Abstract
Protein–protein interactions are crucial for a wide variety of biological processes. These interactions range from high affinity (Kd < nM) to very low affinity (Kd > mM). While much is known about the nature of high affinity protein complexes, our knowledge about structural characteristics of weak protein–protein interactions (wPPIs) remains limited: in addition to the technical difficulties associated with their investigation, historically wPPIs used to be considered physiologically irrelevant. However, emerging evidence suggests that wPPIs, either in the form of intact protein complexes or as part of large molecular machineries, are fundamentally important for promoting rapid on/off switches of signal transduction, reversible cell–cell contacts, transient assembly/disassembly of signaling complexes, and enzyme–substrate recognition. Therefore an atomic-level elucidation of wPPIs is vital to understanding a cornucopia of diverse cellular events. Nuclear magnetic resonance (NMR) is famous for its unique abilities to study wPPIs and, by utilization of the new technical developments combined with sparse data based computational analysis, it now allows rapid identification and structural characterization of wPPIs. Here we present our perspective on the NMR methods employed.
Keywords: Chemical shifts, NMR, NOE, PCS, PRE, RDC
1 Introduction
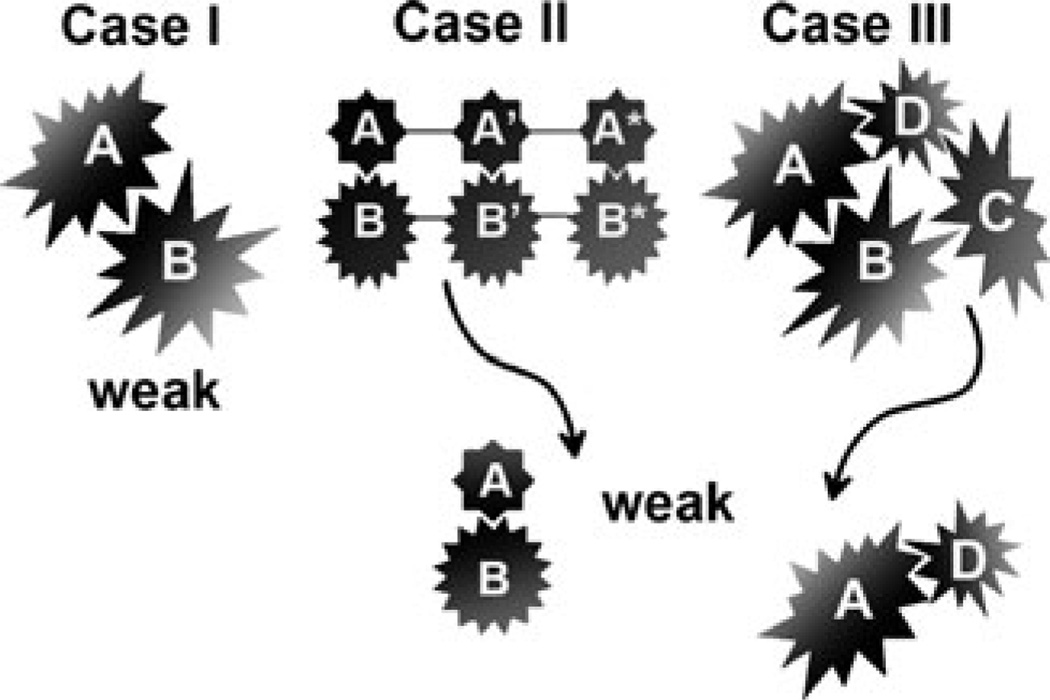
Living organisms are very complex, highly structured, tightly regulated systems with precisely orchestrated communications at every level of organizational hierarchy. These communications are largely mediated by protein–protein interactions (PPIs). The complete genome sequencing now reveals that there are thousands of potential PPIs that may function as building blocks for these communication networks [1,2]. PPIs can be classified based upon the strength of interaction, which is often rendered by the equilibrium dissociation constant (Kd) equal to koff/kon, where koff is the rate constant of the complex dissociation reaction and kon is the rate constant of the association reaction. The window of biologically relevant Kd values is extremely wide and can cover 12 orders of magnitude [3]. PPIs can be very loosely divided into three major subclasses [4]: (1) strong, with Kd < 10−9M, and permanent association, (2) strong and transient, where the change in the quaternary state can be triggered, for example, by ligand binding, and (3) weak, with Kd > 10−4M, and transient association with koff rates of up to 104 s−1, which results in lifetimes as short as 100 µs [5]. Decades of extensive studies have illuminated structural and functional features for the PPIs from the first two subclasses characterized by strong binding with Kd < 10−6 M, which are summarized in numerous reviews [6–8]. The weak PPIs and their physiological importance (wPPIs, with Kd > 10−4 M), on the other hand, are less well understood. This could be attributed in part to the technical difficulties encountered during attempts to characterize them directly in vitro or in vivo. The other reason relates to a common prejudice that wPPIs might not be found in living cells, especially considering low (<10−7 M) protein concentrations estimated by the whole cell volumes. However, it is now being increasingly appreciated that wPPIs are crucial for promoting diverse biologically important processes such as reversible cell–cell contacts, transient assembly and/or disassembly of large signaling complexes, and dynamic regulation of enzymes [9]. Figure 1 provides three possible scenarios of wPPIs: (1) wPPI between two intact proteins, (2) wPPI as part of multi-domain interactions between two intact proteins, and (3) wPPI as part of a multi-protein complex. Conventional methods such as X-ray crystallography, surface Plasmon resonance (SPR), and isothermal titration calorimetry (ITC) often fail to study these wPPIs accurately. In contrast, nuclear magnetic resonance (NMR) has been proven as a particularly powerful tool to examine them at atomic level resolution and at near physiological conditions [7,8,10,11]. In this chapter we present the various NMR approaches to assess and characterize these three types of wPPIs structurally.
