7.5.1 cells. Dotted arrows represent possible cross-talks of the two parallel pathways.
DISCUSSION
Our study demonstrates a hitherto unrecognized function of IKKα in regulating cellular lipid metabolism and hence HCV assembly, both independent of its role in intrinsic innate immunity. We extensively validated and mechanistically investigated this novel function of IKKα. We showed that HCV, through the action of viral 3’UTR that contains a previously identified PAMP, specifically interacts with DDX3X and activates IKKα to mediate an NF-κB-independent function in the nucleus. Recent accumulating evidence indicates an emerging role of DDX3X in innate immunity, although the precise mechanism remains unclear28,29. Other members of the DEAD box family including the well-known RLRs and recently identified DDX1, DDX21 and DHX36 are important intracellular PRRs for RNA viruses27. Although DDX3X has been suggested to function as a PRR28, direct evidence is lacking. Here we show that DDX3X specifically recognizes HCV 3’UTR as a possible PRR. Binding of HCV 3’UTR to DDX3X leads to activation of IKKα. HCV core protein has been shown to bind to DDX3X33, but this interaction is dispensable for HCV-triggered activation of DDX3X-IKKα pathway.
This novel pathway involving DDX3X and IKKα appears to be unique to HCV because genome-wide siRNA screens of several viruses including HIV-1, Dengue, West Nile virus and influenza virus using the same screening platform did not identify this pathway as a crucial host factor for these viruses38–41. Towards this end, a proposed model is generated to illustrate the mechanism of action for DDX3X and IKKα in HCV infection (Fig. 6f). Unlike other factors in the NF-κB activation pathway that are exclusively antiviral genes, IKKα, although capable of modestly inducing ISG expression (Supplementary Fig. 12e), exerts predominantly a proviral effect that targets the assembly stage of HCV life cycle by activating SREBP-mediated lipogenesis and LD biogenesis. It is worth noting here that various HCV proteins have also been implicated to interact with the host lipid metabolism to facilitate HCV assembly22,23, but our data indicate that the primary trigger of LD biogenesis is mediated by the viral 3’UTR and viral proteins mainly play a downstream role in viral assembly22,23.
RNA viruses selectively exploit host specific elements to form specialized organelles where cellular lipids are crucial in enhancing viral production. Dengue virus (DENV) infection launches an autophagy-mediated processing of LDs and triglycerides that is required for efficient viral replication42. West Nile virus infection stimulates cholesterol biosynthesis and redistributes cholesterol to viral RNA replication membranes43. Recently, HCV infection was shown to depend on PI4P lipid-enriched membranes and PI4KIII kinases for RNA replication44. HCV infection also induces LD formation for productive virion assembly24. Cellular lipogenesis is coordinated with LD formation and maturation45.
HCV infection is associated with hepatic steatosis and insulin resistance4, presumably because of HCV-induced dysregulation of lipid metabolism. Hepatic steatosis substantially contributes to the progression of fibrosis in chronic hepatitis C patients46. Interestingly, hepatic activation of IKKβ and NF-κB has been implicated in the etiology of insulin resistance and T2D47,48. Targeted deletion or pharmacological inhibition of IKKβ can restore insulin sensitivity in obese mice or humans49,50. Recent genome-wide association studies have shown that single nucleotide polymorphisms in the CHUK (IKKα) gene locus are associated with nonalcoholic fatty liver diseases51. As we functionally link IKKα to lipogenesis in hepatocytes, it will be interesting to investigate the interaction among the various IKKs in HCV-associated steatosis and insulin resistance as well as in general cellular metabolism.
IKKα, as part of the NF-κB activation pathway, is involved in host recognition and defense against pathogen infection. Here we show that IKKα plays an important role in constitutive maintenance of lipogenic gene expression and LD formation in hepatocytes, and its activation by HCV further stimulates the lipogenic pathways to facilitate viral assembly. We reason that IKKα and its associated pathways have a dual effect on HCV infection – an antiviral effect via the NF-κB pathway and a proviral effect via the lipogenic pathway - with the latter being the dominant pathway in our experimental models. IKKα signaling may therefore represent a pivotal mechanism whereby chronic infection and inflammation leads to dysregulated metabolisms that are implicated in the development of various chronic metabolic disorders52. Our findings offer crucial insights into the inflammatory origin of metabolic disease and pathogenic consequences of chronic viral infection as well as potential novel approaches to treatment14,53.
