Abstract
Matrix metalloproteinases (MMPs) regulate composition of extracellular matrix and play a critical role in cancer, fibrosis and wound healing. This paper describes a novel peptide based electrochemical biosensor for detecting activity of cell-secreted protease MMP9. In this sensing strategy, a peptide specific to MMP9 was modified with a redox label (methylene blue (MB)) and immobilized on microfabricated 300 μm diameter Au electrodes. Challenging the electrodes with known concentrations of MMP9 resulted in the cleavage of the MB containing peptide fragment and caused a decrease in electrical signal measured by square wave voltammetry (SWV). The limit of detection for MMP9 was determined to be 60 pM with linear range extending to 50 nM. In preparation to detect cell-secreted MMP9, glass surfaces with Au electrode arrays were further micropatterned with poly(ethylene glycol) (PEG) gel to define annular cell adhesive regions next to electrodes and render remainder of the surface non-fouling. The surfaces were further modified with CD14 antibody to promote attachment of monocytes. The peptide-modified electrode arrays were integrated into PDMS microfluidic devices and incubated with U-937 cells, transformed monocytes known to produce MMPs. These studies revealed a 3 fold higher electrochemical signal from ~400 activated monocytes after 10 min activation compared to non-activated monocytes. While this paper focuses on MMP9 detection, the general strategy of employing redox-labeled peptides on electrodes should be broadly applicable for detection of other proteases and should have clinical as well as basic science applications.
Keywords: Matrix metalloproteinase (MMP), electrochemical detection, square wave voltammetry (SWV), microfabrication, peptide substrate
INTRODUCTION
Matrix metalloproteinases (MMPs) regulate cell matrix composition and play a key role in a number of pathological and physiological processes including inflammation, arthritis, cardiovascular diseases, pulmonary diseases, embryonic development, morphogenesis, reproduction tissue resorption, and remodeling.1–5 Importantly, matrix digestion and remodeling promoted by MMPs is a key event in cancer cell migration and metastasis.6,7
Matrix metalloproteinase 9 (MMP9) is a 92-kDa gelatinase subgroup of the MMPs. MMP9 is produced by several cell types, including keratinocytes, monocytes, macrophages, granulocytes, and by various malignant cells.4 MMP9 degrades type IV collagen, the main material in basement membranes, thus helping separate epithelial cells from the underlying stroma.8–11 Increased expression of MMP9 in tumors leads to the degradation of basement membranes, a critical step in tumor invasion.12–14
A number of bioanalytical approaches for detection of MMP expression in cancer cells described in the literature include immunoassay,15,16 gelatinase zymography7,17–19 and fluorescence resonance energy transfer (FRET) assay.20–22 While enabling robust and sensitive detection of proteases, the existing approaches have limitations. Although gelatinase zymography is a sensitive assay of proteolytic activity for MMP2 and MMP9, it is a time-consuming process requiring multiple steps for sample preparation and signal readout. In the case of fluorogenic assays, fluorophores may be subject to photobleaching by repetitive excitation and detection over time. Electrochemical detection of proteases is emerging as a viable alternative over other bioassays due to its simplicity, reliability and sensitivity.23–26
There is an increasing emphasis on developing in vitro surrogates or models of tissues and diseases. Our laboratory is interested in integration of miniature biosensors onto microfabricated cell culture surfaces for local monitoring of cell function. The proximity of cells to biosensors is important as it ensures high local concentration of analyte and enables detection from a small number of cells. To ensure cell-to-sensor proximity we have made an extensive use of poly (ethylene glycol) (PEG) hydrogel micropatterning to define sites for cell attachment on sensing surfaces.27–30 Such micropatterned sensing surfaces have been used by us previously in the development of aptasensors (DNA or RNA aptamer based biosensors) for detection of cell secreted cytokines such as IFN-γ and TNF-α.29, 31 In these devices, hairpin containing aptamers were conjugated with redox reporters, and immobilized on micropatterned electrodes through SH-Au chemistry.29,31 Binding of cell-secreted IFN-γ or TNF-α caused a conformational change in the aptamer, resulting in decrease of the redox current.
Building on these previous studies, we sought to develop an electrochemical biosensor modified with peptides for monitoring protease activity of cells. By analogy with aptamer-based biosensors described above, peptides were labeled with redox reporters and designed to contain terminal cysteine for assembly on Au (Figure 1A). These sensing electrodes were packaged in PEG hydrogel and integrated with microfluidics for cell capture, activation and on-chip detection of MMP9. As shown in Figure 1(B,C), cell-secreted MMP9 molecules cleaved redox-labeled peptides modified on the electrode causing a decrease in electrochemical signal measured at the electrode. This signal decrease appeared as early as 10 min after mitogenic activation of ~400 cells. The limit of detection for our biosensor was 60 pM with linear range extending to 50 nM. MMP9 release rate from cells was estimated to be 0.65 pg cell−1 h−1 with PMA activation and 0.036 pg cell−1 h−1 without PMA activation after 4 h incubation. To the best of our knowledge, this is the first report describing local detection of protease production by a small group of cells residing on a micropatterned surface. One future direction will entail integration of protease biosensors into engineered cell microenvironment to study changes in protease release cellular in the context of cancer or fibrosis. In general, the use of peptide-modified electrochemical biosensors may offer a new tool in detection of protease activity and may have a range of clinical and basic science application.
