Abstract
We show that RNA editing sites can be called with high confidence using RNA sequencing data from multiple samples across either individuals or species, without the need for matched genomic DNA sequence. We identified many previously unidentified editing sites in both humans and Drosophila; our results nearly double the known number of human protein recoding events. We also found that human genes harboring conserved editing sites within Alu repeats are enriched for neuronal functions.
RNA editing is the post- or co-transcriptional modification of RNA nucleotides from their genome-encoded sequence. In humans, the most prevalent type is adenosine-to-inosine (A-to-I) editing, catalyzed by the adenosine deaminase acting on RNA (ADAR) family of enzymes1. The ADAR enzymes bind double-stranded RNAs and deaminate adenosine to inosine, which is recognized as guanosine by the cellular machinery. A-to-I editing is pervasive in Alu repeats because of the double-stranded RNA structures formed by inverted Alu repeats in many genes2,3. However, only a few dozen human RNA editing targets that change amino acids in nonrepetitive regions have been identified4, and most of them were identified in nervous system tissues5.
High-throughput RNA sequencing (RNA-seq) has enabled transcriptome-wide identification of A-to-I editing sites. The major challenge in identifying RNA editing sites using RNA-seq data is the discrimination of RNA editing sites from genome-encoded single-nucleotide polymorphisms (SNPs) and technical artifacts caused by sequencing or read-mapping errors. Recently, we and others have developed computational frameworks to identify RNA editing sites by comparing the sequence differences between RNA-seq and matched genomic DNA sequencing from a single individual6–8. This approach is robust in minimizing erroneous variant calls caused by sequencing or read-mapping errors, but it requires deep sequencing of both the transcriptome and the genome from the same sample. Samples with such data are relatively uncommon, and are currently biased toward lymphocyte cell lines, which may not be biologically relevant for RNA editing studies.
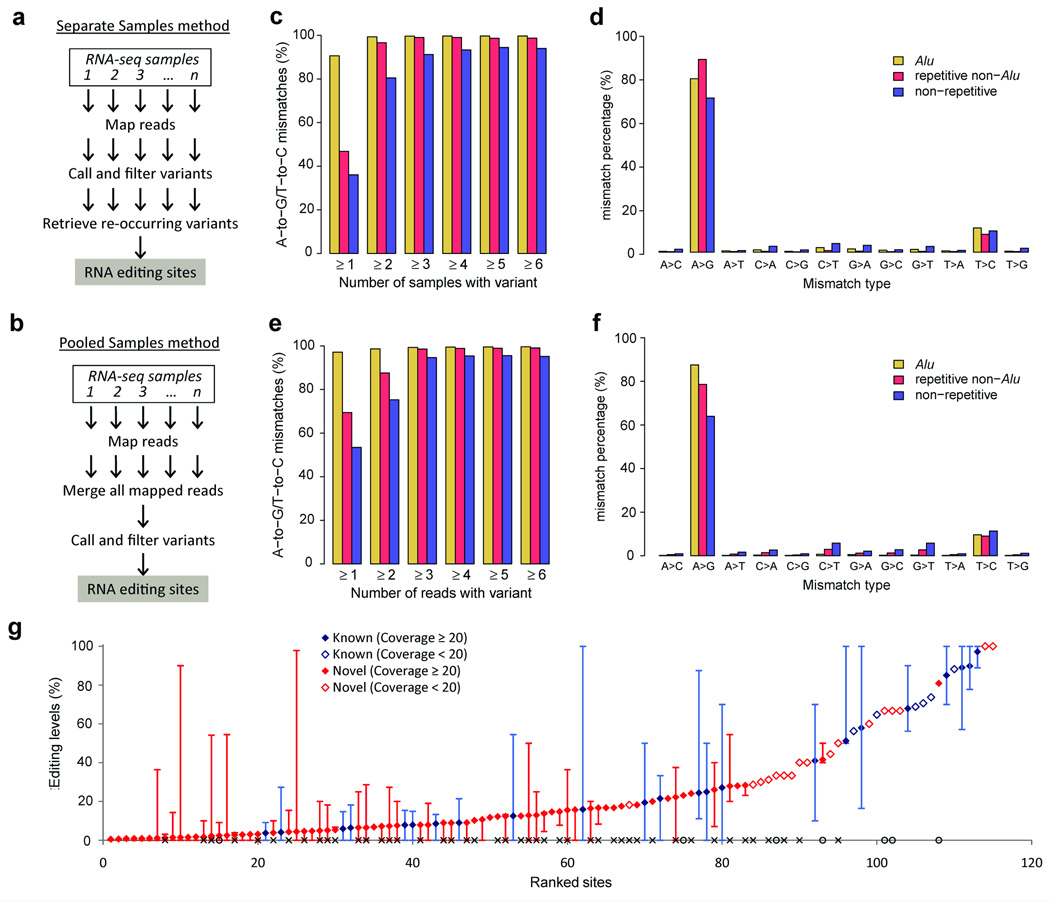
To use the multitude of publicly available RNA-seq data sets for discovery of RNA editing sites, we developed two related and complementary methods to accurately identify RNA editing sites using RNA-seq data from multiple individuals in a single species. In the first method (‘separate samples method’; Fig. 1a), RNA variants are called separately in each RNA-seq sample after mapping sequencing reads to a (nonmatched) genomic reference sequence, and known common genomic SNPs are removed. To distinguish RNA editing sites from rare SNPs in the remaining pool of RNA variants, we took advantage of the fact that the same editing sites are often present in different individuals whereas rare SNPs are most likely not.