A comprehensive understanding of the crosstalk between HCV and host innate response and lipid metabolism may help identify novel and broadly active antiviral targets54. The discovery of a critical proviral function of IKKα offers therapeutic opportunities for IKKα inhibitors in the treatment of HCV infection. The demonstrated efficacy of commercially available inhibitors of IKK activities in blocking HCV infection in this study provides a proof-of-concept approach for novel HCV therapeutic development. Our study also presents the first evidence that HCV usurps intrinsic innate immune pathway for its own advantage and survival. Such an exploitation of host antiviral defense may underlie the mechanism whereby HCV infection has a high propensity for persistent infection in the presence of an active immune response.
ONLINE METHODS
siRNA transfection
siRNAs were transfected into Huh7.5.1 cells at a 50 nM final concentration, using a reverse transfection protocol employing Oligofectamine (Invitrogen) as previously described19. For PHHs, cells were seeded on 12-well plates at 500,000 cells per well and transfected with siRNA at a final concentration of 50 nM using RNAiMAX (Invitrogen). Unless otherwise indicated, further treatments or assays were typically performed 72 h after siRNA transfection, when gene silencing efficiency reaches maximal.
HCV core staining part-one (early stage) and part-two (late stage)
For part one, Huh7.5.1 cells were treated with indicated SMARTpool siRNAs at a concentration of 50 nM for 72 h, then infected with HCV JFH-1 strain. After 48 h, cells were stained and imaged for HCV core production. This part detects host factors involved in the early stages of viral life cycle, from entry to viral protein translation and RNA replication. Culture supernatants of part one cells were transferred and infected naïve Huh7 cells, starting part two, which detects proteins involved in the later stages of viral infection, including virion assembly and release. siRNAs against CD81 and ApoE served as proviral controls for part one and part two respectively. Detailed protocol of core staining is described in the Supplementary Methods.
In Vitro transcription and labeling of HCV RNA and transfection
Plasmids carrying JFH1, J6/JFH1 or Luc-JFH1/P7 genomic RNA or subgenomic replion RNA sequences were linearized with Xba I and purified by phenol-chloroform-isoamyl alcohol extraction. In vitro transcription was performed by using the MEGAscript T7 kit (Ambion), according to the manufacturer’s protocol. The quality and quantity of RNA were evaluated by NanoDrop spectrophotometer (Thermo Scientific). Aliquots (22µl, 1 µg µl−1) of RNA were stored frozen at –80 °C until use. HCV 3’UTR RNA was generated from a plasmid harboring the 3’-UTR of HCV that contains a poly-U/C region previously defined as the viral PAMP25. Cy3 labeling of RNA was conducted using Silencer siRNA labeling kit-Cy3 (Ambion). Biotin-labeled RNA was generated by including biotin-UTP (Roche; 1:6 ratio to UTP) during the synthesis process. RNA transfection was performed using DMRIE-C Reagent (Invitrogen) according to the manufacturer’s instructions.
HCV life cycle assays
HCV life cycle assays were performed using HCV pseudoparticles (for viral entry) and subgenomic replicons (for viral IRES-mediated translation and RNA replication), and single cycle infection assays were conducted using CD81-deficient Huh7-25 cells transfected with JFH1/P7-Luc or J6/JFH1 RNA. Detailed protocols for various assays are described in the Supplementary Methods.
Immunofluorescence, lipid staining, confocal microscopy and quantification of images
Cells grown on Lab-Tek®II borosilicate 4-well chamber coverslips (Nunc) were fixed with 4% paraformaldehyde, permeabilized in 0.3% Triton X-100, and incubated with blocking solution in PBS containing 3% BSA and 10% normal goat serum (Vector Laboratories). Cells were then labeled with appropriate primary antibodies diluted in PBS with 1% BSA, followed by incubation with Alexa Fluor 488, 568 or 647 secondary antibodies (Invitrogen) in PBS with 1% BSA. Nuclei were counterstained with Hoechst 33342 (Invitrogen) at 1:5,000 in PBS. LDs were stained with BODIPY 493/503 (Invitrogen), applied at 1 µg ml−1 for 1 h in PBS with 1% BSA. Each step was followed by three washes with PBS. Confocal laser scanning microscopy analysis was performed with an Axio Observer.Z1 microscope equipped with a Zeiss LSM 5 Live DuoScan System under an oil-immersion 1.4 NA 63× objective lens (Carl Zeiss). Images were acquired using ZEN 2009 software (Carl Zeiss). Dual or triple color images were acquired by consecutive scanning with only one laser line active per scan to avoid cross-excition. Quantification of LDs was performed with ImageJ (National Institutes of Health), ZNF2009, and a set of defined intensity thresholds that were applied to all images.