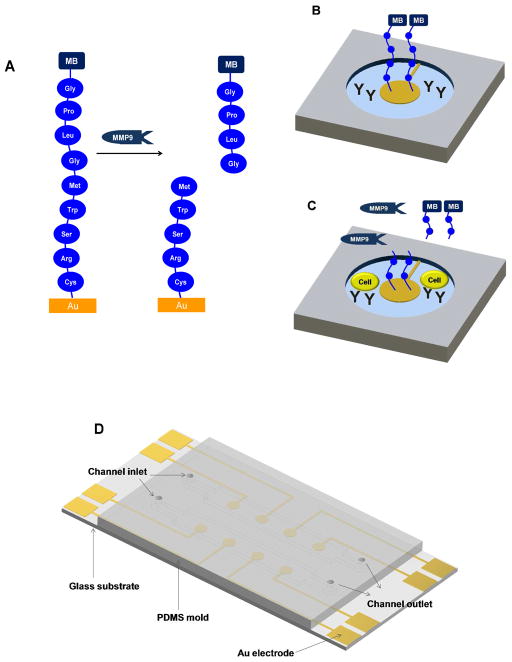
Figure 1.
Detection of MMP9 release from monocytes. (A) Peptide Gly-Pro-Leu-Gly-Met-Trp-Ser-Arg-Cys is assembled on Au surface and is subject to MMP9 mediated cleavage between Gly and Met. (B) An individual cell sensing site, consisting of an electrode with redox-labeled peptide, Abs for cell capture and nonfouling PEG hydrogel. (C) Monocytes become captured on Ab-modified regions and release protease molecules upon activation. Reduction current decreases upon cleavage of peptide and diffusion of fragments carrying redox labels. (D) Layout of the microfluidic devices for sensing.
EXPERIMENTAL SECTION
Materials
Matrix metalloproteinase 9 (MMP9, human, recombinant), poly(ethylene glycol) diacrylate (PEG-DA, MW 575) and 2-hydroxy-2-methyl-propiophenone (photoinitiator) were purchased from Sigma-Aldrich. Chromium etchant (CR-4S) and gold etchant (Au-5) were purchased from Cyantek Corp. (Fremont, CA). Positive photoresist (AZ 5214-E IR) and developer solution (AZ300 MIF) were bought from Mays Chemical (Indianapolis, IN). Phosphate buffered saline (PBS) was purchased from TEKnova (Hollister, CA). Mouse anti-human CD14 antibody (anti-CD14 Ab) were purchased from Beckman Coulter (Miami, FL). Glass slides (75 × 25 mm2) were purchased from Fisher Scientific (Pittsburg, PA). Methylene blue carboxylic acid N-hydroxysuccinimidyl ester (MB-NHS) was purchased from Biosearch Technologies, Inc. (Novato, CA). 3-Acryloxypropyl trichlorosilane was purchased from Gelest (Morrisville, PA). Peptide (Gly-Pro-Leu-Gly-Met-Trp-Ser-Arg-Cys; GPLGMWSRC) was purchased from GL Biochem (Shanghai, China). 11-Mercaptoundecanol hexaethyleneglycol ether (Hydroxy-EG6-undecanethiol) was purchased from Dojindo Molecular Technologies (Rockville, MD). All other chemicals were obtained from Sigma (St. Louis, MO) or Aldrich Chemicals (Milwaukee, WI). MATLAB (MathWorks Inc., Natick, MA) and COMSOL Multiphysics (COMSOL, Inc., Burlington, MA) were used for modeling of peptide cleavage and calculating MMP9 release rate from cells.
Monocytes (U-937 cells) were purchased from American Type Culture Collection (ATCC) and cultured in 10% (v/v) fetal bovine serum (FBS, Invitrogen, Carlsbad, CA), 100 U/mL penicillin and 100 μg/mL streptomycin in RPMI-1640 media (VWR, West Chester, PA) at 37 °C in a humidified 5% CO2 atmosphere.
Surface plasmon resonance and ellipsometry analysis of proteolytic cleavage
Proteolytic cleavage reaction of peptide on Au surface was characterized by surface plasmon resonance (SPR) and ellipsometry. SPR experiments were performed using a BI-3000 instrument (Biosensing Instrument, Tempe, AZ). The SPR instrument was connected to a two-channel programmable syringe pump for uniform continuous flow.
All SPR experiments were performed on bare Au chips obtained from Biosensing Instrument. The peptide, Gly-Pro-Leu-Gly-Met-Trp-Ser-Arg-Cys (5 mM, 0.4 μL) was dissolved in PBS and then mixed with hydroxy-EG6-undecanethiol solution (1 mM, 200 μL) in PBS. This solution was flowed into the SPR instrument for 5 min at a flow rate of 40 μL/min resulting in peptide immobilization. MMP9 (2.5–10 nM) was dissolved in working buffer (50 mM Tris-HCl, 1 mM CaCl2 and 0.05% Triton X-100; pH=7.5) and then perfused through SPR flow cell 5 min at a flow rate of 40 μL/min. The SPR chip was then washed with PBS and exposed to a higher concentration of MMP9.
Fabrication of sensing surfaces
The layout of an electrode array was designed in AutoCAD and converted into plastic transparencies by CAD Art Services (Portland, OR). Glass slides containing 15-nm Cr adhesion layer and 100-nm Au layer from Lance Goddard Associates (Santa Clara, CA) were patterned using photolithography and wet-etching approaches described previously.32 The pattern consisted of eight working microelectrodes (diameter=300 μm) connected to 2 mm × 2 mm square contact pads with 15-μm wide leads (see Figure 1D for layout of electrodes). The photoresist layer was not removed immediately after metal etching to protect Au during silane modification described below.
The glass substrates with photoresist-covered Au electrodes were immersed in a solution of 0.1% (v/v) 3-acryloxypropyltrichlorosilane in toluene for 1 h under nitrogen atmosphere. The slides were rinsed in toluene, sonicated in acetone for 2 min to remove photoresist and then dried using nitrogen gas. The surfaces were then packaged in PEG gel using protocols described by us previously. Briefly, a solution of 2% (v/v) photoinitiator (2-hydroxy-2-methylpropiophenone) in PEG-DA was spin-coated at 800 rpm for 5 sec on acrylated glass slides containing Au electrode patterns. A photomask containing a 500-μm circular pattern was placed on top of the surface and aligned precisely by using fiduciary marks. The slides were irradiated through the photomask for 5 sec by a 365-nm UV light source (80 mW/cm2; OmniCure Series 1000, Lumen Dynamics Group, Mississauga, Ontario, Canada), washed with DI water and dried using nitrogen gas.
