Summary
The protein actin forms filaments that provide cells with mechanical support and driving forces for movement. Actin contributes to biological processes such as sensing environmental forces, internalizing membrane vesicles, moving over surfaces and dividing the cell in two. These cellular activities are complex; they depend on interactions of actin monomers and filaments with numerous other proteins. Here, we present a summary of the key questions in the field and suggest how those questions might be answered. Understanding actin-based biological phenomena will depend on identifying the participating molecules and defining their molecular mechanisms. Comparisons of quantitative measurements of reactions in live cells with computer simulations of mathematical models will also help generate meaningful insights.
Life on earth arose by divergent evolution from a common ancestor that lived ~3 billion years ago. Among its ~400 genes was the ancestral gene encoding actin (1). Actin and its bacterial counterparts polymerize into filaments (Fig. 1) that offer myriad advantages to cells. High cellular concentrations of actin make the protein one of the most abundant on earth. Actin is essential for the survival of most cells: filaments provide internal mechanical support, tracks for movements of intracellular materials and force to drive cell movements. Many modern species of prokaryotes use actin relatives to maintain asymmetrical shapes and to move DNA through the cytoplasm. Essentially all eukaryotes have genes for actin, and most have genes for myosin motor proteins that generate forces on actin filaments (2). In animals actin filaments complement two other cytoskeletal polymers, microtubules and intermediate filaments (Fig. 1B).
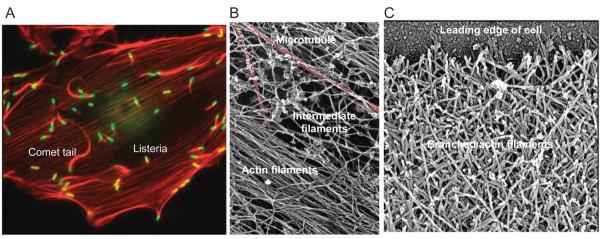
Figure 1.
Micrographs of actin filament structures in cells. A, Fluorescent light micrograph of an animal epithelial cell grown in tissue culture and infected with a bacterium, Listeria monocytogenese. Actin filaments are red and bacteria are green. Actin bundles, called stress fibers, bridge sites of adhesion to the substrate. The bacteria use Arp2/3 complex to assemble comet tails for transport through the cytoplasm. B, Electron micrograph of three types of cytoskeletal polymers in a cell permeabilized to release soluble components. After rapid freezing, the frozen water was sublimed away and cellular components were coated with platinum. Red colorization highlights a microtubule. Bundle of actin filaments and a network of intermediate filaments are labeled. C. Electron micrograph of the network of branched actin filaments at the leading edge (top) of a motile keratocyte. The cell was grown in tissue culture, extracted to release soluble materials, dried and coated with platinum. (All images from T.D. Pollard and W.C. Earnshaw, Cell Biology, W.B. Saunders, 2007. Sources are (A) Matthew Welch, (B) John Heuser and (C) Tatyana Svitkina and Gary Borisy.)
Actin and myosin were discovered during the 1940s in muscle, where the two proteins comprise highly regular arrays of filaments that make up more than half of the total protein. Pioneering research on muscle established general principles that apply to actin assembly and function in all cells, including the mechanism used by myosin to produce force and movement from ATP hydrolysis (3). Two decades later, actin and myosin were discovered in other cells (4) (5), revealing that muscle filaments are a specialized example of a common cellular system. Subsequent research identified numerous proteins that regulate actin, analyzed their mechanisms of action and linked the proteins to cellular processes.
Under physiological conditions, actin monomers (Fig. 2A) spontaneously polymerizes into long stable filaments (Fig. 2B) with a helical arrangement of subunits (for review see (6)). Polymerization starts slowly, because small oligomers are very unstable, but once filaments have been created, then actin polymerizes rapidly and almost completely. Actin filaments are polar, because the subunits in the filament all point in the same direction. One end of the filament grows much faster than the other end. Actin binds an adenine nucleotide (ATP or ADP), and soon after assembly into filaments, actin hydrolyzes the terminal phosphate from the bound ATP and slowly dissociates the phosphate. Subtle changes in the structure of the actin subunits associated with these chemical reactions prepare ADP-actin filaments for disassembly by regulatory proteins.
