Summary
Protein degradation by the ubiquitin-proteasome system is central to cell homeostasis and survival. Defects in this process are associated with diseases such as cancer and neurodegenerative disorders. The 26S proteasome is a large protease complex that degrades ubiquitinated proteins. Here we show that ADP-ribosylation promotes 26S proteasome activity in both Drosophila and human cells. We identify the ADP-ribosyl transferase Tankyrase (TNKS) and the 19S assembly chaperones dp27 and dS5b as direct binding partners of the proteasome regulator PI31. TNKS-mediated ADP-ribosylation of PI31 drastically reduces its affinity for 20S proteasome α-subunits to relieve 20S repression by PI31. Additionally, PI31 modification increases binding to and sequestration of dp27 and dS5b from 19S regulatory particles, promoting 26S assembly. Inhibition of TNKS by either RNAi or a small-molecule inhibitor, XAV939, blocks this process to reduce 26S assembly. These results unravel a mechanism of proteasome regulation that can be targeted with existing small-molecule inhibitors.
Introduction
Selective protein degradation plays a central role for the removal of misfolded, potentially toxic proteins, the control of cell cycle progression, regulation of gene expression, and changes in cell size and morphology (Baumeister et al., 1998; Demartino and Gillette, 2007; Finley, 2009; Glickman and Ciechanover, 2002; Hershko, 2005; Hershko and Ciechanover, 1998; Murata et al., 2009; Tanaka et al., 2012). Moreover, abnormal protein degradation is associated with a wide range of human diseases, such as cancer, muscle wasting diseases and neurodegenerative disorders (Glickman and Ciechanover, 2002; Goldberg, 2007; Hershko and Ciechanover, 1998). The selective degradation of most intracellular proteins is carried out by the ubiquitin-proteasome-system (UPS) (Finley, 2009; Glickman and Ciechanover, 2002; Hershko and Ciechanover, 1998; Varshavsky, 2012). Proteins tagged with poly-ubiquitin chains are hydrolyzed into small peptides by the 26S proteasome in an energy-dependent manner (Baumeister et al., 1998; Besche et al., 2009b; Demartino and Gillette, 2007; Finley, 2009; Tanaka et al., 2012; Tomko and Hochstrasser, 2011).
The 26S proteasome is a large protease complex composed of a catalytic 20S subunit (also known as 20S core particle) and a 19S regulatory particle that caps one or both ends of the 20S proteasome (Baumeister et al., 1998; Besche et al., 2009b; Demartino and Gillette, 2007; Finley, 2009; Lander et al., 2012; Lasker et al., 2012; Murata et al., 2009; Tanaka et al., 2012; Tomko and Hochstrasser, 2011). The assembly and activity of the 26S proteasome is tightly regulated by a large number of loosely associated proteins that function as regulators or cofactors (Besche et al., 2009b; Finley, 2009; Tanaka et al., 2012; Tomko and Hochstrasser, 2011). One such factor is PI31, an evolutionarily conserved regulator of proteasome activity (Bader et al., 2011; Chu-Ping et al., 1992; McCutchen-Maloney et al., 2000; Zaiss et al., 1999). PI31 was initially identified based on its ability to inhibit 20S proteasome activity in vitro (Chu-Ping et al., 1992; McCutchen-Maloney et al., 2000; Zaiss et al., 1999). However, PI31 can also activate the 26S proteasome in vitro, and mutational inactivation of the corresponding gene in Drosophila causes lethality, reduced proteasome activity and defects in protein degradation in vivo (Bader et al., 2011). Therefore, PI31 serves a crucial physiological function as an activator of 26S proteasome activity. The C-terminus of PI31 contains a functionally important HbYX (Hydrophobic residue-Tyrosine-any amino acid) motif, which is commonly found in modulators of proteasome activity, such as Rpt base subunits of the 19S regulatory particle (Gillette et al., 2008; Rabl et al., 2008; Smith et al., 2007). This suggests that PI31 can bind to the 20S particle via its HbYX-motif, and this may cause inhibition by hindering substrate access to the enzymatic core (Bader et al., 2011; McCutchen-Maloney et al., 2000). However, the precise molecular mechanism by which PI31 modulates proteasome activity remains unknown. Previous work also indicated that PI31 function is regulated in vivo to increase 26S proteasome activity under conditions where maximal proteolytic activity is required, for example for the removal of most cellular proteins during the terminal differentiation of sperm (Bader et al., 2011). To gain further insight into the regulation of PI31 activity we looked for novel binding partners of this protein and identified the ADP-ribosyl transferase tankyrase (TNKS) as a direct interactor that modulates PI31 activity. TNKS-mediated ADP-ribosylation of PI31 is necessary for the ability of this protein to stimulate 26S proteasome function, and inhibition of TNKS reduces 26S proteasome activity in both Drosophila and mammalian cells. TNKS-mediated ADP-ribosylation of PI31 drastically reduces the affinity of this protein for binding to 20S proteasome α-subunits and thereby relieves 20S repression by PI31. We also identified the 19S assembly chaperones dp27 and dS5b as binding partners of PI31. In this case, ADP-ribosylation of PI31 causes increased binding to dp27 and dS5b, and sequestration of these assembly chaperones from 19S regulatory particles, which promotes 26S assembly. These results reveal a novel mechanism of proteasome regulation and define TNKS-inhibitors as a new class of proteasome modulators that may have utility in the clinic.
Results
dTNKS interacts with DmPI31
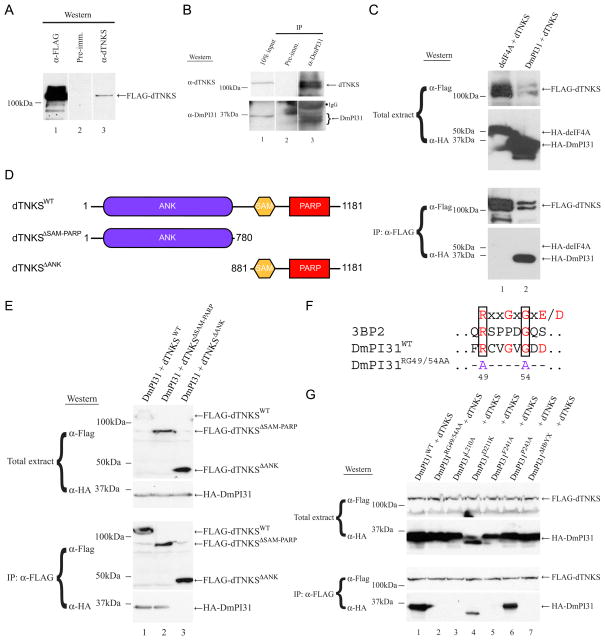
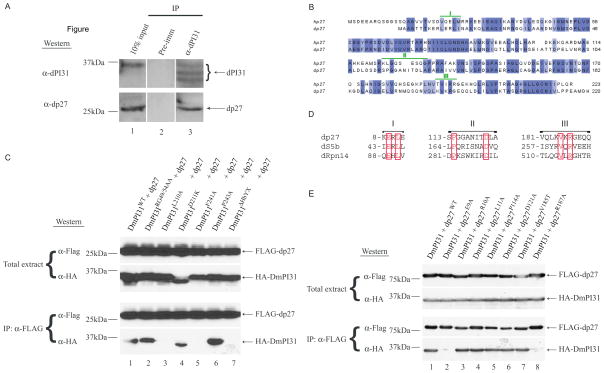
To gain insight into the mechanism of PI31-mediated proteasome regulation, we looked for binding partners of this protein. For this purpose, we screened a Drosophila embryo cDNA library by Far-Western analysis using P32-labeled HMK-tagged Drosophila PI31 (DmPI31) as a probe. This led to the identification of Drosophila tankyrase (dTNKS, CG4719) as a potential interacting protein (data not shown). dTNKS protein is 44% homologous to human tankyrase 1 and 2 (hTNKS1/2; Figure S1A). Next, we used co-IP experiments to investigate endogenous interactions between DmPI31 and dTNKS (Figure 1B). Extracts from wild-type (WT) embryos were immunoprecipitated with anti-DmPI31 and blots were probed with an antibody raised against dTNKS (Figures 1A and S1B). These experiments show that dTNKS and DmPI31 form a complex in vivo (Figure 1B). We also co-expressed FLAG-tagged dTNKS and HA-tagged DmPI31 in human 293 cells to establish a cell culture system for subsequent interaction studies (Figure 1C). Again, co-IP experiments with extracts from these cells showed the presence of a DmPI31:dTNKS complex (Figure 1C).
Figure 1. dTNKS, a novel interacting partner of DmPI31.