Figure 1.
Identification and validation of A-to-I RNA editing sites using RNA-seq data from human brain tissues. (a,b) Overview of the separate samples method (a) and the pooled samples method (b). (c,d) Identification of editing sites using the separate samples method. Minimum number of samples containing each variant relative to the proportion of variants that are either A-to-G or T-to-C mismatches (c). Variants were required to be supported by at least one read in each sample in c. Percentage of all 12 mismatch types (d); variants in Alu and non-Alu regions were required to be present in least one or two samples, respectively. (e,f) Identification of editing sites using the pooled samples method. Minimum number of reads supporting each variant relative to the proportion of variants that are either A-to-G or T-to-C mismatches (e). Percentage of all 12 mismatch types (f); variants in Alu and non-Alu regions were required to be supported by at least one or two reads, respectively. (g) RNA editing levels of 115 nonsynonymous nonrepetitive editing sites measured using the pooled alignments for all 50 brain data samples. Editing sites covered by less than 20 reads are identified as open diamonds. Measurements of RNA editing levels for each site in each sample (where ≥ 10 reads were available) are shown by error bars, with highest and lowest editing levels observed. Previously unidentified sites that were validated are marked with an ‘×’ on the x axis; previously unidentified sites that were not validated by PCR are marked with open circles on the x axis.
In the second method (‘pooled samples method’; Fig. 1b), RNA-seq alignments from different individuals are pooled together to achieve higher read coverage, enhancing the sensitivity for calling RNA variants. RNA variants are called, and common SNPs are removed, similarly to the separate samples method. As rare SNPs are unlikely to be present in multiple individuals, they exist at a very low frequency in the pooled alignment file. The method for mapping RNA-seq reads and calling variants is based on our previously published computational pipeline8 (Online Methods). The hallmark of our pipeline is separate filtering criteria for variants occurring in Alu repeats and variants occurring in non-Alu regions of the genome, resulting in much greater sensitivity in detecting editing sites in Alu repeats (where A-to-I editing is prevalent) and drastically improved specificity for detecting editing sites in non-Alu regions as compared to other methods8. The major modification from our previous pipeline is the use of the Genome Analysis ToolKit (GATK)9 instead of empirically determined parameters for variant calling to provide a uniform statistical framework for variant calling that can be applied to diverse RNA-seq data sets. We noticed that variant calling using empirical parameters instead of using GATK resulted in an abundance of false positive mismatches, especially when the proportion of transcripts being edited, or editing level, is very low (see below).
As a proof of concept, we applied our two methods to identify RNA editing sites using RNA-seq data obtained from 40 human lymphoblastoid cell lines (Supplementary Note 1 and Supplementary Table 1). We found that the majority of mismatches identified using both methods were A-to-G mismatches, indicative of A-to-I editing (Supplementary Fig. 1). We observed a slight enrichment in T-to-C mismatches, the majority of which are incorrectly annotated A-to-G mismatches (Supplementary Note 1 and Supplementary Fig. 2). These same 40 RNA-seq data sets have been used in a previous study10 that provided evidence to support the possibility of noncanonical editing mechanisms. However, more recent studies have shown that these noncanonical mismatches are false positives8,11–16. Our results support the observation that all non–A-to-G mismatches are false positives. Furthermore, we analyzed the same lymphoblastoid RNA-seq data that had been used in the above-mentioned study10, and we only found evidence to support A-to-I editing in these samples. Overall, we identified 303,624 A-to-G variants in Alu repeats, 2,796 A-to-G variants in non-Alu repeats and 2,815 A-to-G variants in nonrepetitive regions using RNA-seq data from lymphoblastoid cell lines (Supplementary Tables 2,3 and Supplementary Data 1,2). We found that more RNA editing sites were called using RNA-seq data only than by comparing sequence differences between RNA and DNA sequencing data using our previous method8 (Supplementary Note 1 and Supplementary Fig. 3). We greatly enhanced sensitivity to detect editing sites by using multiple RNA-seq samples, which allowed us to accurately identify RNA editing sites supported by only one mismatched read in a particular sample (Supplementary Fig. 4).