Fig. 1.
Three representative cases of wPPIs. Case I: a weak protein–protein interaction found in a locally highly crowded manner. Case II: a weak domain–domain interaction, exemplified by A–B pair, as part of a tight multi-domain complex. Such weak binary domain–domain interaction may be undetectable by many conventional methods including deletion mapping, yeast-hybrid approach, immunoprecipitation, etc., but become apparent when the tertiary structure of the tight complex is challengingly determined. However, NMR may be able to pick this interaction at early stage of the characterization. Case III: a weak protein–protein interaction as a part of multi-protein complex. Similar to (II), a weak A–D pair may not be detectable in isolated manner by any conventional techniques except NMR
1.1 Chemical Shift Perturbation Mapping
The resonance frequencies, also known as chemical shifts, of individual atoms in a particular protein strongly depend on the local environment and, because of that, are often considered as fingerprints of its structure. The chemical shift patterns of 15N and directly attached amide 1H are especially sensitive in this respect. Thus their perturbation, as the result of complex formation, provides a highly sensitive tool for mapping the binding interface. Binding surface on the target protein is identified by titrating the unlabeled target into the solution of the 15N-labeled protein and monitoring the resultant spectral changes in its 1H–15N HSQC (heteronuclear single quantum correlation) spectrum or, for the larger proteins, its TROSY-based (transverse relaxation-optimized spectroscopy) version [12]. The utility and popularity of this experiment are based upon its straightforward nature and high sensitivity – the spectrum can be recorded in 5–40 min on a typical protein sample (~0.1–1 mM). The spectral changes, also denoted as chemical shift perturbations (CSP), are usually associated with a particular set of amino acids that either (1) directly participate in interaction or are situated very closely to the binding site or (2) reflect binding-induced conformational rearrangements (e.g., a disorder–order transition). The former happens more frequently for wPPIs, which have characteristic small CSP with little or no conformational change, at least within the backbone of well folded domains. Hence the binding interface can be qualitatively deduced from the spectrally perturbed residues. In addition, for wPPIs with fast exchange, characterized by high koff rates, Kd can be estimated from concentration-dependent titrations [13]. However, the potential problem associated with this analysis relates to the low affinity of the complex: an interfacial residue might not necessarily undergo a significant CSP upon binding, meaning that interfaces derived from CSP data alone are not always complete [6]. One way to avoid this caveat is to increase the ratio of the unlabeled titrant, which can drive the equilibrium towards the bound form with bigger CSP. However, the ratio cannot be too high since it may cause some non-specific effects. The weakest PPI analyzed by CSP demonstrated a Kd of 10−2 M for the flavodoxin/flavodoxin reductase complex [14]. Both (1) and (2) might occur for strong PPIs, which often undergo significant conformational rearrangements upon binding, especially if a disorder–order transition occurs. In such cases, analysis of binding interfaces is more challenging and less straightforward, and one has to rely on additional techniques, such as incorporation of a cross-saturation [15] or inter-molecular Nuclear Overhauser Effects/Enhancements (NOEs) (described in detail in the next section). To conclude, although for wPPIs the CSP method provides fast and robust assessment of the residues forming an intermolecular interface, the mutual orientation of the two partners remains elusive. Thus, if the goal is to generate the structural model of the complex, additional experiments have to be performed and/or novel computational approaches have to be employed.