Attaching biomolecules onto micropatterned electrode surfaces
Prior to assembly on electrodes, peptides were labeled with redox reporter (MB) by reacting peptide (Gly-Pro-Leu-Gly-Met-Trp-Ser-Arg-Cys, 1 μmol, 1.0 mg) in DMF (100 μL) with MB-NHS (2.5 μmol, 1.5 mg) and DIPEA (5 μmol, 0.87 μL) at room temperature for 2 h. The completion of labeling reaction was confirmed by the peptide disappearance on thin layer chromatography (TLC). Unreacted MB-NHS was deactivated by adding ethanolamine (1 M) in DMF (5 μL) and the mixture solution was used without further separation. Subsequently, glass substrates containing Au electrode arrays and PEG hydrogel layer were incubated for 2 h in 1 mM peptide solution in 1× PBS and then rinsed with water. The MB-tagged peptide molecules self-assembled on Au via thiolate bonds of terminal Cys amino acid. Peptide coverage on Au electrode was optimized and the details are described in Figure S1 (see Supporting Information for detailed description). After peptide assembly the surfaces were blocked by incubation in 3 mM 6-mercapto-1-hexanol (MCH). In the final step, designed to promote cell attachment next to electrodes, surfaces were incubated for 1 h with monocyte-specific CD14-Ab (50 μg/mL) in 1× PBS washed with DI water and dried using nitrogen gas.
Electrochemical detection of MMP9 production by monocytes
A double-channel poly(dimethylsiloxane) (PDMS) mold (3 mm (width)× 10 mm (length)× 0.1 mm (height)) was integrated to the glass substrates with Au electrodes for real time detection of MMP9 release from monocytes. The microfluidic devices were fabricated by using standard soft lithography approaches as reported previously.33,34 Electrochemical measurements were performed using a CHI 842B Electrochemical Workstation (CHInstruments, Austin, TX) with a three electrode system consisting of a Ag/AgCl (3 M KCl) reference electrode at the outlet, Pt wire counter electrode in the inlet of the channel and Au working electrodes. Electrochemical signals were obtained in serum-free and Phenol Red-free RPMI-1640 media using square wave voltammetry (SWV) with a 40 mV amplitude signal at a frequency of 60 Hz, over the range from 0 to −0.50 V versus Ag/AgCl references.
SWV responses of peptide-functionalized electrodes were measured by varying concentrations of MMP9 and incubating each solution over time. To analyze MMP9 release from cells, U-937 cells suspended in PBS were incubated on anti-CD14 Ab immobilized PEG-patterned glass surface with Au electrodes described above for 20 min and then unbound cells were removed by washing with PBS. The microchannel was filled with 100 ng/mL phorbol 12-myristate 13-acetate (PMA) in serum-free and Phenol Red-free RPMI-1640 media and the electrochemical signals were monitored over time for up to 4 h.
RESULTS AND DISCUSSION
This paper describes a bioanalytical method for monitoring protease release from a small group of cells. In this method, electroactive peptides are assembled on electrodes and then, become cleaved by protease molecules released from neighboring cells. Peptide digestion is monitored electrochemically to gauge cellular activity (Figure 1).
SPR analysis of peptide assembly and cleavage
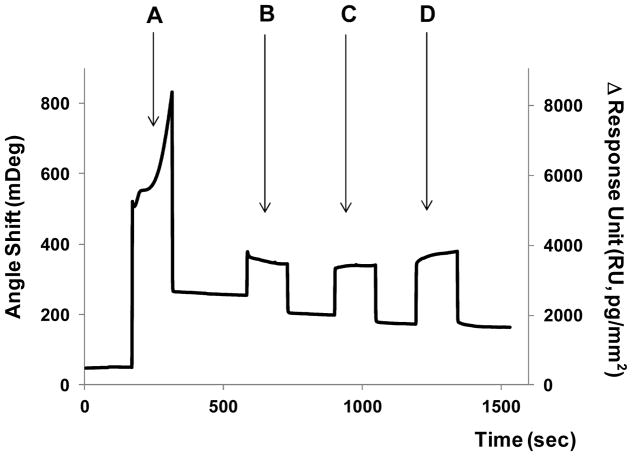
The peptide used for MMP9 detection ((Gly-Pro-Leu-Gly-Met-Trp-Ser-Arg-Cys) was designed to contain terminal Cys amino acid to form thiolate bonds on Au substrates (Figure 1A). Assembly of peptide molecules was verified by SPR analysis described in Figure 2. As seen from these data, infusion of peptide/EG6 mixture resulted in 200 mDeg SPR signal change. Using methodology described by Shumaker-Parry et al. this SPR signal may be converted into a surface coverage value of 2.1×10−10 mol/cm2.35 Subsequent to peptide assembly, surfaces were exposed to varying concentrations of MMP9. As seen from Figure 2 (steps B to D) protease molecules caused decrease in SPR signal (90 mDeg) suggesting that peptide cleavage took place. It is of note that SPR signal decrease was larger for higher MMP9 concentrations. In addition to SPR analysis, ellipsometry was employed to measure the thickness change after peptide cleavage by protease. Peptide adsorption onto Au surface resulted in thickness increase of 2.33±0.03 nm. Subsequent cleavage by varying concentrations of MMP9 decreases to 2.29±0.03 nm for 5 nM and 2.27±0.03 nm for 10 nM. Therefore, surface analysis points to cleavage of hydrolysis of peptide molecules assembled on Au surfaces by MMP9.