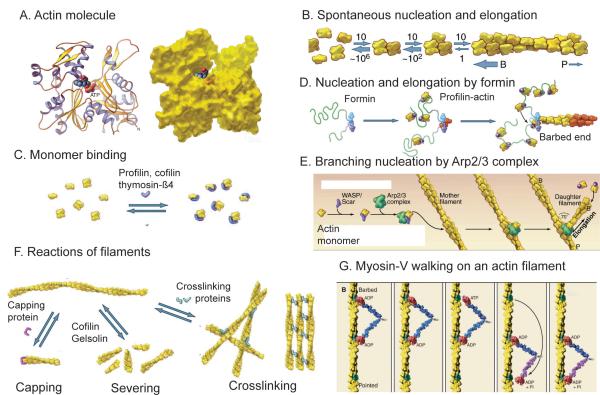
Figure 2.
Structures of actin and diagrams of fundamental reactions. A, Ribbon and space filling models of the actin molecule (pdb:1ATN). B, Spontaneous nucleation and elongation. Dimers and trimers are unstable. Longer polymers grow rapidly at the barbed end (B) and slowly at the pointed end (P). C, Actin monomer binding proteins. Thymosin-ß1 blocks all assembly reactions; profilin promotes nucleotide exchange and inhibits pointed end elongation and nucleation but not barbed end elongation; cofilin inhibits nucleotide exchange and promotes nucleation. D, Nucleation and elongation by formins. Formins initiate polymerization from free actin monomers and remain associated with the growing barbed end. Profilin-actin binds to formin and transfers actin onto the barbed end of the filament. E, Nucleation by Arp2/3 complex. Nucleation promoting factors such as WASp bind an actin monomer and Arp2/3 complex. Binding to the side of a filament completes activation, and the barbed end of the daughter filament grows from Arp2/3 complex. F, Reactions of actin filaments. Capping proteins bind to and block barbed ends; cofilin and gelsolin sever filaments; crosslinking proteins assemble networks and bundles of actin filaments. G, Myosin motors, such as myosin-V, use cycles of ATP hydrolysis to walk along actin filaments, generally toward the barbed end. (Redrawn from images in T.D. Pollard and W.C. Earnshaw, Cell Biology, W.B. Saunders, 2007.)
Eukaryotic cells use >100 accessory proteins to maintain a pool of actin monomers, initiate polymerization, restrict the length of actin filaments, regulate the assembly and turnover of actin filaments and crosslink filaments into networks or bundles (Fig. 2C-F). The mechanisms forming new filaments include growing a branch on the side of an existing filament, severing a filament to create two new ends or starting up a filament from monomers. Genes for most of these accessory proteins were in place about 1 billion years ago when the top branches formed on the phylogenetic tree, so amoebas, fungi and animals share many molecular mechanisms that run their actin systems (2). Some species, such as the intestinal parasite Giardia, lack genes for myosin and many actin-binding proteins. These organisms may have diverged before these genes emerged (7), or they may have lost these genes, just as plants lost more than 200 genes required for the assembly of cilia and flagella (8).
Polymerization of actin filaments drives the crawling locomotion of eukaryotic cells, a characteristic feature of amoebae and animal cells (Fig. 3G). Actin polymerization also contributes to the internalization of membrane vesicles to control the composition of the cell membrane and the interface of the cell with the environment.