(A) dTNKS antiserum detects purified dTNKS. Purified FLAG-tagged dTNKS is detected in a Western blot with a dTNKS antiserum (lane 3), but not with pre-immune serum (lane 2). (B) 0–2h yw (WT) embryo extract was used in an anti-DmPI31 immunoprecipitation (IP) to demonstrate that DmPI31 interacts with dTNKS in vivo. (C) A DmPI31:dTNKS complex can form in mammalian 293 cells. Anti-FLAG IP experiment were performed using extracts from 293 cells expressing FLAG-tagged dTNKS together with HA-tagged deIF4A (negative control) or DmPI31. (D) Schematic representation of WT and mutant dTNKS with their respective domains. Numbers denote amino acid positions. (E) dTNKS recruits DmPI31 through its ANK domain as demonstrated via a Co-IP experiment using 293 cell extracts expressing HA-tagged DmPI31 with FLAG-tagged dTNKSWT, dTNKS ΔSAM-PARP and dTNKS ΔANK. (F) Alignments of the TNKS-binding motif with those found in human 3BP2 (Guettler et al., 2011; Levaot et al., 2011) and DmPI31 reveal the presence of a putative motif in DmPI31. Numbers indicate the position of residues within DmPI31. (G) dTNKS interacts with multiple DmPI31 surfaces: the putative DmPI31 TNKS-binding motif (RG49/54AA) and conserved C-terminal residues (D211 and P243) including the HbYX-domain. See also Figure S1.
TNKS belongs to the poly(ADP-ribose) polymerase (PARP) super-family (D’Amours et al., 1999; Hsiao and Smith, 2008; Smith et al., 1998). TNKSs play diverse roles in telomere maintenance, centrosome maturation, Wnt signaling, embryonic development and the pathogenesis of Cherubism (Guettler et al., 2011; Hsiao and Smith, 2008; Huang et al., 2009; Levaot et al., 2011). TNKSs recruit and modify target proteins by ADP-ribosylation using their Ankyrin (ANK) and PARP domains (Figure S1A) (Guettler et al., 2011; Hsiao and Smith, 2008; Huang et al., 2009; Levaot et al., 2011; Morrone et al., 2012; Smith et al., 1998). Unlike in mammals, where two isoforms of TNKS with partially redundant function are present, the Drosophila genome contains only one TNKS (Chiang et al., 2008; Chiang et al., 2006; Hsiao et al., 2006; Hsiao and Smith, 2008; Yeh et al., 2009). In order to identify which region of dTNKS is responsible for interaction with DmPI31, we generated truncations of dTNKS that lack either the ANK or SAM-PARP domains and tested them for binding to DmPI31 (Figures 1D and 1E). Whereas the SAM-PARP domains were dispensable for DmPI31-binding, deletion of the ANK domain in the dTNKS ΔANK mutant prevented formation of the DmPI31:dTNKS complex (Figure 1E). This suggests that dTNKS binds DmPI31 via its ANK-domain.
A hallmark of TNKS-binding partners is the presence of a canonical TNKS-binding motif (RxxGxGxE/D) (Guettler et al., 2011; Hsiao and Smith, 2008; Levaot et al., 2011). DmPI31 contains a good fit to this consensus motif at its N-terminus (Figure 1F). To investigate the functional importance of this motif, we mutated this sequence by altering two amino acids (RG49/54AA) and tested binding to dTNKS. The mutant protein was no longer able to form a complex with dTNKS, indicating that this motif is required for interaction with dTNKS (Figure 1G, lane 2). To further characterize the DmPI31-dTNKS interaction, we generated DmPI31 mutants in which the C-terminal residues, including HbYX motif, are either mutated or deleted (L210A, D211K, F241A and P243A and ΔHbYX - Figure 1G; lanes 3–7). Whereas DmPI31D211K and DmPI31P243A mutants retained their ability to bind dTNKS, DmPI31L210A and DmPI31F241A were unable to form a complex with dTNKS (Figure 1G). Furthermore, deletion of the HbYX motif in DmPI31 also abrogated the interaction between DmPI31 and dTNKS (Figure 1G, lane 7). This suggests that dTNKS uses its ANK-domain to bind DmPI31, and that both the N-terminal TNKS-binding motif and the C-terminal HbYX domain of DmPI31 are required for interaction with dTNKS.
dTNKS ADP-ribosylates DmPI31
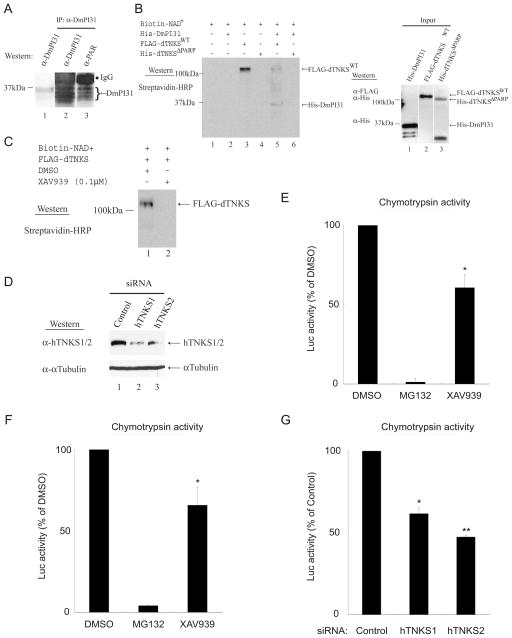
TNKSs modulate the activity of target proteins by ADP-ribosylation (Hsiao and Smith, 2008; Huang et al., 2009; Smith et al., 1998). Therefore, we investigated whether DmPI31 is a substrate for dTNKS-mediated ADP-ribosylation. First, we used Western blot analysis with a monoclonal antibody recognizing poly ADP-ribosylation (PAR) and found that DmPI31 is ADP-ribosylated in vivo (Figure 2A, lane 3). Next, we performed an in vitro ADP-ribosylation assay by incubating Biotin-labeled NAD+ and His-DmPI31 together with either FLAG-dTNKSWT or His-dTNKS ΔPARP recombinant proteins (Figure 2B). These experiments demonstrated that dTNKS can directly ADP-ribosylate DmPI31 in vitro, and that this activity requires its PARP domain.
Figure 2. dTNKS post-translationally modifies PI31 via ADP-ribosylation to regulate 26S proteasome activity.
(A) DmPI31 is poly ADP-ribosylated in vivo. Endogenous DmPI31 ADP-ribosylation was detected using 0–2h WT embryo extracts immuno-precipitated with anti-DmPI31. Eluted proteins were analyzed by Western blotting using an anti-poly ADP-ribose (PAR) antibody. (B) dTNKS directly ADP-ribosylates DmPI31 in vitro and this is dependent on its PARP domain. Samples containing Biotin-NAD+, His-DmPI31, and FLAG-dTNKSWT or His-dTNKS ΔPARP were analyzed for ADP-ribosylation by Western analysis. Input proteins were also analyzed by Western blotting (right panel). (C and D) Tools used to investigate the role of TNKS in 26S proteasome regulation. (C) XAV939 inhibits Drosophila TNKS (dTNKS) in vitro, indicating that the activity of this drug is conserved across species. (D) siRNAs targeting hTNKS1 and hTNKS2 efficiently knocked-down expressions in 293 cells. α-tubulin was used as a loading control. (E and F) TNKS inhibition by XAV939 reduces proteasome activity in Drosophila embryo (E) and 293 cell (F) extracts. (G) Reduced hTNKS1 and hTNKS2 levels inhibit 26S proteasome activity. (E–G) Data are presented as mean ± standard deviation from three independent experiments. Values obtained for DMSO (E–F) and control siRNA (G) were set as 100%. (E) *, p=0.01; (F) *, p=0.05; (G) *, p=0.04 and **, p=0.006. Statistical analysis was performed with a two-tailed, paired, t-test. See also Figure S2.
We investigated the significance of TNKS-mediated ADP-ribosylation for PI31 function and proteasome regulation, first in Drosophila and subsequently in mammalian cells. For this purpose, we took advantage of a small molecule inhibitor of hTNKS1/2, XAV939 (Huang et al., 2009). XAV939 was discovered in a screen for small molecules affecting the Wnt/β-catenin signaling pathway (Huang et al., 2009). XAV939 is a highly specific inhibitor of hTNKS1/2 (Kd = 0.09 μM) and has anti-cancer effects towards APC-deficient colorectal cancer cells (Huang et al., 2009). We tested the ability of XAV939 to inhibit dTNKS and found that it was able to inhibit the auto-ADP-ribosylation activity of dTNKS, but not PARP (Figures 2C and S2A) (Gibson and Kraus, 2012; Smith et al., 1998). Therefore, XAV939 is an effective and specific TNKS-inhibitor in Drosophila.
