Abstract
Breast cancer is the leading cause of cancer mortality in women between the ages of 15 and 54. During mammography screening, radiologists use a strict lexicon (BI-RADS) to describe and report their findings. Mammography records are then stored in a well-defined database format (NMD). Lately, researchers have applied data mining and machine learning techniques to these databases. They successfully built breast cancer classifiers that can help in early detection of malignancy. However, the validity of these models depends on the quality of the underlying databases. Unfortunately, most databases suffer from inconsistencies, missing data, inter-observer variability and inappropriate term usage. In addition, many databases are not compliant with the NMD format and/or solely consist of text reports. BI-RADS feature extraction from free text and consistency checks between recorded predictive variables and text reports are crucial to addressing this problem.
We describe a general scheme for concept information retrieval from free text given a lexicon, and present a BI-RADS features extraction algorithm for clinical data mining. It consists of a syntax analyzer, a concept finder and a negation detector. The syntax analyzer preprocesses the input into individual sentences. The concept finder uses a semantic grammar based on the BI-RADS lexicon and the experts’ input. It parses sentences detecting BI-RADS concepts. Once a concept is located, a lexical scanner checks for negation. Our method can handle multiple latent concepts within the text, filtering out ultrasound concepts. On our dataset, our algorithm achieves 97.7% precision, 95.5% recall and an F1-score of 0.97. It outperforms manual feature extraction at the 5% statistical significance level.
Keywords: BI-RADS, free text, lexicon, mammography, clinical data mining
I. Introduction
Breast cancer is the most common type of cancer among women. Researchers estimated that 636, 000 cases occurred in developed countries and 514, 000 in developing countries during 2002 [1]. Currently, a woman living in the US has a 12.3% lifetime risk of developing breast cancer [2]. There is considerable evidence that mammography screening is effective at reducing mortality from breast cancer [3].
The American College of Radiology (ACR) developed a specific lexicon to homogenize mammographic findings and reports. The BI-RADS (Breast Imaging Reporting and Data System [4]) lexicon consists of 43 descriptors organized in a hierarchy (Fig. 1).
Fig. 1.
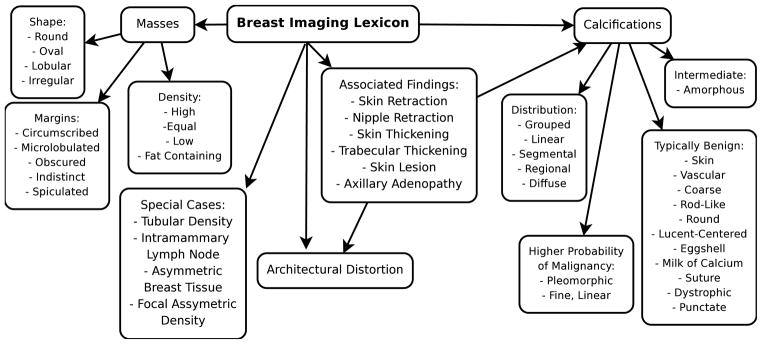
BI-RADS lexicon
Mammography practice is heavily regulated and mandates quality assurance audits over the generated data. Radiologists often use structured reporting software to support required audits. The ACR developed a database format, the National Mammography Database (NMD), which standardizes data collection [5]. NMD is a structured database that combines BI-RADS features with various demographic variables. Radiologists can describe and record their mammography interpretations directly into a NMD-compliant form. Databases containing BI-RADS and NMD features have been used to build successful breast-cancer models and classifiers [6]–[9].
Nevertheless, NMD databases suffer from inconsistencies. There is still a substantial inter-observer variability in the application of the BI-RADS lexicon [10], including inappropriate term usage and missing data. Consistency checks between recorded predictive variables and text reports are necessary before the data can be used for decision support [6]. Natural language processing techniques can parse textual records and recover missing data. Extracting BI-RADS features from free-text can help address these problems.