Microarray analysis
Huh7.5.1 cells were transfected with NT or IKKα siRNA for 72 h, then were mock-infected or infected with HCV at an MOI of 1. 48 h later, cellular RNA was extracted and purified using RNeasy Mini Kit (Qiagen). RNA was quantified with a spectrophotometer, and the RNA quality was analyzed with an Agilent Bioanalyzer according to the manufacturer’s instructions. RNA was then amplified with an Agilent Enzo kit. Amplified complementary RNA was hybridized to an Affymetrix Human 133 Plus 2.0 microarray chip containing 54,675 gene transcripts. Microarray analyses were performed at the NIDDK Microarray Core Facility. The bioinformatics and statistical analysis were described previously55. A >1.5-fold change in expression combining a greater than 95% probability of being differentially expressed (P < 0.05) was considered to be biologically significant. The infectivity of JFH-1 virus for Huh7.5.1 cells was tested prior to the microarray experiment.
Gene expression assay
Total cellular RNA from a replica experiment was prepared with RNeasy Mini Kit (Qiagen). Complementary DNA (cDNA) was synthesized from total RNA with First Strand cDNA Synthesis Kit (Roche). The mRNA expression levels of target genes were quantified by quantitative PCR using gene-specific primers and probes (IDT) and TaqMan Gene Express Master Mix (Applied Biosystems) on an ABI 7500 Real Time PCR System. Relative transcript levels were calculated using the ΔΔCT method, with 18S rRNA as the normalizing control gene.
ChIP assays
ChIP assays were performed using SimpleChIP® Enzymatic Chromatin IP Kit (#9003, Cell Signaling) as described by manufacturers. In brief, after cross-linking, nuclei were purified and chromatin was sheared by sonication (three times, 20 s each). Chromatin was incubated overnight with specific antibodies against IKKα (ab4111, Abcam), CBP (ab2832, Abcam), and RNA-Polymerase II CTD (MA1-46093, Thermo Scientific). Isotype IgG were used as a negative control for the immunoprecipitation. Immunoprecipitated chromatin was then incubated with protein G magnetic beads, washed and eluted. After reversal of the cross-links and purification of DNA, precipitated DNAs were analysed by quantitative real-time PCR (40 cycles) with specific primers to the human actin promoter as a negative control, the IL-8 promoter as a positive control17, and SREBP-1 promoter (5’-GCTGTCCCGTGTTAGCCCTT-3’ and 5’-TCTACCCGGGAGGTAGGGA-3’). Quantitative real-time PCR was performed in duplicate on duplicate ChIP on duplicate assays by using 500 nM of the above oligonucleotide primers and input DNA standards diluted in 5-fold increments from 5% to 0.008% with SYBR® Green PCR Master Mix (#4309155, Applied Biosystems) and Applied Biosystems 7500 Sequence Detection System. Fold-increase of association of IKKα, CBP and Pol II to IL-8 and SREBP-1 promoters were normalized to actin signals and calculated for HCV-infected and HCV PAMP RNA-transfected cells as compared to untreated cells. P value was calculated using the Student’s t test.
Chemical inhibitor studies
IKK inhibitors II (Wedelolactone), XII, and III (BMS-345541) were purchased from Merck (EMD) Chemicals. Serial dilutions of the IKK inhibitors were made in 100% dimethyl sulfoxide (DMSO) immediately prior to the assay so that the final concentration of DMSO in each reaction was identical. Cells were infected with JFH-1 at an MOI of 0.5 for 6 h, and then treated with fresh DMEM containing DMSO with or without IKK inhibitors. Cultures were incubated for 48 h, and HCV RNA was extracted from cell lysate or culture medium and quantified by TaqMan real-time PCR. The experiments were done in triplicate, and each dose was tested at least twice.
Additional methods
Detailed methodology is described in the Supplementary Methods.
Supplementary Material
ACKNOWLEDGMENTS
We thank Z. Hu, Y.-Y. Zhang, Y.-M. Li, E. Thomas, V. Gonzalez-Munoz and L. Holz for technical assistance, Y.-P. Wu of the NIDDK Confocal Microscopy Core for helping with confocal imaging, W.-P. Chen of the NIDDK Microarray Core Facility for DNA microarray analysis. We also thank A. Patel of University of Glasgow for providing the HCV core mutant (F24Y) virus. This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, US National Institutes of Health. Primary human hepatocytes were provided by the NIH funded Liver Tissue Procurement and Cell Distribution System (N01-DK-7-0004/HHSN26700700004C, PI-S. Strom, University of Pittsburgh).
