Abstract
Prebiotic peptide formation under aqueous conditions in the presence of metal ions is one of the plausible triggers of the emergence of life. The salt-induced peptide formation reaction has been suggested as being prebiotically relevant and was examined for the formation of peptides in NaCl solutions. In previous work we have argued that the first protocell could have emerged in KCl solution. Using HPLC-MS/MS analysis, we found that K+ is more than an order of magnitude more effective in the L-glutamic acid oligomerization with 1,1'-carbonyldiimidazole in aqueous solutions than the same concentration of Na+, which is consistent with the diffusion theory calculations. We anticipate that prebiotic peptides could have formed with K+ as the driving force, not Na+, as commonly believed.
Keywords: Potassium, Sodium, Prebiotic, Peptide formation, Origin of life
Introduction
Since Oparin’s ideas (1924; 1938) and Miller-Urey’s famous experiment (1953) on the prebiotic synthesis of amino acids, one of the main problems of prebiotic chemistry is to “re-invent” the plausible range of indispensable physical-chemical boundary requirements that would enable the emergence of stable and replicable molecules on the primordial Earth (Eschenmoser 2003). According to almost all discussions of the prebiotic soup theory, biological precursor molecules might have formed and evolved in the context of the sodium chloride ocean on primordial Earth (Fox 1960; Rode 1999; Zimmer 2009). Salt-induced peptide formation reaction has been suggested to be prebiotically relevant for the very first steps of chemical evolution (Schwendinger and Rode 1989). Based on Monte Carlo computer simulations, Rode and co-workers found that sodium chloride at concentrations above 3 M effectively acts as a dehydrating agent to overcome the thermodynamic barrier of peptide bond formation in aqueous solutions, and the first hydration shell of the sodium ion was assumed to no longer be saturated with water molecules (Jakschitz and Rode 2012). Furthermore, using HPLC-MS/MS analysis, a high concentration of sodium chloride was found to significantly enhance the formation of peptides from L-glutamic acid (L-Glu) in homogenous water solutions (Wang et al. 2005).
All the references we have found that discuss the presence of other mono- and divalent inorganic cations in prebiotic peptide formation speculate that these ions support the dehydrating effect of sodium chloride. However, the level of potassium exceeds that of sodium by more than an order of magnitude inside all living cells (Aronson et al. 2009), and the ion ratio is actively preserved with Na+/K+ pumps in the cell membrane, which suggests that potassium is more essential for life. The physical-chemical differences between Na+ and K+ are small (Freedman 1995), although the bio-directed activity of these ions differs dramatically; for example, K+ is required for ribosomal peptide synthesis (Spirin and Gavrilova 1971) and the amplification of DNA with thermostable Taq polymerase (Saiki et al. 1988), whereas Na+ attenuates these processes. The contradiction between the Na+ and K+ compositions of seawater and living cell cytoplasm led to the hypothesis that the first protocell could have emerged in KCl solution (Natochin 2007; Natochin 2010). However, the hypothesis of the K+-driven emergence of prebiotic peptides remains to be tested. Here we investigate the relative effects of Na+ and K+ in a model peptide synthesis reaction.
Methods
L-glutamic acid and 1,1′-carbonyldiimidazole (CDI) were obtained from Sigma-Aldrich Co. LLC (St. Louis, USA). In total, 10 mmol KCl or 10 mmol NaCl was added to reaction mixtures containing 3 mmol L-Glu in 5 ml distilled water. The mixture was diluted to 10 ml and cooled on a crashed ice-NaCl mixture, and 6 mmol CDI was added into each mixture and incubated at room temperature for 24 h. A 10 μl sample was loaded onto a Zorbax SAX (4.6 mm × 250 mm, 5 μm) column using an autosampler.
Peptide separation was performed at a flow rate of 0.5 ml/min using an NaCl gradient (2–80 % B for 80 min; buffer A: 20 % acetonitrile in 0.020 M NaH2PO4 at pH 7.0; buffer B: 2.0 M NaCl in buffer A) using an Agilent 1100 nano-HPLC system (Agilent Technologies Inc., USA). LC analysis of the peptides was performed by an established procedure (Ishihama et al. 2002) on a homemade 12-cm reverse-phase spraying fused silica capillary column (75 μm i.d. × 12 cm) with a 3-μm ReproSil-Pur Basic-C18 (Dr. Maisch HPLC GmbH, Germany). Peptide fractions were collected for further analysis.
