Abstract
Unlike many localized infections, the development and resolution of bacteremia involves physical and immunological interactions between many anatomic sites. In an effort to better understand these interactions, we developed a computational model of bacteremia as a dynamical system fashioned after multicompartmental pharmacodynamic models, incorporating bacterial proliferation and clearance in the blood, liver, spleen, and lungs, and the transport of pathogens between these sites. A system of four first-order homogeneous ODEs was developed. Blood and organ bacterial burdens were measured at various time points from 3 to 48 h postinoculation using an LD25 murine model of Staphylococcus epidermidis bacteremia. Using these empiric data, solutions to the mathematical model were recovered. A bootstrap resampling method was used to generate 95% confidence intervals around the solved parameters. The validity of the model was examined in parallel experiments using animals acutely immunocompromised with cyclophosphamide; the model captured abnormalities in bacterial partitioning previously described with this antineoplastic agent. Lastly, the approach was used to explore possible benefits to clinically observed hyperdynamic blood flow during sepsis: in simulation, normal mice, but not those treated with cyclophosphamide, enjoyed significantly more rapid bacterial clearance from the bloodstream under hyperdynamic conditions.
Keywords: Bacteremia, immunocompromise, mathematical models, statistical methods
INTRODUCTION
Staphylococcus epidermidis, a member of the coagulase-negative staphylococci, is among the most frequently identified causes of bloodstream infections in the United States, a problem related to increasing use of short- and long-term implantable devices (1, 2). In recent clinical series, this group of organisms causes between 11% and 33% of nosocomial bloodstream infections, with an incidence of 15.8 per 10,000 hospital admissions (3, 4), and is a disproportionately frequent cause of bloodstream infection in intensive care units (5). Perhaps the strongest risk factor for developing a bloodstream infection is neutropenia. Short-term crude mortality after a neutropenic bloodstream infection exceeds 30%, and attributable mortality has been estimated to be approximately 12% in patients with hematological malignancy (6, 7).
Surviving a bloodstream infection requires effective filtration of bacteria from the blood and elimination of pathogens from organs that have been seeded during episodes of bacteremia. Host defense in this context seems similar to the pharmacokinetic clearance of drugs, a phenomenon that has been successfully conceptualized as a series of compartments into which agents may enter, between which agents may be transferred, and within which they may be degraded. However, the analogy is not perfect; unlike drugs, bacterial burden can increase in either the early hours of infection when host defenses are coming online or in the event of unsuccessful host defense. Numerically analyzing complex experimental data taken from multiple compartments at multiple time points in a manner that imposes physiological and immunological relationships is a significant departure from standard statistical techniques, where the most common analyses conclude at the rejection of a null hypothesis.
Considering bacterial clearance from the bloodstream in a physiologically based pharmacokinetic (PBPK) modeling paradigm was first described in 1983 (8). In that report, a limited 2-compartment model was fit to measures of blood and mesenteric lymph node bacterial content in mice over time. The authors assumed that bacteria did not proliferate during the experiment and computationally had the capacity to fit only the mean of the measured data from multiple animals. Accordingly, the conclusions of the work were limited to the observation that bacteria are cleared from the bloodstream in a manner approximated more closely by functions of a higher order than linear or single-exponential forms.
Improved methods of quantitatively thinking about host-pathogen interactions during life-threatening bloodstream infection could provide a useful framework for both experimental and analytical work in this area. Therefore, in the current work, we substantially expanded the concept of bacteremia as a PBPK phenomenon. A four-compartment mathematical model that related bacterial growth, clearance, and intercompartmental transfer between the blood, lung, liver, and spleen was used to characterize a murine LD25 S. epidermidis bacteremia model. This model was used to study two questions. First, what are the effects of acute immunocompromise induced by the antineoplastic agent cyclophosphamide as they relate to the resolution of an episode of bacteremia? Second, how does hyperdynamic blood flow affect the conclusions drawn from this computational model? Although more technically demanding than traditional statistical methods, analyzing experimental results from an animal model of multiorgan infection with a PBPK mathematical approach provides an opportunity to pursue novel questions the answers to which would otherwise be inaccessible.
MATERIALS AND METHODS
Organisms and culture conditions
Staphylococcus epidermidis RP62A, a strain originally isolated from an infected intravascular catheter, was obtained from American Type Culture Collection (ATCC 35984) (9, 10). Klebsiella pneumoniae Xen 39 is an engineered strain with a chromosomally inserted constitutively expressed lux operon and therefore can be visualized in vivo using high-sensitivity cameras (Xenogen, Inc, Hopkinton, Mass). Bacteria were grown from cryopreserved stock in tryptic soy broth or on solid (TSA) media (MP Biomedicals, Solon, Ohio) and incubated at 37°C. To obtain midlog phase cells, overnight cultures from single colonies were subcultured in fresh tryptic soy broth by 1:100 dilution and grown for 2 to 3 h until optical density (OD600nm) readings reached 0.4. Cultures were washed, resuspended in sterile 0.9% NaCl, and quantified turbidometrically (11). Six- to 10-week-old outbred male ICR mice (Harlan Sprague-Dawley, Indianapolis, Ind) were used in all experiments. Animal protocols were approved by the University of Michigan’s Animal Use Committee. The experiments were perfomed in adherence to the National Institutes of Health Guidelines on the Use of Laboratory Animals.
Murine bacteremia model
Mice were anesthetized using 2.5% inhaled isoflurane (Vedco, Inc, St Joseph, Mo) and injected into the deep dorsal vein of the penis with 200 μL of the bacterial suspension containing 109 colony-forming units (CFUs) of S. epidermidis. The magnitudes of bacterial inocula were confirmed by plating serial dilutions. In preliminary experiments, this dose was determined to produce a 7-day LD25 infection. At specified time points, animals were euthanized by exsanguination and cervical dislocation under deep isoflurane anesthesia. The entire liver, spleen, and lungs, and 1 mL of blood were harvested in a sterile fashion. Portions of each organ were weighed in tared tubes, homogenized in 3 mL of 0.9% NaCl, and then quantitatively cultured on TSA. For survival analysis, a separate cohort of animals was injected with the same dose of bacteria and monitored daily for 9 days.
Leukopenia was induced in some animals via intraperitoneal injection of cyclophosphamide (200 mg/kg; Fluka Biochemika 29875, Switzerland) 72 h before the study (12). Neutropenia was confirmed by automated complete blood cell count and by peripheral blood smear analysis. Neutropenia was maintained by giving a second injection of cyclophosphamide at the time of bacterial inoculation.