1.2 Nuclear Overhauser Effect
NOE, a relaxation mechanism based upon magnetic dipole–dipole interactions of the nuclei, allows measurement of interproton distances with the basic r−6 distance proportionality. This provides major distance restraints for structural calculations. Supplemented with additional data, such as original dihedral angle restraints obtained from J-couplings or more recent information about the orientation of the bond vectors connecting magnetically active nuclei with respect to the external magnetic field, this approach has been the foundation for NMR-based protein structure determination since its dawn in 1984 [11].
1.2.1 NOE in the Determination of the Structure of wPPIs
From the perspective of wPPIs structural characterization, two particular applications of NOE are proven most fruitful.
Transferred NOE Experiments
Transferred NOE Experiments (trNOESY) is a quick two-dimensional 1H NMR experiment that allows the observation of intramolecular proton contacts (<5 Å) for the small peptide bound to its target protein [16]. In a nutshell, if the affinity of the interaction in question is low due to the fast dissociation rate, cross relaxation between protons of the peptide in the bound state, which is governed by the large correlation time of the complex, is transferred to the free state through chemical exchange. This phenomenon is manifested by the appearance of additional peaks at the original (corresponding to free state) frequencies in the NOESY spectrum of the peptide when it is mixed with a small molar portion of the large, typically over 50 kDa in molecular weight, target protein. Protein–peptide ratios for trNOESY may vary from 1:10 to 1:200 with mixing times ranging from 50 to 500 ms. Both parameters need to be optimized for each particular case. Substantially increased number of NOEs should be observed for the peptide in the presence of its target protein comparatively to the peptide free form. The method requires no isotopelabeling and is suitable for examination of protein–ligand interactions over a wide range of Kds (micromolar–millimolar) [7]. The method can be applied to study initial lead compounds weakly bound to the target protein, which allows the structure determination of the bound compounds for further optimization leading to high affinity binding.
Half-Filtered NOESY (Intermolecular NOEs)
Half-filtered NOESY approach was developed to detect inter-molecular contacts in the form of NOEs only between protons pairs in which one of the protons is attached to 15N or 13C nuclei while the other is attached to magnetically inactive 14N or 12C nuclei. Thus it requires the preparation of 15N–13C-labeled protein mixed with its unlabeled binding partner, and/or vice versa. Two types of half-filtered NOESY experiment are commonly used [17]: (1) three dimensional 13C-separated–15N,13C-filtered NOESY, which detects NOEs between protons attached to 13C atoms of the doubly labeled protein and those attached to 12C and 14N on the unlabeled protein, and (2) three-dimensional 15N-separated-15N,13C-filtered NOESY, which detects NOEs between 15N-attached and 12C-,14N-attached protons. However, there are pulse sequences available with smart combinations of both, when separation in 13C and 15N dimensions can be achieved simultaneously, significantly reducing the experimental time. The sensitivity of this experiment crucially depends on the lifetime of a protein complex. For weak PPIs characterized by high koff the actual portion of the complex within the sample might not be high enough for observing the intermolecular NOEs. However, in favorable cases, when the concentrations of both binding partners are high [18], the complexed state could be detectible with the help of high-sensitivity NMR instruments, such as those equipped with cryo-probes. Another relatively more sensitive experiment is the 15N-edited NOESY on a 15N/100% deuterated protein bound to the target, which will detect the NOEs between the amide proton of the 15N-labeled protein and any nearby protons of the target [19]. This experiment can be complementary to those in (1) and (2) but provides a very unambiguous assignment of the amide protons, which are usually well resolved in the HSQC spectrum, to the protons of the unlabeled target. This experiment can detect very weak NOEs, possibly up to 7 Å in distance, due to the deuteration effect.