Next, we applied our approaches to identify RNA editing sites using RNA-seq data obtained from brain tissues of 50 human individuals (Fig. 1 and Supplementary Table 4). Using the separate samples method, we found that RNA variants present in one or more samples in Alu repeats and RNA variants present in two or more samples in non-Alu regions were highly enriched for potential A-to-I editing sites (Fig. 1c,d). Using the pooled samples method, we found that RNA variants with one or more variant reads in Alu repeats and RNA variants with two or more variant reads in non-Alu regions were highly enriched for potential A-to-I editing sites (Fig. 1e,f). We identified 612,573, 13,724, and 12,160 A-to-G variants in Alu repeats, non-Alu repeats, and nonrepetitive regions, repectively using RNA-seq data from human brain tissues (Supplementary Fig. 5, Supplementary Table 2 and Supplementary Data 3,4). As expected for A-to-I editing sites17, these A-to-G variants spanned a wide spectrum of editing levels (Supplementary Fig. 6) and were associated with an under- and overrepresentation of guanosines immediately 5′ and 3′ of the edited adenosine, respectively, although the sequence preferences at these two positions were not completely independent (Supplementary Fig. 7). We also identified RNA editing sites from other human tissues (Supplementary Note 2, Supplementary Fig. 8, Supplementary Tables 2,5 and Supplementary Data 5,6). Altogether, from human RNA-seq data alone we identified 996,012, 16,622 and 15,020 A-to-I RNA editing sites in Alu, repetitive non-Alu and non-repetitive regions, respectively, most of which we identified in the brain samples only (Supplementary Fig. 9).
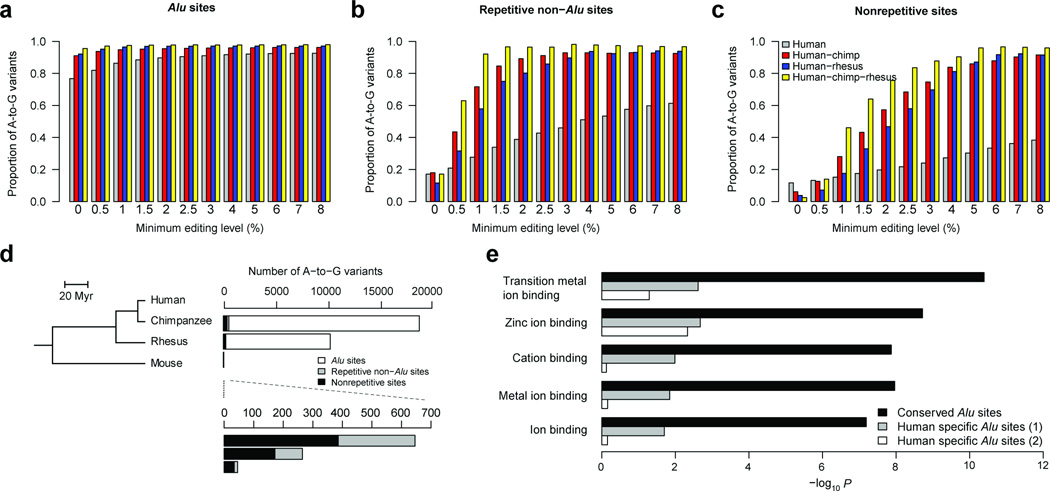
As large numbers of RNA editing sites are identified, it is difficult to pinpoint the functionally important ones. Additionally, the accuracy (proportion of total variants that are A-to-G type, or A-to-G fraction) of the two methods described above in functionally important regions, such as in nonrepetitive coding regions, is not as good as in intronic or untranslated regions (Supplementary Table 3), most likely because of challenges in mapping reads to spliced exons. To address these challenges, we developed a cross-species transcriptome comparison method based on the fact that functionally relevant RNA editing events tend to be conserved between related species, whereas SNPs or false positives, mainly from errors in DNA sequencing and computational mapping, are unlikely to be common to unrelated species (Supplementary Fig. 10). To enrich for functionally relevant editing sites, we focused on identifying conserved RNA variants in exonic regions. We first applied this method to the primate lineage to identify human RNA editing sites conserved in chimpanzee and rhesus macaque (Fig. 2), which diverged from humans ~6 million and ~25 million years ago, respectively18. As RNA editing is implicated in neuronal functions, we used RNA-seq data from primate brains (Supplementary Table 6). In this method, we used empirically determined parameters instead of GATK for variant calling to increase the sensitivity of variant detection especially at low editing levels. In contrast to what we observed with human-only RNA-seq data, we could accurately find conserved editing sites with very low editing levels and we tuned the thresholds of editing levels to maximize the identification of editing sites with tolerable false-discovery rates (Fig. 2a–c). To achieve high accuracy in calling A-to-I edits without a substantial reduction in sensitivity, we chose editing level cutoffs such that the proportion of A-to-G variants to total variants was at least 80% . Assuming that all non–A-to-G mismatches are false and the error rate for all 12 mismatch types is equal, the false discovery rate at this cutoff is (20% / 11) / 80% = 2.3%. However, this rate is conservative because many T-to-C mismatches are actually incorrectly annotated A-to-G mismatches (Supplementary Fig. 2).