TNKS regulates proteasome activity by ADP-ribosylation
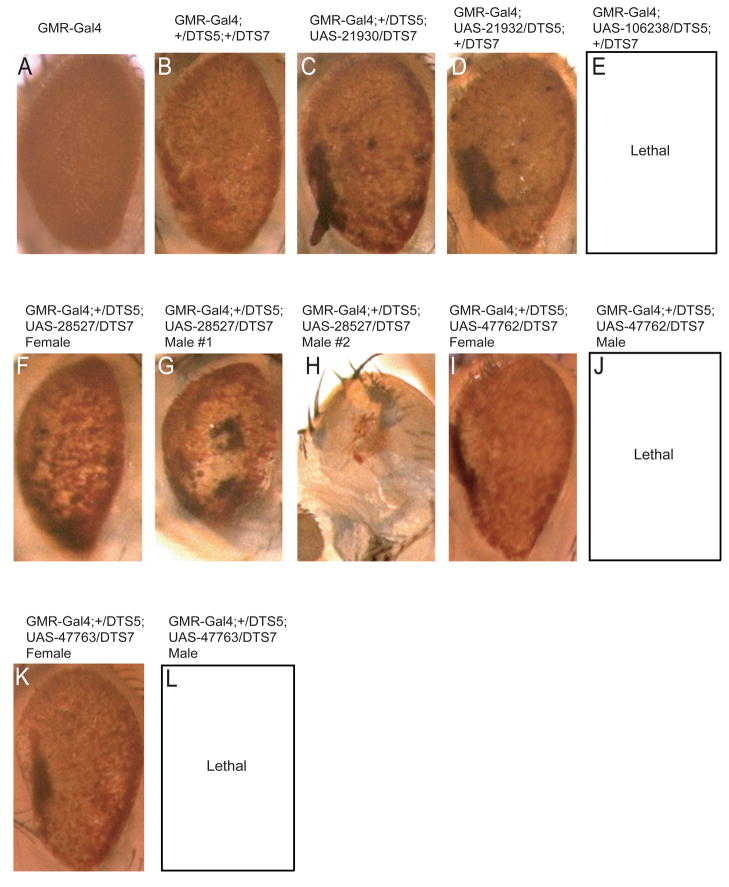
Because DmPI31 is required for optimal proteasome activity in vivo, and because dTNKS can post-translationally modulate DmPI31, we explored the possibility that inhibition of TNKS affects proteasome function. First, we examined the effect of the TNKS-inhibitor XAV939 on 26S proteasome activity in Drosophila. Extracts from 0–2h WT embryo were treated with different compounds and assayed for changes in proteasome activity (Figure 2E). Treatment with XAV939 significantly reduced 26S proteasome activity (Figure 2E). Although treatment with XAV939 did not cause complete inhibition of proteasome activity as observed for MG312, the effect was very similar to inactivation of DmPI31 (Bader et al., 2011). These results are consistent with a requirement of dTNKS for DmPI31-mediated proteasome activation. Next we asked whether XAV939 could also inhibit proteasome activity in mammalian cells. For this purpose, we treated 293 cell extracts with XAV939 and measured 26S proteasome activity (Figure 2F). Again, we found that exposure to this compound decreased proteasome activity. Similar results were obtained with IWR-1, another TNKS inhibitor that is structurally unrelated to XAV939 (Figure S2B) (Chen et al., 2009; Narwal et al., 2012). This supports the conclusion that the observed decrease in proteasome activity is caused by TNKS inhibition, and not an unrelated off-target effect of XAV939. To further corroborate this idea, we also used RNAi to target TNKS in both Drosophila and mammalian cells. First, we expressed RNAi against dTNKS in the Drosophila retina in a background that is compromised for proteasome activity (Figure 3). Specifically, expression of temperature sensitive dominant negative mutant for the 20S proteasome β2 and β6 subunits causes a rough, reduced eye (Belote and Fortier, 2002). Expression of RNAi against dTNKS further enhanced this phenotype, similarly to what was previously observed for reduction of DmPI31 function (Figures 3C–E) (Bader et al., 2011). Therefore, down-regulation of dTNKS reduces proteasome activity in vivo. Finally, we used siRNA to knock-down hTNKS1 and hTNKS2 in human 293 cells (Figure 2D). Again, a significant reduction of 26S proteasome activities was observed (Figure 2G). Taken together, these results show that TNKS stimulates proteasome activity by ADP-ribosylation of PI31.
Figure 3. dTNKS and dp27 RNAi enhances compromised proteasome activity.
To test dTNKS and dp27 role in modulating proteasome activity in vivo, we expressed RNAi against dTNKS (21930, 21932 and 106238; C–E) and dp27 (28527, 47762 and 47763; F–L) in the background of dominant-negative temperature sensitive mutants (UAS-DTS5 and UAS-DTS7) at 29°C. As expected, down-regulation of these factors resulted in a significant enhancement of the mutant rough reduced eye phenotype (compare panels B to C–L) and in some dp27 RNAi lines, caused male lethality. These observations are indicative of reduced proteasome activities in these tissues. GMR-Gal4 expression caused no discernable eye defects (panel A). Orientation of eyes is anterior left and dorsal up.
ADP-ribosylation blocks DmPI31 inhibition of 20S proteasomes
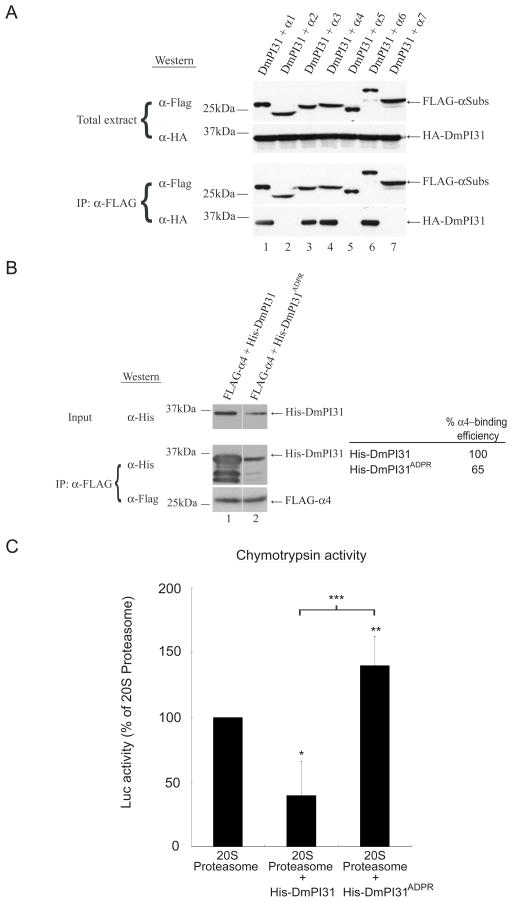
Best known for its role in DNA damage, ADP-ribosylation is a transient post-translational modification that can drastically alter the physical properties of target proteins (Chambers et al., 2012; Gagne et al., 2006; Gibson and Kraus, 2012; Wang et al., 2009). In order to understand the biochemical consequences of TNKS-mediated ADP-ribosylation of PI31, we investigated whether this modification affects the binding properties of this protein. In mammalian cells, PI31 can bind to 20S proteasomes (McCutchen-Maloney et al., 2000; Zaiss et al., 1999). Likewise, we found that DmPI31 can bind to 20S proteasomes in Drosophila (Figure 4A). In particular, DmPI31 bound selectively to several α-subunits: α1, α3, α4 and α6, but not α2 and α5 (Figure 4A). Next, we used co-immunoprecipitation experiments to evaluate the role of ADP-ribosylation on the DmPI31: α 4-complex. Whereas unmodified His-PI31 strongly interacted with α4, binding of the ADP-ribosylated version was significantly diminished (Figure 4B). Consistent with the observed change in DmPI31 affinity for α-subunits, we found that ADP-ribosylation interfered with the ability of PI31 to inhibit 20S proteasome particles (Figure 4C). These results demonstrate that ADP-ribosylation blocks binding of DmPI31 to 20S subunits and relieves repression of their proteolytic activity by this protein.
Figure 4. ADP-ribosylation relieves 20S inhibition by PI31.
(A) DmPI31 interacts with 20S proteasomes via select α-subunits: α1, α3, α4 and α6. (B) ADP-ribosylation significantly decreases the affinity of DmPI31 for α4. FLAG- α4 expressing 293 cell extract was incubated with modified DmPI31 (His-DmPI31ADPR) to test its effect on the α4:DmPI31 complex. Chemoluminescence was quantitated using ImageQuant LAS-4000 (GE Healthcare). α4-binding is reported as relative amount of immunoprecipitated His-DmPI31 compared to the level of input His-DmPI31. (C) Modification of DmPI31 relieves its inhibitory effect on 20S proteasomes. Purified bovine 20S proteasomes (0.1 μg) were incubated with His-DmPI31 and His-DmPI31ADPR proteins (2 μg each), and chymotrypsin-like proteasome activity was measured. Data are presented as mean ± standard deviation from three independent experiments. The value obtained for 20S proteasome was set as 100%. *, p=0.04, **, p=0.6 and ***, p=0.045. Statistical analysis was performed with a two-tailed, paired, t-test.
dTNKS and DmPI31 actively participates in proteasome assembly
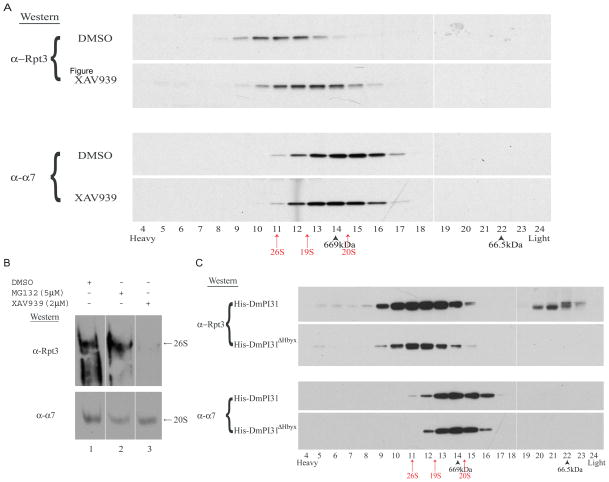
Since TNKS-mediated ADP-ribosylation of PI31 affects both 20S and 26S proteasome activities, we investigated a possible role of TNKS and PI31 in 26S proteasome assembly. For this, we performed Superose 6 gel filtration (FPLC) assays and native gel analyses to study the effect of TNKS-inhibitor XAV939 on proteasome assembly. Drosophila embryos are an ideal system to study effects on 26S assembly because ~65% of proteasomes exist as 20S particles (Nickell et al., 2007). XAV939 treatment of Drosophila embryo extracts led to a significant shift in Rpt bands towards lighter fractions when compared to controls, and it prevented the formation of 26S proteasomes when analyzed on native gels (Figures 5A, 5B, S3A and S3B). This indicates that TNKS activity promotes 26S proteasome assembly.