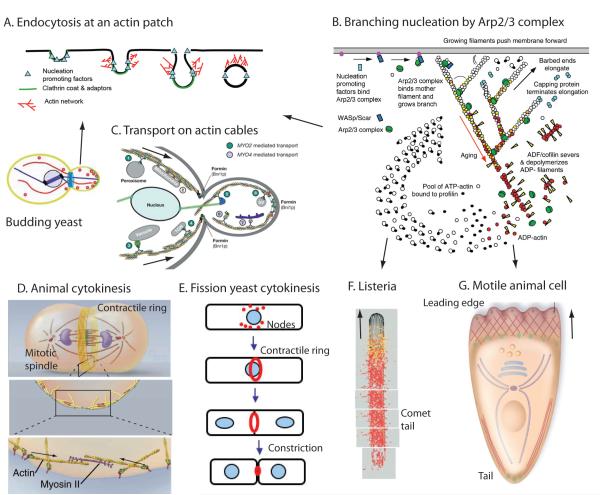
Figure 3.
Cartoons showing actin based movements. (A) Clathrin-mediated endocytosis at fungal actin patches. (B) Formation of branched filament networks nucleated by Arp2/3 complex, which is used for three of these types of movement, those depicted in panels A, F and G. (C) Transport of membrane-bound vesicles, organelles and RNAs from the mother cell to the daughter cell by class V myosins in budding yeast. (D) Cytokinesis in animal cells by constriction of a contractile ring of actin filaments and myosin-II. (E) Cytokinesis in fission yeast. The contractile ring of actin filaments and myosin-II forms by condensation of nodes. (F) The Listeria bacterium stimulates the assembly of an actin filament comet tail to push it through the cytoplasm of a host animal cell. (G) Locomotion of an animal cell by assembly of actin filaments at the leading edge and retraction of the tail. (A, Redrawn from Ref. 13. B, From Ref. 32. C, Redrawn from Ref. 25. D, Redrawn from T.D. Pollard and W.C. Earnshaw, Cell Biology, W.B. Saunders, 2007. E, From T.D. Pollard. F, Redrawn from Ref. 18. G, Redrawn from Ref. 30.)
Interactions of myosin motor proteins with actin filaments (Fig. 2G) produce two types of movements. First, myosin generates force between actin filaments, producing contractions that pull up the rear of moving cells (Fig. 3G), pinch dividing cells in two (Fig. 3D,E) and change cellular shapes to form tissues. A similar mechanism contracts muscle cells. Second, myosins associated with subcellular organelles and macromolecular complexes of proteins and RNA move these cargos along actin filaments over short distances (Fig. 3C). In budding yeast cells, which are small, actin filament tracks are responsible for distributing nearly all of the organelles and secretory vesicles to daughter cells before cell division. Movement of cargo over longer distances in many other cell types involves microtubules and their motors.
Questions to ask
First, researchers need a complete list of parts to understand how the actin system or any biological system works. Do we know all the parts of the actin system? We know many, but the list is far from complete. Second, which parts interact with other parts? Third, how does this system of connected parts work as a whole? Most biological systems are so complicated that their operations are not intuitively obvious, so mathematical models and simulations are needed to connect hypotheses with experimental observations (9).
Examples of Biological Processes that Depend on Actin
Making connections between molecules and biology can be challenging. On one hand, any cellular process depends on many different proteins. On the other hand, a given molecule will contribute many processes, as shown by doing a PubMed search on any protein mentioned in this article. For example, recent publications provide evidence that cofilin participates in cancer, embryonic development, HIV infection, path finding by nerve cell axons, learning and memory, programmed cell death, Alzheimer’s disease, traffic of intracellular membranes, mitosis, cytokinesis, tight junctions and immune reactions of T-lymphocytes! In each of the following examples, the inventories of participating molecules are more advanced than our understanding of the molecular mechanisms or the operations of these processes at the system level.