Figure 2.
SPR analysis of peptide cleavage. Each step showed the signal increase upon injection of reagent solution and decreased to a certain baseline by surface change after washing the surface. (A) When peptide molecules were immobilized on Au SPR chip, baseline moved up. The immobilized peptide was cleaved upon the injection of MMP9 solution ((B) 2.5, (C) 5 and (D) 10 nM, subsequently). The decrease in SPR signal observed in steps (B) to (C) suggests that peptide cleavage took place. (C) The peptide cleavage reaction was completed at 5 nM and (D) there was no further decrease in base line at 10 nM.
Fabrication and characterization of peptide-modified electrodes
The surfaces used for cell detection consisted of 300 μm diameter Au electrodes residing in the center of 500 μm diameter PEG hydrogel wells. This design, shown in Figure 1D, was used to position cells inside the hydrogel microwells in the immediate vicinity of the Au electrodes. Prior to cell analysis electrodes were functionalized with MB-labeled peptides and challenged with known concentrations of MMP9 to establish detection limit and linear range of the biosensor. The peptide, MB-Gly-Pro-Leu-Gly-Met-Trp-Ser-Arg-Cys, is the substrate for MMP9 and was expected to become cleaved between Gly and Met amino acids,22 resulting in loss of the peptide fragment carrying redox reporter MB (see Figure 1A).
Surface density of the MB moiety (Γ) is described as Γ= Q/nFA, where Q is the total charge passed (1.81×10−4 C), n is the number of electrons transferred per MB moiety (=2), A is the electrode surface area (7.07×10−4 cm2) and F is the Faraday constant (9.65×104 C/mol).25–27 The initial MB-peptide surface density of our sensor was estimated to be 1.33×10−10 mol/cm2. The surface density is comparable to other reports employing redox reporter-tagged peptides.25,26
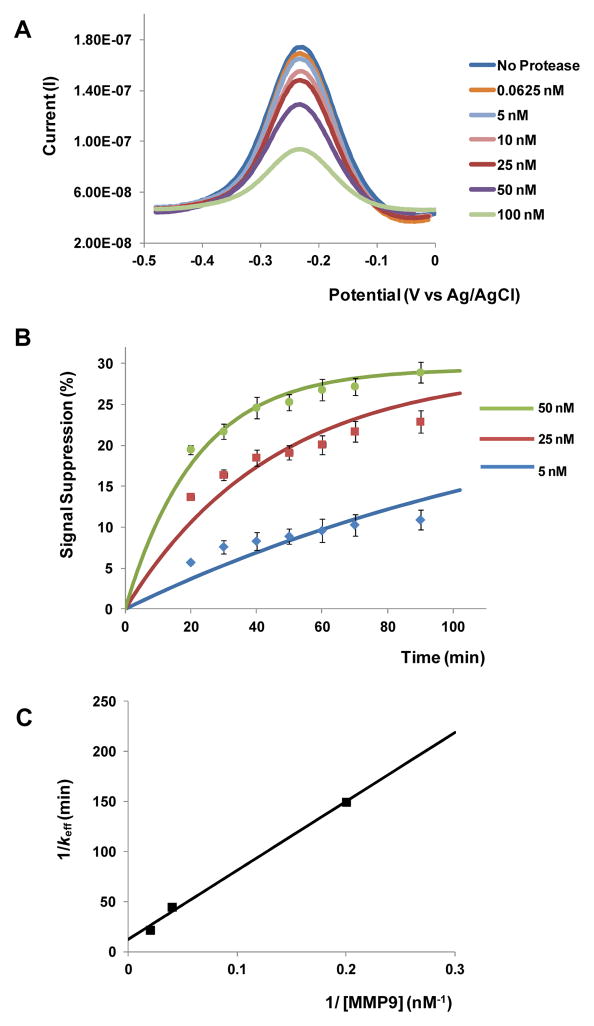
MMP9 molecules cleave the redox reporters from the peptide layer, causing a decrease in the redox current measured at the electrode. This change in electrical signal is termed “signal suppression” and is defined as the ratio of final to initial values of SWV peak current.29 Figure 3A shows SWV responses of peptide-functionalized electrodes to varying concentrations of MMP9. As seen from these data, redox current measured at the electrode decreases upon addition of protease due to cleavage and diffusion of redox-labeled peptide fragments away from the electrode. The SWV measurements in Figure 3A obtained 30 min after adding protease show limit of detection of ca. 60 pM. We determined the limit of detection estimated to be a certain protease concentration based on signal-to-noise characteristics (S/N = 3).25,29 It is worth noting that sensitivity is similar to that of fluorescent assays for MMP detection reported in the literature.36,37 Unlike affinity biosensors where analyte binding to the surface may reach equilibrium rapidly, MMP9 is capable of turning over and digesting multiple peptide molecules per enzyme molecule, hence the signal suppression increases over time (dots in Figure 3B).
Figure 3.
Electrochemical detection of recombinant MMP9. (A) Square wave voltammetry (SWV) results after incubating peptide-functionalized surfaces with different concentrations of MMP9 for 30 min. (B) Monitoring signal suppression in various MMP9 solutions over time: dots represent experimental data and lines represent simulated data. 100 % signal suppression indicates complete peptide cleavage. (C) The reaction rate, keff, is a function of the concentration of enzyme [E]. A plot of 1/keff vs. 1/[E] was used to determine kcat and Km.