Figure 5. TNKS and PI31 regulate proteasome assembly.
(A) Inhibition of dTNKS by XAV939 led to a drastic shift of the proteasome profile to “lighter” fractions, indicative of a reduced amount of 26S particles. To assess the effect of TNKS activity on 26S assembly, embryo extracts treated with XAV939 were subjected to Superose 6 gel filtration chromatography. (B) TNKS inhibition severely affects 26S proteasome assembly. The effect of XAV939 on proteasome assembly in embryo extracts was assessed by native gel analysis. (C) Similar to XAV939, surplus DmPI31 dramatically affected proteasome assembly. Furthermore, it also caused an increase in levels of Rpt3 monomers (fractions 20–23), as visualized by gel filtration chromatography. (A–C) Western blot analyses were performed to detect the presence of the proteasomal subunits Rpt3 (a 19S component that can be used to identify 26S) and α7 (to identify 20S). (A and C) Arrows and arrowheads indicate the position of proteasome components (red arrows: 26S, 19S and 20S) and standards (black arrowheads: Thyroglobulin-660 kDa and BSA-66.5 kDa), respectively. See also Figure S3.
Next we examined the effect of PI31 on 26S proteasome assembly. Again, we used gel filtration assays to analyze embryo extracts supplemented with high concentrations of His-DmPI31 recombinant protein (Figure 5C). Again, addition of His-DmPI31 caused a significant change in the proteasome architectural landscape by redistributing Rpt3 bands (Figure 5C). We also observed an increase in Rpt3 monomers (Figure 5C; top panel - fractions 20–23). These effects were seen for WT DmPI31, but not for a mutant lacking the HbYX motif (Figure 5C). The increase in Rpt3 monomers can be explained by the ability of PI31 to bind 19S assembly chaperones (see below) that are known to stabilize the assembly of both 19S regulatory particle and 26S proteasome (Funakoshi et al., 2009; Kaneko et al., 2009; Le Tallec et al., 2009; Park et al., 2009; Roelofs et al., 2009; Saeki et al., 2009). Collectively, these results show that both TNKS activity and PI31 can modulate 26S proteasome assembly, and suggest a role of PI31 in promoting 19S stability.
19S assembly chaperones interact with DmPI31
To better understand the role of DmPI31 in proteasome assembly, we looked for additional DmPI31 binding partners. In the same screen that revealed dTNKS as a DmPI31 interactor, we discovered two other proteasome-associated proteins, Drosophila p27 (dp27; CG9588) and Drosophila S5b (dS5b; CG12096). p27 and S5b play a known role as 19S assembly chaperones and, along with Rpn14 and Nas6, bind to 19S Rpt subunits to foster orderly proteasome assembly (Funakoshi et al., 2009; Kaneko et al., 2009; Le Tallec et al., 2009; Park et al., 2009; Roelofs et al., 2009; Saeki et al., 2009). As in yeast and human, the Drosophila homologs of the 19S assembly chaperones (dp27: 41% and dS5b: 23.3% homologous; Figures 6B and S5A) are essential genes and required for organismal viability (Figures 3F–L, S4A and S4B). IP experiments with anti-DmPI31 using Drosophila embryo extracts followed by Western analysis using anti-dp27 reveal that DmPI31 binds dp27 in vivo (Figures 6A and S4C). To identify DmPI31 residues involved in dp27 and dS5b binding, we used point mutants previously employed to map the DmPI31:dTNKS interaction (Figure 1G). While mutants defective for TNKS-binding (RG49/54AA) failed to destabilize the DmPI31:assembly chaperone complex, mutating the conserved C-terminus of DmPI31 (L210, F241 and HbYX motif) disrupted these interactions (Figures 6C and S5B). These results show that the C-terminal HbYX motif is required for the formation of a complex between DmPI31 and the assembly chaperones dp27 and dS5b.
Figure 6. The 19S assembly chaperone dp27 interacts with DmPI31.
(A) Endogenous dp27:DmPI31 interaction was detected in an anti-DmPI31 IP experiment using embryo extracts. (B) Sequence alignment of p27 from human (hp27) and D. melanogaster (dp27). dp27 is 41% homologous to hp27. Identical amino acids are colored in violet and the DmPI31-binding motifs (I–III; refer to panel D) are denoted in green. (C) DmPI31 recruits dp27 via its C-terminal residues: L210, F241 and HbYX-domain. Similar observations were made for TNKS (Figure 1G). (D) dp27, dS5b and dRpn14 protein sequence alignments reveal three putative DmPI31-binding motifs (I–III) with unique conserved residues (in red boxes). Numbers denote N-terminal end amino acid positions of each motif in respective proteins. (E) Multiple dp27 surfaces (motifs I (E9A) and III (R187A)) make contacts with DmPI31 to stabilize the complex. See also Figures S4 and S5.
Despite their structural differences, 19S assembly chaperones use a common mechanism to bind to 19S Rpt subunits (Barrault et al., 2012; Funakoshi et al., 2009; Lee et al., 2011; Park et al., 2009; Roelofs et al., 2009; Saeki et al., 2009; Takagi et al., 2012). Since dp27 and dS5b both interact with the DmPI31 C-terminus, we performed protein sequence alignments of dp27, dS5b and dRpn14. This identified three putative DmPI31-binding motifs (I–III; Figure 6D). In order to test the functional relevance of these motifs, we mutated these sites and examined the consequences for DmPI31 interaction (Figures 6E and S5C). Point mutations that replaced key residues in motifs I and III of dp27 and dS5b (dp27: E9A and R187A; dS5b: E44A and R263A) failed to recruit DmPI31, indicating a requirement of these motifs for interaction with DmPI31 (Figures 6E and S5C). These results suggest that dp27 and dS5b are novel interacting partners of DmPI31.
ADP-ribosylation promotes 26S proteasome assembly by increasing the affinity of DmPI31 for assembly chaperones
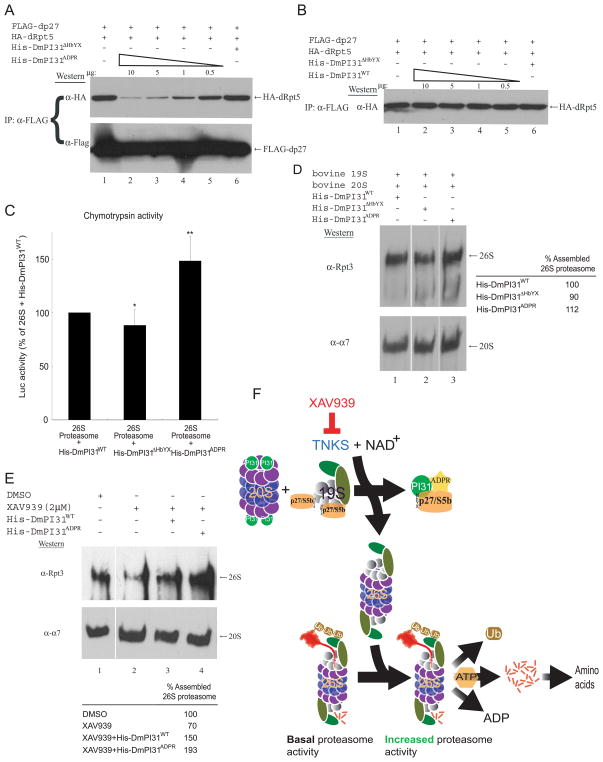
Since ADP-ribosylation of PI31 modulates 20S proteasome activity by altering its affinity to α-subunits (Figure 4), we wondered if this post-translational modification may also affect interaction with the 19S assembly chaperones p27 and S5b. To examine this possibility, we first performed an anti-FLAG IP experiment using 293 cell extracts standardized for FLAG-dp27 and HA-dRpt5 expression, followed by rigorous washing and the addition of incremental amounts of in vitro modified DmPI31. The effects of ADP-ribosylated DmPI31 on the stability of dp27:dRpt5 and dS5b:dRpt2 complexes were visualized via Western blot (Figures 7A and S6A). Countering its effect on α4 subunit, when compared to its unmodified version, ADP-ribosylation considerably increased the ability of DmPI31 to recruit the assembly chaperones, sequestering them away from the Rpt base subunits in a dose-dependent manner (compare Figures 7A and S6A to Figures 7B and S6B, respectively). These results, together with those presented earlier indicate that ADP-ribosylation directly modifies DmPI31 affinity for proteasome-associated proteins.