MS/MS analysis of the samples was performed using a 7-Tesla LTQ-FT Ultra mass spectrometer and Xcalibur software in data-dependent mode (Thermo Fisher Scientific Inc., USA). The precursor ion MS spectra were acquired in the ICR trap with a resolution of 50,000 at m/z 400. The three most intense ions were isolated from MS/MS spectra and fragmented in LTQ. Oligomers from 2- to 9-mers were identified with ESI-MS. Other oligomers were assigned based on the one-charge increase in oligomers on HPLC traces.
We used the basic theories of catalytic reactions and nucleation (Dubrovskii and Nazarenko 2010) to model the ion-mediated condensation of amino acids in the liquid phase.
Results
Liquid Chromatography and Mass Spectrometry
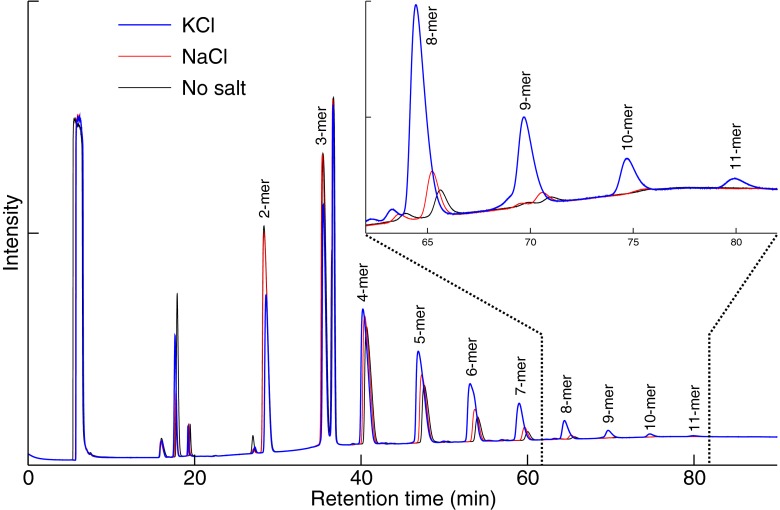
We first prepared L-Glu oligomerization reactions in the presence of 1.0 M KCl based on an established procedure using CDI, followed by HPLC-MS/MS analysis. CDI is an efficient dehydrating agent that can be used to produce homooligopeptides or random oligopeptides in water via a carboxyanhydride intermediate as a route for the prebiotic activation of amino acids to form oligopeptides (Brack 1987; Hill and Orgel 1996). In the control reaction, we added 1.0 M NaCl, which is the most effective salt concentration for the CDI-mediated formation of peptides (Wang et al. 2005). The chromatograms of the reactions with 1.0 M KCl or 1.0 M NaCl or no salts are shown in Fig. 1.
Fig. 1.
Chromatograms of the K+- and Na+-mediated oligomerization of peptides. Each peak matched specific CDI-induced L-Glu peptides in 1.0 M KCl or 1.0 M NaCl solution or water without any salts
We found that the lengths of the oligomers increased up to 11-mer in the presence of K+ compared to 9-mer in the presence of Na+. For the mass spectra of the oligomers, see Table 1. We then studied L-Glu oligomerization in the presence of 0.5 M and 2.0 M KCl and NaCl. We found that ion concentrations below and above 1.0 M reduced L-Glu peptide yields. K+ predominance was found in all the reactions.
Table 1.