Interestingly, as seen from Figure 3B, only 30% signal suppression was achieved when challenging electrodes with 50 nM MMP, suggesting that only a fraction of peptides become cleaved. A further enhancement in signal could not be achieved even after immersing peptide-functionalized electrodes in a high concentration of MMP9 solution. A possible explanation for partial cleavage of peptide layer is provided by Anne et al.,38 who report that the extent of peptide cleavage by proteases was significantly higher on ultra-flat Au surfaces compared to regular Au electrodes. It is therefore conceivable that the roughness of our micropatterned Au electrodes may need to be decreased in the future to improve peptide cleavage and by extension redox signal.
Modeling of peptide cleavage
The empirical measurements of peptide cleavage on Au electrodes (dots in Figure 3B) were used to determine rate constants and Michaelis-Menten constant for this reaction. Assuming Michaelis-Menten kinetics, the proteolytic cleavage may be described by the following equation:
The enzyme (E), MMP9, first interacts with the substrate (S), peptide, on the Au surface and forms an intermediate complex (C), which subsequently dissociates to release both MMP9 and the peptide fragment carrying redox reporter (P) in bulk solution. Unlike standard expressions where enzyme-substrate interactions occur in solution, our reaction is heterogeneous with surface bound peptide and soluble enzyme. Such reaction may be described by the equation below,39
| (1) |
where [S]0 is the initial substrate (peptide) concentration on the electrode surface and Km = (kcat+k−1)/k1 is the Michaelis-Menten constant.
In order to solve eq. (1), we defined the initial peptide concentration, [S]0 = ε[S]T, where [S]T is the total peptide on the electrode and ε is defined as the maximum fraction of peptides cleaved from the electrode. The [S]T is 1.33×10−10 mol/cm2 represents peptide coverage on the electrode and was determined earlier in this paper by integrating SWV curves of redox-labeled peptides. The value for ε was determined to be 0.3 based on the data in Figure 3B. The signal suppression, defined as the ratio of final to initial values of SWV peak current, had a linear relationship with [P]/[S]T, which could be calculated by solving eq. (1),
| (2) |
where
| (3) |
As shown in Figure 3B (dots in the graph), addition of known concentrations of MMP9 resulted in redox signal changing in a concentration dependent and time dependent manner. MATLAB was used to fit eq. (2) to the experimental data and then determine values for reaction rate denoted keff. Solid lines in Figure 3B represent curve fitting to the experimental data. With keff values in hand, eq. (3) was linearized and solved for kcat and Michaelis-Menten constant (Km). Figure 3C shows a plot of 1/keff vs. 1/[E], with kcat = 0.0809 min−1 (derived from the intercept) and Km = 55.75 nM (obtained from the slope). The kcat/Km value (slope of the line) for our peptide GPLGMWSRC was 2.4×104 M−1 sec−1, which is close to the value of 1.03×105 M−1 sec−1 reported for a similar sequence GPLGLWAR.40 The enzyme rate constants derived in this section were used to solve diffusion-reaction equation in order to determine rate of protease release from cells. This discussion is presented in the section on cell release monitoring.
Specificity of peptide-modified electrodes
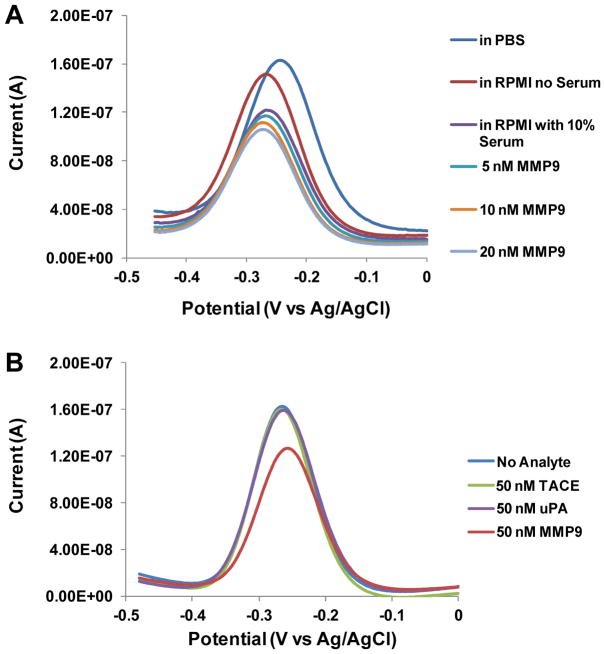
To test performance of the biosensor, experiments were carried out in complex media and in the presence of non-specific proteases. To demonstrate feasibility of our sensor to cell-related experiments, we wanted to detect signal responses in serum-containing media. As shown in Figure 4A, addition of 10% serum into RPMI media resulted in signal loss of ~20% compared to media alone. However, despite this loss in signal, spiking MMP9 into serum-containing media resulted in measurable changes in redox current as demonstrated in Figure 4A. When the sensor was challenged with varying concentrations of MMP9 in serum-containing media, 5 nM, 10 nM and 20 nM of MMP9 caused 4%, 11% and 17% signal suppression, respectively. The ability to selectively detect MMP9 molecules in the presence of overabundant non-specific proteins confirmed the suitability of this sensor for the detection of cell secreted proteins under physiological condition.
Figure 4.
Peptide-based biosensor responses in complex media and in the presence of non-specific proteases. (A) SWV signals from MB-peptide on Au electrodes using recombinant MMP9 in serum containing RPMI-1640 media. Even though the signal suppression was reduced, the response to varying MMP9 concentration was still discernible. (B) Responses of peptide-functionlized electrodes to nonspecific proteases TACE and uPA as well as to specific protease MMP9.
To demonstrate substrate specificity of MMP9, the peptide sensor was challenged with non-specific proteases including tumor necrosis factor-α-converting enzyme (TACE) and urokinase-type plasminogen activator (uPA). We observed that minimal signal suppression (2–3%) was shown with TACE (50 nM) and uPA (50 nM), respectively, while 25% signal suppression was shown with MMP9 at the identical condition (Figure 4B). This result demonstrates that our peptide sensor can detect MMP9 in the presence of non-specific proteases.