Figure 7. ADP-ribosylation increases DmPI31 affinity for dp27 to regulate proteasome assembly, and model for proteasome regulation by ADP-ribosylation.
(A and B) ADP-ribosylation increased the affinity of DmPI31 for dp27, allowing it to compete with dRpt5 for chaperones in a dose-dependent manner. Anti-FLAG IP using 293 cell extracts standardized for FLAG-dp27 and HA-dRpt5 expressions were supplemented with in vitro modified (A) and unmodified (B) DmPI31 to assess their ability to compete with dRpt5 for dp27. (C) ADP-ribosylated DmPI31 activated 26S proteasome activity in vitro to levels beyond that seen for its unmodified control. Purified bovine 26S proteasome (0.1μg) was incubated with His-DmPI31, His-DmPI31ΔHbYX and His-DmPI31ADPR (2 μg each), and chymotrypsin-like proteasome activity was assayed. Data are presented as mean ± standard deviation from three independent experiments. The value obtained for “26S + His-DmPI31WT” was set as 100%. *, p=0.46 and **, p=0.007. Statistical analysis was performed with a two-tailed, paired, t-test. (D) DmPI31 modification promotes de novo assembly of the 26S proteasome. Purified bovine 19S and 20S proteasome particles (1 μg each) were incubated with His-DmPI31, His-DmPI31ΔHbYX and His-DmPI31ADPR (5 μg each) in the presence of ATP, followed by native gel analysis to assess the effect of modified DmPI31 on proteasome assembly. (E) Modified DmPI31 rescues XAV939-induced defects in 26S proteasome assembly to a greater extent than its unmodified control. 200 μg of embryo extracts treated with the TNKS-inhibitor were supplemented with 5 μg of respective proteins followed by native gel analysis. (D and E) Native gels were analyzed via Western blot using anti-Rpt3 (for 26S proteasome) and anti- α7 (for 20S proteasome). Chemoluminescence was quantitated using ImageQuant LAS-4000 (GE Healthcare). 26S proteasome assembly status is reported as relative intensity of Rpt3 bands. (F) Model for proteasome regulation by ADP-ribosylation. PI31 is a conserved proteasome regulatory protein that is required for normal 26S proteasome activity in vivo and organismal viability (Bader et al., 2011). ADP-ribosylation of PI31 by TNKS, which transfers the ADP-ribose group from NAD+, activates PI31 and leads to increased proteolytic activity of the 26S proteasome by promoting its assembly. Specifically, ADP-ribosylation of PI31 causes decreased affinity for 20S proteasome α-subunits and dislodges PI31 from 20S particles. On the other hand, ADP-ribosylation increases binding of PI31 to the assembly chaperones dp27 and dS5b and thereby sequesters them away from 19S regulatory particles. Taken together, this promotes 26S proteasome assembly and facilitates the breakdown of intracellular proteins. Inhibition of TNKS by small-molecule compounds, such as XAV939, blocks this process and reduces 26S proteasome activity. Therefore, TNKS-inhibitors represent a novel class of proteasome inhibitors that target the assembly of 26S proteasomes. See also Figure S6.
Evidence for a role of PI31 as an activator of the 26S proteasome was previously based on the requirement of this protein for normal proteasome activity in vivo, and the ability of this protein to stimulate 26S proteasome activity in vitro (Bader et al., 2011). Since modification by ADP-ribosylation differentially affects interaction of DmPI31 with different proteasome proteins, we investigated the consequences of PI31-modification on 26S proteasome activity in vitro. ADP-ribosylated DmPI31 was significantly more potent in stimulating the chymotrypsin-like activity of purified 26S proteasomes in vitro (Figure 7C). Furthermore, when modified DmPI31 was incubated with purified 20S and 19S particles and analyzed on native gels, we saw that ADP-ribosylation increased the ability of DmPI31 to promote de novo 26S assembly in vitro (Figure 7D). Finally, the addition of ADP-ribosylated DmPI31 to extracts pre-treated with XAV939 reversed the inhibitory effect of this compound on 26S proteasome assembly and was even able to further increase 26S proteasome assembly (Figure 7E). We also found that surplus PI31 can overcome the effects of XAV939, albeit less efficiently than its modified counterpart. This is consistent with our observation that unmodified PI31 can bind and sequester 19S assembly chaperones (Figure S6B). Taken together, our data show that dTNKS-dependent ADP-ribosylation of DmPI31 stimulates 26S proteasome activity by promoting 26S assembly.
Discussion
It is becoming increasingly clear that the proteasome, often thought to be a constitutively active protease complex, is dynamically regulated to meet the changing proteolytic needs of a cell (Demartino and Gillette, 2007; Glickman and Ciechanover, 2002; Hershko and Ciechanover, 1998). Although poly-ubiquitination of substrates is central to select specific proteins for degradation, additional mechanisms must exist to account for the plasticity of proteasome activity (Crosas et al., 2006; Demartino and Gillette, 2007; Hanna et al., 2007; Kurucz et al., 2002; Murata et al., 2009; Peth et al., 2009; Peth et al., 2013; Peth et al., 2010; Princiotta et al., 2001). Here, we identified TNKS as a novel regulator of proteasome activity in both Drosophila and mammalian cells. Specifically, we show that TNKS-mediated ADP-ribosylation of PI31 alters the affinity of this protein for 20S proteasome α-subunits and 19S assembly chaperones and thereby stimulates 26S proteasome assembly. Our results support a model in which TNKS activates an evolutionarily conserved proteasome regulatory protein, PI31, by ADP-ribosylation (Figure 7F). PI31 is physiologically required for optimal 26S proteasome activity in vivo, and inactivation of the corresponding gene in Drosophila causes reduced protein breakdown and organismal lethality (Bader et al., 2011). Inhibition of TNKS by either RNAi or with a specific small-molecule inhibitor, XAV939, blocked ADP-ribosylation of PI31 and impaired 26S proteasome activity similar to inactivation of PI31. It was suggested that PI31 may act as a modulator of proteasome assembly by reversibly associating with 20S proteasome (Besche et al., 2009a; McCutchen-Maloney et al., 2000; Tai et al., 2010; Tanahashi et al., 1999; Zaiss et al., 1999; Zaiss et al., 2002). In support of this idea, we show that PI31 modification by TNKS facilitates the assembly of the 26S proteasome by acting at two crucial stages (Figure 7F). First, TNKS-mediated ADP-ribosylation decreases the affinity of PI31 for α4 subunits of the 20S particle. This dislodges PI31 from 20S proteasomes and prevents PI31 from inhibiting 20S activity. Once free, modified PI31 competes with Rpt subunits to sequester the assembly chaperones p27 and S5b away from 19S regulatory particles, thereby promoting the capping of 20S with 19S particles. This is consistent with previous observations where overexpression of p27, S5b and Rpn14/PAAF1 proteins negatively affected proteasome assembly (Kaneko et al., 2009; Lassot et al., 2007; Park et al., 2005; Shim et al., 2012). By acting on both 20S particles and assembly chaperones, PI31 appears to function as a central regulator of 26S proteasome assembly (Figure 7F). Significantly, this process can be disrupted with existing small-molecule compounds: TNKS-inhibitors represent a novel class of compounds that inhibit 26S activity by interfering with proteasome assembly.
PI31 is required for normal proteasome activity in vivo, and it can also stimulate the activity of purified 26S proteasome in vitro (Bader et al., 2011). In order to explain the latter finding, we invoke the possibility that purified 26S proteasomes used in these experiments contain associated proteins, including 19S assembly chaperones. This is consistent with observations showing p27 and S5b in association with purified 26S proteasome (Besche et al., 2009a; Deveraux et al., 1995; Gomes et al., 2006; Tai et al., 2010; Watanabe et al., 1998). Furthermore, unmodified PI31 can still bind to assembly chaperones, albeit with much lower efficiency (Figure S6B). Taken together, these results suggest that PI31 stimulates proteasome activity in vitro by increasing 26S particles.
