Abstract
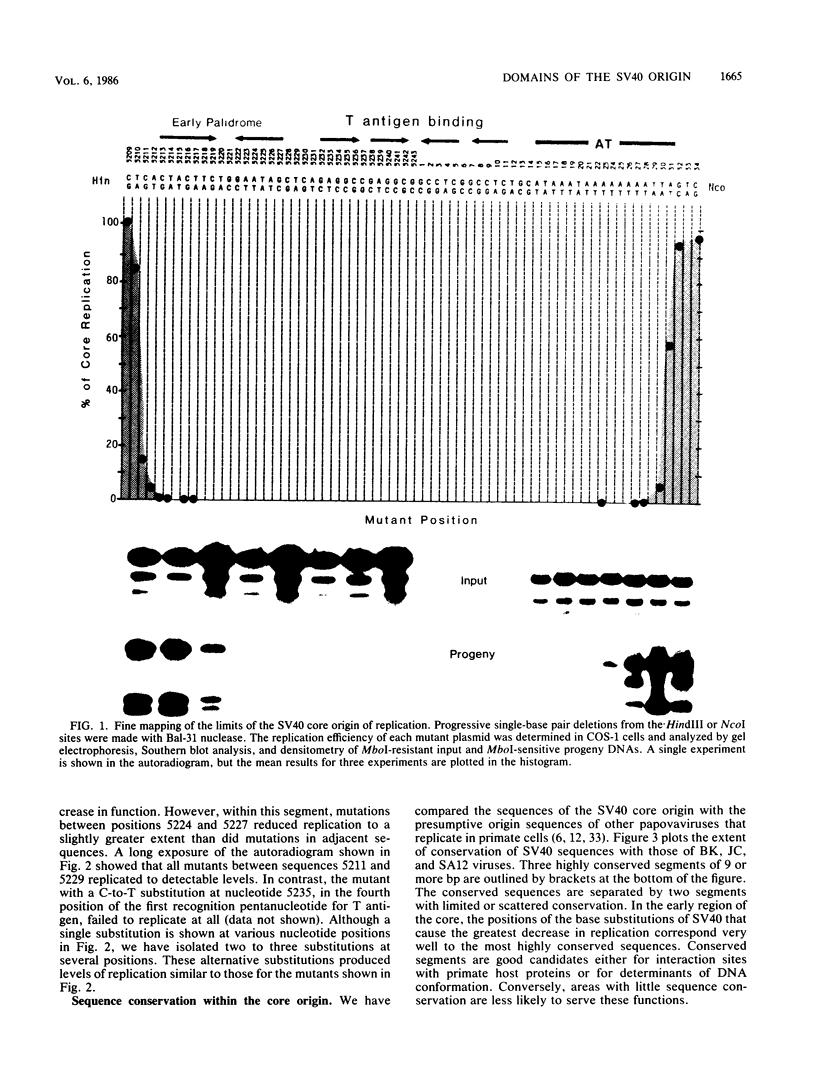
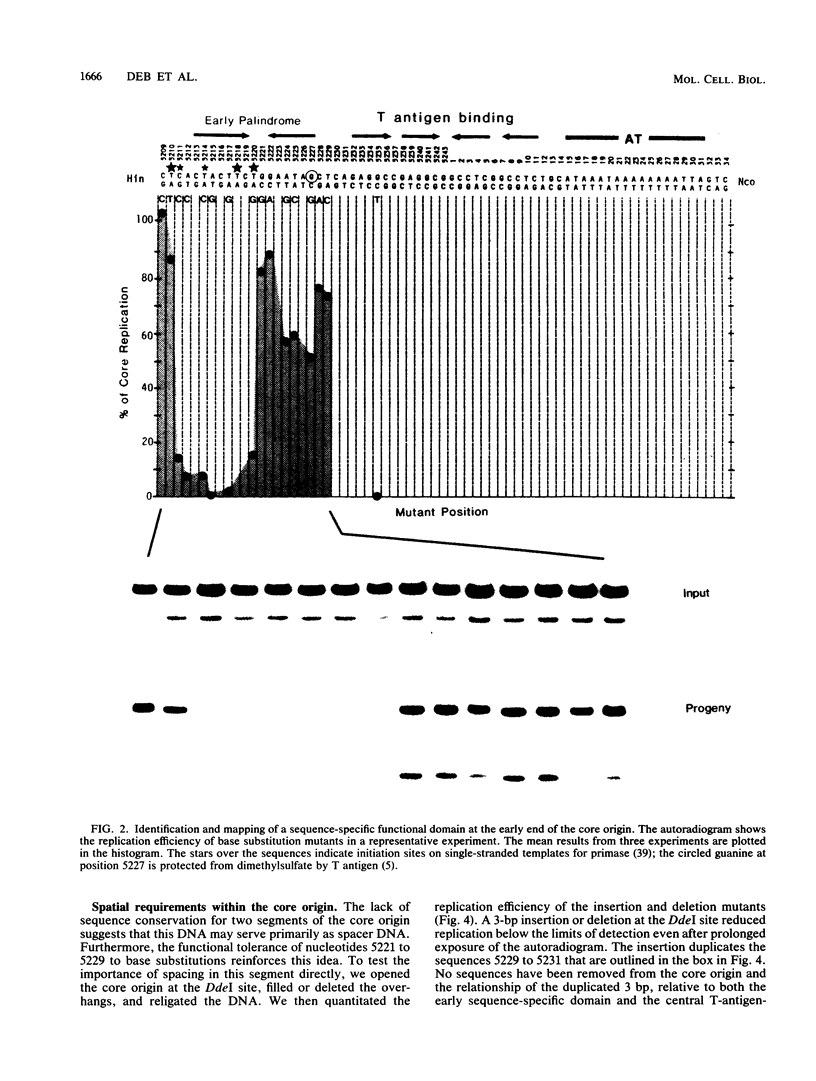
The simian virus 40 core origin of replication consists of nucleotides 5211 through 31. These 64 base pairs contain three functional domains with strict sequence requirements and two spacer regions with relaxed sequence specificity but precise positional constraints. The early domain extends for 10 contiguous base pairs between nucleotides 5211 and 5220. A 9-base pair spacer from sequences 5221 through 5229 separates the early domain from the 23-base pair central palindrome that directs the binding of T antigen. The late end of the core between nucleotides 12 and 31 also contains spacer and sequence-specific functions that are not yet completely mapped. We propose that the sequence-specific domains are interaction sites for viral and cellular proteins, determinants of DNA conformation, or both. The spacers would position these signals at required distances and rotations relative to one another.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abarzúa P., Marians K. J. Enzymatic techniques for the isolation of random single-base substitutions in vitro at high frequency. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2030–2034. doi: 10.1073/pnas.81.7.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsma D. J., Olive D. M., Hartzell S. W., Subramanian K. N. Territorial limits and functional anatomy of the simian virus 40 replication origin. Proc Natl Acad Sci U S A. 1982 Jan;79(2):381–385. doi: 10.1073/pnas.79.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereghini S., Yaniv M. Assembly of transfected DNA into chromatin: structural changes in the origin-promoter-enhancer region upon replication. EMBO J. 1984 Jun;3(6):1243–1253. doi: 10.1002/j.1460-2075.1984.tb01959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen G. L., Wright P. J., DeLucia A. L., Lewton B. A., Anderson M. E., Tegtmeyer P. Critical spatial requirement within the origin of simian virus 40 DNA replication. J Virol. 1984 Jul;51(1):91–96. doi: 10.1128/jvi.51.1.91-96.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham T. P., Pipas J. M. Simian agent 12 is a BK virus-like papovavirus which replicates in monkey cells. J Virol. 1985 May;54(2):483–492. doi: 10.1128/jvi.54.2.483-492.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLucia A. L., Deb S., Partin K., Tegtmeyer P. Functional interactions of the simian virus 40 core origin of replication with flanking regulatory sequences. J Virol. 1986 Jan;57(1):138–144. doi: 10.1128/jvi.57.1.138-144.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLucia A. L., Lewton B. A., Tjian R., Tegtmeyer P. Topography of simian virus 40 A protein-DNA complexes: arrangement of pentanucleotide interaction sites at the origin of replication. J Virol. 1983 Apr;46(1):143–150. doi: 10.1128/jvi.46.1.143-150.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMaio D., Nathans D. Cold-sensitive regulatory mutants of simian virus 40. J Mol Biol. 1980 Jun 15;140(1):129–142. doi: 10.1016/0022-2836(80)90359-9. [DOI] [PubMed] [Google Scholar]