Monitoring MMP9 release from monocytes
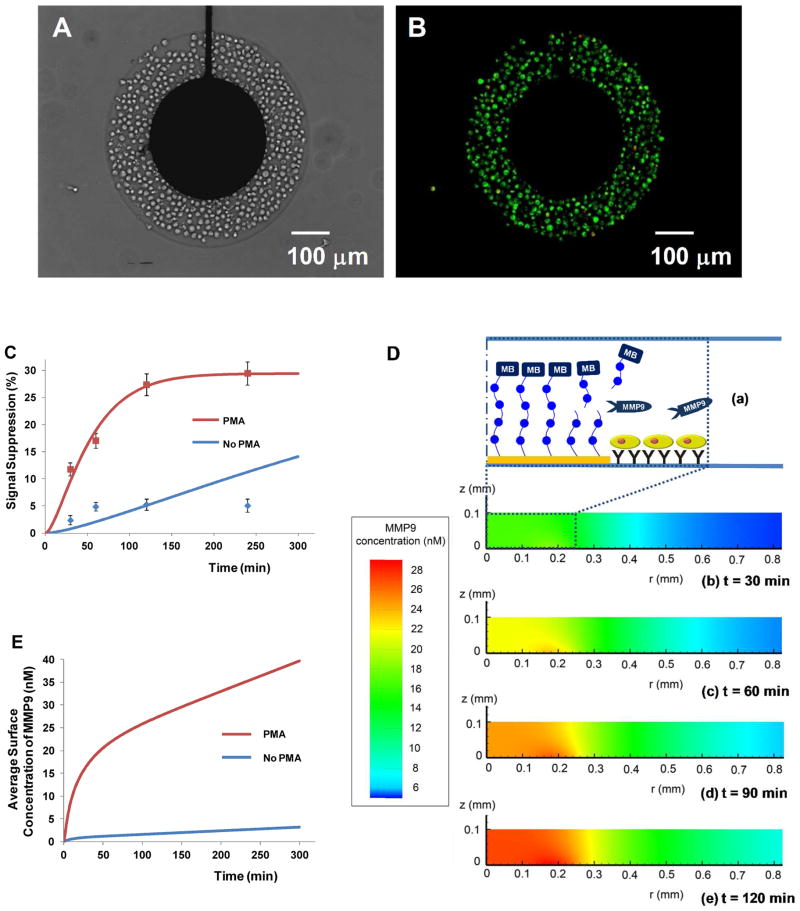
In the final set of experiments, we wanted to demonstrate real time detection of MMP9 release from monocytes using peptide-functionalized electrodes. U-937 cells were used as model monocytes secreting MMP9 under mitogenic stimulation by PMA.7 The miniature peptide-modified electrodes were placed inside microfluidic PDMS channels and exposed to monocyte solution. As shown in Figure 5A, monocytes were captured in the annular Ab-modified regions of glass, formed between 300 μm diameter Au electrodes and 500 μm diameter hydrogel wells. Approximately ~400 cells attached near each electrode. Cell viability after immobilization was over 95% as determined by live/dead assay (Figure 5B). Upon mitogenic stimulation, MMP9 activity from the monocytes was monitored by SWV measurements for up to 4 h (dots in Figure 5C). Monocytes captured in a parallel channel of a microfluidic device and incubated in media without PMA were used as a negative control. Figure 5C shows that as early as 10 min post stimulation, signal from activated monocytes was 3-fold higher than that from non-activated cells. The signal suppression from U-937 cells with PMA activation was 32% after 4 h incubation, whereas that without activation was 8% (Figure 5C).
Figure 5.
Monitoring cellular production of MMP9. (A) U-937 monocytes captured next to sensing electrodes: The dimensions of Au electrodes (diameter= 300 μm) and PEG hydrogel (diameter= 500 μm) allowed for attachment of ~400 cells. (B) Live/dead staining (green: live; red: dead) shows that over 95% of cells were alive. (C) Change in signal suppression over time for mitogenically activated vs. quiescent cells. Dots are experimental measurements whereas solid curves are theoretical results obtained from reaction-diffusion model based on the assumption of cellular production rate. 100 % signal suppression indicates complete peptide cleavage. (D) Modeling change in MMP9 concentration over time. (a) Schematic to orient the reader as to the location of cells electrode in the z- and r-directions. (b–e) The simulation domain was 0.1 mm in z-direction and 1.5 mm in r-direction. MMP9 molecules being released from the cells diffused to the electrode, which increased the concentration of MMP9 on the electrode surface. (E) Change in concentration over time based on the numerical simulation that used cellular production rates determined in part (C).
In order to determine the rate of MMP9 production by the cells we needed to consider that the enzyme concentration in the proximity of the electrode changes as a function of time and that the rate of reaction on the electrode surface is dependent on the enzyme concentration (see Figure 5D). The enzyme reaction constants kcat and Km were determined in Figure 3C. The reaction-diffusion model was then set-up to determine the change in [E] at the electrode over time and to then solve Michaelis-Mention expression (eq. (1)) for [P] – the concentration of cleaved peptide fragment carrying redox reporter. Diffusion of enzyme molecules is described by the expression below:
| (4) |
where D = 8.18×10−7 cm2/s is the diffusion coefficient of MMP9.41 The boundary conditions are
| (5) |
where Nc is the cell density in the vicinity of the electrode and σ̇ is the protease secretion rate per cell. These boundary conditions state that the rate of protease release is constant and only occurs next to cells. Enzyme kinetics on electrode is described by eq. (1) with the initial condition for cleaved peptide fragment [P]t=0 = 0. The system of reaction and diffusion equations was solved using COMSOL Multiphysics.