Figure 2.
Accurate identification of RNA editing sites in the primate lineage. (a–c) Relationship between the A-to-G proportion of detected mismatches and the minimum editing level in Alu (a), repetitive non-Alu (b) and nonrepetitive regions (c). (d) Phylogenetic relationships among the four analyzed mammalian species (left); Myr: million years ago. Number of conserved A-to-G variants that were identified in human and each of the other three selected species (right), with a magnification for variants in non-Alu regions (bottom). (e) Functional enrichment in transcripts with edited Alu repeats. The conserved editing sites in Alu repeats, sites edited in human and chimpanzee and/or rhesus macaque brains, occur in 1,400 genes. As controls, we collected two groups of genes (1,065 and 831, respectively) with editing sites in Alu repeats that were edited in human only (Online Methods). The P values (Expression Analysis Systematic Explorer (EASE) scores22) shown on the x axis were corrected for multiple hypotheses testing using the Benjamini-Hochberg method.
For variants common to both human and chimpanzee, we identified 17,800 A-to-G edited sites in Alu regions, 308 in repetitive non-Alu regions and 464 in nonrepetitive regions (Supplementary Table 2). For these three types of regions, we used 0%, 1.5% and 4% editing level cutoffs. Similarly, we identified variants present in both human and rhesus macaque and variants present in all three species (Fig. 2a–c and Supplementary Fig. 11). Combining all common sites identified through two- or three-species comparisons, we identified 21,108, 334 and 542 exonic A-to-G edit sites in Alu, repetitive non-Alu and nonrepetitive regions, respectively (Supplementary Data 7,8). Compared to the separate samples and pooled samples methods, we observed an improvement in the accuracy (A-to-G fraction), especially in nonrepetitive coding regions (Supplementary Table 3).
As expected, the vast majority of editing sites in human samples occurred in Alu repeats (Supplementary Table 2), but their function has been largely unexplored. We found that genes harboring conserved Alu sites edited in human as well as other primate species were highly enriched in various ion-binding activities. In contrast, there was no obvious enrichment for genes harboring Alu sites edited only in humans (Fig. 2e), despite a previous observation that genes with human-specific Alu insertions are enriched for neuronal functions19. Thus, Alu sites that are edited in multiple primate species may have been integrated into the neuronal function of RNA editing in primates. Our cross-species comparison method will facilitate the identification of these functionally relevant editing sites in Alu repeats.
From the brain data set, we identified 115 nonrepetitive nonsynonymous editing sites, 87 of which have not been previously identified (Fig. 1g). Genes with amino acid–recoding sites were highly enriched for ion transporter and ion channel activities (Supplementary Fig. 12), consistent with the known neuronal functions of RNA editing5. We validated 47 of these previously unidentified sites using Sanger sequencing and targeted deep sequencing of three human brain samples, nearly doubling the total number of validated human recoding sites (Supplementary Note 3, Supplementary Figs. 13,14 and Supplementary Table 7). Given the spatiotemporal dynamics of RNA editing, it is possible that more sites can be validated in additional human samples.