The rate of intracellular protein degradation is dramatically affected by various signals, including the metabolic state of the cell (Glickman and Ciechanover, 2002). For example, caloric restriction can cause a severe reduction of skeletal muscles, and humans loose a large fraction of their muscle mass during aging. Although this appears to have evolved as a protective mechanism to cope with food shortage, muscle atrophy can be a highly debilitating process that also occurs in various diseases, such as cancer cachexia, AIDS, renal failure, and in neuro-degenerative diseases. In all these cases, proteasome activity is responsible for protein breakdown and cellular atrophy, but a direct connection between metabolism and proteasome activity has not yet been established. Since NAD+ is the source of the ADP-ribose group, proteasome regulation by TNKS provides a potential new link between cell metabolism and the regulation of protein degradation (Gibson and Kraus, 2012). Because ADP-ribosylation has a very short half-life (1–2 min), it is ideally suited to rapidly signal transient changes in cell physiology (Chambers et al., 2012; Gibson and Kraus, 2012; Lindahl et al., 1995; Wang et al., 2009). We speculate that TNKS-mediated activation of PI31 may serve to stimulate proteasome activity when cells need to dynamically boost their proteolytic capacity to meet changing demands, such as during cellular remodeling, upon stress and during caloric restriction. It is also possible that activation of PI31 occurs only in specific subcellular compartments, for example within the nucleus, in specific neuronal processes undergoing pruning, or in synapses during remodeling. In this way, it may be possible to locally fine-tune the assembly and activity of 26S proteasomes to meet a cell’s changing needs for controlled proteolysis.
The proteasome is a validated drug target for cancer therapy and bortezomib (“Velcade”), which inhibits the chymotrypsin-like activity of the proteasome, is approved in the US for the treatment of multiple myeloma and mantle cell lymphoma (Goldberg, 2012; Kisselev et al., 2012; Raab et al., 2009). Problems associated with bortezomib include notable side effects, such as peripheral neuropathy and drug resistance (Goldberg, 2012; Kisselev et al., 2012; Raab et al., 2009). Therefore, drugs that inhibit proteasome activity by a separate mechanism may have considerable clinical value. Our findings suggest that TNKS-inhibitors, such as XAV939 and IWR-1, may be useful for the treatment of multiple myeloma, mantle cell lymphoma and other cancers sensitive to proteasome inhibition. Consistent with this idea, we have preliminary evidence that XAV939 can block the growth of multiple myeloma cells (Figure S6C). Besides inhibiting proteasome activity by an entirely distinct mechanism, TNKS-inhibitors are expected to only block maximal activation of 26S function, but not the basal activity of the proteasome and hence may have fewer side effects (Figure 7F). Interestingly, XAV939 has already been shown to be effective against colorectal cancer cells and is thought to antagonize Wnt-signaling by preventing the degradation of axin (Huang et al., 2009). However, the contribution of proteasome inhibition in this paradigm has not yet been determined. Proteasome regulation by tankyrase-mediated ADP-ribosylation provides an unexpected new mechanism to regulate protein degradation that can be targeted with small-molecule inhibitors.
Experimental Procedures
Recombinant Protein Purification and Far-Western screening of cDNA library
For the purification of His-DmPI31, His-DmPI31ΔHbYX, His-HMK-DmPI31 and His-dTNKSΔPARP fusion proteins, E.coli BL21 (Invitrogen) was transformed with the pET28-His-DmPI31, pET28-His-DmPI31ΔHbYX, pET101-His-HMK-DmPI31 and pET101-His-dTNKSΔPARP constructs. Following a 2h induction at 37°C with 0.1 mM IPTG, the fusion proteins were purified on a TALON™ Metal Affinity resin (BD Bioscience) according to the manufacturer’s instructions. For the purification of FLAG-dTNKS and FLAG-dPARP, HEK293 cell extracts expressing the pcDNA3.2-FLAG-dTNKS and pcDNA3.2-FLAG-dPARP constructs were incubated with anti- FLAG®M2-Affinity Gel (Sigma) for 3h and purified using FLAG® peptide (Sigma) according to the manufacturer’s instructions. Far-Western screening of cDNA library was performed as previous described (Pause et al., 1994) using Drosophila λ cDNA library (Stratagene).
Cell Culture/Growth assay
Cationic lipid reagent (20 μl of Lipofectamine 2000; Invitrogen) was diluted in serum free media (Opti-MEM; Invitrogen) for transfection in Human Embryonic Kidney 293 cells (100mm dish). Following a 5h incubation, the medium was replaced with Dulbecco’s Modified Eagle Medium(DMEM; Invitrogen) supplemented with 10% Fetal Bovine Serum (FBS). Transfected cells were harvested in PBS 48h following the addition of serum containing media. The cells were then lysed by repeated freeze/thaw cycles in 600 μl of lysis buffer (20mM Hepes-KOH, pH 7.6, 200mM KCl, 0.5mM EDTA, 10% glycerol, 1% Triton X-100 and Protease Inhibitor Cocktail (Complete™; Roche)) that contains RNAse A (50 μg/ml; Sigma). Cell debris was pelleted by centrifugation, and the protein concentration in the supernatant was determined using the Bio-Rad assay. The siRNAs purchased from Invitrogen (sequences shown in Table S1) were transfected in 293 cells at a final concentration of 50nM/100mm dishes using Lipofectamine 2000. Effects of drug treatment on U266 multiple myeloma cell growth were performed using 0.1 μM of bortezomib and 5 μM of XAV939 in low FBS (0.5%) RPMI media, and 96 well plates seeded with ~8000 cells/well. Samples were assayed for growth/viability using PrestoBlue® Cell Viability Reagent (Invitrogen) in a Spectramax M2 reader (Molecular Devices) per manufacturer’s instructions.
Co-Immunoprecipitation
For co-immunoprecipitation, 293 cell extract (200 μl; 6–10 μg/ μl) was brought up to 1ml with the lysis buffer and pre-cleared for 1h at 4°C with 25 μl of Protein A Sepharose (GE Healthcare). The supernatant was immunoprecipitated for 1h at 4°C with 25 μl of anti-FLAG®M2-Affinity Gel (Sigma). The resin was washed twice with lysis buffer and once with lysis buffer containing 300mM KCl. Immunoprecipitates were eluted in 3X red sample buffer (NEB). For anti-HA and anti-DmPI31 immunoprecipitations, 25 μl of Protein A Sepharose were pre-incubated for 2h with anti-HA (3 μl) and anti-DmPI31 (5 μl). The resins were washed three times with the lysis buffer prior to immunoprecipitation as described above. Embryo extracts were used at a concentration of 5 μg/ μl.
Proteasome activity Assay
50 μg of 0–2h WT (yw) embryo or 3 μg of 293 cell extracts in PIPES buffer (50mM PIPES, 1mMMgCl2, 50mM NaCl, 2mM EGTA and 2mM ATP) were programmed with 1% DMSO (Sigma), 5 μM MG132 (Calbiochem), 2 μM XAV939 (Sigma) and 2 μM IWR-1 (Sigma), respectively, and incubated for 20 min at room temperature. Samples were assayed for proteasome activity using the Proteasome-Glo™ Chymotrypsin-like Cell-Based assay (Promega) in a Spectramax M2 reader (Molecular Devices). To assess the effect of ADP-ribosylation on 20S and the 26S proteasome activities purified bovine 20S and 26S proteasomes (0.1 μg; UBPBio) in PIPES buffer + 2mM ATP were programmed with 2 μg of His-DmPI31WT, His-DmPI31ΔHbYX and His-DmPI31ADPR, and incubated for 20 min at room temperature. Samples were assayed for proteasome activity as described above.
Gel Filtration assay
2mg of 0–2h WT embryo extracts prepared using the PIPES buffer + 2mM ATP were centrifuged twice at 14 000 rpm for 20 min at 4°C prior to being treated with 50 μg His-DmPI31, 50 μg His-DmPI31ΔHbYX, 50 μg His-DmPI31ADP-ribosylation, 1% DMSO, 5 μM MG132, 2μM XAV939 and 2 μM Olaparib (LC laboratories) for 30 min. Gel filtration assay was subsequently performed using Superose 6 10/300GL FPLC column (AKTA; GE Healthcare). 500 μl fractions were collected using 0.250 ml/min flow rate. Purified bovine 19S, 20S and 26S proteasomes (UBPBio), thyroglobulin, and BSA were used as standards.
In vitro ADP-ribosylation Assay and Purification of His-DmPI31ADPR
Recombinant His-DmPI31 (1 μg), FLAG-dTNKS (0.1 μg) and His-dTNKSΔPARP (0.1 μg), together with biotin-NAD+ (3.5 μM; Trevigen) were incubated for 5h at 37°C and subjected to SDS-PAGE followed by Western Blotting using Streptavidin-HRP (Thermo-Scientific; 1:10000). Effect of XAV939 on dTNKS and dPARP were assessed by incubating FLAG-dTNKS or FLAG-dPARP/Biotin-NAD+ mixture with either 0.1% DMSO or 0.1 μM XAV939. For the purification of His-DmPI31ADPR protein, in vitro ADP-ribosylation of recombinant His-DmPI31 protein was performed as above. This was followed by serial purifications using TALON Metal Affinity resin (ClonTech) and SoftLink™ Soft Release Avidin resin (Promega) per manufactures’ instructions.