1.3 Residual Dipolar Couplings
Although NMR-based applications on dipolar coupling have been mainly associated with solid-state NMR or NMR of oriented samples, they have been recently applied to solution NMR for studying macromolecular structure and function in an aqueous solution [20,21]. The dipolar coupling describes a through-space interaction that arises between any two magnetically active nuclei. It depends upon the distance between two atoms, which is constant for the nuclei connected by covalent bonds such as 1H–15N or 1H–13C, and the orientation of the connecting vector with respect to the external magnetic field. In solution, dipolar interactions are averaged because of fast isotropic tumbling. However, if the macromolecules experience obstacles in some directions due to partial alignment with orienting media, for example, bacteriophage, bicelles or polyacrylamide gels, the dipolar couplings are not completely canceled out and whatever is left is designated as residual dipolar couplings (RDC). By measuring RDC the orientation of the molecular alignment tensor could be defined, providing the information about mutual orientation between the domains within single macromolecule or between binary units of the complex. Thus the quest began to find robust ways to orient the media weakly without significant increase in viscosity of the system or generation nucleation points for aggregation. The general idea is based upon a fact that certain media, possessing sufficiently large magnetic susceptibility anisotropy, can be aligned spontaneously by high magnetic field. In earlier 1990s, bicelles, disk-shaped particles made from the lipid/detergent mixtures, were introduced [22] for this purpose, followed by rod-shaped viruses [23], mechanically orientable systems [24,25], and G-tetrad DNA [26]. As compared to the more conventional NOEs approach, RDC carry complementary information: while NOEs provide only local distance restraints, RDC contain long range orientational information (e.g., see review by [27]), thus delivering powerful long-range geometric constraints for proper subunits orientation during the structure determination of the complex. In the case of wPPIs that may undergo fast exchange between the bound and free forms of the binding partners, measured RDC will represent the population weighted average values of those in bound and free form. Theoretically, knowing the molar ratio of bound and free forms (from Kd and molar concentrations), and after measuring RDC in the free form, one can calculate RDC in bound form [27]. From the RDC of weakly bound subunits, their alignment tensors can be calculated and matched for defining the structure of a weak complex. One example using this strategy to determine the structure of weak complex is α-methyl mannose bound to mannose-binding protein with a Kd ~ 1 mM [28]. In practice, however, it is not always straightforward for small ligands to determine the accurate fraction of bound form and, thus, full saturation could be beneficial to utilize the RDCs of the fully bound form.
1.4 Paramagnetic NMR
As NMR spectroscopists are constantly on the quest to improve the line-shape and to reduce the width of the peaks in their spectra, elimination of the paramagnetic species, often causing significant line-broadening effects, has been considered as paramount in sample preparations. However, the usually undesirable line-broadening effect can provide unique structural long range information [29] when the effect is specific and paramagnetic center is localized to a particular site of the macromolecules. Historically, this understanding has been mainly applied to proteins containing metal-binding sites (reviewed in [30]). The idea to utilize surface exposed cysteines for introduction of paramagnetic tags came later with a cornucopia of chemical agents and procedures developed for reliable conjugation at specific sites (reviewed in [31]). This approach not only provides information about intramolecular distances but can also help in defining alignment of binding subunits within complexes characterized by wPPIs [32], although actual quantification of the distance dependence in such systems is not always straightforward as we discuss below. Two distinguished NMR phenomena, based upon the specific nature of the magnetic moment of an attached paramagnetic center, present the basis for structural investigation. These are paramagnetic relaxation enhancement (PRE) characteristic for all paramagnetic moieties and the pseudocontact shifts (PCS) effect specific for the subclass of paramagnetic ions with an anisotropic electron g-tensor.