Chromatography and mass spectrometry data for Na+- or K+ - catalyzed peptides
| Number of residues | L-Glu oligomers + 1.0 M NaCl | L-Glu oligomers + 1.0 M KCl | ||||||
|---|---|---|---|---|---|---|---|---|
| Mass spectrometry [M + H]+ ([M + Na]+) | Chromatography | Mass spectrometry [M + H]+ ([M + K]+) | Chromatography | |||||
| Calculated, Da | Found, Da | Peak area | Relative area, % | Calculated, Da | Found, Da | Peak area | Relative area, % | |
| 2 | C10H17O7N2 277.104 C10H16O7N2Na (299.086) | 277.101 (299.085) | 963 | 100.0 | C10H17O7N2 277.104 C10H16O7N2K (315.059) | 277.103 (315.089) | 534 | 100.0 |
| 3 | C15H24O10N3 406.146 C15H23O10N3Na (428.128) | 406.146 (428.127) | 1060 | 110.1 | C15H24O10N3 406.146 C15H23O10N3K (444.102) | 406.146 (444.101) | 709 | 132.8 |
| 4 | C20H31O13N4 535.189 C20H30O13N4Na (557.171) | 535.187 (557.172) | 770 | 80.0 | C20H31O13N4 535.189 C20H30O13N4K (573.145) | 535.187 (573.145) | 833 | 156.0 |
| 5 | C25H38O16N5 664.231 C25H37O16N5Na (686.213) | 664.230 (686.212) | 408 | 42.4 | C25H38O16N5 664.231 C25H37O16N5K (702.187) | 664.231 (702.187) | 651 | 121.9 |
| 6 | C30H45O19N6 793.274 C30H44O19N6Na (815.256) | 793.272 (815.252) | 174 | 18.1 | C30H45O19N6 793.274 C30H44O19N6K (831.230) | 793.272 (831.229) | 411 | 77.0 |
| 7 | C35H52O22N7 922.317 C35H51O22N7Na (944.298) | 922.315 (944.285) | 61 | 6.3 | C35H52O22N7 922.317 C35H51O22N7K (960.272) | 922.315 (960.273) | 223 | 41.8 |
| 8 | C40H59O25N8 1051.359 | 1051.352 | 18 | 1.9 | C40H59O25N8 1051.359 C40H58O25N8K (1089.315) | 1051.352 (1089.311) | 99 | 18.5 |
| 9 | – | – | 4 | 0.4 | C45H66O28N9 1180.401 | 1180.394 | 45 | 8.4 |
| 10 | – | – | – | – | – | – | 17 | 3.2 |
| 11 | – | – | – | – | – | – | 6 | 1.1 |
Physical Model
To provide theoretical evidence in favour of the difference between the peptide formation reactions in the presence of K+ and Na+, we modelled the ion-mediated condensation of amino acids in the liquid phase. In general, the reaction chain producing the complexes A n with n monomers in presence of a catalyst B can be put in the form
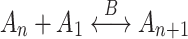 |
1 |
This assumes the effective absence of interactions between the complexes as well as three-body interactions, the properties that should pertain for a dilute solution in water. The catalyst is assumed to promote the monomer attachment via one of the following heterogeneous reactions
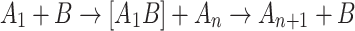 |
2 |
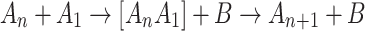 |
3 |
In scheme (2), the heterogeneous complex [A 1 B] is long-lived, and the growth is controlled by the diffusion transport of the reactants. Scheme (3) assumes that the homogeneous complex [A n A 1] is long-lived, where the growth should be limited by the diffusion transport of the catalyst. We considered the conventional quasi-chemical nucleation model for the concentrations C n of complexes containing n monomers at time t
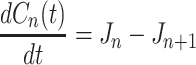 |
4 |
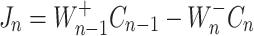 |
5 |
whereas, 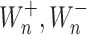 denote the B − dependent rate constants for the monomer attachment and detachment, respectively, and J
n represents the corresponding flux. The monomer concentration is generally obtained from the mass conservation
denote the B − dependent rate constants for the monomer attachment and detachment, respectively, and J
n represents the corresponding flux. The monomer concentration is generally obtained from the mass conservation 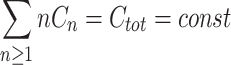 at any time, where C
tot is the total concentration of monomers in the system. In according to the nucleation theory (Dubrovskii and Nazarenko 2010) the time scale hierarchy of the entire agglomeration process results in a rather slow time dependence of the monomer concentration C
1(t), while the concentrations of differently sized complexes depend on time only through C
1(t). For small enough n, the C
n can be obtained within the quasi-equilibrium approximation relating to J
n = 0. This yields the size distribution of the form
at any time, where C
tot is the total concentration of monomers in the system. In according to the nucleation theory (Dubrovskii and Nazarenko 2010) the time scale hierarchy of the entire agglomeration process results in a rather slow time dependence of the monomer concentration C
1(t), while the concentrations of differently sized complexes depend on time only through C
1(t). For small enough n, the C
n can be obtained within the quasi-equilibrium approximation relating to J
n = 0. This yields the size distribution of the form
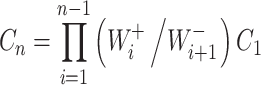 |
6 |
Now we assume that the equilibrium constant 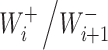 is proportional to a certain characteristic b that depends on the catalyst type
is proportional to a certain characteristic b that depends on the catalyst type
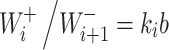 |
7 |
Substitution of equation (7) into equation (6) readily gives
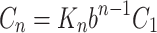 |
8 |
whereas, dependence of the complexes concentration C
n on the catalyst is described by the b
n−1 and 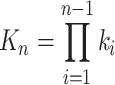 can be considered as being catalyst-independent.