Footnotes
AUTHOR CONTRIBUTIONS
Q.L. and T.J.L. conceived and designed the study. Q.L., V.P., S.K. and H.C. conducted experiments. Q.L., V.P. and T.J.L. analyzed data. Q.L. and T.J.L. wrote the paper with the input from V.P. T.J.L. supervised the studies.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Liang TJ, Rehermann B, Seeff LB, Hoofnagle JH. Pathogenesis, natural history, treatment, and prevention of hepatitis C. Ann Intern Med. 2000;132:296–305. doi: 10.7326/0003-4819-132-4-200002150-00008. [DOI] [PubMed] [Google Scholar]
- 2.Liang TJ, Ghany MG. N Engl J Med. 2013 in press. [Google Scholar]
- 3.Rehermann B. Hepatitis C virus versus innate and adaptive immune responses: a tale of coevolution and coexistence. J Clin Invest. 2009;119:1745–1754. doi: 10.1172/JCI39133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Syed GH, Amako Y, Siddiqui A. Hepatitis C virus hijacks host lipid metabolism. Trends Endocrinol Metab. 2010;21:33–40. doi: 10.1016/j.tem.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herker E, Ott M. Unique ties between hepatitis C virus replication and intracellular lipids. Trends Endocrinol Metab. 2011;22:241–248. doi: 10.1016/j.tem.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diamond DL, et al. Temporal proteome and lipidome profiles reveal hepatitis C virus-associated reprogramming of hepatocellular metabolism and bioenergetics. PLoS Pathog. 2010;6:e1000719. doi: 10.1371/journal.ppat.1000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waris G, Felmlee DJ, Negro F, Siddiqui A. Hepatitis C virus induces proteolytic cleavage of sterol regulatory element binding proteins and stimulates their phosphorylation via oxidative stress. J Virol. 2007;81:8122–8130. doi: 10.1128/JVI.00125-07. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Lerat H, et al. Hepatitis C virus proteins induce lipogenesis and defective triglyceride secretion in transgenic mice. J Biol Chem. 2009;284:33466–33474. doi: 10.1074/jbc.M109.019810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowie AG, Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol. 2008;8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 12.Perkins ND. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat Rev Mol Cell Biol. 2007;8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 13.Senftleben U, et al. Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signaling pathway. Science. 2001;293:1495–1499. doi: 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
- 14.Pasparakis M. Regulation of tissue homeostasis by NF-kappaB signalling: implications for inflammatory diseases. Nat Rev Immunol. 2009;9:778–788. doi: 10.1038/nri2655. [DOI] [PubMed] [Google Scholar]
- 15.Anest V, et al. A nucleosomal function for IkappaB kinase-alpha in NF-kappaB-dependent gene expression. Nature. 2003;423:659–663. doi: 10.1038/nature01648. [DOI] [PubMed] [Google Scholar]
- 16.Birbach A, et al. Signaling molecules of the NF-kappa B pathway shuttle constitutively between cytoplasm and nucleus. J Biol Chem. 2002;277:10842–10851. doi: 10.1074/jbc.M112475200. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto Y, Verma UN, Prajapati S, Kwak YT, Gaynor RB. Histone H3 phosphorylation by IKK-alpha is critical for cytokine-induced gene expression. Nature. 2003;423:655–659. doi: 10.1038/nature01576. [DOI] [PubMed] [Google Scholar]
- 18.Huang WC, Ju TK, Hung MC, Chen CC. Phosphorylation of CBP by IKKalpha promotes cell growth by switching the binding preference of CBP from p53 to NF-kappaB. Mol Cell. 2007;26:75–87. doi: 10.1016/j.molcel.2007.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Q, et al. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc Natl Acad Sci U S A. 2009;106:16410–16415. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zandi E, Rothwarf DM, Delhase M, Hayakawa M, Karin M. The IkappaB kinase complex (IKK) contains two kinase subunits, IKKalpha and IKKbeta, necessary for IkappaB phosphorylation and NF-kappaB activation. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 21.Akazawa D, et al. CD81 expression is important for the permissiveness of Huh7 cell clones for heterogeneous hepatitis C virus infection. J Virol. 2007;81:5036–5045. doi: 10.1128/JVI.01573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyanari Y, et al. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 23.Herker E, et al. Efficient hepatitis C virus particle formation requires diacylglycerol acyltransferase-1. Nat Med. 2010;16:1295–1298. doi: 10.1038/nm.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLauchlan J. Lipid droplets and hepatitis C virus infection. Biochim Biophys Acta. 2009;1791:552–559. doi: 10.1016/j.bbalip.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 25.Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rothenfusser S, et al. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J Immunol. 2005;175:5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Z, et al. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity. 2011;34:866–878. doi: 10.1016/j.immuni.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oshiumi H, Sakai K, Matsumoto M, Seya T. DEAD/H BOX 3 (DDX3) helicase binds the RIG-I adaptor IPS-1 to up-regulate IFN-beta-inducing potential. Eur J Immunol. 2010;40:940–948. doi: 10.1002/eji.200940203. [DOI] [PubMed] [Google Scholar]