Acute bacterial uptake studies
For analysis of very early postinoculation uptake, we used luminescent bacteria injected i.v. into isoflurane-anesthetized mice positioned in a dedicated imaging system (IVIS Imaging System, Xenogen/Caliper Life Sciences, Hopkinton, Mass). Image series were recorded using default instrument settings and 30-s exposures. Five animals were studied under identical experimental conditions. Unfortunately, luminescent S. epidermidis was not available at the time of this writing. Therefore, we alternatively chose K. pneumoniae Xen 39.
Bacterial proliferation in blood ex vivo
To establish the upper bound for the bacterial growth rate in whole blood, midlog phase bacteria were diluted to various concentrations, added to 200 μL whole mouse blood (1:10 by volume), and incubated at 37°C. At specified time points, 20-μL samples were taken from each tube and used for quantitative cultures on TSA. Growth constants (as h−1) were determined assuming first-order growth kinetics.
PBPK mathematical model
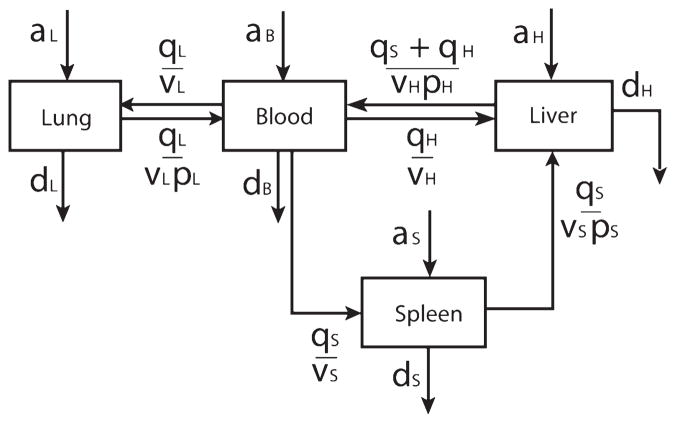
Bacteria within infected mice were assumed to exist as a well-mixed suspension of CFUs with diameter = 0 (i.e., no explicit relationship between bacterial and capillary dimensions was included) and to reside only within four compartments: the blood, liver, spleen, and lung (Fig. 1). Within each compartment, bacteria proliferated at rate a (a ≥ 0) and were eliminated at rate d (d ≥ 0), both as per hour (h−1), in proportion to the number of bacteria in the compartment. As there was no experimental means to distinguish a from d in vivo, the two terms were treated as a net bacterial growth rate (a–d) in the model. The upper bounds of a, and thus a–d, was established experimentally to be 0.92 h−1 in the whole blood bacterial growth assay described previously. Transfer between blood and other compartments was described as a function of the rate of blood flow, q, to each compartment, the volume, v, of each compartment, and a partitioning coefficient, p, where p is greater than 1 when bacteria were being filtered from the blood, and 0 is less than p is less than 1 when bacteria were being shed into the bloodstream from the compartment. Like proliferation or elimination, partitioning from one compartment into another was proportional to the number of bacteria present in the donor compartment.
Fig. 1. Schematic outline of the mathematical model.
Bacteria were assumed to exist in one of four compartments. Within each compartment, bacteria could proliferate (at rate a, in h−1), be eliminated (at rate d, in h−1), or transfer to another compartment. Transfer was a function of blood flow per compartment volume (q/v, in mL/h) and a compartment-specific partitioning coefficient (p, dimensionless). Portal circulation through the spleen into the liver was included.
These features were assembled into a system of four autonomous ordinary differential equations (ODEs) describing the instantaneous rate of change of bacterial burden in each compartment,
| (1) |
| (2) |
| (3) |
| (4) |
where l, s, h, and b are the bacterial concentrations, as CFU/mL of tissue, in the lung, spleen, liver (h, hepatic), and blood, respectively. Whereas circulating blood could transfer bacteria into the spleen, the spleen transferred bacteria only into the liver via the portal circulation. Hepatic circulation was divided into two components: splenic portal blood flow and the combination of hepatic arterial flow and all nonsplenic portal flow. Values for organ volume, v, were taken from organ mass measurements in the current experiments with an assumption that tissue density in each case was 1 g/mL. Values for organ blood flow, q, were linearly scaled by body mass from previously published microsphere-based flow measurements in rats with bacterial peritonitis (13). Modeling was performed using both normodynamic and hyperdynamic blood flow assumptions (Table 1).
Table 1.
Parameter definitions and values
| Parameter | Normodynamic | Hyperdynamic |
|---|---|---|
| Blood flow, mL/min per g | ||
| Liver* | 1.5 | 2.4 |
| Spleen | 1.2 | 1.9 |
| Lung† | 6.84 | 10.05 |
| Organ mass or volume | ||
| Liver, g | 1.99 | — |
| Spleen, g | 0.15 | — |
| Lung, g | 2.10 | — |
| Blood, mL | 1.8 | — |
Total hepatic blood flow, including arterial and portal venous.
Taken as cardiac output.
Model fitting and bootstrap resampling for parameter estimation
Software for model fitting and all analysis was written in Matlab R2006b (The Mathworks, Inc, Natick, Mass). To generate median and 95% confidence interval (CI) estimates of each fitted parameter, a bootstrapping method was used as has been discussed elsewhere (14). For each bootstrap replicate, the liver, spleen, lung, and blood bacterial content of one mouse was selected randomly, with replacement, from each time point to generate a time series of multicompartment bacterial burden. For this time course of measurements, the model equations were solved using the appropriate ODE solver in Matlab. The parameters were estimated by fitting the solution of the differential equations to the bootstrap data using least squares optimization. In this way, each bootstrap replicate produced one estimate of all net bacterial growth rates and intercompartmental partitioning coefficients. One thousand replicates of this procedure were performed. An identical process was undertaken for neutropenic animals. Importantly, the model’s initial conditions were taken to be the first postinoculation time point (i.e., 3 h postinfection). This decision was made to avoid early intercompartmental redistribution that presumably occurred at rates too fast to be reliably measured by our intermittent culture technique.
Stability analysis
To determine whether parameter estimates produced a stable solution (i.e., one which would ultimately lead to a fixed point, in this case, zero bacteria in all compartments) or an unstable one (i.e., in which the bacterial content increased in a nonsurvivable fashion), stability analysis was conducted using the Routh-Hurwitz criteria as has been described previously (15). Details of this procedure are presented in the appendix.