- DiMaio D., Nathans D. Regulatory mutants of simian virus 40. Effect of mutations at a T antigen binding site on DNA replication and expression of viral genes. J Mol Biol. 1982 Apr 15;156(3):531–548. doi: 10.1016/0022-2836(82)90265-0. [DOI] [PubMed] [Google Scholar]
- Frisque R. J., Bream G. L., Cannella M. T. Human polyomavirus JC virus genome. J Virol. 1984 Aug;51(2):458–469. doi: 10.1128/jvi.51.2.458-469.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm M., Berg P. Deletion mapping of DNA regions required for SV40 early region promoter function in vivo. J Mol Appl Genet. 1982;1(5):457–481. [PubMed] [Google Scholar]
- Gerard R. D., Montelone B. A., Walter C. F., Innis J. W., Scott W. A. Role of specific simian virus 40 sequences in the nuclease-sensitive structure in viral chromatin. Mol Cell Biol. 1985 Jan;5(1):52–58. doi: 10.1128/mcb.5.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluzman Y., Frisque R. J., Sambrook J. Origin-defective mutants of SV40. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):293–300. doi: 10.1101/sqb.1980.044.01.033. [DOI] [PubMed] [Google Scholar]
- Hay R. T., DePamphilis M. L. Initiation of SV40 DNA replication in vivo: location and structure of 5' ends of DNA synthesized in the ori region. Cell. 1982 Apr;28(4):767–779. doi: 10.1016/0092-8674(82)90056-3. [DOI] [PubMed] [Google Scholar]
- Innis J. W., Scott W. A. DNA replication and chromatin structure of simian virus 40 insertion mutants. Mol Cell Biol. 1984 Aug;4(8):1499–1507. doi: 10.1128/mcb.4.8.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K. A., Myers R. M., Tjian R. Mutational analysis of simian virus 40 large T antigen DNA binding sites. EMBO J. 1984 Dec 20;3(13):3247–3255. doi: 10.1002/j.1460-2075.1984.tb02286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongstra J., Reudelhuber T. L., Oudet P., Benoist C., Chae C. B., Jeltsch J. M., Mathis D. J., Chambon P. Induction of altered chromatin structures by simian virus 40 enhancer and promoter elements. Nature. 1984 Feb 23;307(5953):708–714. doi: 10.1038/307708a0. [DOI] [PubMed] [Google Scholar]
- Kalderon D., Richardson W. D., Markham A. F., Smith A. E. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature. 1984 Sep 6;311(5981):33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- Li J. J., Kelly T. J. Simian virus 40 DNA replication in vitro: specificity of initiation and evidence for bidirectional replication. Mol Cell Biol. 1985 Jun;5(6):1238–1246. doi: 10.1128/mcb.5.6.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers R. M., Tjian R. Construction and analysis of simian virus 40 origins defective in tumor antigen binding and DNA replication. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6491–6495. doi: 10.1073/pnas.77.11.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder K., Silver S., DeLucia A. L., Fanning E., Tegtmeyer P. An altered DNA conformation in origin region I is a determinant for the binding of SV40 large T antigen. Cell. 1986 Mar 14;44(5):719–725. doi: 10.1016/0092-8674(86)90838-x. [DOI] [PubMed] [Google Scholar]