Actin filaments as part of the cytoskeleton
The protein polymers forming the cytoskeleton are responsible for the mechanical properties and shapes of cells, which are often critical to their functions. If the membranes of an human cell are dissolved away to release soluble components, a ghost-like meshwork of cytoskeletal polymers remains (Fig. 1B) (10). The polymers include actin filaments, microtubules and intermediate filaments in various proportions and geometries. Actin filaments provide mechanical structure and motility for amoeboid and animal cells. Microtubules are responsible for separating chromosomes and long-range transport of large particles in all eukaryotes. Intermediate filaments in vertebrates function as intracellular ligaments and tendons to resist mechanical forces.
Interactions among the three cytoskeletal polymers reinforce the cytoskeleton, although some crosslinking proteins exchange rapidly and the polymers themselves turn over on time scales of seconds to minutes. These features give the cytoplasm useful properties, such as being stiff when deformed rapidly and malleable when deformed slowly. Even the cells of plants and fungi, despite being encased in a cell wall, use cytoskeletal polymers to direct the shape of their compartments (11). In addition, the cytoskeleton is part of a system that senses external forces applied to the cell along with the mechanical properties of the cell’s environment. This system can influence diverse aspects of cell function including gene expression and differentiation (12).
Actin Patches and Endocytosis
Actin filaments assemble occurs at sites of plasma membrane internalization in budding and fission yeast (13) (14). In these “actin patches” filaments assemble de novo, provide force to form and internalize an endocytic vesicle from the plasma membrane and then disassemble in a process that is self-limited in space and time (Fig. 3A). While powerful molecular and genetic tools have identified ~30 to 50 participating proteins, the parts list still appears to be incomplete. Actin is associated with endocytosis in many cells besides yeast, with an apparently similar set of molecular players (15).
The process of endocytosis begins at multiple independent sites with the spontaneous assembly of membrane proteins along with clathrin and adaptor proteins. Next to be recruited are proteins including WASp family proteins and certain class-I myosins that bind to and/or activate Arp 2/3 complex, which creates new filaments as branches on older filaments (Fig. 2E). The source of the very first filaments remains unclear. Capping protein limits the growth of actin filaments, and filaments are linked together along their sides by fimbrin, among other proteins. While assembly of such a network of filaments alone can create force sufficient to deform a membrane, specialized proteins associate with membrane to induce curvature. The density of actin filaments decreases rapidly as the endocytic vesicle moves into the cytoplasm, a process that depends on the filament severing protein cofilin, perhaps aided by proteins Aip1 and coronin (16). Although actin patches are one of the best characterized actin systems, our understanding of these reactions is limited, and some apparently contradictory observations exist, illustrating just how little we know about the process.
Bacterial comet tails
After invading a eukaryotic cell, some bacteria usurp cellular proteins to assemble a comet tail of actin filaments for propulsion through the cytoplasm (Figs. 1A, 3B,F). Nucleation-promoting proteins on the surface of the bacterium recruit Arp2/3 complex to polymerize actin filaments. Growth of those filaments pushes the bacterium forward. The whole process can be reconstituted with the bacterial nucleation-promoting protein on the surface of a bead or lipid vesicle in a solution with purified actin, profilin, Arp2/3 complex, a capping protein and the severing protein cofilin (17) and simulated with a computer model (18).
Cytokinesis
The physical separation of two daughter cells is the last step in the cell cycle (Fig. 3D,E). Amoebae, fungi and animals use a contractile ring of actin filaments and myosin-II to pinch themselves in two. Myosin-II polymerizes into bipolar filaments, which can produce a contraction by pulling actin filaments together. Multi-cellular animals adapted the contractile ring machinery in specialized cells that evolved into muscle. Organisms on the other branch of the tree (including algae, plants and ciliates) lack myosin-II, so cytokinesis depends primarily on membrane fusion (plants) or on mechanisms that are not understood. Remarkably, prokayotes use a protein related to the microtubule subunit tubulin to assemble a ring of filaments that pinches these cells in two, much like a contractile ring but without the obvious participation of a motor protein (19).