RESULTS
Bacterial clearance dynamics in normal mice
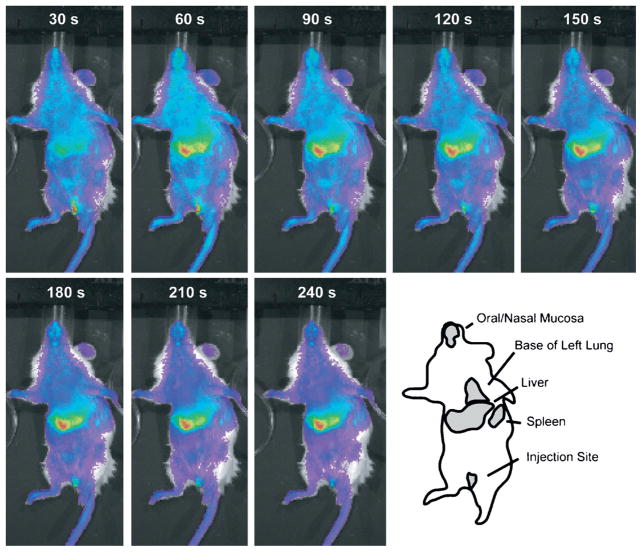
To establish the very early dynamics of bacterial distribution and clearance in this model, we used a luminescent strain of K. pneumoniae to conduct real-time imaging studies. Strain differences notwithstanding, sequential whole-body imaging revealed that within 30 s, bacteria began concentrating in the region of the liver. Within 4 min, the lung, liver, and spleen all demonstrated intense uptake of blood-borne bacteria (Fig. 2). Based on these data, the blood and these three organs were the targets of our subsequent modeling. In addition, given the very rapid distribution of bacteria in the opening minutes of a bacteremia episode, we concentrated our experimental measures and modeling on the slower process of bacterial removal.
Fig. 2. Initial time course of bacterial distribution into anatomic compartments.
Luminescent bacteria were administered by penile vein and imaged in 30-s intervals for the first 5 min of bacteremia. Almost immediately, partitioning into the liver was detected, and a generally stable steady state was achieved in each case within 3 to 4 min of administration. The rapidity of initial dynamics was such that attention was focused on subsequent events related to further clearance and eradication of bacterial burden. Image series is representative of five animals studied.
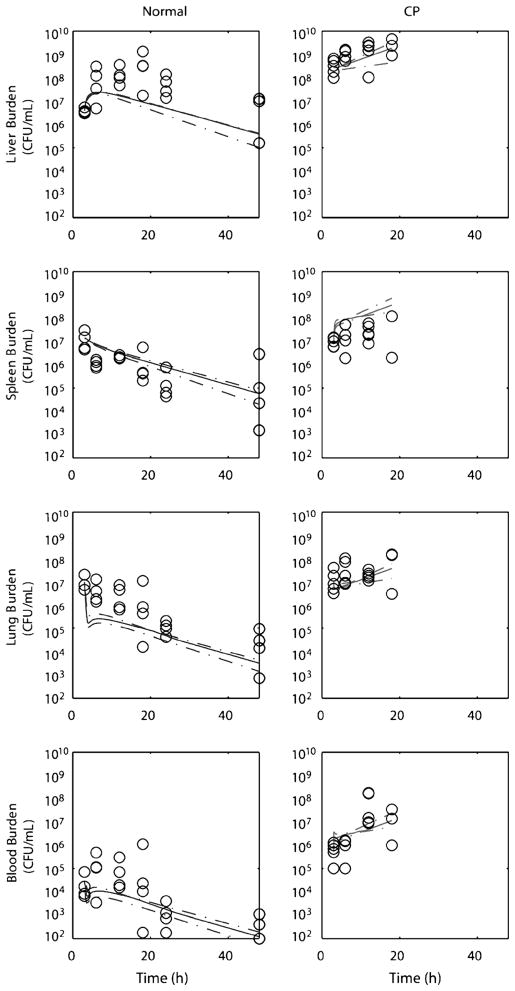
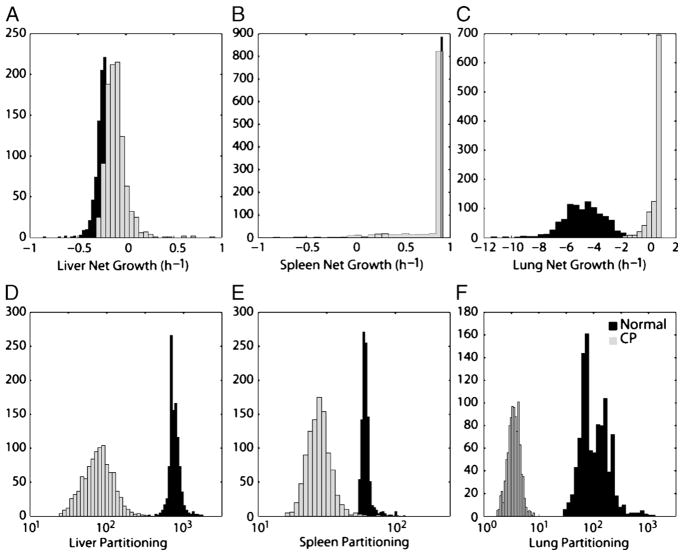
When later time points (3–48 h) were studied by quantitative culture rather than bacterial luminescence, pathogen burden was found to decrease over time in all compartments, although at different rates (Fig. 3). In particular, liver bacterial burden increased over the first 24 h, whereas it decreased in other compartments. Clearance dynamics were captured by estimated growth rates and intercompartmental partitioning coefficients; bootstrap estimates of these parameter values and their 95% CIs are shown in Figure 4 and in Table 2. Of note, the model suggests that the lung, and not the liver, had the highest bacterial killing rate. A bacterial growth rate of −4.70 h−1 for the lung indicates that 90% of the accumulated burden would be expected to be cleared every 30 min in the lung, whereas the liver rate of −0.24 h−1 suggests that 90% of accumulated bacteria would be cleared in roughly 9 h. However, the bacterial partitioning in the liver was significantly more efficient (×8-fold), supporting the claim that despite differences in organ perfusion, the liver is the main site of bacterial clearance during a bacteremic episode, a conclusion in line with extensive previous experimental observations (16–18).