1.4.1 The PRE Effect
The large magnetic dipolar interaction of the unpaired electron from a paramagnetic atom with the neighboring NMR-active nucleus results in an increase of the relaxation rate of the above nucleus [33]. Similar to NOE, this effect has basic r−6 distance proportionality, but, because of the larger magnetic moment of the electron, PRE effect is observable at longer, up to 25–35 Å, distances depending upon the nature of the particular paramagnetic group [34,35]. Thus PRE measurements can provide much longer range distance restraints for structural calculations in comparison to the classical NOE approach. The caveat of PRE application for short distances determination comes from the same original source and relates to the fact that nuclei situated very close to a paramagnetic center are often broadened beyond detection. However, the data sets acquired by NOE and PRE approaches are at least complementary. The PRE measurements are based on the correlation of the increased transverse relaxation rate with the distance between the introduced paramagnetic moiety and the affected nucleus [36]. Simply speaking, we are measuring the distance-dependent reduction in peak intensities in a 15N-HSQC spectrum of the target protein when a single paramagnetic tag is attached to it at the specific site, usually through a thioether bond formed with the side chain of the cysteine residue. Nitroxide stable radicals or metal chelators, such as EDTA-Mn2+, which are characterized by an unpaired electron with an isotropic g-tensor, are especially useful since they do not give rise to pseudo-contact shifts, and Curie-spin relaxation is negligible [37]. The advantage of their employment for pure PRE measurements is the fast and straightforward nature of the method: the resonance assignments in the correlation spectra, known from through-bond scalar triple resonance experiments, are not perturbed by paramagnetic modification and the high sensitivity results in an experimental time of 5–40 min on a typical protein sample (the same advantages we discussed for CSP method, as both are based on the same types of data acquisition – HSQC experiments). The potential problems are associated with intrinsic flexibility/rotation of either the paramagnetic tag itself or its attachment to the protein, resulting in the time average distances sampled over all possible conformations. Thus certain caution is required for incorporation of the derived distances as the restraints for structure calculations, where a paramagnetic center, for example, can be treated as an ensemble average rather than a fixed point [38]. The other possibility for highly flexible systems is to use PRE data loosely, as a guide, rather than major geometric restraints, for example, in structure determination of the complex when the orientation of the peptide, which could be labeled by a paramagnetic tag, in a particular binding site needs to be addressed [39,40]. The potential ability to study transient low population intermediates in macromolecular interactions is conceivably one of the most exciting PRE implementations in structural biology. These illusive species are rarely accessible by other than NMR biophysical techniques. In an exchanging system the observed PRE measured on the resonance of the major species can be modulated by the minor species to the extent depending upon the rate of exchange [41], with the fast exchange allowing one to characterize structurally populations comprising as low as 1%. The example illustrating PRE potential to demonstrate the existence and visualize the distribution of an ensemble of transient non-specific intermediates in addition to specific complex formation has been presented by Clore and colleagues for a bacterial phosphotransferase system [42]. Thus, it has been proven that PRE data is highly sensitive asserting weak interactions characterized by large koff rate and is salutary for structural analysis of weak PPIs.
1.4.2 The PCS Effect
PCS is a phenomenon that is only observable for paramagnetic systems with anisotropic unpaired electrons such as those found in Dy3+, Tb3+, and Fe3+ lanthanide ions, characterized by an anisotropic electron g-tensor. In general, if the g-tensor is anisotropic, than the magnetic susceptibility tensor (usually referred to as the χ-tensor) is anisotropic as well. The magnitude of the PCS depends on the orientations of the vectors connecting the lanthanide ion and affected nuclei with respect to direction of the external magnetic field. These orientations are not averaged because tumbling in aqueous solution appears to be non-isotropic due to the effect of large χ-tensor intrinsic for these paramagnetic species. This large magnetic susceptibility tensor provides enough energy to overcome random Brownian motion and to generate preferable orientation or alignment of the macromolecule containing the paramagnetic tag. This phenomenon causes changes in chemical shift of the affected nuclei which are sufficiently close to the paramagnetic center (reviewed in [43]). Importantly, the PCS displays basic r−3 distance proportionality in contrast to the r−6 dependence for the PRE or NOE. In theory, this will define a relatively longer experimentally attainable distance range, extending it up to, for example, ~40 Å for Dy3+in metalloproteins. In practice, the principal axis of the χ-tensor is not rigidly fixed within the molecular frame when an extrinsic metal ion is attached to a macromolecule using a chelator with a flexible linker, causing significant reduction in the magnitude of the PCS because of χ-tensor principal axes fluctuations within the frame of the macromolecule. From the perspective of studying wPPIs, PCS restraints can be generated using a 15N-labeled and/or 13C-labeled protein bound to an unlabeled but paramagnetically tagged partner. 15N and/or 13C-HSQC experiments then need to be recorded for both the paramagnetic and diamagnetic states of a sample and chemical shift changes should be extracted from the spectra [31]. However, to use PCS data, one first has to define the tensor describing the anisotropic magnetic moment of the paramagnetic center [44]. When the structures of the individual proteins are known, PCS data can be combined with rigid-body docking to produce a model for a protein complex. This approach has been proven successful in determination of a 30-kDa complex between the θ and ε subunits of Escherichia coli polymerase III [45], where the active-site bound Mg2+/Mn2+ pairs were exchanged with paramagnetic Dy3+ or Er3+ and corresponding 15N-HSQC spectra of the diamagnetic apo-complex and paramagnetic-ion-bound complexes were compared. An analogous approach taking advantage of the intrinsic iron-binding capability of cytochrome f has been used earlier to define the structure of its transient complex with plastocyanin: conveniently, iron in its oxidized Fe3+ form is paramagnetic while in the Fe2+ form it displays a diamagnetic nature [46].