Native Gel analysis
To analyze proteasome assembly status, 200 μg of embryo extracts exposed to different experimental conditions + 2mM ATP were resolved using 5% Tris-HCl acrylamide gels (Bio-rad) under non-denaturing conditions and transferred onto a 0.22 μm PVDF membrane. Western blot analysis using specific antibodies were performed as described above. Concentrations of various reagents were: 1% DMSO, 5 μM MG132, 2 μM XAV939, purified bovine 19S and 20S (1 μg), and 5μg of His-DmPI31WT, His-DmPI31ΔHbYX and His-DmPI31ADPR proteins.
Fly Strains
UAS-DTS5 and UAS-DTS7 lines were obtained from J. Belote, dTNKS (21930, 21932 and 106238), dp27 (28527, 47762 and 47763), dS5b (104492) and dRpn14 (32697 and 32698) RNAi lines were obtained from the Vienna Drosophila RNAi Center (VDRC) and Bloomington Drosophila Stock Center.
Supplementary Material
Highlights.
TNKS modifies the proteasome regulator PI31 by ADP -ribosylation
ADP-ribosylation alters PI31 affinity for proteasome proteins
TNKS regulates the 26S proteasome activity by controlling its assembly
TNKS-inhibitors represent a new class of agents that block 26S assembly
Acknowledgments
In alphabetical order, we thank J. Belote for providing fly strains; G. DeMartino, A. Goldberg and S. Larisch for critical comments on the manuscript; S. Benjamin-Hong, O. Cho, I.B. Cho-Park, Y.A. Cho-Park, D. Ferres-Marco, N. Gandagar, T. Hsiao, A. Kelkar, L.I. Park, C. Pham, J. Rodriguez, I. Sachrai, C. Sandu and members of our lab for valuable discussions; J. Gee-Esposito, T. Gorenc, O. Matthew and A. Persaud for logistical support. H.S. is an Investigator of the Howard Hughes Medical Institute. This work was supported by NIH grant RO1GM60124 to H.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bader M, Benjamin S, Wapinski OL, Smith DM, Goldberg AL, Steller H. A conserved F box regulatory complex controls proteasome activity in Drosophila. Cell. 2011;145:371–382. doi: 10.1016/j.cell.2011.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrault MB, Richet N, Godard C, Murciano B, Le Tallec B, Rousseau E, Legrand P, Charbonnier JB, Le Du MH, Guerois R, et al. Dual functions of the Hsm3 protein in chaperoning and scaffolding regulatory particle subunits during the proteasome assembly. Proc Natl Acad Sci U S A. 2012;109:E1001–1010. doi: 10.1073/pnas.1116538109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister W, Walz J, Zuhl F, Seemuller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- Belote JM, Fortier E. Targeted expression of dominant negative proteasome mutants in Drosophila melanogaster. Genesis. 2002;34:80–82. doi: 10.1002/gene.10131. [DOI] [PubMed] [Google Scholar]
- Besche HC, Haas W, Gygi SP, Goldberg AL. Isolation of mammalian 26S proteasomes and p97/VCP complexes using the ubiquitin-like domain from HHR23B reveals novel proteasome-associated proteins. Biochemistry. 2009a;48:2538–2549. doi: 10.1021/bi802198q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besche HC, Peth A, Goldberg AL. Getting to first base in proteasome assembly. Cell. 2009b;138:25–28. doi: 10.1016/j.cell.2009.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers JE, Petrova K, Tomba G, Vendruscolo M, Ron D. ADP ribosylation adapts an ER chaperone response to short-term fluctuations in unfolded protein load. J Cell Biol. 2012;198:371–385. doi: 10.1083/jcb.201202005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Dodge ME, Tang W, Lu J, Ma Z, Fan CW, Wei S, Hao W, Kilgore J, Williams NS, et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol. 2009;5:100–107. doi: 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang YJ, Hsiao SJ, Yver D, Cushman SW, Tessarollo L, Smith S, Hodes RJ. Tankyrase 1 and tankyrase 2 are essential but redundant for mouse embryonic development. PLoS One. 2008;3:e2639. doi: 10.1371/journal.pone.0002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang YJ, Nguyen ML, Gurunathan S, Kaminker P, Tessarollo L, Campisi J, Hodes RJ. Generation and characterization of telomere length maintenance in tankyrase 2-deficient mice. Mol Cell Biol. 2006;26:2037–2043. doi: 10.1128/MCB.26.6.2037-2043.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu-Ping M, Slaughter CA, DeMartino GN. Purification and characterization of a protein inhibitor of the 20S proteasome (macropain) Biochim Biophys Acta. 1992;1119:303–311. doi: 10.1016/0167-4838(92)90218-3. [DOI] [PubMed] [Google Scholar]
- Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, Buecker C, Leggett DS, Schmidt M, King RW, et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127:1401–1413. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
- D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342(Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- Demartino GN, Gillette TG. Proteasomes: machines for all reasons. Cell. 2007;129:659–662. doi: 10.1016/j.cell.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Deveraux Q, Jensen C, Rechsteiner M. Molecular cloning and expression of a 26 S protease subunit enriched in dileucine repeats. J Biol Chem. 1995;270:23726–23729. doi: 10.1074/jbc.270.40.23726. [DOI] [PubMed] [Google Scholar]
- Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi M, Tomko RJ, Jr, Kobayashi H, Hochstrasser M. Multiple assembly chaperones govern biogenesis of the proteasome regulatory particle base. Cell. 2009;137:887–899. doi: 10.1016/j.cell.2009.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne JP, Hendzel MJ, Droit A, Poirier GG. The expanding role of poly(ADP-ribose) metabolism: current challenges and new perspectives. Curr Opin Cell Biol. 2006;18:145–151. doi: 10.1016/j.ceb.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- Gillette TG, Kumar B, Thompson D, Slaughter CA, DeMartino GN. Differential roles of the COOH termini of AAA subunits of PA700 (19 S regulator) in asymmetric assembly and activation of the 26 S proteasome. J Biol Chem. 2008;283:31813–31822. doi: 10.1074/jbc.M805935200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Goldberg AL. On prions, proteasomes, and mad cows. N Engl J Med. 2007;357:1150–1152. doi: 10.1056/NEJMcibr073962. [DOI] [PubMed] [Google Scholar]
- Goldberg AL. Development of proteasome inhibitors as research tools and cancer drugs. J Cell Biol. 2012;199:583–588. doi: 10.1083/jcb.201210077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes AV, Zong C, Edmondson RD, Li X, Stefani E, Zhang J, Jones RC, Thyparambil S, Wang GW, Qiao X, et al. Mapping the murine cardiac 26S proteasome complexes. Circ Res. 2006;99:362–371. doi: 10.1161/01.RES.0000237386.98506.f7. [DOI] [PubMed] [Google Scholar]
- Guettler S, LaRose J, Petsalaki E, Gish G, Scotter A, Pawson T, Rottapel R, Sicheri F. Structural basis and sequence rules for substrate recognition by Tankyrase explain the basis for cherubism disease. Cell. 2011;147:1340–1354. doi: 10.1016/j.cell.2011.10.046. [DOI] [PubMed] [Google Scholar]
- Hanna J, Meides A, Zhang DP, Finley D. A ubiquitin stress response induces altered proteasome composition. Cell. 2007;129:747–759. doi: 10.1016/j.cell.2007.03.042. [DOI] [PubMed] [Google Scholar]
- Hershko A. The ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle. Cell Death Differ. 2005;12:1191–1197. doi: 10.1038/sj.cdd.4401702. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hsiao SJ, Poitras MF, Cook BD, Liu Y, Smith S. Tankyrase 2 poly(ADP-ribose) polymerase domain-deleted mice exhibit growth defects but have normal telomere length and capping. Mol Cell Biol. 2006;26:2044–2054. doi: 10.1128/MCB.26.6.2044-2054.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao SJ, Smith S. Tankyrase function at telomeres, spindle poles, and beyond. Biochimie. 2008;90:83–92. doi: 10.1016/j.biochi.2007.07.012. [DOI] [PubMed] [Google Scholar]
- Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Hamazaki J, Iemura S, Sasaki K, Furuyama K, Natsume T, Tanaka K, Murata S. Assembly pathway of the Mammalian proteasome base subcomplex is mediated by multiple specific chaperones. Cell. 2009;137:914–925. doi: 10.1016/j.cell.2009.05.008. [DOI] [PubMed] [Google Scholar]