Having started with a common genetic toolbox for cytokinesis, one expects amoebas, fungi and animals to use similar mechanisms for cytokinesis (20). Nevertheless, the general principles are still cloudy for two reasons. First, the process is very complicated, involving the products of >130 genes in fission yeast, a molecular inventory shared at least in part by other less well-characterized organisms. Second, some cells emphasize certain features more than other cells, because certain cytokinesis genes were added or lost from various lineages over the past billion years.
Successful cytokinesis depends on (i) placing the cleavage furrow in the right place between the separated chromosomes, (ii) assembling, constricting and disassembling the contractile ring and (iii) fusing the plasma membrane between the daughter cells. In animal cells the information to place the cleavage furrow comes from the mitotic spindle, part from the poles of the spindle and part from the center of the spindle, where the chromosomes initially congregate. The cleavage site around the equator is marked with active signaling proteins called Rho-GTPases (21). The mechanisms linking these GTPases to the assembly of the contractile ring are still being investigated. In fission yeast negative signals from the ends of the cell and positive signals from the centrally placed nucleus localize contractile ring precursors called nodes to the middle of the cell (20). Nodes accumulate myosin-II along with a formin protein that nucleates the growth of actin filaments. Computer simulations established the feasibility of one hypothesis for the assembly of the ring: myosin molecules capture actin filaments growing randomly from adjacent nodes and pull the nodes together into a ring over 10 minutes (22). Contractile ring assembly is less well understood in other cells (23).
After the mitotic apparatus separates the two daughter nuclei, the contractile ring constricts, pulling the cell membrane into a cleavage furrow. Remarkably, the proteins of the contractile ring disperse as it constricts. Membrane fusion resolves the membranes of the two daughter cells (24).
Actin Cables and Organelle Transport
Many, perhaps all, eukaryotic cells, use myosin motors to transport organelles along actin filaments (Fig. 3C). Budding yeast replicate by directing secretion of cell-wall materials to grow a bud from a particular location on the plasma membrane of a mother cell. At a pre-determined bud site, molecular polarity cues activate formins to nucleate actin filaments. A formin remains associated with each fast-growing barbed end to promote elongation (at 200 subunits per second) and prevent capping. The uniformly polarized filaments form bundles that serve as tracks for the movement of organelles (25). Class-V myosin walks toward the barbed ends of these filaments (Fig. 2G) and moves secretory vesicles and intracellular organelles to the bud (26). Tropomyosin lies along the actin filaments to stabilize the bundles; it may also influence the action of the myosin motor. Myosin-V also moves certain mRNAs on cables into the daughter, to influence cell fate and fitness.
Fission yeast (27) and plant cells (28) also depend on formins to assemble uniformly polarized actin filament cables as tracks for transport of materials for polarized growth. In animal cells and in elongated fungal hyphae, long-range movements depend largely on microtubules, and actin filaments do not appear to be organized into cables of uniform polarity. However, myosins coordinate with microtubule motors to move organelles over short distances along the actin filaments (29).
Cellular motility
Actin filaments are essential for cell locomotion, a defining feature of animal cells (30). For example, cells of the immune system migrate to search and destroy pathogens or cancer cells. During development of animal embryos, some cells move from one location in the body to another, by crawling between neighboring cells and through the extracellular matrix. Cancer cells use similar mechanisms to spread through the body. Nerve cells provide spectacular examples of both cell migration and cell process extension. Neurons destined to control the intestine migrate large distances as neural crest cells during development (31), nerve cells grow processes up to 1 meter long to find their targets and the human brain has 1.5 million km of such cellular processes.
Assembly of actin filaments from their monomeric subunits can suffice to change the shape of the cell and produce a protrusion, which is often the first step in cell locomotion. Arp2/3 complex assembles a dense network of short, branched actin filaments (Figs. 1C, 3B) that grow in successive generations like the twigs of a bush (32). Each filament can produce picoNewton forces (33), allowing the front end of cells to move at rates up to about 1 μm per second (34). Most filaments are capped before growing longer than 0.5 μm; longer filaments would presumably buckle under force. Computer simulations reveal that the components have the capacity for self-organization into networks (35).