can be considered as being catalyst-independent.
The theoretical model above can be used to obtain dependence of the L-Glu peptides concentration on the peptide length in presence of ions, if we consider the monomer is L-Glu and the catalyst B is K+ or Na+. In case of reaction (2), the dependence might be explained with different ion adsorption probabilities onto the surface of the amino acid. For the reaction (3), the equilibrium constant 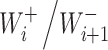 should be proportional to the diffusion coefficient
should be proportional to the diffusion coefficient 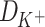 or
or 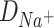 of the corresponding ion in water. The diffusion limit gives the equation (9) for the ratio of peptide concentrations in the presence of K+ or Na+ in water solutions
of the corresponding ion in water. The diffusion limit gives the equation (9) for the ratio of peptide concentrations in the presence of K+ or Na+ in water solutions
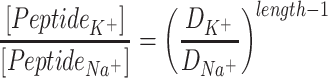 |
9 |
whereas, 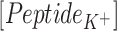 and
and 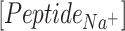 are concentrations of the peptides,
are concentrations of the peptides, 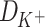 and
and 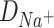 are diffusion coefficients of the ions in water and length is the number of L-Glu residues in the peptide.
are diffusion coefficients of the ions in water and length is the number of L-Glu residues in the peptide.
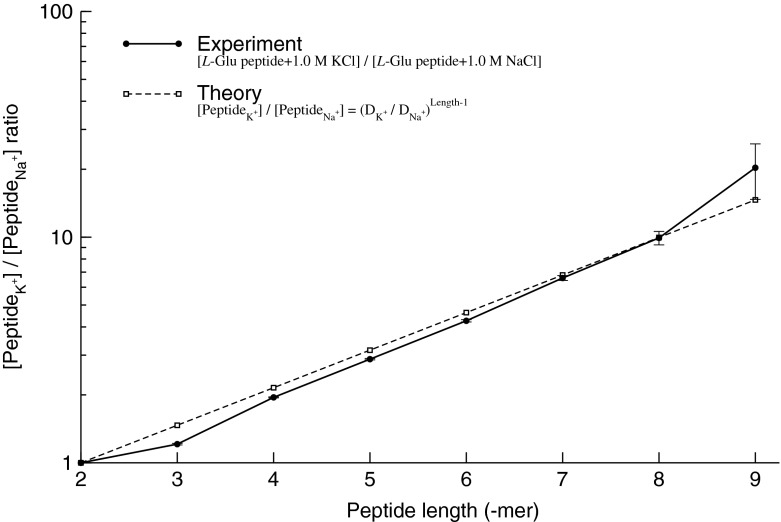
Thus, the equation (9) above, with the diffusion coefficients of K+ (DK + = 1.957 × 10−5 cm2/s) and Na+ (DNa + = 1.334 × 10−5 cm2/s) in water solutions (Lide and David, 1998), clearly corresponds to the K+/Na+ ratio of the salt-mediated formation of L-Glu peptides (Fig. 2), which was calculated as the peak area of each oligomer on the chromatogram divided by the peak area of the dipeptide in the same reaction (Table 1).
Fig. 2.