The need for BI-RADS feature extraction is further amplified by the fact that many databases are not compliant with the NMD format and/or solely consist of text reports. Radiologists variably follow the BI-RADS guidelines to write semi-structured free-text reports, and any further analysis of such databases needs mammography terminology indexing using words or concepts.
This paper presents a general method for information extraction from loosely structured free-text as a prerequisite for clinical data mining. Our method takes full advantage of the available lexicon and incorporates expert knowledge. We apply this method to a free-text mammography database. We compare our method to a 100-record subset manually indexed by a practicing radiologist, who is one of the authors, fellowship-trained in breast imaging [11].
II. Background
Only one prior study addresses BI-RADS information extraction from compliant radiology reports. This research used a Linear Least Squares Fit to create a mapping between mammography report words-frequency and BI-RADS terms [11]. It makes minimal use of lexical techniques. However, several researchers tackled the similar problem of clinical information extraction from medical discharge summaries.
Most approaches to processing clinical reports heavily rely on natural language processing techniques. For instance, the MedLEE processor [12], [13] is capable of complex concept extraction in clinical reports. It first parses the text, using a semantic grammar to identify its structure. It then standardizes the semantic terms and maps them to a controlled vocabulary.
In parallel, the emergence of medical dictionaries emphasizes a phrase-match approach. The National Library of Medicine’s (NLM) Unified Medical Language System [14] (UMLS) compiles a large number of medical dictionaries and controlled vocabulary into a metathesaurus, a thesaurus of thesauri, which provides a comprehensive coverage of biomedical concepts. The UMLS metathesaurus was used to index concepts and perform information extraction on medical texts [15], [16]. Similar approaches have been used with more specialized terminology metathesauri, like caTIES and SNOMED CT [17]. The BI-RADS lexicon can be seen as a metathesaurus for our task.
Negation presents another substantial challenge for information extraction from free text. In fact, pertinent negative observations often comprise the majority of the content in medical reports [18]. Fortunately, medical narrative is a sublanguage limited in its purpose, and its documents are lexically less ambiguous than unrestricted documents [19]. Clinical negations thus tend to be much more direct and straightforward, especially in radiology reports [20]. A very small set of negation words (“no”, “not”, “without”, “denies”) accounts for the large majority of clinical negations [20], [21].
Negation detection systems first identify propositions, or concepts, and then determine whether the concepts are negated. Basic negation detection methods are based on regular expression matching [20], [21]. More recent approaches add grammatical parsing [22], triggers [23] and recursion [24].
Finally, most clinical reports are dictated. They contain a high number of grammatically incorrect sentences, misspellings, errors in phraseology, transcription errors, acronyms and abbreviations. Very few of these abbreviations and acronyms can be found in a dictionary, and they are highly idiosyncratic to the domain and local practice [25]. For this reason, expert knowledge can contribute to effective data extraction.
III. Materials and Methods
We next present our algorithm and further describe the dataset on which we evaluated it.
A. Algorithm Overview
The BI-RADS lexicon clearly depicts 43 distinct mammography features. In radiology reports, these concepts are not uniformly described. Radiologists use different words to refer to the same concept. Some of these synonyms are identified in the lexicon (e.g. “equal density” and “isodense”), while others are provided by experts (e.g. “oval” and “ovoid”). Some lexicon words are ambiguous, referring to more than one concept, or to no concept at all. The word “indistinct” may refer to the “indistinct margin” or to the “amorphous/indistinct calcification” concepts. Or it may be used in a non-mammography context, like “the image is blurred and indistinct”.
Therefore, to map words and phrases in the text into concepts, we cannot solely rely on the lexicon. We supplement it by a semantic grammar. The grammar consists of rules specifying well-defined semantic patterns and the underlying BI-RADS categories into which they are mapped.
Our algorithm has three main modules (Fig. 2). Given the free-text BI-RADS reports, it applies a syntax preprocessor. Then the semantic parser maps subsentences to concepts. Finally a lexical scanner detects negated concepts and outputs the BI-RADS features.