A short distance behind the leading edge, the network of branched filaments turns over within a few seconds, replaced by one composed of longer unbranched filaments (34). Formins are logical candidates to help remodel the network in this manner, because formins are known to create long unbranched actin filaments in thin protrusions called filopodia (36). The relative contributions of formins and Arp2/3 complex to motility have been difficult to sort out, in part because Arp2/3 complex is essential for viability and active at very low concentrations (making depletion experiments difficult). Specific pharmacologic inhibitors for Arp2/3 complex only recently became available (37). During cell locomotion, myosin interacts with actin filaments to pull up the rear of the cell, working as structural elements with myosin motor proteins.
Conclusions
The big questions for the actin system are not so much about “what happens”, but rather “how these systems work” at the molecular level. Given the common origin of the genes for the actin system, evolution should be able to help sort out the complicated mechanisms. Continuing focus on tractable model systems should help to establish the molecular basis for each actin-based function and general principles that apply more broadly. Research with systems-level genomics approaches based on genetic and physical interactions in model organisms continues to grow the parts list and to reveal new interactions. The missing rate and equilibrium constants required to trace pathways and explain how molecules work together still need to be defined. The regulation of actin by signaling mechanisms and the interaction of actin with other cellular systems, such as membranes requires further attention.
Technical advances should prove critical. For example, we can now image the behavior of single molecules over time, in vitro and in cells, and we can reconstitute complex processes using mixtures of purified components. Furthermore, advances in light and electron microscopy allow for nanometer-level localization of protein components and for measurement of global and local concentrations of molecules inside living cells. These advances should prove critical to advance our understanding of these actin systems, especially how and where filaments are created and assembled into networks of varying geometry.
The field has only recently started to create mathematical models, at the microscopic, mesoscopic and macroscopic scales. Still, great progress has been made in several areas, such as understanding how the actin filaments in a protrusion assemble and create force on the plasma membrane (9)(18)(35). Quantitative measurements in live cells aided by genetics, specific drugs and depletion strategies should provide the data required to test hypotheses embodied in mathematical models.
Acknowledgments
This work was supported by NIH research grants GM-026132, GM026338, GM066311, GM38542 and GM47337.
References and Notes
- 1.Erickson HP. Bioessays. 2007;29:668. doi: 10.1002/bies.20601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richards TA, Cavalier-Smith T. Nature. 2005;436:1113. doi: 10.1038/nature03949. [DOI] [PubMed] [Google Scholar]
- 3.Geeves MA, Holmes KC. Adv Protein Chem. 2005;71:161. doi: 10.1016/S0065-3233(04)71005-0. [DOI] [PubMed] [Google Scholar]
- 4.Hatano S, Oosawa F. Biochim. Biophys. Acta. 1966;127:488. doi: 10.1016/0304-4165(66)90402-8. [DOI] [PubMed] [Google Scholar]
- 5.Adelman MR, Taylor EW. Biochemistry. 1969 Dec;8:4964. doi: 10.1021/bi00840a046. [DOI] [PubMed] [Google Scholar]
- 6.Pollard TD. Annu Rev Biophys Biomol Struct. 2007;36:451. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- 7.Morrison HG, et al. Science. 2007;317:1875. [Google Scholar]