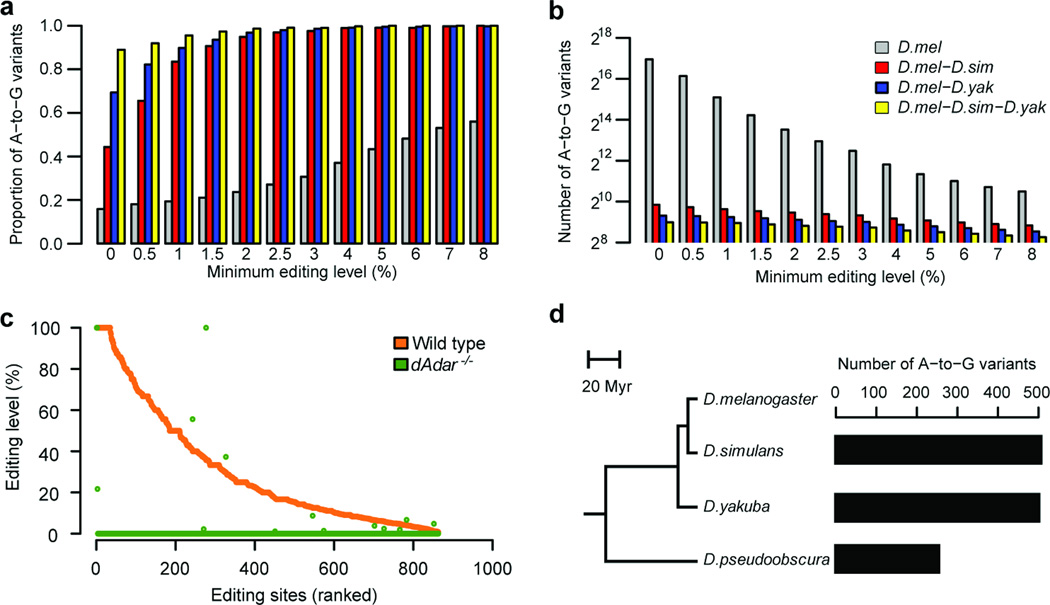
We extended our analysis beyond the primate lineage by applying our cross-species comparative transcriptome method to identify exonic RNA editing sites in Drosophila melanogaster. We obtained RNA-seq data from adult whole bodies of D. melanogaster, D. simulans and D. yakuba; the latter two species diverged from D. melanogaster ~5 million and ~11 million years ago, respectively20 (Supplementary Table 8). We then identified variants present in D. melanogaster that were also present in D. simulans, D. yakuba or both. Similar to the primate lineage, we found that common RNA variants were highly enriched for A-to-G mismatches (Fig. 3a). The A-to-G mismatch proportion approached 100% with increasing thresholds of editing level requirement, suggesting that A-to-I RNA editing is the only conserved editing type in Drosophila.
Figure 3.
RNA editing site identification in Drosophila. (a) A-to-G proportion of detected mismatches relative to the minimum editing level for the species labeled in b. (b) Number of A-to-G variants relative to the minimum editing level. (c) RNA editing levels of 863 editing sites measured from the heads of male wild-type and Adar5G1 mutant flies. (d) Phylogenetic relationships among the four analyzed Drosophila species (left). Myr: Million years ago. Numbers of A-to-G variants identified in D. melanogaster and each of the other three selected Drosophila species analyzed (right).
We chose the minimal editing levels for comparisons of D. melanogaster and D. simulans (1%), D. melanogaster and D. yakuba (0.5%), and D. melanogaster and D. simulans-D. yakuba (0%) by requiring the proportion of A-to-G mismatches to be at least 80%, and identified 793, 628 and 508 exonic sites, respectively (Fig. 3a,b). To identify more editing sites, we analyzed two additional data sets from D. melanogaster: the whole-body transcriptome of 1-d-old y1;cn bw1 sp1 flies and the head transcriptome of another D. melanogaster strain, OregonR. We identified 1,038 and 937 exonic A-to-G sites with high specificity, respectively (Supplementary Fig. 15). In total we identified 1,327 A-to-I editing sites in D. melanogaster, including 847 newly identified sites (Supplementary Fig. 16a and Supplementary Data 9,10) and 452 amino acid–recoding sites (Supplementary Fig. 16b). Genes with newly identified sites were highly enriched in various channel activity, ion transport and neurotransmitter transport functions (Supplementary Fig. 16c), consistent with the known neuronal function of RNA editing events in Drosophila21.
To validate whether the identified A-to-G sites were bona fide A-to-I editing events, we performed RNA-seq for the D. melanogaster wild-type strain (w1118) and for the Adar5G1 mutant that eliminates RNA editing (Online Methods). Of all 1,327 identified A-to-G sites, we examined 863 that were edited in the wild-type head RNA sample. As expected, we achieved high accuracy; 98.2% of all A-to-G sites showed only adenosine in the Adar5G1 sample (Fig. 3c).
Our cross-species comparisons in both primate and Drosophila lineages allowed us to investigate the relationship between the number of identified editing events and genetic distance. The number of identified sites inversely correlated with the genetic distance between two species under comparison (Figs. 2d,3d, Supplementary Note 4 and Supplementary Fig. 17). In the primate lineage, human had almost twice as many editing sites in common with chimpanzee as it did with rhesus macaque, suggesting a recent origin of many editing sites (mostly in Alu sites) in the great ape lineage. Whether this rapid turnover of RNA editing is due to lineage-specific adaptation or the lack of evolutionary constraint needs further investigation.