Fig. 2.
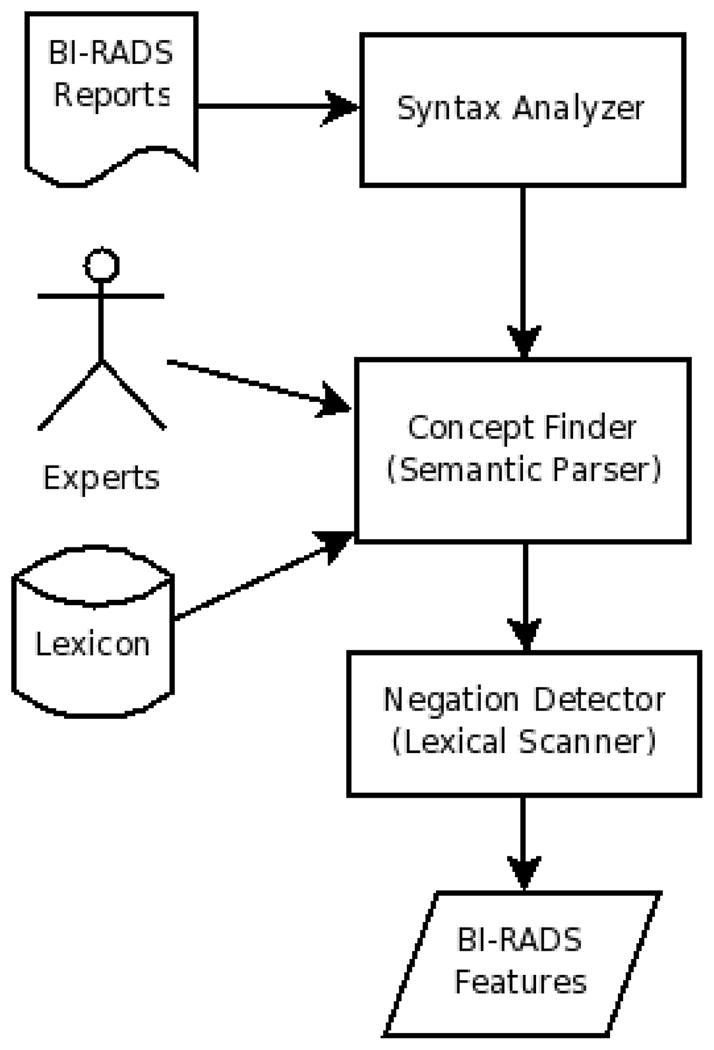
Algorithm flowchart
B. Syntax Analyzer
The first module in our system is a preprocessing step that performs syntactic analysis. Since BI-RADS concepts do not cross sentence boundaries, we process the reports by individual sentences. We simply assume that punctuation delimits sentence boundaries. This is not perfect—for example “St. Jude’s Hospital” would be improperly partitioned—but we have not found this to be an issue in our testbed. We then remove all remaining punctuation. We keep stop words because some of them are used in the negation detection phase.
C. Concept Finder
The concept finder module takes the syntactic token (a sentence) and applies grammar rules to search for concepts. We base the semantic grammar on the lexicon and augment it using manual scanning and semiautomated learning with experts’ input to closely capture the clinical practice. Experts provide, among other things, domain synonyms, acronyms and idiosyncrasies. We formulate the rules as a context free grammar, and express them using Perl’s pattern matching capacities [26].
We found that a clearly defined list of terms (describing important, domain-specific patterns of usage) was critical for our data extraction task. For example, the “regional distribution” concept requires the presence of the word “regional” without being followed by the words “medical” or “hospital”, while the concept of “skin lesion” is shown by the presence of both words “skin” and “lesion” within close proximity. We established the order, if any, of these words and their proximity degree by monitoring the rule performance over the training set.
Due to different word forms and misspellings, we use stem-words and ease our matching constraints. For example, we map the words “pleomorph”, “pleomorphic”, “plimorph”, “plemorph” and “plmorfic”, among others, to the “pleomorphic calcifications” concept.