Experimental and theoretical evidence of the K+- versus Na+-mediated formation of peptides The experimental data for the K+/Na+ ratio of L-Glu peptides was calculated from Fig. 1 as the peak area of each oligomer on the chromatogram divided by the peak area of the dipeptide in the same reaction
Discussion
Our experimental results demonstrate that K+ has a 3-fold to 10-fold greater catalytic effect than the same concentration of Na+ on the reaction peak of 5-mer to 8-mer L-Glu condensation in aqueous solutions. Computations and blackbody infrared radioactive dissociations have shown that Na+ is coordinated to the nitrogen and carbonyl oxygen atoms (NO coordination) of amino acids, whereas K+ is coordinated to both oxygen atoms (OO coordination), with lower binding energy (Jockusch et al. 2001). This finding allows us to suggest that NO coordination decreases the reactivity of amino acids for each subsequent peptide bond formation, as it begins with a nucleophilic attack on the lone electron pair of the N atom, in full agreement with our obtained experimental data showing the weaker Na+-mediated formation of L-Glu peptides.
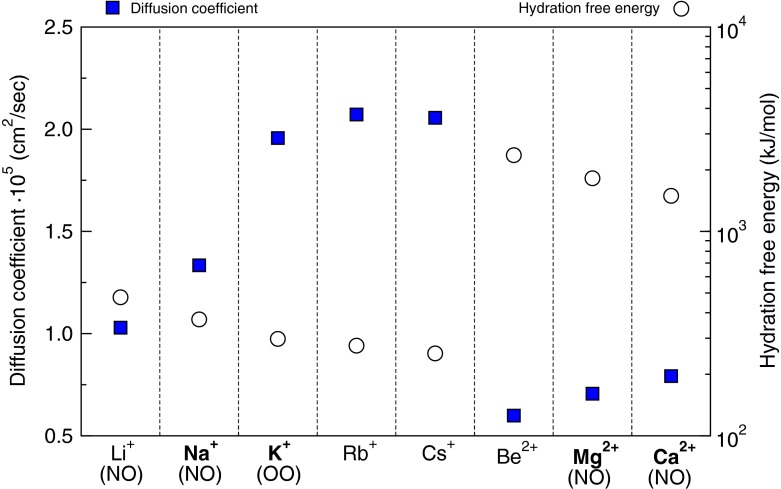
Although the mechanism described above explains the results of the experimental L-Glu peptide formations in the presence of K+ and Na+, this interpretation of the mechanism is not exhaustive. Our data on the calculated difference between the K+ and Na+ diffusion-controlled condensation of amino acids is fully consistent with the experimental data (Fig. 2). Using the model above for other mono- and divalent ions, we summarised in Fig. 3 the available data on diffusion coefficients, hydration energy of the ions and their coordination to the amino acids in aqueous solutions (Lide and David 1998; Schmid et al. 2000; Jockusch et al. 2001; Remko and Rode 2006). We found that Rb+ and Cs+ might be similar to K+ in mediating peptide formation in the OO coordination to amino acids, which has not yet been modelled, to the best of our knowledge.
Fig. 3.
Metal ion diffusion, hydration and coordination to amino acids. The coordination of the ions to amino acids in aqueous solutions is shown in parentheses. The most abundant ions are shown in bold
Taken together, our experimental and theoretical evidences show that K+ predominates over Na+ ions in the formation of peptides. This allows us to suggest that the high K+/Na+ ratio in any prebiotic water reservoir could accelerate the first step in the chemical evolution of self-assembling organic molecules. Geochemically, a high K+/Na+ ratio in aqueous solution could also have formed during the differentiation of primary chondritic material into the Earth’s core and mantle (Galimov et al. 2011). It was also suggested that the ion composition required for the initial environment for the first cells could have emerged in inland geothermal ponds (Mulkidjanian et al. 2012). Although this assumption has been criticised (Switek 2012), from a biological point of view, the “modern” cytoplasm of the living cells might represent the same functional conditions that determined the first protocell’s chemical content. Thus, if the emergence of the ancient metabolic and information systems of the protocells occurred in potassium-rich habitats, it seems evident that all the living cells would have evolved to preserve the initial ion gradients by using energy-dependent membrane pumps in sodium aqueous media.