In contrast to previous methods that rely on coupled RNA and DNA sequencing6–8, we identified RNA editing sites using RNA sequencing data by itself. This allowed us to explore RNA editing in a wide variety of human, primate and Drosophila samples where RNA-seq data are widely available. Summing human A-to-I editing sites identified in this and previous work yields a total of 1,319,602, 24,322 and 20,622 sites in Alu regions, repetitive non-Alu regions and nonrepetitive regions, respectively (Supplementary Fig. 18), a notable expansion in the catalog of human RNA editing sites. This public database of human editing sites can also be used to identify RNA editing sites in a single RNA-seq sample of interest (Supplementary Note 5 and Supplementary Fig. 19). We anticipate that our methods will be even more effective in the future as additional RNA-seq data become available.
ONLINE METHODS
RNA-seq data collection
We obtained unstranded Illumina RNA-seq data from the US National Center for Biotechnology Information Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra/) and the modENCODE project (http://www.modencode.org/). Details about the data samples are available in Supplementary Tables 1,4,5,6,8.
Mapping of RNA-seq reads
We adopted our previously published pipeline to accurately map RNA-seq reads onto the genome. In brief, we used the Burrows-Wheeler Algorithm (BWA)23 to align RNA-seq reads to a combination of the reference genome and exonic sequences surrounding known splice junctions from available gene models. We chose the length of the splice junction regions to be slightly shorter than the RNA-seq reads to prevent redundant hits. The reference genomes used were: human, hg19; chimpanzee, ptr2; rhesus monkey, rhe2; mouse, mm9; D. melanogaster, dm3; D. simulans, dsim1; D. yakuba, dyak2; and D. pseudoobscura, dps3. We obtained gene models from University of California Santa Cruz (UCSC) genome browser: D. melanogaster, FlyBase genes; D. simulans, Genscan genes; D. yakuba, Genscan genes; D. pseudoobscura, Genscan genes; human, a combination of Gencode, RefSeq, Ensembl and UCSC genes; chimpanzee, Ensembl genes; rhesus macaque, Ensembl genes; and mouse, Ensembl genes. We used the MarkDuplicates tool from Picard (http://picard.sourceforge.net/) to remove identical reads (PCR duplicates) that mapped to the same location. For human RNA-seq alignments, GATK tools IndelRealigner and TableRecalibration were used to perform local realignment around insertion and/or deletion polymorphisms and to recalibrate base quality scores.
Variant calling and filtering
For the human RNA-seq–only methods (separate samples and pooled samples), we called variants using the GATK9 UnifiedGenotyper tool with options stand_call_conf of 0 and stand_emit_conf of 0. We required variants to be supported by at least one mismatched read with a base quality score ≥ 25 and a mapping quality score ≥ 20. We removed all known SNPs present in dbSNP (except SNPs of molecular type “cDNA”; database version 135; http://www.ncbi.nlm.nih.gov/SNP/), the 1000 Genomes Project, and the University of Washington Exome Sequencing Project (http://evs.gs.washington.edu/EVS/). Additional filters were used to remove false positive mismatches as previously described8. In brief, we discarded mismatches in the first six bases of each read to avoid artificial mismatches derived from random-hexamer priming. In non-Alu regions, we removed intronic candidates if they were located within 4 base pairs of a known splice junction, removed sites in homopolymer runs of ≥ 5 base pairs and removed sites in simple repeats. We also removed sites in regions that were highly similar to other parts of the genome using the BLAST-Like Alignment Tool (BLAT). Finally, we excluded variant sites in hypervariable regions of the genome (UCSC transcripts named as ‘abParts’). We inferred the editing type of each site based on the strand of overlapping annotated genes. Regions with bidirectional transcription (sense and antisense gene pairs) were discarded.
For the cross-species comparative method, we took variant positions where the mismatch was supported by two or more reads with a base quality score of ≥ 20 and a mapping quality score ≥ 20. We used additional filters to remove false positive mismatches as described above. For the mouse data, we removed all known SNPs present in dbSNP (except SNPs of molecular type “cDNA”; database version 135) and all known SNPs identified by the Sanger Mouse Genomes Project (http://www.sanger.ac.uk/resources/mouse/genomes/). For Drosophila species, we excluded variant sites with an extreme degree of variation (100%), which are likely genomic SNPs.
Cross-species position conversion
The LiftOver tool was used to convert genomic positions between different species. As LiftOver does not provide strand information between two species (for example, a sense strand in one species corresponds to a reverse stand in another species), pairwise alignment files downloaded from UCSC genome browser were used to extract the strand information.