2 Conclusions
While tight protein interactions can be addressed experimentally by many techniques, including X-ray crystallography, the vast majority of these approaches fail or become unreliable when the interactions are weak. Solution NMR spectroscopy is unique among the structural techniques, permitting the characterizing of weak interactions and providing structures of weak protein-target complexes. If such interactions involve small molecules, NMR can be employed for optimization and development of drug-leads. In the current post-genomic era, the NMR methods we have highlighted in combination with functional and computational approaches hold significant promise for characterizing the plethora of weak protein complexes that regulate cellular events, thereby providing an unbiased and comprehensive view of how proteins function in living cells.
Contributor Information
Olga Vinogradova, Department of Pharmaceutical Sciences, University of Connecticut, 69 North Eagleville Rd, Unit 3092, Storrs, CT 06269-3092, USA, olga.vinogradova@uconn.edu.
Jun Qin, Department of Molecular Cardiology, Cleveland Clinic, NB20, 9500 Euclid Ave., Cleveland, OH 44195, USA, qinj@ccf.org.
References
- 1.Gavin AC, Bosche M, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415(6868):141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 2.Ho Y, Gruhler A, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415(6868):180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 3.Nooren IM, Thornton JM. Diversity of protein-protein interactions. EMBO J. 2003;22(14):3486–3492. doi: 10.1093/emboj/cdg359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perkins JR, Diboun I, et al. Transient protein-protein interactions: structural, functional, and network properties. Structure. 2010;18(10):1233–1243. doi: 10.1016/j.str.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Prudencio M, Ubbink M. Transient complexes of redox proteins: structural and dynamic details from NMR studies. J Mol Recognit. 2004;17(6):524–539. doi: 10.1002/jmr.686. [DOI] [PubMed] [Google Scholar]
- 6.O’Connell MR, Gamsjaeger R, et al. The structural analysis of protein-protein interactions by NMR spectroscopy. Proteomics. 2009;9(23):5224–5232. doi: 10.1002/pmic.200900303. [DOI] [PubMed] [Google Scholar]
- 7.Qin J, Vinogradova O, et al. Protein-protein interactions probed by nuclear magnetic resonance spectroscopy. Meth Enzymol. 2001;339:377–389. doi: 10.1016/s0076-6879(01)39323-0. [DOI] [PubMed] [Google Scholar]
- 8.Zuiderweg ER. Mapping protein-protein interactions in solution by NMR spectroscopy. Biochemistry. 2002;41(1):1–7. doi: 10.1021/bi011870b. [DOI] [PubMed] [Google Scholar]
- 9.Vaynberg J, Qin J. Weak protein-protein interactions as probed by NMR spectroscopy. Trends Biotechnol. 2006;24(1):22–27. doi: 10.1016/j.tibtech.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 10.Clore GM, Gronenborn AM. Determining the structures of large proteins and protein complexes by NMR. Trends Biotechnol. 1998;16(1):22–34. doi: 10.1016/S0167-7799(97)01135-9. [DOI] [PubMed] [Google Scholar]
- 11.Wuthrich K. NMR of proteins and nucleic acids. New York: Wiley; 1986. [Google Scholar]
- 12.Pervushin K, Riek R, et al. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci USA. 1997;94(23):12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Velyvis A, Vaynberg J, et al. Structural and functional insights into PINCH LIM4 domain-mediated integrin signaling. Nat Struct Biol. 2003;10(7):558–564. doi: 10.1038/nsb938. [DOI] [PubMed] [Google Scholar]