- Ryder K., Vakalopoulou E., Mertz R., Mastrangelo I., Hough P., Tegtmeyer P., Fanning E. Seventeen base pairs of region I encode a novel tripartite binding signal for SV40 T antigen. Cell. 1985 Sep;42(2):539–548. doi: 10.1016/0092-8674(85)90111-4. [DOI] [PubMed] [Google Scholar]
- Shortle D. R., Margolskee R. F., Nathans D. Mutational analysis of the simian virus 40 replicon: pseudorevertants of mutants with a defective replication origin. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6128–6131. doi: 10.1073/pnas.76.12.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortle D., Nathans D. Local mutagenesis: a method for generating viral mutants with base substitutions in preselected regions of the viral genome. Proc Natl Acad Sci U S A. 1978 May;75(5):2170–2174. doi: 10.1073/pnas.75.5.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. W., Gluzman Y. Replication and supercoiling of simian virus 40 DNA in cell extracts from human cells. Mol Cell Biol. 1985 Aug;5(8):2051–2060. doi: 10.1128/mcb.5.8.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B., Gerard R. D., Guggenheimer R. A., Gluzman Y. T antigen and template requirements for SV40 DNA replication in vitro. EMBO J. 1985 Nov;4(11):2933–2939. doi: 10.1002/j.1460-2075.1985.tb04026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian K. N., Shenk T. Definition of the boundaries of the origin of DNA replication in simian virus 40. Nucleic Acids Res. 1978 Oct;5(10):3635–3642. doi: 10.1093/nar/5.10.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegtmeyer P., Lewton B. A., DeLucia A. L., Wilson V. G., Ryder K. Topography of simian virus 40 A protein-DNA complexes: arrangement of protein bound to the origin of replication. J Virol. 1983 Apr;46(1):151–161. doi: 10.1128/jvi.46.1.151-161.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegtmeyer P. Simian virus 40 deoxyribonucleic acid synthesis: the viral replicon. J Virol. 1972 Oct;10(4):591–598. doi: 10.1128/jvi.10.4.591-598.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace R. B., Johnson M. J., Suggs S. V., Miyoshi K., Bhatt R., Itakura K. A set of synthetic oligodeoxyribonucleotide primers for DNA sequencing in the plasmid vector pBR322. Gene. 1981 Dec;16(1-3):21–26. doi: 10.1016/0378-1119(81)90057-3. [DOI] [PubMed] [Google Scholar]
- Wilson V. G., Tevethia M. J., Lewton B. A., Tegtmeyer P. DNA binding properties of simian virus 40 temperature-sensitive A proteins. J Virol. 1982 Nov;44(2):458–466. doi: 10.1128/jvi.44.2.458-466.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobbe C. R., Dean F., Weissbach L., Hurwitz J. In vitro replication of duplex circular DNA containing the simian virus 40 DNA origin site. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5710–5714. doi: 10.1073/pnas.82.17.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Hendrickson E. A., DePamphilis M. L. DNA primase-DNA polymerase alpha from simian cells: sequence specificity of initiation sites on simian virus 40 DNA. Mol Cell Biol. 1985 May;5(5):1170–1183. doi: 10.1128/mcb.5.5.1170. [DOI] [PMC free article] [PubMed] [Google Scholar]