The solid lines in Figure 5C were obtained by iteratively assuming a rate of release σ̇ and fitting the resultant theoretical curve from diffusion-reaction model to the experimental data. Based on this, the MMP9 release rate was determined to be 0.65 pg cell−1 h−1 for activated cells and 0.036 pg cell−1 h−1 for quiescent cells over the course of 4 h. In particular, a poor curve fitting was obtained from resting cells in performing the numerical simulation. A more sophisticated model may need to be developed to predict protease release from resting cells. Based on the detection limit and the MMP9 secretion rate per cell, electrochemical signals from ca. 25 cells will be detectable within an hour using our system.
Eqs. (3) and (4) were solved numerically to determine concentration profile over time for activated vs. resting monocytes. Simulations were performed in cylindrical coordinates with the origin at the center of the electrode. The simulation domain was 0.1 mm in z-direction and 1.5 mm in r-direction. Time-dependent profiles of MMP9 diffusion from the activated cells are described in Figure 5D. We could observe that MMP9 molecules being released from the cells diffused to the electrode, which increased the concentration of MMP9 on the electrode surface over time. These data presented in Figure 5E show the average MMP9 concentration on the electrode surface, which demonstrate significant differences between activated and non-activated cells and may be used to quantify protease production from a small cell population.
Additional experiments were carried to verify MMP9 production from activated monocytes using conventional methods.7 As described in Figure S2 (see Supporting Information for detailed description), fluorescence-based assay revealed that mitogenic activation of U937 cells did indeed cause an upregulation in MMP9 production.
CONCLUSIONS
In this study, we developed micropatterned peptide-functionalized sensing surfaces for detecting protease release from cells. The biosensing surfaces were designed so as to place small groups of cells (~400) next to Au electrodes inside microfluidic devices. MMP9 molecules secreted from cells diffused rapidly to the nearby electrode, cleaving MB-labeled peptides and causing a change in redox signal at the electrode. This biosensor was specific to MMP9 when tested in mixtures with nonspecific proteases and showed 60 pM limit of detection. Reaction-diffusion modeling was used to determine protease release rate to be 0.65 pg cell−1 h−1 for activated cells and 0.036 pg cell−1 h−1 for quiescent monocytes – an 18 fold difference. The difference in response could be detected as early as 10 min post activation. Significantly, the signal was monitored in real-time during the cell stimulation experiments. Using this sensing system, electrochemical signal from ca. 25 cells is measurable within an hour. Miniature protease sensors described here may be particularly useful when integrated into in vitro models of tissues to address questions related to local protease activity in developmental biology, cancer metastasis and injury remodeling.
Supplementary Material
Acknowledgments
This work was supported by NSF EFRI grant awarded to AR.
Footnotes
Additional information as noted in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Nagase H, Woessner JF., Jr J Biol Chem. 1999;274:21491–21494. doi: 10.1074/jbc.274.31.21491. [DOI] [PubMed] [Google Scholar]
- 2.Zitka O, Kukacka J, Krizkova S, Huska D, Adam V, Masarik M, Prusa R, Kizek R. Curr Med Chem. 2010;17:3751–3768. doi: 10.2174/092986710793213724. [DOI] [PubMed] [Google Scholar]
- 3.Murphy G, Nagase H. Mol Asp Med. 2008;29:290–308. doi: 10.1016/j.mam.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ram M, Sherer Y, Shoenfeld Y. J Clin Immunol. 2006;26:299–307. doi: 10.1007/s10875-006-9022-6. [DOI] [PubMed] [Google Scholar]
- 5.Matache C, Stefanescu M, Dragomir C, Tanaseanu S, Onu A, Ofiteru A, Szegli G. J Autoimmun. 2003;20:323–331. doi: 10.1016/s0896-8411(03)00037-4. [DOI] [PubMed] [Google Scholar]
- 6.Deryugina EI, Quigley JP. Cancer Metastasis Rev. 2006;25:9–34. doi: 10.1007/s10555-006-7886-9. [DOI] [PubMed] [Google Scholar]
- 7.Roomi MW, Monterrey JC, Kalinovsky T, Rath M, Niedzwiecki A. Oncol Rep. 2009;21:1323–1333. doi: 10.3892/or_00000358. [DOI] [PubMed] [Google Scholar]
- 8.Kohrmann A, Kammerer U, Kapp M, Dietl J, Anacker J. BMC cancer. 2009;9:188. doi: 10.1186/1471-2407-9-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato Y, Yamashita T, Ishikawa M. Oncol Rep. 2002;9:565–569. [PubMed] [Google Scholar]
- 10.Iwasaki M, Nishikawa A, Fujimoto T, Akutagawa N, Manase K, Endo T, Yoshida K, Maekawa R, Yoshioka T, Kudo R. Gynecol Oncol. 2002;85:103–107. doi: 10.1006/gyno.2001.6573. [DOI] [PubMed] [Google Scholar]