- 14.Hall DA, Vander Kooi CW, et al. Mapping the interactions between flavodoxin and its physiological partners flavodoxin reductase and cobalamin-dependent methionine synthase. Proc Natl Acad Sci USA. 2001;98(17):9521–9526. doi: 10.1073/pnas.171168898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi H, Nakanishi T, et al. A novel NMR method for determining the interfaces of large protein-protein complexes. Nat Struct Biol. 2000;7(3):220–223. doi: 10.1038/73331. [DOI] [PubMed] [Google Scholar]
- 16.Clore GM, Gronenborn AM. The two-dimensional transferred nuclear Overhauser effect. J Magn Reson. 1982;48:402–417. [Google Scholar]
- 17.Zwahlen C, Legault P, et al. Methods for measurement of intermolecular NOEs by multinuclear NMR spectroscopy: application to a bacterio-phage l N-peptide/boxB RNA complex. J Am Chem Soc. 1997;119:711–721. [Google Scholar]
- 18.Vaynberg J, Fukuda T, et al. Structure of an ultraweak protein-protein complex and its crucial role in regulation of cell morphology and motility. Mol Cell. 2005;17(4):513–523. doi: 10.1016/j.molcel.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 19.Walters KJ, Matsuo H, et al. A simple method to distinguish intermonomer nuclear Overhauser effects in homodimeric proteins with C2 symmetry. J Am Chem Soc. 1997;119:5958–5959. [Google Scholar]
- 20.Bax A, Kontaxis G, et al. Dipolar couplings in macromolecular structure determination. Meth Enzymol. 2001;339:127–174. doi: 10.1016/s0076-6879(01)39313-8. [DOI] [PubMed] [Google Scholar]
- 21.Prestegard JH, Bougault CM, et al. Residual dipolar couplings in structure determination of biomolecules. Chem Rev. 2004;104(8):3519–3540. doi: 10.1021/cr030419i. [DOI] [PubMed] [Google Scholar]
- 22.Sanders CR, Hare BJ, et al. Magnetically-oriented phospholipid micelles as a tool for the study of membrane-associated molecules. Prog Nucl Magn Reson Spectrosc. 1994;26:421–444. [Google Scholar]
- 23.Clore GM, Starich MR, et al. Measurement of residual dipolar couplings of macromolecules aligned in the nematic phase of a colloidal suspension of rod-shaped viruses. J Am Chem Soc. 1998;120:10571–10572. [Google Scholar]
- 24.Chou JJ, Kaufman JD, et al. Micelle-induced curvature in a water-insoluble HIV-1 Env peptide revealed by NMR dipolar coupling measurement in stretched polyacrylamide gel. J Am Chem Soc. 2002;124(11):2450–2451. doi: 10.1021/ja017875d. [DOI] [PubMed] [Google Scholar]
- 25.Ishii Y, Markus MA, et al. Controlling residual dipolar couplings in high-resolution NMR of proteins by strain induced alignment in a gel. J Biomol NMR. 2001;21(2):141–151. doi: 10.1023/a:1012417721455. [DOI] [PubMed] [Google Scholar]
- 26.Lorieau J, Yao L, et al. Liquid crystalline phase of G-tetrad DNA for NMR study of detergent-solubilized proteins. J Am Chem Soc. 2008;130(24):7536–7537. doi: 10.1021/ja801729f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lipsitz RS, Tjandra N. Residual dipolar couplings in NMR structure analysis. Annu Rev Biophys Biomol Struct. 2004;33:387–413. doi: 10.1146/annurev.biophys.33.110502.140306. [DOI] [PubMed] [Google Scholar]
- 28.Bolon PJ, Al-Hashimi HM, et al. Residual dipolar coupling derived orientational constraints on ligand geometry in a 53 kDa protein-ligand complex. J Mol Biol. 1999;293(1):107–115. doi: 10.1006/jmbi.1999.3133. [DOI] [PubMed] [Google Scholar]
- 29.Kosen PA. Spin labeling of proteins. Meth Enzymol. 1989;177:86–121. doi: 10.1016/0076-6879(89)77007-5. [DOI] [PubMed] [Google Scholar]
- 30.Bertini I, Luchinat C, et al. NMR spectroscopy of paramagnetic metalloproteins. Chembiochem. 2005;6(9):1536–1549. doi: 10.1002/cbic.200500124. [DOI] [PubMed] [Google Scholar]