For example, the “oval shape” concept is defined by two rules. The first is the word “oval” or “ovoid” followed, within a ten words span, by words containing “dens”, “mass”, “struc”, “asym” or “nodul”. The second rule is a word containing “mass” or “nodul”, followed by the word “oval” or “ovoid” within a five words span. The experts provided the synonym “ovoid” as well as the delimiting words. Representing “density” as a word containing “dens” allows us to match “isodense”, “dense” and “densities”. We varied the proximity degrees and opted for the ones with the best accuracy over the training set (ten and five, respectively).
Using the grammar rules, we parse the whole report searching for concepts. The concept finder outputs extracted subsentences that are mapped to concepts. These subsentences can overlap. For example, the “oval shape” concept subsentence, “oval 12 × 18 mm circumscribed density”, contains a “circumscribed margin” concept formed by the “circumscribed” subsentence. Each of these two subsentences, within the same sentence, is a different token. If the same concept occurs repeatedly, we treat each occurrence individually. We can thus report features for multiple findings in a single mammogram.
D. Negation Detector
Once the semantic grammar detects a concept occurrence, it hands the subsentence token to the negation detection module. The negation detection module is a lexical scanner that searches for negation signals using regular expressions. It analyzes their negation scope to determine if they apply over the concept.
Following the approach of Gindl et al. [23], we identify adverbial (“not”, if not preceded by “where”) and intra-phrase (“no”, “without”) negation triggers. Similar to previous findings [20], we find that negation triggers usually precede, but sometimes succeed, the concepts they act upon. In addition, we report negation from within the concept. Since our approach maps a concept to a subsentence, the negation trigger may appear within the concept’s underlying indexed text structure. For instance, the word “mass” followed by “oval” within 5 words, is a rule for the “oval shape” concept. The subsentence “mass is not oval” is a negation within the concept.
We also note that there may be several words between the negation trigger and the concept it negates, and a single trigger may negate several concepts. The maximum degree of word separation between a trigger and its concept, referred to as the negation scope, differs among concepts. Accurate analysis of scope may involve lexical, syntactic, or even semantic analysis. We establish each concept’s negation scope by counting and looking at a subset of the trigger’s hits over the unlabeled training set. Starting with a high scope, we assess the number of false positives we get. With smaller scopes, we can assess the number of false negatives. We choose the scope that minimizes the error ratio. For example, we allow a maximum of 5 words between a negation trigger and the “round shape” concept’s subsentence; while the scope is 8 words for the “grouped distribution” concept.
Since we treat each concept occurrence individually, we can correctly detect a concept in a sentence containing both the concept and its negation. We hence avoid the pitfall of erroneously rejecting a concept encountered by Chapman et al. [21], who negated the entire concept if a single instance of that concept was negated.
While analyzing negation errors, Mutalik et al. [20] reported errors caused by double negatives. We address this issue using the same approach to detect negation triggers. We identify a set of double-negation triggers which, when coupled with negation triggers, deactivate them. These signals are: “change”, “all”, “correlation”, “differ” and “other”. Therefore “there is no change in rounded density” does not negate the concept “round shape”.
As a working example, the concept finder detects the “round shape” concept and passes the subsentence “rounded density” as a token to the negation detector. The negation detector searches for a set of negation triggers before and within the subsentence, and finds the trigger “no”. The trigger is located two words before the subsentence, well within its negation scope of five words. The subsentence token becomes “no change in rounded density”. The negation detector now searches for a set of double-negation triggers within the subsentence, and finds the trigger “change”. It concludes that the concept is not negated.
Given a concept subsentence token, the negation detector outputs a Boolean value: 0 for a negated subsentence, and 1 for a non-negated subsentence. For each mammography report, our algorithm sums the Boolean outputs into a feature vector, which depicts the number of times a concept occurred in a certain report.