- 11.Giambernardi TA, Grant GM, Taylor GP, Hay RJ, Maher VM, McCormick JJ, Klebe RJ. Matrix Biol. 1998;16:483–496. doi: 10.1016/s0945-053x(98)90019-1. [DOI] [PubMed] [Google Scholar]
- 12.Sato T, Sakai T, Noguchi Y, Takita M, Hirakawa S, Ito A. Gynecol Oncol. 2004;92:47–56. doi: 10.1016/j.ygyno.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 13.Berube M, Deschambeault A, Boucher M, Germain L, Petitclerc E, Guerin SL. Mol Vis. 2005;11:1101–1111. [PubMed] [Google Scholar]
- 14.Di Nezza LA, Misajon A, Zhang J, Jobling T, Quinn MA, Ostor AG, Nie G, Lopata A, Salamonsen LA. Cancer. 2002;94:1466–1475. doi: 10.1002/cncr.10355. [DOI] [PubMed] [Google Scholar]
- 15.Yan AT, Yan RT, Spinale FG, Afzal R, Gunasinghe HR, Stroud RE, McKelvie RS, Liu PP. Eur J Heart Fail. 2008;10:125–128. doi: 10.1016/j.ejheart.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banfi C, Cavalca V, Veglia F, Brioschi M, Barcella S, Mussoni L, Boccotti L, Tremoli E, Biglioli P, Agostoni P. Eur Heart J. 2005;26:481–488. doi: 10.1093/eurheartj/ehi073. [DOI] [PubMed] [Google Scholar]
- 17.Kodera T, Nakagawa T, Kubota T, Kabuto M, Sato K, Kobayashi H. J Neurooncol. 2000;46:105–114. doi: 10.1023/a:1006387600909. [DOI] [PubMed] [Google Scholar]
- 18.Sawaya RE, Yamamoto M, Gokaslan ZL, Wang SW, Mohanam S, Fuller GN, McCutcheon IE, Stetler-Stevenson WG, Nicolson GL, Rao JS. Clin Exp Metastasis. 1996;14:35–42. doi: 10.1007/BF00157684. [DOI] [PubMed] [Google Scholar]
- 19.Rao JS, Yamamoto M, Mohaman S, Gokaslan ZL, Fuller GN, Stetler-Stevenson WG, Rao VH, Liotta LA, Nicolson GL, Sawaya RE. Clin Exp Metastasis. 1996;14:12–18. doi: 10.1007/BF00157681. [DOI] [PubMed] [Google Scholar]
- 20.Miller MA, Barkal L, Jeng K, Herrlich A, Moss M, Griffith LG, Lauffenburger DA. Integr Biol. 2011;3:422–438. doi: 10.1039/c0ib00083c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knight CG, Willenbrock F, Murphy G. FEBS Lett. 1992;296:263–266. doi: 10.1016/0014-5793(92)80300-6. [DOI] [PubMed] [Google Scholar]
- 22.Netzel-Arnett S, Mallya SK, Nagase H, Birkedal-Hansen H, Van Wart HE. Anal Biochem. 1991;195:86–92. doi: 10.1016/0003-2697(91)90299-9. [DOI] [PubMed] [Google Scholar]
- 23.Munge BS, Fisher J, Millord LN, Krause CE, Dowd RS, Rusling JF. Analyst. 2010;135:1345–1350. doi: 10.1039/c0an00028k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abd-Rabboh HS, Nevins SA, Durust N, Meyerhoff ME. Biosens Bioelectron. 2003;18:229–236. doi: 10.1016/s0956-5663(02)00181-1. [DOI] [PubMed] [Google Scholar]
- 25.Zhao N, He Y, Mao X, Sun Y, Zhang X, Li C-z, Lin Y, Liu G. Electrochem Commun. 2010;12:471–474. [Google Scholar]
- 26.Liu G, Wang J, Wunschel DS, Lin Y. J Am Chem Soc. 2006;128:12382–12383. doi: 10.1021/ja0626638. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y, Tuleouva N, Ramanculov E, Revzin A. Anal Chem. 2010;82:8131–8136. doi: 10.1021/ac101409t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan J, Pedrosa VA, Simonian AL, Revzin A. ACS Appl Mater Interfaces. 2010;2:748–755. doi: 10.1021/am9007819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, Yan J, Howland MC, Kwa T, Revzin A. Anal Chem. 2011;83:8286–8292. doi: 10.1021/ac202117g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan J, Pedrosa VA, Enomoto J, Simonian AL, Revzin A. Biomicrofluidics. 2011;5:032008. doi: 10.1063/1.3624739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, Kwa T, Revzin A. Biomaterials. 2012;33:7347–7355. doi: 10.1016/j.biomaterials.2012.06.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Revzin A, Tompkins RG, Toner M. Langmuir. 2003;19:9855–9862. [Google Scholar]
- 33.Stybayeva G, Mudanyali O, Seo S, Silangcruz J, Macal M, Ramanculov E, Dandekar S, Erlinger A, Ozcan A, Revzin A. Anal Chem. 2010;82:3736–3744. doi: 10.1021/ac100142a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu H, Stybayeva G, Silangcruz J, Yan J, Ramanculov E, Dandekar S, George MD, Revzin A. Anal Chem. 2009;81:8150–8156. doi: 10.1021/ac901390j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shumaker-Parry JS, Campbell CT. Anal Chem. 2004;76:907–917. doi: 10.1021/ac034962a. [DOI] [PubMed] [Google Scholar]
- 36.Xia Z, Xing Y, So MK, Koh AL, Sinclair R, Rao J. Anal Chem. 2008;80:8649–8655. doi: 10.1021/ac801562f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng D, Zhang Y, Feng T, Shi W, Li X, Ma H. Chem Commun. 2011;47:10680–10682. doi: 10.1039/c1cc13975d. [DOI] [PubMed] [Google Scholar]
- 38.Anne A, Chovin A, Demaille C. Langmuir. 2012;28:8804–8813. doi: 10.1021/la301316r. [DOI] [PubMed] [Google Scholar]
- 39.Gutierrez OA, Chavez M, Lissi E. Anal Chem. 2004;76:2664–2668. doi: 10.1021/ac049885d. [DOI] [PubMed] [Google Scholar]
- 40.Patterson J, Hubbell JA. Biomaterials. 2010;31:7836–7845. doi: 10.1016/j.biomaterials.2010.06.061. [DOI] [PubMed] [Google Scholar]
- 41.Tyn MT, Gusek TW. Biotechnol Bioeng. 1990;35:327–338. doi: 10.1002/bit.260350402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.