Conclusion
In conclusion, we hypothesise that the following conditions could have led to the prebiotic polymerisation of amino acids: (1) aqueous media contained the building blocks of organic matter and positive inorganic ions, which are geochemically abundant; (2) binding reversibility to amino acids and the moderate hydration energy of the ions in liquid phase at 0–100 °C; and (3) high diffusion and specific ion coordination to oxygen atoms of amino acids in zwitterion form, which enhances the ion-dependent yields of oligomerization. We propose that K+ complies with all the above-listed requirements, which is unique in contrast to other mono- and divalent metallic ions (Fig. 3). Further peptide evolution at later stages could have occurred due to the presence of other abundant cations, e.g., Na+, Mg2+ and Ca2+, which may have resulted from their lower diffusion and higher hydration energy. The elongation and functionalization of the peptides might also have been driven by other inorganic cations or clays or minerals (Ferris et al. 1996; Hill and Orgel 1999; Rode et al. 1999; Rees and Howard 2003) because they form more stable complexes with biomolecules.
We assume that our findings could be useful not only for discussions of the origin of life but also for more sophisticated research on the role of the physical-chemical properties of inorganic ions, biomolecules and nanoparticles in molecular physiology. The data on the difference in K+ versus Na+ coordination- and diffusion-controlled condensation of amino acids may be of particular interest in understanding ion-exchange regulation by the membrane Na+/K+-ATPase pump.
Acknowledgments
We are grateful to Prof. Yuri V. Trushin and Prof. Vladimir G. Dubrovskii for helpful discussions of the physics of diffusion, Dr. Viktor G. Zgoda for his discussions of mass spectrometry and PhD student Ivan N. Terterov for his technical assistance. This work was performed under a grant from the Presidium of the Russian Academy of Sciences.
Footnotes
Author Contributions
MVD and YuVN developed the concept and supervised the project, MVD designed the experiments, interpreted the data, proposed conclusions and wrote the manuscript, YuVN provided conceptual advice; SYuV and VMB performed the experiments, analysed the data of liquid chromatography and mass spectrometry; IEE designed the theoretical model; and ENN, IAP and ASK gathered the HPLC-MS/MS data.
References
- Aronson PS, Boron WF, Boulpaep EL. Transport of solutes and water. In: Boron WF, Boulpaep EL, editors. Medical physiology. 2. Philadelphia: Saunders Elsevier; 2009. pp. 106–146. [Google Scholar]
- Brack A. Selective emergence and survival of early polypeptides in water. Orig Life Evol Biosph. 1987;17:367–379. doi: 10.1007/BF02386475. [DOI] [PubMed] [Google Scholar]
- Dubrovskii VG, Nazarenko MV. Nucleation theory beyond the deterministic limit. I. The nucleation stage. J Chem Phys. 2010;132:114507. doi: 10.1063/1.3354118. [DOI] [PubMed] [Google Scholar]
- Eschenmoser A. The search for the chemistry of life's origin. Tetrahedron. 2003;63:12821–12844. doi: 10.1016/j.tet.2007.10.012. [DOI] [Google Scholar]
- Ferris LP, Hill AR, Jr, Liu R, Orgel LE. Synthesis of long prebiotic oligomers on mineral surfaces. Nature. 1996;381:59–61. doi: 10.1038/381059a0. [DOI] [PubMed] [Google Scholar]
- Fox SW. How did life begin? Science. 1960;132:200–208. doi: 10.1126/science.132.3421.200. [DOI] [PubMed] [Google Scholar]
- Freedman J. In: Cell physiology, source book. Biophysical chemistry of cellular electrolytes. Sperelakys N, editor. San Diego: Academic; 1995. pp. 3–17. [Google Scholar]
- Galimov EM, Ryzhenko BN, Cherkasova EV. Estimation of the composition of the Earth's primary aqueous phase. 2. Synthesis from the mantle and igneous rock material. Comparison with synthesis from the carbonaceous chondrite material. Geochem Int. 2011;49:1057–1071. doi: 10.1134/S0016702911110036. [DOI] [Google Scholar]