Sequence preferences and GO analyses
The ADAR-binding sequence preferences were plotted using two-sample Logo tool24. Background nucleotides were chosen as random adenosines in genes harboring editing sites. The Database for Annotation, Visualization and Integrated Discovery (DAVID)22 was used to perform Gene Ontology analysis. The list of all human genes was used as background for enrichment analyses. We collected two different groups of genes with editing sites Alu repeats that are edited in human only. For group 1 genes, we first collected all genes with exonic Alu editing sites in the brain data set. We then selected genes that are edited in humans only by excluding genes that have Alu editing sites conserved in either chimpanzee or rhesus macaque. For group 2 genes, we first collected all genes with previously known exonic Alu editing sites from the Database of RNa Editing (DARNED)25 database and two recent genome-wide RNA editing identification publications7,8. We then selected genes that are observed in humans only by excluding genes that have Alu editing sites conserved in either chimpanzee or rhesus macaque.
Validation of Drosophila A-to-I editing sites using wild-type and male head Adar5G1 RNA-seq data
We collected heads of 5-d-old male Adar−/− mutant (y, 5G1 allele, w)26 and wild-type (w1118) flies. Poly(A)+ RNA was used to prepare RNA-seq libraries, which were subsequently single-end sequenced by an Illumina GAII. Sequences were mapped as described above. We examined all identified A-to-G sites that are edited in the wild type strain. Sites that are not edited in the Adar−/− mutant were considered to be genuine A-to-I RNA editing sites.
Edited gene conservation analysis
Orthologous gene relationship between human and D. melanogaster was obtained via Ensmbl Biomart (http://www.ensembl.org/biomart/martview). ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/) was used to align orthologous proteins.
The relationship between the number of common editing sites and genetic distance
For mammalian species, rhesus macaque has the lowest sequencing coverage with ~8,000 million mappable bases. For a fair comparison, RNA-seq data with similar number of mappable bases to rhesus macaque in chimpanzee and mouse were used. Common variants for human and one of these species were identified.
For Drosophila species, D. pseudoobscura has the lowest sequencing coverage with ~1,300 million mappable bases. For a fair comparison, RNA-seq data with similar number of mappable bases to D. pseudoobscura in D. simulans and D. yakuba were used. Variants common to one of these species above and the same variant set (5-d-old y1; cn bw1 sp1 RNA-seq data set) were identified.
Validation of sites with PCR and deep sequencing
For each selected site, we designed PCR primers for both cDNA and genomic DNA (gDNA). Primer sequences are listed in Supplementary Table 9. We obtained three RNA samples: (1) frontal lobe, (2) equal amount RNA from cerebellum, corpus callosum, diencephalon, frontal lobe, parietal lobe and temporal lobe, and (3) a Brain Reference Total RNA sample pooled from brain samples of 23 individuals (Ambion, 6050). Samples 1 and 2 were from the same individual (a 26-year-old male), and gDNA from the frontal lobe of the same individual was also obtained (all from Biochain Institute). The gDNA of sample 3 was unavailable to us. PCRs were set up as described below. All amplicons of each sample were pooled together, and the four samples were barcoded by a secondary round of PCR. All four pools were then combined and purified via QIAquick Gel Extraction Kit (Qiagen). The resulting library was loaded onto an Illumina MiSeq instrument and analyzed by 50-base-pair single-end read with index sequencing (all editing sites were designed to be within 50 base pairs from the sequencing primer). Reads were mapped as described above. For each site, we only considered reads with a mapping quality score ≥20 and a base quality score ≥20. Rates of sequencing errors of A-to-G and T-to-C were estimated using the gDNA sequencing data (of all 50 base pairs). Statistically significant editing sites were determined by applying Fisher’s exact test to compare the observed and expected A-to-G or T-to-C occurrences in each editing site27. P values were corrected using the Benjamini-Hochberg method, and a confidence level of 0.05 was used as the cutoff.
Validation of sites with PCR analysis and Sanger sequencing
We used Sanger sequencing to validate whether a subset of candidate sites are edited in vivo. We obtained cDNA and gDNA from the cerebellum of a 26-year-old human male (Biochain Institute). Typically, a 12-µl PCR was assembled with 1 × iQ SYBR Green Supermix (Bio-Rad), ~10 ng of gDNA (or ~5 ng of cDNA) template, and 125 nM each of the forward and reverse primers. We used the following touch-down PCR program: 95 °C for 5 min, 24 cycles of 95 °C for 30 s, 72 °C for 30 s with a decrement of 0.7 °C every cycle, and 72 °C for 45 s, then 40 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 45 s. PCR amplicons were sequenced by Eurofins MWG Operon. Primer sequences are listed in Supplementary Table 10.