Fig. 3. Organ and blood culture time course during bacteremia.
Normal or CP-treated mice inoculated with 109 CFU S. epidermidis underwent quantitative culture of liver, spleen, lung, and blood at various times. Experimental data are shown as open circles. Median and 95% CI values for predictions based on the mathematical model are shown as solid and dashed lines, respectively. CP–cyclophosphamide.
Fig. 4. Bootstrap distributions of the six fitted model parameters for normal (black bars) and CP-treated mice (gray bars).
For each group of animals, bootstrap resampling was used to generate 1,000 solutions (i.e., 1,000 sets of estimates for each of the six parameters shown). The net bacterial growth rate (equal to the rate of bacterial proliferation minus the rate of bacterial elimination) for the liver, spleen, and lung are shown in A to C. In each instance, the upper limit for net growth rate was set at 0.92 h−1, the rate at which S. epidermidis was found to proliferate in mouse blood ex vivo. Only in the lung were net growth rates found to be significantly impaired by treatment with CP (P < 0.01). In D to F, the partitioning coefficients between blood and the three organ compartments are shown. In each case, CP treatment significantly reduced apparent filtration of bacteria from the bloodstream (P < 0.01 in each case).
Table 2.
Fitted model at 18 h
| Parameter | Control (median, 95% CI) | Cyclophosphamide-treated |
|---|---|---|
| Net growth rate* | ||
| Liver (ah–dh), h−1 | −0.24 (−0.42 to −0.13) | −0.12 (−0.26 to 0.13) |
| Spleen (as–ds), h− 1 | 0.92 (0.68 to 0.92) | 0.92 (0.04 to 0.92)† |
| Lung (al –dl), h−1 | −4.70 (−8.02 to −2.01) | 0.78 (−1.07 to 0.92) |
| Partitioning coefficient | ||
| Liver (ph) | 746 (575 to 1153) | 79 (33 to 181) |
| Spleen (ps) | 59 (54 to 78) | 28 (19 to 43) |
| Lung (pl) | 93 (43 to 355) | 3 (2 to 6) |
Convention in compartmental modeling holds that all rates be greater than or equal to 0, therefore net growth rate is listed here as the difference between intracompartmental growth a and killing d.
0.92 h−1 is the fastest growth achievable by S. epidermidis in normal mouse blood in vitro in our laboratory and is considered the theoretical maximum for both a and (a–d) in the in vivo model.
Impact of cyclophosphamide
As expected, cyclophosphamide administration 72 h in advance of inoculation produced significant reductions in circulating leukocytes, particularly neutrophils (Figs. 5, A–C). Seventy percent (14/20) of normal mice survived 7 days after bacterial inoculation, whereas no bacteremic animals pre-treated with cyclophosphamide survived 24 h (0/22, Fig. 5D). Changes in compartmental bacterial burden and model parameter estimates for immunocompromised animals are shown alongside their normal counterparts in Figures 3 and 4 and in Table 2. Interestingly, the most prominent differences between normal and cyclophosphamide-treated animals were among intercompartmental partitioning coefficients, rather than bacterial clearance rates. This finding supports previous reports that neutrophils participate in bacterial clearance from the bloodstream by more than bactericide alone, and that the immune lesion produced by cyclophosphamide extends to the Kupffer cells and vascular endothelium (16, 18, 19).
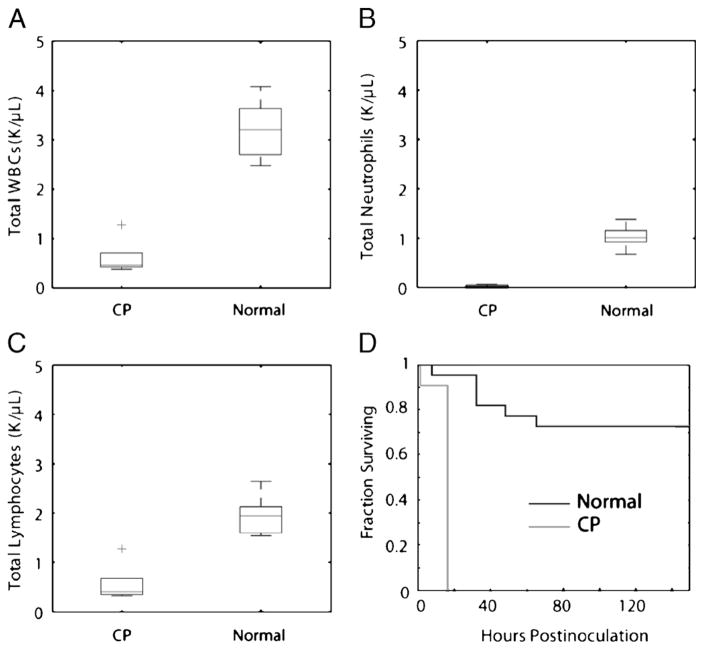
Fig. 5. Effect of CP treatment on S. epidermidis bloodstream infection.
Mice received either saline (normal) or 200 mg/kg CP intraperitoneally 72 h before measurement. As expected, treatment resulted in a significant decrease in total circulating leukocytes (A), a profound decrease in neutrophils (B), and a lesser decrease in circulating lymphocytes (C), n = 5 per group. Box plots represent the values for the five animals in each group. In D, 1-week survival curves of normal and CP-treated animals subjected to an i.v. inoculation of 109 CFU S. epidermidis are shown, P < 0.001, n = 20 per treatment group. WBCs–white blood cells.
Stability analysis of model results
An important feature of systems of ODEs is stability, the existence of a range of model parameters that, over time, will drive the state of the system toward a fixed point. In the current context, stable solutions were those that would ultimately result in complete eradication of bacterial burden and thus were a prerequisite for host survival. Conversely, unstable solutions were those that would over time produce bacterial burdens approaching infinity and necessarily would be fatal to the host. In the current model, the boundaries between stable and unstable solutions were a family of five membranes that existed in the 6-dimensional space whose axes were the three net growth rates (ah − dh, as − ds, and al − dl ) and three partition coefficients (ph, ps, and pl). As described in the appendix, the system of equations can be rewritten in matrix-vector form, with parameter coefficient matrix M. Based on our analysis, one of the membranes, along which the determinant of the matrix M was equal to zero (det[M] = 0), was found to separate the solutions of normal and cyclophosphamide-treated mice, and therefore defined the cyclophosphamide-treated parameters as necessarily unstable and not survivable. In Figure 6, 2-dimensional sections of the 6-dimensional parameter space are shown with density contour plots of the 1,000 numerical solutions from normal and cyclophosphamide-treated mice and the calculated boundary between stable and unstable solutions. The solutions for the two groups lay on opposite sides of the stability boundary det(M) = 0, indicating that the solutions lead in the case of the normal animals to complete elimination and, in the case of chemotherapy-treated animals, to uncontrolled bacterial proliferation.