E. Handling Latent Concepts
Multiple latent concepts may exist in a given report. For instance, our mammography reports often contain ultrasound concepts. Ultrasound and mammography concepts can have common underlying words, thus the need to discriminate them. A “round mass” is a BI-RADS feature, while a “round hypoechoic mass” is an ultrasound feature. We use an ultrasound lexicon, composed of the concepts “echoic” and “sonogram” and apply the same approach (Fig. 2) to detect ultrasound concepts. We require that a BI-RADS concept not share common subsentences with an ultrasound concept. Our method is thus able to handle multiple latent concepts within the text.
F. Dataset
Our database consists of 146 972 consecutive mammograms recorded at the University of California San Francisco Medical Center (UCSFMC) between January 6, 1997 and June 27, 2007. This database does not follow the NMD format and contains BI-RADS free-text reports. As a preprocessing step, we wrote a program to match the mammograms to their reports and remove redundancies. We were left with 146 198 reports for our analysis. An information extraction step is crucial for any subsequent clinical data mining or modeling of the UCSFMC database.
To test our method, we compare our algorithm’s results to manual information extraction performed by radiologists. Our testing set consists of 100 records from the database that a radiologist on our team manually indexed in 1999 [11]. Each record has a Boolean feature vector of 43 elements representing the BI-RADS lexicon categories (see Fig. 1). The information extraction task is to correctly populate the 43 × 100 = 4300 elements matrix by assigning an element to 1 if its corresponding BI-RADS feature is present in the report, and to 0 otherwise. The manual method extracted a total of 203 BI-RADS features, leaving 4097 empty slots.
IV. Results
A. First Run
We first perform a double-blind run. We manually altered the algorithm using the UCSFMC database except for the 100 hand-curated records, which are solely used for testing. The algorithm extracts a total of 216 BI-RADS features, out of which 188 are in agreement with the manual extraction. In 43 cases, only one of the methods claims the presence of a BI-RADS feature. Upon review of these disparate results, a radiologist determined that our algorithm correctly classified 28 cases while the manual method correctly classified 15.
Clearly the manual method, applied in 1999, does not constitute ground truth. In fact, correctly labeling a text corpus is complicated enough that even experts need several passes to reduce labeling errors [27]. Due to the high labeling cost, in practice one must rely on the imperfect judgments of experts [28]. Since time spent cleaning labels is often not as effective as time spent labeling extra samples [29], our reviewing radiologist reexamined only the diverging cases.
We consider as ground truth the features that both computational and manual methods agree on, in addition to the relabeling of diverging cases by experts. This approach is likely underestimating the number of true features. The omission error of a method is bounded by the number of diverging cases correctly labeled by the other method. We assume that the classifier and the labelers make errors independently, since humans and computers generally classify samples using different methodologies. We use Lam and Stork’s method of handling noisy labels [29]: we treat the classification differences between the two methods as apparent errors, and the classification differences between each method and ground truth as labeling errors. We factor both error terms to get the true classification errors and the confusion matrices for both our algorithm and the manual method (Table I).
TABLE I.
Automated and manual extraction, 1st run
| Method | Predicted | Actual
|
|
|---|---|---|---|
| Feature present | Feature absent | ||
| Automated | Feature present | 211 | 5 |
| Feature absent | 10 | 4074 | |
| Manual | Feature present | 198 | 5 |
| Feature absent | 23 | 4074 | |
To compute test statistics, we treat the present features as positives and the absent features as negatives. Our data being highly skewed, we employ precision-recall analysis instead of accuracy. For the double-blind run, the manual method achieves a 97.5% precision, a 89.6% recall rates and a 0.93 F1-score. Our algorithm achieves a much better recall (95.5%) and F1-score (0.97) for a similar precision (97.7%). It correctly classifies 65.1% of the disputed cases.
To compare both methods, we use the probabilistic interpretation of precision, recall and F-score [30]. Using a Laplace prior, the probability that the computational method is superior to the manual method is 97.6%. Our result is statistically significant at the 5% level (p-value = 0.024).