- 29.Schroder M, Baran M, Bowie AG. Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon-mediated IRF activation. EMBO J. 2008;27:2147–2157. doi: 10.1038/emboj.2008.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ariumi Y, et al. DDX3 DEAD-box RNA helicase is required for hepatitis C virus RNA replication. J Virol. 2007;81:13922–13926. doi: 10.1128/JVI.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Randall G, et al. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci U S A. 2007;104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oshiumi H, et al. Hepatitis C virus core protein abrogates the DDX3 function that enhances IPS-1-mediated IFN-beta induction. PLoS One. 2010;5:e14258. doi: 10.1371/journal.pone.0014258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Angus AG, et al. Requirement of cellular DDX3 for hepatitis C virus replication is unrelated to its interaction with the viral core protein. J Gen Virol. 2010;91:122–132. doi: 10.1099/vir.0.015909-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reed BD, Charos AE, Szekely AM, Weissman SM, Snyder M. Genome-wide occupancy of SREBP1 and its partners NFY and SP1 reveals novel functional roles and combinatorial regulation of distinct classes of genes. PLoS Genet. 2008;4:e1000133. doi: 10.1371/journal.pgen.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ericsson J, Edwards PA. CBP is required for sterol-regulated and sterol regulatory element-binding protein-regulated transcription. J Biol Chem. 1998;273:17865–17870. doi: 10.1074/jbc.273.28.17865. [DOI] [PubMed] [Google Scholar]
- 36.Oliner JD, Andresen JM, Hansen SK, Zhou S, Tjian R. SREBP transcriptional activity is mediated through an interaction with the CREB-binding protein. Genes Dev. 1996;10:2903–2911. doi: 10.1101/gad.10.22.2903. [DOI] [PubMed] [Google Scholar]
- 37.Giandomenico V, Simonsson M, Gronroos E, Ericsson J. Coactivator-dependent acetylation stabilizes members of the SREBP family of transcription factors. Mol Cell Biol. 2003;23:2587–2599. doi: 10.1128/MCB.23.7.2587-2599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brass AL, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 39.Sessions OM, et al. Discovery of insect and human dengue virus host factors. Nature. 2009;458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krishnan MN, et al. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455:242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brass AL, et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heaton NS, Randall G. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe. 2010;8:422–432. doi: 10.1016/j.chom.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mackenzie JM, Khromykh AA, Parton RG. Cholesterol manipulation by West Nile virus perturbs the cellular immune response. Cell Host Microbe. 2007;2:229–239. doi: 10.1016/j.chom.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 44.Hsu NY, et al. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farese RV, Jr, Walther TC. Lipid droplets finally get a little R-E-S-P-E-C-T. Cell. 2009;139:855–860. doi: 10.1016/j.cell.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Asselah T, Rubbia-Brandt L, Marcellin P, Negro F. Steatosis in chronic hepatitis C: why does it really matter? Gut. 2006;55:123–130. doi: 10.1136/gut.2005.069757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cai D, et al. Local and systemic insulin resistance resulting from hepatic activation of IKK-beta and NF-kappaB. Nat Med. 2005;11:183–190. doi: 10.1038/nm1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuan M, et al. Reversal of obesity- and diet-induced insulin resistance with salicylates or targeted disruption of Ikkbeta. Science. 2001;293:1673–1677. doi: 10.1126/science.1061620. [DOI] [PubMed] [Google Scholar]
- 49.Arkan MC, et al. IKK-beta links inflammation to obesity-induced insulin resistance. Nat Med. 2005;11:191–198. doi: 10.1038/nm1185. [DOI] [PubMed] [Google Scholar]
- 50.Zhang X, et al. Hypothalamic IKKbeta/NF-kappaB and ER stress link overnutrition to energy imbalance and obesity. Cell. 2008;135:61–73. doi: 10.1016/j.cell.2008.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Romeo S, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 53.Baker RG, Hayden MS, Ghosh S. NF-kappaB, inflammation, and metabolic disease. Cell Metab. 2011;13:11–22. doi: 10.1016/j.cmet.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Munger J, et al. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol. 2008;26:1179–1186. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feld JJ, et al. Hepatic gene expression during treatment with peginterferon and ribavirin: Identifying molecular pathways for treatment response. Hepatology. 2007;46:1548–1563. doi: 10.1002/hep.21853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.