- Kisselev AF, van der Linden WA, Overkleeft HS. Proteasome inhibitors: an expanding army attacking a unique target. Chem Biol. 2012;19:99–115. doi: 10.1016/j.chembiol.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurucz E, Ando I, Sumegi M, Holzl H, Kapelari B, Baumeister W, Udvardy A. Assembly of the Drosophila 26 S proteasome is accompanied by extensive subunit rearrangements. Biochem J. 2002;365:527–536. doi: 10.1042/BJ20011520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasker K, Forster F, Bohn S, Walzthoeni T, Villa E, Unverdorben P, Beck F, Aebersold R, Sali A, Baumeister W. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc Natl Acad Sci U S A. 2012;109:1380–1387. doi: 10.1073/pnas.1120559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassot I, Latreille D, Rousset E, Sourisseau M, Linares LK, Chable-Bessia C, Coux O, Benkirane M, Kiernan RE. The proteasome regulates HIV-1 transcription by both proteolytic and nonproteolytic mechanisms. Mol Cell. 2007;25:369–383. doi: 10.1016/j.molcel.2006.12.020. [DOI] [PubMed] [Google Scholar]
- Le Tallec B, Barrault MB, Guerois R, Carre T, Peyroche A. Hsm3/S5b participates in the assembly pathway of the 19S regulatory particle of the proteasome. Mol Cell. 2009;33:389–399. doi: 10.1016/j.molcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- Lee SY, De la Mota-Peynado A, Roelofs J. Loss of Rpt5 protein interactions with the core particle and Nas2 protein causes the formation of faulty proteasomes that are inhibited by Ecm29 protein. J Biol Chem. 2011;286:36641–36651. doi: 10.1074/jbc.M111.280875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levaot N, Voytyuk O, Dimitriou I, Sircoulomb F, Chandrakumar A, Deckert M, Krzyzanowski PM, Scotter A, Gu S, Janmohamed S, et al. Loss of Tankyrase-mediated destruction of 3BP2 is the underlying pathogenic mechanism of cherubism. Cell. 2011;147:1324–1339. doi: 10.1016/j.cell.2011.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T, Satoh MS, Poirier GG, Klungland A. Post-translational modification of poly(ADP-ribose) polymerase induced by DNA strand breaks. Trends Biochem Sci. 1995;20:405–411. doi: 10.1016/s0968-0004(00)89089-1. [DOI] [PubMed] [Google Scholar]
- McCutchen-Maloney SL, Matsuda K, Shimbara N, Binns DD, Tanaka K, Slaughter CA, DeMartino GN. cDNA cloning, expression, and functional characterization of PI31, a proline-rich inhibitor of the proteasome. J Biol Chem. 2000;275:18557–18565. doi: 10.1074/jbc.M001697200. [DOI] [PubMed] [Google Scholar]
- Morrone S, Cheng Z, Moon RT, Cong F, Xu W. Crystal structure of a Tankyrase-Axin complex and its implications for Axin turnover and Tankyrase substrate recruitment. Proc Natl Acad Sci U S A. 2012;109:1500–1505. doi: 10.1073/pnas.1116618109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata S, Yashiroda H, Tanaka K. Molecular mechanisms of proteasome assembly. Nat Rev Mol Cell Biol. 2009;10:104–115. doi: 10.1038/nrm2630. [DOI] [PubMed] [Google Scholar]
- Narwal M, Venkannagari H, Lehtio L. Structural basis of selective inhibition of human tankyrases. J Med Chem. 2012;55:1360–1367. doi: 10.1021/jm201510p. [DOI] [PubMed] [Google Scholar]
- Nickell S, Mihalache O, Beck F, Hegerl R, Korinek A, Baumeister W. Structural analysis of the 26S proteasome by cryoelectron tomography. Biochem Biophys Res Commun. 2007;353:115–120. doi: 10.1016/j.bbrc.2006.11.141. [DOI] [PubMed] [Google Scholar]
- Park S, Roelofs J, Kim W, Robert J, Schmidt M, Gygi SP, Finley D. Hexameric assembly of the proteasomal ATPases is templated through their C termini. Nature. 2009;459:866–870. doi: 10.1038/nature08065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y, Hwang YP, Lee JS, Seo SH, Yoon SK, Yoon JB. Proteasomal ATPase-associated factor 1 negatively regulates proteasome activity by interacting with proteasomal ATPases. Mol Cell Biol. 2005;25:3842–3853. doi: 10.1128/MCB.25.9.3842-3853.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC, Jr, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- Peth A, Besche HC, Goldberg AL. Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening. Mol Cell. 2009;36:794–804. doi: 10.1016/j.molcel.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peth A, Kukushkin N, Bosse M, Goldberg AL. Ubiquitinated proteins activate the proteasomal ATPases by binding to Usp14 or Uch37. J Biol Chem. 2013 doi: 10.1074/jbc.M112.441907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peth A, Uchiki T, Goldberg AL. ATP-dependent steps in the binding of ubiquitin conjugates to the 26S proteasome that commit to degradation. Mol Cell. 2010;40:671–681. doi: 10.1016/j.molcel.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Princiotta MF, Schubert U, Chen W, Bennink JR, Myung J, Crews CM, Yewdell JW. Cells adapted to the proteasome inhibitor 4-hydroxy- 5-iodo-3-nitrophenylacetyl-Leu-Leu-leucinal-vinyl sulfone require enzymatically active proteasomes for continued survival. Proc Natl Acad Sci U S A. 2001;98:513–518. doi: 10.1073/pnas.021132398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab MS, Podar K, Breitkreutz I, Richardson PG, Anderson KC. Multiple myeloma. Lancet. 2009;374:324–339. doi: 10.1016/S0140-6736(09)60221-X. [DOI] [PubMed] [Google Scholar]
- Rabl J, Smith DM, Yu Y, Chang SC, Goldberg AL, Cheng Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol Cell. 2008;30:360–368. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelofs J, Park S, Haas W, Tian G, McAllister FE, Huo Y, Lee BH, Zhang F, Shi Y, Gygi SP, Finley D. Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature. 2009;459:861–865. doi: 10.1038/nature08063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki Y, Toh EA, Kudo T, Kawamura H, Tanaka K. Multiple proteasome-interacting proteins assist the assembly of the yeast 19S regulatory particle. Cell. 2009;137:900–913. doi: 10.1016/j.cell.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Shim SM, Lee WJ, Kim Y, Chang JW, Song S, Jung YK. Role of S5b/PSMD5 in proteasome inhibition caused by TNF-alpha/NFkappaB in higher eukaryotes. Cell Rep. 2012;2:603–615. doi: 10.1016/j.celrep.2012.07.013. [DOI] [PubMed] [Google Scholar]
- Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Giriat I, Schmitt A, de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998;282:1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- Tai HC, Besche H, Goldberg AL, Schuman EM. Characterization of the Brain 26S Proteasome and its Interacting Proteins. Front Mol Neurosci. 2010:3. doi: 10.3389/fnmol.2010.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi K, Kim S, Yukii H, Ueno M, Morishita R, Endo Y, Kato K, Tanaka K, Saeki Y, Mizushima T. Structural basis for specific recognition of Rpt1p, an ATPase subunit of 26 S proteasome, by proteasome-dedicated chaperone Hsm3p. J Biol Chem. 2012;287:12172–12182. doi: 10.1074/jbc.M112.345876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanahashi N, Kawahara H, Murakami Y, Tanaka K. The proteasome-dependent proteolytic system. Mol Biol Rep. 1999;26:3–9. doi: 10.1023/a:1006909522731. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Mizushima T, Saeki Y. The proteasome: molecular machinery and pathophysiological roles. Biol Chem. 2012;393:217–234. doi: 10.1515/hsz-2011-0285. [DOI] [PubMed] [Google Scholar]
- Tomko RJ, Jr, Hochstrasser M. Order of the proteasomal ATPases and eukaryotic proteasome assembly. Cell Biochem Biophys. 2011;60:13–20. doi: 10.1007/s12013-011-9178-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. The ubiquitin system, an immense realm. Annu Rev Biochem. 2012;81:167–176. doi: 10.1146/annurev-biochem-051910-094049. [DOI] [PubMed] [Google Scholar]
- Wang Y, Dawson VL, Dawson TM. Poly(ADP-ribose) signals to mitochondrial AIF: a key event in parthanatos. Exp Neurol. 2009;218:193–202. doi: 10.1016/j.expneurol.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe TK, Saito A, Suzuki M, Fujiwara T, Takahashi E, Slaughter CA, DeMartino GN, Hendil KB, Chung CH, Tanahashi N, Tanaka K. cDNA cloning and characterization of a human proteasomal modulator subunit, p27 (PSMD9) Genomics. 1998;50:241–250. doi: 10.1006/geno.1998.5301. [DOI] [PubMed] [Google Scholar]
- Yeh TY, Beiswenger KK, Li P, Bolin KE, Lee RM, Tsao TS, Murphy AN, Hevener AL, Chi NW. Hypermetabolism, hyperphagia, and reduced adiposity in tankyrase-deficient mice. Diabetes. 2009;58:2476–2485. doi: 10.2337/db08-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaiss DM, Standera S, Holzhutter H, Kloetzel P, Sijts AJ. The proteasome inhibitor PI31 competes with PA28 for binding to 20S proteasomes. FEBS Lett. 1999;457:333–338. doi: 10.1016/s0014-5793(99)01072-8. [DOI] [PubMed] [Google Scholar]
- Zaiss DM, Standera S, Kloetzel PM, Sijts AJ. PI31 is a modulator of proteasome formation and antigen processing. Proc Natl Acad Sci U S A. 2002;99:14344–14349. doi: 10.1073/pnas.212257299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.