B. Second Run
Before the first run, we only adjusted the algorithm using unlabeled data. After performing the first run on labeled data, the experts suggested slight changes to some of the rules. We consider this modified version our final algorithm and use it for extracting terms from the UCSFMC database. This approach can be viewed as utilizing both labeled and unlabeled data to modify the algorithm [31]. Using the final version of the algorithm, we perform a second run over the test data (Table II). Note that the test set is no longer a valid test set, since we looked at it to modify the algorithm. We are showing the results as a confirmation step, due to the lack of ground truth and the small number of labeled data.
TABLE II.
Automated and manual extraction, 2nd run
| Method | Predicted | Actual
|
|
|---|---|---|---|
| Feature present | Feature absent | ||
| Automated | Feature present | 219 | 2 |
| Feature absent | 4 | 4075 | |
| Manual | Feature present | 198 | 5 |
| Feature absent | 25 | 4072 | |
During the second run, the algorithm correctly classifies some of its previous mismatches, dropping its false positive and false negative counts. It now achieves a precision of 99.1%, a recall of 98.2% and an F1-score of 0.99. In addition, the algorithm discovers two more previously unrecognized true positives, which increases the manual method’s false negative count.
V. Discussion
As in most clinical data, false negative mammograms are critical and often more costly than false positive ones [32]. Many technical or human errors cause missed or delayed diagnosis of breast cancer. Among the several reasons are observer error, unreasonable diagnostic evaluation, and problems in communication [33]. Therefore, it is notable that the main gain of our algorithm is in recall, by achieving low false negative counts. The algorithm’s recall rate of 95.5% is higher than the manual method’s 89.6% and the Linear Least Squares Fit method’s reported 35.4% recall rate [11].
To account for higher false negative costs, we use the generalized F-score statistic. By attaching β times as much importance to recall (r) as precision (p), the general Fβ-score becomes:
| (1) |
As β increases, the difference between the computational and manual method’s Fβ-scores increases. Taking into account the relative weight of false negatives further improves the algorithm’s performance.
These results show that the algorithm may match or surpass the manual method for information extraction from free text mammography reports. Our algorithm can thus be used, with high confidence, for consistency checks, data preprocessing and information extraction for clinical data mining. We applied the second version of the algorithm to the UCSFMC data and generated a BI-RADS features database. We intend to use it to improve our current breast-cancer classifier [6].
In addition to information extraction, our algorithm allows the assessment of radiologist’s labeling of mammography reports. By comparing the features extracted by the radiologist to the algorithm’s output, we can detect repeatedly missed concepts and suggest areas for improvement. This may be useful for radiology trainees.
In an effort to increase the accuracy of mammography interpretation, the Institute of Medicine notes that data collection is inadequate without resources for accurate and uniform analysis [34]. It points at double reading and computer-aided detection (CAD) as potential methods for increasing recall. Given a manually indexed report, our algorithm may act as a double reader. For partly-labeled or missing data, it may act as a CAD method. In both events, it may be able to provide decision support for physicians, which helps decrease medical errors. Further tests regarding our algorithm’s decision support capacities are needed to assess this claim.
VI. Future Work
Compared to state-of-the-art procedures, our syntax, semantic and lexical scanners are simple. Achieving high recall (95.5%) and precision (97.7%) values, it can be argued that a more complex natural language processor would add little performance for a high complexity price. Nevertheless, we plan on refining our parser by adding a part-of-speech tagger.
Another concern is the small number of the labeled dataset (100 records). Manually indexing reports is a laborious time-consuming task. Although many studies in the medical diagnostics domain have similar data ranges [11], [12], [16], [20], [22], [23], we plan on expanding our testing set.
Finally, it would be interesting to study the impact of inter-observer variability on our method. We can have multiple test sets each indexed by a different radiologist, and compare our algorithm’s performance on each. We can also train our algorithm on reports written by one radiologist and train on a test set indexed by another.