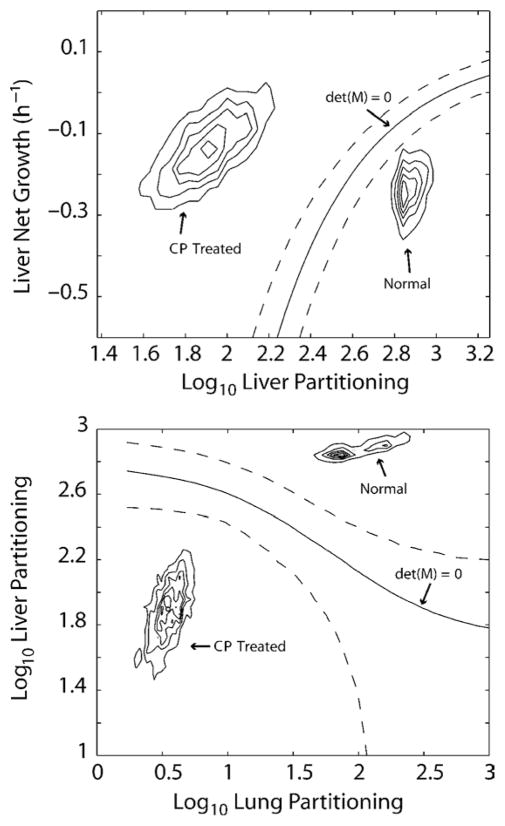
Fig. 6. Stability analysis of the model.
Panels represent sections through parameter space, plots of one estimated parameter in the model versus another estimated parameter and should not be confused with plots of measured data. Shown are density contours of the estimated parameter values for normal and CP-treated mice for liver partitioning (ph) and liver net bacterial growth rate (ah–dh) in the upper panel and lung partitioning (pl) and liver partitioning (ph) in the lower panel. The solid curves labeled det(M) = 0 are locations of the predominant stability boundaries of the model (the determinant of the coefficient matrix M), and the dotted lines are the estimated 95% CIs for those boundaries. In both plots, the parameter estimates for normal animals fall on the side of the boundary in which bacterial elimination from all compartments will occur. Cyclophosphamide-treated mice fall on the opposite side of the boundary, where bacterial burdens over time would increase exponentially until the death of the animal. A discussion of the location of this boundary is provided in the appendix.
Role of hyperdynamic blood flow
In Figure 7, the median parameters for normal and cyclophosphamide-treated mice were applied with either normodynamic or hyperdynamic blood flow values q (Table 1). For both experimental conditions, the model was provided with mean compartmental bacterial burdens measured at 3 h as its initial conditions. The bacterial burden over the ensuing 15 h (18 h postinoculation) was then forecast. Hyperdynamic blood flow offered a significant advantage to normal mice, accelerating the elimination of bacteria from the bloodstream. However, given the impaired partitioning described by the mathematical model, there was little quantitative difference when these blood flow values were applied to the cyclophosphamide-treated group.
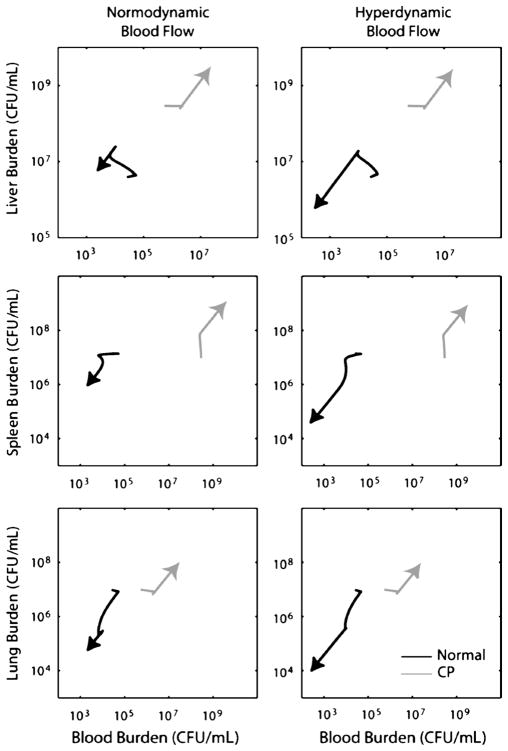
Fig. 7. Trajectories of normal and CP-treated mice from h 3 to h 18 postinoculation.
The median numbers of bacteria in each compartment observed experimentally at 3 h postinoculation were used as the initial condition for these simulations, and trajectories were generated using the median parameter estimates shown in Table 2. Plots are of the expected burden over time in the bloodstream (the horizontal axis in each panel) versus the burden in one of the three compartments. Hyperdynamic blood flow (values for which are shown in Table 1) significantly increased the rate of bacterial clearance in normal mice, but provided no meaningful benefit in CP-treated animals.
DISCUSSION
In the current work, a physiologically based pharmacokinetic model was used to characterize S. epidermidis bacteremia in mice. The fitted models agreed well with experimental data, and the use of bootstrap resampling of empiric measurements allowed statistical comparisons to be made between normal animals and those with a clinically important test condition, cyclophosphamide-induced leukopenia. Formal stability analysis of the differential equations used to describe host-pathogen interactions demonstrated that model parameters in cyclophosphamide-treated mice were not only quantitatively different from those of normal mice, but lay on the far side of an analytical boundary beyond which bacterial proliferation would progress unchecked. Surprisingly, the parameters most altered by cyclophosphamide were not the rates of bacterial clearing, but of blood:organ partitioning, indicating defects in the shuttling of bacteria out of the bloodstream and not just in bacterial killing.