Supplementary Material
ACKNOWLEDGMENTS
We thank E. Levanon, A. Fire and members of Li lab for constructive discussions, S. Blair for assistance with data set curation, And modENCODE Consortium for the use of Drosophila RNA-seq data sets. G.R. and P.D. were supported by the Stanford Genome Training Program funded by the US National Institutes of Health. R.Z. was partially supported by a Dean’s fellowship from Stanford University School of Medicine. R.P. was supported by a fellowship from the German Academic Exchange Service. This work was supported by startup funds from Stanford University Department of Genetics and Ellison Medical Foundation (to J.B.L.) and Medical Research Council, UK (to M.A.O.).
Footnotes
Accession codes. Gene Expression Omnibus: GSE42815 (sequencing data for wild-type and Adar5G1 Drosophila strains).
Note: Supplementary information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS
G.R. and R.Z. performed computational analyses with help from R.P., P.D. and J.B.L.; R.Z. and G.R. carried out the validation experiments; L.P.K. and M.A.O. generated RNA-seq data for wild-type and Adar−/− flies; and G.R., R.Z. and J.B.L. wrote the paper with input from other authors.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Nishikura K. Annu. Rev. Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. Proc. Natl. Acad. Sci. USA. 1994;91:11457–11461. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levanon EY, et al. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 4.Eisenberg E, Li JB, Levanon EY. RNA Biol. 2010;7:248–252. doi: 10.4161/rna.7.2.11565. [DOI] [PubMed] [Google Scholar]
- 5.Rosenthal JJ, Seeburg PH. Neuron. 2012;74:432–439. doi: 10.1016/j.neuron.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bahn JH, et al. Genome Res. 2012;22:142–150. doi: 10.1101/gr.124107.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peng Z, et al. Nat. Biotechnol. 2012;30:253–260. doi: 10.1038/nbt.2122. [DOI] [PubMed] [Google Scholar]
- 8.Ramaswami G, et al. Nat. Methods. 2012;9:579–581. doi: 10.1038/nmeth.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DePristo MA, et al. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li M, et al. Science. 2011;333:53–58. doi: 10.1126/science.1207018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kleinman CL, Adoue V, Majewski J. RNA. 2012;18:1586–1596. doi: 10.1261/rna.033233.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kleinman CL, Majewski J. Science. 2012;335:1302. doi: 10.1126/science.1209658. [DOI] [PubMed] [Google Scholar]
- 13.Lin W, Piskol R, Tan MH, Li JB. Science. 2012;335:1302. doi: 10.1126/science.1210419. [DOI] [PubMed] [Google Scholar]
- 14.Pickrell JK, Gilad Y, Pritchard JK. Science. 2012;335:1302. doi: 10.1126/science.1210484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schrider DR, Gout JF, Hahn MW. PLoS ONE. 2011;6:e25842. doi: 10.1371/journal.pone.0025842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piskol R, Peng Z, Wang J, Li JB. Nat. Biotechnol. 2013;31:19–20. doi: 10.1038/nbt.2472. [DOI] [PubMed] [Google Scholar]
- 17.Li JB, et al. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 18.Goodman M. Am. J. Hum. Genet. 1999;64:31–39. doi: 10.1086/302218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paz-Yaacov N, et al. Proc. Natl. Acad. Sci. USA. 2010;107:12174–12179. doi: 10.1073/pnas.1006183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamura K, Subramanian S, Kumar S. Mol. Biol. Evol. 2004;21:36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]
- 21.Hoopengardner B, Bhalla T, Staber C, Reenan R. Science. 2003;301:832–836. doi: 10.1126/science.1086763. [DOI] [PubMed] [Google Scholar]
- 22.Huang da W. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 23.Li H, Durbin R. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vacic V, Iakoucheva LM, Radivojac P. Bioinformatics. 2006;22:1536–1537. doi: 10.1093/bioinformatics/btl151. [DOI] [PubMed] [Google Scholar]
- 25.Kiran A, Baranov PV. Bioinformatics. 2010;26:1772–1776. doi: 10.1093/bioinformatics/btq285. [DOI] [PubMed] [Google Scholar]
- 26.Palladino MJ, Keegan LP, O'Connell MA, Reenan RA. Cell. 2000;102:437–449. doi: 10.1016/s0092-8674(00)00049-0. [DOI] [PubMed] [Google Scholar]
- 27.Picardi E, et al. Nucleic Acids Res. 2010;38:4755–4767. doi: 10.1093/nar/gkq202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.