VII. Conclusion
We describe a general scheme for concept information retrieval from free text given a lexicon, and present a BI-RADS features extraction algorithm for clinical data mining. On our dataset, our algorithm achieves 97.7% precision, 95.5% recall and an F1-score of 0.97. It outperforms manual feature extraction at the 5% statistical significance level. It particularly achieves a high recall gain over manual indexing. We stipulate that our method can help avoid clinical false negatives by performing consistency checks and providing physicians with decision support.
Contributor Information
Houssam Nassif, Email: nassif@biostat.wisc.edu.
David Page, Email: page@biostat.wisc.edu.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA-Cancer J Clin. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.American Cancer Society. Breast Cancer Facts & Figures 2007–2008. Atlanta, GA: American Cancer Society, Inc; 2007. [Google Scholar]
- 3.Boyle P, Levin B. World Cancer Report 2008. Lyon, France: International Agency for Research on Cancer; 2008. [Google Scholar]
- 4.Breast Imaging Reporting and Data System (BI-RADS™) American College of Radiology; Reston, VA: 1998. [Google Scholar]
- 5.National Mammography Database. American College of Radiology; 2001. [Google Scholar]
- 6.Burnside ES, Davis J, Chhatwal J, Alagoz O, Lindstrom MJ, Geller BM, Littenberg B, Shaffer KA, Kahn CE, Page D. Probabilistic computer model developed from clinical data in national mammography database format to classify mammographic findings. Radiology. 2009;251:663–672. doi: 10.1148/radiol.2513081346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burnside ES, Davis J, Costa VS, de Castro Dutra I, Kahn CE, Fine J, Page D. Knowledge discovery from structured mammography reports using inductive logic programming. American Medical Informatics Association Annual Symposium Proceedings; Washington, DC. 2005. pp. 96–100. [PMC free article] [PubMed] [Google Scholar]
- 8.Chhatwal J, Alagoz O, Kahn C, Burnside E. A logistic regression model based on the national mammography database format to aid breast cancer diagnosis. Am J of Roentgenol. 2009;192(4):1117–1127. doi: 10.2214/AJR.07.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis J, Burnside E, Dutra I, Page D, Ramakrishnan R, Costa VS, Shavlik J. View learning for statistical relational learning: With an application to mammography. Proc. of the 19th International Joint Conference on Artificial Intelligence; Edinburgh, Scotland. 2005. pp. 677–683. [Google Scholar]
- 10.Liberman L, Menell JH. Breast imaging reporting and data system (BI-RADS) Radiol Clin N Am. 2002;40(3):409–430. doi: 10.1016/s0033-8389(01)00017-3. [DOI] [PubMed] [Google Scholar]
- 11.Burnside B, Strasberg H, Rubin D. Automated indexing of mammography reports using linear least squares fit. Proc. of the 14th International Congress and Exhibition on Computer Assisted Radiology and Surgery; San Francisco, CA. 2000. pp. 449–454. [Google Scholar]
- 12.Friedman C, Alderson P, Austin J, Cimino J, Johnson S. A general natural-language text processor for clinical radiology. J Am Med Inform Assn. 1994;1(2):161–174. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assn. 2004;11(5):392–402. doi: 10.1197/jamia.M1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindberg DA, Humphreys BL, McCray AT. The unified medical language system. Method Inform Med. 1993;32:281–291. doi: 10.1055/s-0038-1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aronson AR. Effective mapping of biomedical text to the UMLS metathesaurus: The MetaMap program. Proc. of the American Medical Informatics Association Symposium; Washington, DC. 2001. pp. 17–21. [PMC free article] [PubMed] [Google Scholar]
- 16.Long W. Lessons extracting diseases from discharge summaries. American Medical Informatics Association Annual Symposium Proceedings; Chicago, IL. 2007. pp. 478–482. [PMC free article] [PubMed] [Google Scholar]