That bacterial disappearance from the bloodstream follows a pattern exponential decay has been recognized since the 1950s after classic studies using radiolabeled heat-killed Escherichia coli and Staphylococcus aureus (20). In those studies, serial measures of blood radioactivity were tracked and found to follow first-order kinetics. Because of the nature of those experiments, no multicompartmental modeling was possible. Mathematical limitations also existed at the time-the first description of a 2-compartment model for any pharmacological application (intercompartmental movement of inulin) had only been reported in 1949, and until the 1960s, pharmacokinetics was largely a nascent field (21, 22). In 1983, Cheewatrakoolpong and colleagues (8) used a 2-compartment model to explain the disappearance of K. pneumoniae from the bloodstream and from mesenteric lymph nodes in mice. They did not treat these anatomic sites as two interrelated compartments, but modeled bacterial counts from each site independently using equations of the form:
| (5) |
where B1 (analogous to our a – d) was the rate constant for the measured compartment (the blood or lymph nodes), and B2 was the rate constant for a hidden unmeasured second compartment. This approach allowed better fitting of experimental data than did a single exponential form but could not provide mechanistic insight into the underlying dynamics. In addition, because of the destructive nature of sampling (an issue in the current work as well), those investigators chose to fit only the mean values of replicate mice killed at each time point. As a result, their numerical approach provided estimates of A and B in the previous equation but no variance around these estimates.
The modeling strategy in the current work sought to overcome these shortcomings by requiring that all numerical solutions to fit simultaneous measurements of bacterial burden in each of the modeled compartments and by generating a large number of bacterial time courses using a resampling method such that parameter estimates, their variances, and joint probabilities could all be generated. The result is a model that can shed light on the process of bacterial clearance during an episode of bacteremia.
An important test of validity is the ability of a mathematical model to demonstrate behavior for which it has not explicitly been designed. In the case of this multicompartmental model, solutions obtained for cyclophosphamide-treated mice support the validity of our approach; pretreatment with this agent seemed to affect the shuttling of bacteria out of the bloodstream, not just to reduce the rate at which bacteria were killed. This was most notable in the liver and lung, where the difference was approximately an order of magnitude. Our experimental data, and our necessarily simplified model of intercompartmental transfer, do not reveal details about the blood-organ interface. However, in the liver, cooperativity between resident Kupffer cells and recruited circulating neutrophils against bacteria in the portal circulation is well described (23). Failure to compartmentalize circulating bacteria because of a lack of available neutrophils for this partnership is consistent with our results. To our knowledge, a similar situation in the lung intravascular compartment, perhaps between marginated monocytes and neutrophils taking place on an ad hoc basis, has not been described. Particularly intriguing are recent descriptions of neutrophil extracellular traps, tangles of neutrophil DNA in which bacteria may become embedded. Recent evidence indicates that after LPS stimulation, neutrophils that marginate in the lung and liver may deploy extracellular DNA as a means of increasing the efficiency of filtration of blood-borne bacteria (24). Our model predictions seem consistent with this experimental observation.
Another benefit of the current model is its ability to draw a clear distinction between normal and cyclophosphamide-treated mice, in an analytical, rather than purely statistical, way. We were able to take advantage of stability analysis to dichotomize model behavior into two outcomes after acute infection: progressive elimination, where total bacterial burden over time approaches zero, and progressive overwhelming proliferation, where the burden approaches infinity. These two outcomes are associated with two distinct sets of model parameters that lay on opposite sides of a multidimensional boundary defined by the Routh-Hurwitz criterion for system stability. The existence of distinctly survivable and lethal trajectories in a model system allows one to pose experimentally testable questions such as what therapies might shift a host’s model parameters from the lethal side of this boundary to the surviving side.
Lastly, by including blood flow in our model, we were able to consider the contribution of hyperdynamic blood flow during severe infection. Hyperdynamic sepsis, in which cardiac output and regional blood flow are increased over uninfected levels, is a common clinical occurrence. Using scaled blood flow values from rats with bacterial peritonitis, we explored the impact of increased organ blood flow on the multicompartmental behavior of bacteria during systemic infection. As might be predicted, elevated blood flow rates accelerated the disappearance of bacteria in normal animals. Because partitioning parameters in cyclophosphamide-treated mice were so much lower than in normal mice, increased blood flow in this context did not significantly benefit the host.
All models of illness, whether mathematical or experimental, fall short of fully reproducing clinical human experience, and our model of disseminated bacterial infection has several noteworthy limitations. The first is the nature and intensity of our bacterial challenge. We chose an i.v. inoculation system for this work because we wanted to capture the behavior of a host exposed to a single episode of bacterial showering into the bloodstream. This model is admittedly artificial but allowed us to develop a mathematical approach that did not require the inclusion of additional “black box” terms that could account for ongoing bacteria seeding that could not be measured experimentally.
A second limitation is that the mathematical model is autonomous or time invariant. It does not have a means of capturing host or pathogen behaviors that change over time; a bacterium entering the system 12 h after infection experiences the same host as one present in the initial inoculum. In reality, infected hosts recruit humoral and cellular defenses that require time to come on line, and hemodynamic changes, beneficial or harmful, also occur with some delay. We have shown previously in a murine model of pneumonia that the rate of bacterial elimination within the lung increases in the opening hours of infection as neutrophils are recruited into the airspaces (25). A reasonable next step in developing the theoretical model will be to incorporate a reliably measurable phenomenon, such as organ neutrophil content, that presumably will track temporally changing bactericidal or partitioning events.
Another unavoidable limitation of the current strategy is the destructive nature of measuring bacterial burden in four compartments. Linear dynamical modeling attempts to estimate a trajectory of future behavior based on the current state of a system. In our experimental system, organ and blood cultures were obtained by killing of the experimental animal, and as such, it was not possible for any mouse to contribute data at two time points. It was therefore impossible to know exactly an individual’s trajectory. We chose to overcome this problem by generating in essence a simulated repeated measures design by bootstrap resampling the available experimental data. This strategy provided the additional benefit of producing estimates of parameter variance, a crucial issue for future work that aims to test more complicated hypotheses or to make sample size calculations for work in involving advanced mathematical techniques. The validity of this bootstrapping is well documented, but the larger statistical implications of this choice of numerical method for data sets such as the one used here deserve, in our view, considerable additional study and development (14).
Lastly, we would point out that the modeling in this article, both experimental and computational, seeks to address only one aspect of clinical bacteremia. In humans, the downstream inflammatory response to a bacterial challenge is frequently more devastating than the bacterial exposure itself. Considerable work is underway to better characterize all aspects of this response in an analytical way (26–28). The current work is one additional step toward that goal.