- Hill AR, Jr, Orgel LE. Oligomerization of negatively-charged amino acids by carbonyldiimidazole. Orig Life Evol Biosph. 1996;26:539–545. doi: 10.1007/BF01808219. [DOI] [PubMed] [Google Scholar]
- Hill AR, Jr, Orgel LE. Oligomerization of L-gamma-carboxyglutamic acid. Orig Life Evol Biosph. 1999;29:115–122. doi: 10.1023/A:1006512304332. [DOI] [PubMed] [Google Scholar]
- Ishihama Y, Rappsilber J, Andersen JS, Mann M. Microcolumns with self-assembled particle frits for proteomics. J Chromatogr A. 2002;979:233–239. doi: 10.1016/S0021-9673(02)01402-4. [DOI] [PubMed] [Google Scholar]
- Jakschitz TA, Rode BM. Chemical evolution from simple inorganic compounds to chiral peptides. Chem Soc Rev. 2012;41:5484–5489. doi: 10.1039/c2cs35073d. [DOI] [PubMed] [Google Scholar]
- Jockusch RA, Lemoff AS, Williams ER. Effect of metal ion and water coordination on the structure of a gas-phase amino acid. J Am Chem Soc. 2001;123:12255–12265. doi: 10.1021/ja0106873. [DOI] [PubMed] [Google Scholar]
- Lide DR, David R. CRC handbook of chemistry and physics. 87. FL: CRC Press; 1998. pp. 76–78. [Google Scholar]
- Miller SL. A production of amino acids under possible primitive Earth conditions. Science. 1953;117:527–528. doi: 10.1126/science.117.3046.528. [DOI] [PubMed] [Google Scholar]
- Mulkidjanian AY, Bychkov AY, Dibrova DV, Galperin MY, Koonin EV. Origin of first cells at terrestrial, anoxic geothermal fields. Proc Natl Acad Sci USA. 2012;109:E821–E830. doi: 10.1073/pnas.1117774109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natochin YV. The physiological evolution of animals: sodium is the clue to resolving contradictions. Her Russ Acad Sci. 2007;77:581–591. doi: 10.1134/S1019331607060068. [DOI] [Google Scholar]
- Natochin YV. The origin of membranes. Paleontol J. 2010;44:860–869. doi: 10.1134/S0031030110070142. [DOI] [Google Scholar]
- Oparin AI. Proiskhozhdenie Zhizny. Moscow: Moskovski Rabochii; 1924. [Google Scholar]
- Oparin AI. The origin of life. New York: Macmillan; 1938. [Google Scholar]
- Rees DC, Howard JB. The interface between the biological and inorganic worlds: iron-sulfur metalloclusters. Science. 2003;300:929–931. doi: 10.1126/science.1083075. [DOI] [PubMed] [Google Scholar]
- Remko M, Rode BM. Effect of metal ions (Li+, Na+, K+, Mg2+, Ca2+, Ni2+, Cu2+, and Zn2+) and water coordination on the structure of glycine and zwitterionic glycine. J Phys Chem A. 2006;110:1960–1967. doi: 10.1021/jp054119b. [DOI] [PubMed] [Google Scholar]
- Rode BM. Peptides and the origin of life. Peptides. 1999;20:773–786. doi: 10.1016/S0196-9781(99)00062-5. [DOI] [PubMed] [Google Scholar]
- Rode BM, Son HL, Suwannachot Y. The combination of salt induced peptide formation reaction and clay catalysis: a way to higher peptides under primitive conditions. Orig Life Evol Biosph. 1999;29:273–286. doi: 10.1023/A:1006540101290. [DOI] [PubMed] [Google Scholar]
- Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Schmid R, Miah AM, Sapunov VN. A new table of the thermodynamic quantities of ionic hydration: values and some applications (enthalpy–entropy compensation and Born radii) Phys Chem Chem Phys. 2000;2:97–102. doi: 10.1039/a907160a. [DOI] [Google Scholar]
- Schwendinger MG, Rode BM. Possible role of copper and sodium chloride in prebiotic evolution of peptides. Analyt Sci. 1989;5:411–414. doi: 10.2116/analsci.5.411. [DOI] [Google Scholar]
- Spirin AS, Gavrilova LP. Ribosome. 2. Moscow: Nauka; 1971. [Google Scholar]
- Switek B (2012) Debate bubbles over the origin of life. Nature. doi:10.1038/nature.2012.10024
- Wang K-J, Yao N, Li C. Sodium chloride enhanced oligomerization of L-glutamic acid in aqueous solution. Orig Life Evol Biosph. 2005;35:313–322. doi: 10.1007/s11084-005-2041-0. [DOI] [PubMed] [Google Scholar]
- Zimmer C. Evolutionary roots. On the origin of life on earth. Science. 2009;323:198–199. doi: 10.1126/science.323.5911.198. [DOI] [PubMed] [Google Scholar]