- 17.Carrell D, Miglioretti D, Smith-Bindman R. Coding free text radiology reports using the cancer text information extraction system (caTIES). American Medical Informatics Association Annual Symposium Proceedings; Chicago, IL. 2007. p. 889. [PubMed] [Google Scholar]
- 18.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. Evaluation of negation phrases in narrative clinical reports. Proc. of the American Medical Informatics Association Symposium; Washington, DC. 2001. pp. 105–109. [PMC free article] [PubMed] [Google Scholar]
- 19.Ruch P, Baud R, Geissbuhler A, Rassinoux AM. Comparing general and medical texts for information retrieval based on natural language processing: an inquiry into lexical disambiguation. Proc. of the 10th World Congress on Medical Informatics; London, UK. 2001. pp. 261–265. [PubMed] [Google Scholar]
- 20.Mutalik PG, Deshpande A, Nadkarni PM. Use of general-purpose negation detection to augment concept indexing of medical documents: A quantitative study using the UMLS. J Am Med Inform Assn. 2001;8(6):598–609. doi: 10.1136/jamia.2001.0080598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001;34:301–310. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 22.Huang Y, Lowe H. A novel hybrid approach to automated negation detection in clinical radiology reports. J Am Med Inform Assn. 2007;14(3):304–311. doi: 10.1197/jamia.M2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gindl S, Kaiser K, Miksch S. Syntactical negation detection in clinical practice guidelines. Proc. of the 21st International Congress of the European Federation for Medical Informatics; Göteborg, Sweden. 2008. pp. 187–192. [PMC free article] [PubMed] [Google Scholar]
- 24.Romano R, Rokach L, Maimon O. Cascaded data mining methods for text understanding, with medical case study. Proc. of the 6th IEEE International Conference on Data Mining - Workshops; Hong Kong, China. 2006. [Google Scholar]
- 25.Rokach L, Maimon O, Averbuch M. Information retrieval system for medical narrative reports. Proc. of the 6th International Conference on Flexible Query Answering Systems; Lyon, France. 2004. pp. 217–228. [Google Scholar]
- 26.Wall L, Schwartz RL. Programming Perl. Sebastopol, CA, United States of America: O’Reilly & Associates; 1992. [Google Scholar]
- 27.Eskin E. Detecting errors within a corpus using anomaly detection. Proc. of the 1st North American chapter of the Association for Computational Linguistics Conference; San Francisco, CA. 2000. pp. 148–153. [Google Scholar]
- 28.Smyth P. Bounds on the mean classification error rate of multiple experts. Pattern Recogn Lett. 1996;17(12):1253–1257. [Google Scholar]
- 29.Lam CP, Stork DG. Evaluating classifiers by means of test data with noisy labels. Proc. of the 18th International Joint Conference on Artificial Intelligence; Acapulco, Mexico. 2003. pp. 513–518. [Google Scholar]
- 30.Goutte C, Gaussier E. A probabilistic interpretation of precision, recall and F-score, with implication for evaluation. Proc. of the 27th European Conference on IR Research; Santiago de Compostela, Spain. 2005. pp. 345–359. [Google Scholar]
- 31.Nigam K, McCallum A, Thrun S, Mitchell T. Learning to classify text from labeled and unlabeled documents. Proc. of the 15th National Conference on Artificial Intelligence; 1998. pp. 792–799. [Google Scholar]
- 32.Petticrew M, Sowden A, Lister-Sharp D. False-negative results in screening programs: Medical, psychological, and other implications. Int J Technol Assess. 2001;17(2):164–170. doi: 10.1017/s0266462300105021. [DOI] [PubMed] [Google Scholar]
- 33.Brenner RJ. False-negative mammograms: Medical, legal, and risk management implications. Radiol Clin N Am. 2000;38(4):741–757. doi: 10.1016/s0033-8389(05)70198-6. [DOI] [PubMed] [Google Scholar]
- 34.Nass SJ, Ball J. Improving Breast Imaging Quality Standards. Washington, DC: National Academies Press; 2005. [Google Scholar]