Acknowledgments
The authors thank Adam P. Thompson for his assistance in the laboratory and the staff of the Center for Advanced Computing at the University of Michigan. The code used to generate Figure 6 was modified from a routine written by Peter Perkins, Mathworks, Inc, and is available at the Mathworks File Exchange, www.mathworks.com/matlabcentral/fileexchange. This work was supported by National Institute of General Medical Sciences grant R01 GM069438 and an associated supplement for computational biology. Additional funds were provided by a pilot grant from the University of Michigan Center for Computational Medicine and Biology. Ms. Chung was sponsored by a Society for Academic Emergency Medicine/Emergency Medicine Foundation student grant.
GLOSSARY
- Dynamical system
A mathematical formulation that describes the evolution of the state of a system over time
- Ordinary differential equation
- An equation that relates the state of a system to its rate of change. For example, an ODE is frequently used to relate the rate of growth of bacteria with the number of bacteria. It is important to realize that there is a distinction between an ODE, with which one studies rates of change, and an ODE’s solution, with which one can predict the later state of a system given an initial condition. In the simplest model of bacterial growth, one might write:
where B is the number of bacteria, dB/dt is the growth rate, and k is a growth constant(10) - Solution
- Whereas in algebra, the solution to an equation is typically a number, the solution to a differential equation is a function. The solution to an ODE allows one to comment on the state of a system at a moment in time rather than its rate of change at that time. In the previous example, if one wanted to know the number of bacteria present 1 h after the initiation of growth, one could use the solution to ODE (1),
where B(t) is the number of bacteria present at time = t, B0 is the number present at time = 0 (sometimes called the initial condition), and t = 1 h, and thus B(1) = B0ek. Although in this simple example, an exact solution to ODE (1) is known, frequently, no explicit solution can be written down for complicated ODEs. In such instances, the solution can only be estimated using a computer(11) - Stability
It is often important to understand if the solution to an ODE will, over time, converge to a constant value (such as zero) or diverge toward infinity. When solutions approach a constant value, that value is known as asymptotically stable. Conversely, if a solution tends to leave the neighborhood of a constant value (often toward infinity), that value is known as unstable. In the current context, a stable solution is one in which the constant value is zero, and an effective host will drive the bacterial numbers toward zero. An unstable solution is one in which the host does not control proliferation, and the bacterial number increases without bound
- Bootstrapping
Bootstrapping is a statistical method used when an experimental design produces relatively few data points that may come from a very nonnormal distribution. It is carried out by repeatedly and randomly drawing (with replacement) a number of data points from a set of observations to estimate that data set’s mean and variance. The method is surprisingly simple and refers interested readers to reference 14
APPENDIX
Stability analysis
To compare the predicted fates of normal and cyclophosphamide-treated mice, we determined the conditions for stability of the model using standard analytical methods (15, 29). Our 4-compartment system can be expressed in matrix form as:
| (6) |
or in abbreviated form as:
| (7) |
where the 4 × 4 matrix M is the system’s coefficient matrix (e.g., m11 = [a−d]l−ql/vlpl). When this model is fit to experimental data, the output of the fitting procedure are quantities for each of the variables m11 through m44. To establish stability, the characteristic equation, p(λ), of the coefficient matrix was determined to be:
| (8) |
The Routh array for this equation was generated using the Symbolics Toolbox in Matlab. The first column of the Routh array was found to be:
| (9) |
where α1 and α2 are more complicated algebraic terms. For a solution to be stable, all five of these elements must be greater than zero. Numerical estimates of the second through fifth elements for normal and chemotherapy-treated mice, and for normal and chemotherapy-treated mice with hypothetical hyperdynamic blood flow, are shown in Table A1.
Table A1.
Numerical estimates of the Routh array for the experimental and hypothetical conditions examined
| RH Criterion | Normodynamic |
Hyperdynamic |
||
|---|---|---|---|---|
| Normal | Chemotherapy-treated | Normal | Chemotherapy-treated | |
| tr(A) | 358 | 418 | 544 | 635 |
| α1 | 2,195 | 10,895 | 4,027 | 26,113 |
| α2 | 1,324 | 23,033 | 4,691 | 97,753 |
| det(A) | 118 | −332 | 895 | −11,178 |
These results suggested that det(M) = 0 was the dominant stability boundary in our system. We therefore solved det(M) = 0 for various model parameters to locate the boundary between stability and instability in relationship to our fitted parameters. As 1,000 bootstrap solutions from both the normal and cyclophosphamide-treated animals were available, the boundary was calculated for all 2,000 sets of parameters. The median and 95% confidence bounds were used for the plots in Figure 5 in the main text.
References
- 1.Diekema D, Beekman S, Chapin K, Morel K, Munson E, Doern G. Epidemiology and outcome of nosocomial and community-onset bloodstream infection. J Clin Microbiol. 2003;41:3655–3660. doi: 10.1128/JCM.41.8.3655-3660.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laupland K, Gregson D, Zygun D, Doig C, Mortis G, Church D. Severe bloodstream infections: a population-based assessment. Crit Care Med. 2004;32:992–997. doi: 10.1097/01.ccm.0000119424.31648.1e. [DOI] [PubMed] [Google Scholar]
- 3.Lark R, Chenoweth C, Saint S, Zemencuk J, Lipsky B, Plorde J. Four year prospective evaluation of nosocomial bacteremia: epidemiology, microbiology, and patient outcome. Diagn Microbiol Infect Dis. 2000;38:131–140. doi: 10.1016/s0732-8893(00)00192-9. [DOI] [PubMed] [Google Scholar]