- 8.Lang D, D. ZA, Rensing SA, Reski R. Trends Plant Sci. 2008;10:542. doi: 10.1016/j.tplants.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Mogilner A, Wollman R, Marshall WF. Dev Cell. 2006 Sep;11:279. doi: 10.1016/j.devcel.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 10.Heuser JE, Kirschner MW. J. Cell Biol. 1980;86:212. doi: 10.1083/jcb.86.1.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hussey PJ, Ketelaar T, Deeks MJ. Annu Rev Plant Biol. 2006;57:109. doi: 10.1146/annurev.arplant.57.032905.105206. [DOI] [PubMed] [Google Scholar]
- 12.Discher DE, Mooney DJ, Zandstra PW. Science. 2009;324:1673. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galletta BJ, Cooper JA. Curr Opin Cell Biol. 2009;21:20. doi: 10.1016/j.ceb.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaksonen M, Toret CP, Drubin DG. Nat Rev Mol Cell Biol. 2006 Jun;7:404. doi: 10.1038/nrm1940. [DOI] [PubMed] [Google Scholar]
- 15.Doherty GJ, McMahon HT. Annu Rev Biochem. 2009;78:857. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 16.Brieher WM, Kueh HY, Ballif BA, Mitchison TJ. J Cell Biol. 2006;175:315. doi: 10.1083/jcb.200603149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. Nature. 1999;401:613. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- 18.Alberts JB, Odell GM. PLoS Biol. 2004 Dec;2:e412. doi: 10.1371/journal.pbio.0020412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osawa M, Anderson DE, Erickson HP. Science. 2008;320:792. doi: 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oliferenko S, Chew TG, Balasubramanian MK. Genes Dev. 2009;23:660. doi: 10.1101/gad.1772009. [DOI] [PubMed] [Google Scholar]
- 21.Miller AL, Bement WM. Nat Cell Biol. 2009;11:71. doi: 10.1038/ncb1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vavylonis D, Wu JQ, Hao S, O’Shaughnessy B, Pollard TD. Science. 2008 Jan 4;319:97. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- 23.Zhou M, Wang YL. Mol Biol Cell. 2008;19:318. doi: 10.1091/mbc.E07-08-0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montagnac G, Echard A, Chavrier P. Curr Opin Cell Biol. 2008;20:454. doi: 10.1016/j.ceb.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 25.Pruyne D, Legesse-Miller A, Gao L, Dong Y, Bretscher A. Annu Rev Cell Dev Biol. 2004;20:559. doi: 10.1146/annurev.cellbio.20.010403.103108. [DOI] [PubMed] [Google Scholar]
- 26.Fagarasanu A, Rachubinski RA. Curr Opin Microbiol. 2007;10:528. doi: 10.1016/j.mib.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Martin SG, Chang F. Curr Biol. 2006 Jun 20;16:1161. doi: 10.1016/j.cub.2006.04.040. [DOI] [PubMed] [Google Scholar]
- 28.Vidali L, et al. Proc Natl Acad Sci U S A. 2009;106:13341. doi: 10.1073/pnas.0901170106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levi V, Serpinskaya AS, Gratton E, Gelfand V. Biophys J. 2006;90:318. doi: 10.1529/biophysj.105.067843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ridley AJ, et al. Science. 2003;302:1704. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 31.Anderson RB, Newgreen DF, Young HM. Adv Exp Med Biol. 2006;589:181. doi: 10.1007/978-0-387-46954-6_11. [DOI] [PubMed] [Google Scholar]
- 32.Pollard TD, Borisy GG. Cell. 2003 Feb 21;112:453. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 33.Kovar DR, Pollard TD. Proc Natl Acad Sci U S A. 2004 Oct 12;101:14725. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Svitkina TM, Verkhovsky AB, McQuade KM, Borisy GG. J. Cell Biol. 1997;139:397. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaus TE, Taylor EW, Borisy GG. Proc Natl Acad Sci U S A. 2007 Apr 24;104:7086. doi: 10.1073/pnas.0701943104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang C, et al. PLoS Biol. 2007;5:e317. doi: 10.1371/journal.pbio.0050317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nolen BJ, et al. Nature. 2009;460:1031. doi: 10.1038/nature08231. [DOI] [PMC free article] [PubMed] [Google Scholar]