- 31.Otting G. Prospects for lanthanides in structural biology by NMR. J Biomol NMR. 2008;42(1):1–9. doi: 10.1007/s10858-008-9256-0. [DOI] [PubMed] [Google Scholar]
- 32.Mahoney NM, Rastogi VK, et al. Binding orientation of proline-rich peptides in solution: polarity of the profilin–ligand interaction. J Am Chem Soc. 2000;122:7851–7852. [Google Scholar]
- 33.Bloembergen N, Morgan LO. Proton relaxation times in paramagnetic solutions. Effects of electron spin relaxation. J Chem Phys. 1961;34:842–850. [Google Scholar]
- 34.Battiste JL, Wagner G. Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear Overhauser effect data. Biochemistry. 2000;39(18):5355–5365. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]
- 35.Iwahara J, Anderson DE, et al. EDTA-derivatized deoxythymidine as a tool for rapid determination of protein binding polarity to DNA by intermolecular paramagnetic relaxation enhancement. J Am Chem Soc. 2003;125(22):6634–6635. doi: 10.1021/ja034488q. [DOI] [PubMed] [Google Scholar]
- 36.Iwahara J, Tang C, et al. Practical aspects of (1)H transverse paramagnetic relaxation enhancement measurements on macromolecules. J Magn Reson. 2007;184(2):185–195. doi: 10.1016/j.jmr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clore GM, Tang C, et al. Elucidating transient macromolecular interactions using paramagnetic relaxation enhancement. Curr Opin Struct Biol. 2007;17(5):603–616. doi: 10.1016/j.sbi.2007.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iwahara J, Schwieters CD, et al. Ensemble approach for NMR structure refinement against (1)H paramagnetic relaxation enhancement data arising from a flexible paramagnetic group attached to a macromolecule. J Am Chem Soc. 2004;126(18):5879–5896. doi: 10.1021/ja031580d. [DOI] [PubMed] [Google Scholar]
- 39.Deshmukh L, Gorbatyuk V, et al. Integrin beta3 phosphorylation dictates its complex with Shc PTB domain. J Biol Chem. 2010;285:34875–34884. doi: 10.1074/jbc.M110.159087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, Fukuda K, et al. The structure of alpha-parvin CH2-paxillin LD1 complex reveals a novel modular recognition for focal adhesion assembly. J Biol Chem. 2008;283(30):21113–21119. doi: 10.1074/jbc.M801270200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iwahara J, Clore GM. Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature. 2006;440(7088):1227–1230. doi: 10.1038/nature04673. [DOI] [PubMed] [Google Scholar]
- 42.Tang C, Iwahara J, et al. Visualization of transient encounter complexes in protein-protein association. Nature. 2006;444(7117):383–386. doi: 10.1038/nature05201. [DOI] [PubMed] [Google Scholar]
- 43.Clore GM, Iwahara J. Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem Rev. 2009;109(9):4108–4139. doi: 10.1021/cr900033p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmitz C, John M, et al. Efficient chi-tensor determination and NH assignment of paramagnetic proteins. J Biomol NMR. 2006;35(2):79–87. doi: 10.1007/s10858-006-9002-4. [DOI] [PubMed] [Google Scholar]
- 45.Keniry MA, Park AY, et al. Structure of the theta subunit of Escherichia coli DNA polymerase III in complex with the epsilon subunit. J Bacteriol. 2006;188(12):4464–4473. doi: 10.1128/JB.01992-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ubbink M, Ejdeback M, et al. The structure of the complex of plastocyanin and cytochrome f, determined by paramagnetic NMR and restrained rigid-body molecular dynamics. Structure. 1998;6(3):323–335. doi: 10.1016/s0969-2126(98)00035-5. [DOI] [PubMed] [Google Scholar]