- 4.Lark R, Saint S, Chenoweth C, Zemencuk J, Lipsky B, Plorde J. Four-year prospective evaluation of community-acquired bacteremia: epidemiology, microbiology, and patient outcome. Diagn Microbiol Infect Dis. 2001;41:15–22. doi: 10.1016/s0732-8893(01)00284-x. [DOI] [PubMed] [Google Scholar]
- 5.Wisplinghoff H, Bischoof T, Tallen S, Seifert H, Wenzel R, Edmond M. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nation-wide surveillance study. Clin Infect Dis. 2004;39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 6.Garrouste-Orgeas M, Chevret S, Mainardi J, Timsit J, Misset B, Carlet J. A one-year prospective study of nosocomial bacteremia in ICU and non-ICU patients and its impact on patient outcome. J Hosp Infect. 2000;44:206–213. doi: 10.1053/jhin.1999.0681. [DOI] [PubMed] [Google Scholar]
- 7.Wisplinghoff H, Cornely O, Moser S, Bethe U, Stutzer H, Salzberger B, Fatkenheuer G, Seifert H. Outcomes of nosocomial bloodstream infections in adult neutropenic patients: a prospective cohort and matched case-control study. Infect Control Hosp Epidemiol. 2003;24:884–886. doi: 10.1086/502158. [DOI] [PubMed] [Google Scholar]
- 8.Cheewatrakoolpong B, Steffen E, Brown R, Berg R. Kinetic analysis of bacterial clearance in mice using ESTRIPc and KINET microcomputer programs. J Immunol Methods. 1983;58:375–381. doi: 10.1016/0022-1759(83)90365-4. [DOI] [PubMed] [Google Scholar]
- 9.Christensen G, Bisno A, Parisi J, McLaughlin B, Hester M, Luther R. Nosocomial septicemia due to multiply antibiotic-resistant Staphylococcus epidermidis. Ann Intern Med. 1982;96:1–10. doi: 10.7326/0003-4819-96-1-1. [DOI] [PubMed] [Google Scholar]
- 10.Christensen G, Simpson W, Bisno A, Beachey E. Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infect Immun. 1982;37:318–326. doi: 10.1128/iai.37.1.318-326.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lugo J, Miller J, Price S, Merrill V, Ben-David I, Weinberg J, Mancuso P, Younger J. Lipopolysaccharide O-antigen promotes persistent murine bacteremia. Shock. 2006;27:186–191. doi: 10.1097/01.shk.0000238058.23837.21. [DOI] [PubMed] [Google Scholar]
- 12.Zuluaga A, Salazar B, Rodriguez C, Zapata A, Agudelo M, Vesga O. Neutropenic induced in outbred mice by a simplified low-dose cyclophosphamide regimen: characterization and applicability to diverse experimental models of infectious diseases. BMC Infect Dis. 2006;6:55. doi: 10.1186/1471-2334-6-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lang C, Bagby G, Ferguson J, Spitzer J. Cardiac output and redistribution of organ blood flow in hypermetabolic sepsis. Am J Physiol Regul Integr Comp Physiol. 1984;246:331–337. doi: 10.1152/ajpregu.1984.246.3.R331. [DOI] [PubMed] [Google Scholar]
- 14.Efron B, Tibshirani R. An Introduction to the Bootstrap. New York: Chapman & Hall; 1993. [Google Scholar]
- 15.Bobisud L. Cannibalism as an evolutionary strategy. Bull Math Biol. 1976;38:359–368. doi: 10.1007/BF02462211. [DOI] [PubMed] [Google Scholar]
- 16.Ashare A, Monick M, Powers L, Yarovinsky T, Hunninghake G. Severe bacteremia results in a loss of hepatic bacterial clearance. Am J Respir Crit Care Med. 2006;173:644–652. doi: 10.1164/rccm.200509-1470OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gregory S, Cousens L, van Rooijen N, Dopp E, Carlos T, Wing E. Complementary adhesion molecules promote neutrophil-Kupffer cell interaction and the elimination of bacteria taken up by the liver. J Immunol. 2002;168:308–315. doi: 10.4049/jimmunol.168.1.308. [DOI] [PubMed] [Google Scholar]
- 18.Gregory S, Sagnimeni A, Wing E. Bacteria in the bloodstream are trapped in the liver and killed by immigrating neutrophils. J Immunol. 1996;157:2514–2520. [PubMed] [Google Scholar]
- 19.Anton E. Ultrastructural changes of stromal cells of bone marrow and liver after cyclophosphamide treatment in mice. Tissue Cell. 1997;29:1–9. doi: 10.1016/s0040-8166(97)80066-3. [DOI] [PubMed] [Google Scholar]
- 20.Benacerraf B, Sebestyen M, Schlossman S. A quantitative study of the kinetics of blood clearance of P32-labeled Escherichia coli and staphylococci by the reticuloendothelial system. J Exp Med. 1959;110:27–48. doi: 10.1084/jem.110.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaudino M. Kinetics of distribution of inulin between two body water compartments. Proc Soc Exp Biol Med. 1949;70:672–674. doi: 10.3181/00379727-70-17029. [DOI] [PubMed] [Google Scholar]
- 22.Wagner J. History of pharmacokinetics. Pharmacol Ther. 1981;12:537–562. doi: 10.1016/0163-7258(81)90097-8. [DOI] [PubMed] [Google Scholar]
- 23.Gregory S, Wing E. Neutrophil-Kupffer cell interaction in host defenses to systemic infection. Immunol Today. 1998;19:507–510. doi: 10.1016/s0167-5699(98)01319-x. [DOI] [PubMed] [Google Scholar]
- 24.Clark S, Ma A, Tavener S, McDonald B, Goodarzi Z, Kelly M, Patel K, Chakrabarti S, McAvoy E, Sinclar G, et al. Platelet TLR4 activates neutrophil extracellular traps to ensnare bacteria in septic blood. Nat Med. 2007;13:463–469. doi: 10.1038/nm1565. [DOI] [PubMed] [Google Scholar]
- 25.Ben-David I, Price S, Bortz D, Greineder C, Cohen S, Bauer A, Jackson T, Younger J. Dynamics of intrapulmonary growth in a murine model of repeated microaspiration. Am J Respir Cell Mol Biol. 2005;33:476–482. doi: 10.1165/rcmb.2005-0053OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Day J, Rubin J, vodovotz Y, Chow C, Reynolds A, Clermont G. A reduced model of the acute inflammatory response. II. Capture scenarios of repeated endotoxin administration. J Theor Biol. 2006;242:237–256. doi: 10.1016/j.jtbi.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 27.Lagoa C, Bartels J, Baratt T, Tseng G, Clermont G, Fink M, Billiar T, Vodovotz Y. The role of initial trauma in the host’s response to injury and hemorrhage: insights from a correlation of mathematical simulations and hepatic transcriptomic analysis. Shock. 2007;26:592–600. doi: 10.1097/01.shk.0000232272.03602.0a. [DOI] [PubMed] [Google Scholar]
- 28.Reynolds A, Rubin J, Clermont G, Day J, Vodovotz Y, Ermentrout G. A reduced mathematical model of the acute inflammatory response: I. Derivation of model and analysis of anti-inflammation. J Theor Biol. 2006;242:220–236. doi: 10.1016/j.jtbi.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 29.Coppel W. Stability and Asymptotic Behavior of Differential Equations. Boston: D. C. Heath and Company; 1965. [Google Scholar]