Abstract
The first successful rabbit SCNT was achieved more than one decade ago, yet rabbits remain one of the most difficult species to clone. The present study was designed to evaluate the effects of two histone deacetylase inhibitors (HDACi), namely trichostatin A (TSA) and scriptaid (SCP), on cloning efficiency in rabbits. The in vitro development, acetylation levels of histone H4 lysine 5 (H4K5ac), and Oct-4 protein expression patterns of cloned embryos were systemically examined after various HDACi treatments. Supplementation of TSA (50 nM) or SCP (250 nM) in the culture medium for 6 h improved blastocyst development rates of cloned embryos compared to treatment without HDACi. The combined treatment with TSA (50 nM) and SCP (250 nM) further enhanced morula (58.6%) and blastocyst (49.4%) rates in vitro. More importantly, compared to single HDACi treatments, embryos with the combined treatment had a higher level of H4K5ac and an increased total cell number (203.7±14.4 vs 158.9±9.0 or 162.1±8.2, P<0.05) with better Oct-4 expression pattern in hatching blastocysts, indicating substantially improved embryo quality. This was apparently the first report regarding Oct-4 expression in cloned rabbit embryos. We inferred that the majority of cloned rabbit embryos had an aberrant ICM structure accompanied with abnormal spatial distribution of Oct-4 signals. This study demonstrated a synergistic effect of TSA and SCP treatments on cloned rabbit embryos, which may be useful to improve cloning efficiency in rabbits.
Keywords: Rabbit, SCNT, HDACi, TSA, SCP
1. Introduction
Somatic cell nuclear transfer (SCNT) technology has been a useful tool for reprogramming terminally differentiated cells in agriculture, as well as in research in biomedicine and regenerative biology. Using this technology, cloned animals have been born in many species; however, the success rates remain low. Improper gene expression patterns, likely from the inheritance of epigenetic marks of differentiated donor nuclei, can lead to developmental arrest or abnormalities [1–6]. Recently, a key enzyme, oxidase TET3 (tet methylcytosine dioxygenase 3) in oocytes, was reported to promote active demethylation of the paternal genome through a base excision repair pathway [7–10]. In addition, TET3–mediated DNA demethylation was also reported in cloned embryos [9,10]. However, it is noted that the somatic genome is much different from the sperm, in which high acetylation levels and lack of normal repressive marks on histone tails are observed after protamine-histone exchange [11]. This discrepancy likely resulted in inefficient demethylation in SCNT embryos and produced a methylation pattern more similar to the donor cell rather than a zygote [12]. Treatments of chromatin modifiers to moderate epigenetic settings are thus suggested to improve SCNT efficiency [13]. Among these approaches, the use of a histone deacetylation inhibitor (HDACi) to alter histone acetylation patterns greatly improved nuclear reprogramming, gene expression, blastocyst quality and full-term development in various species [14–19].
Applying TSA, an HDACi, on cloned rabbit embryos has advantageous effects on histone modification patterns, cell number, and embryo development during preimplantation stages in rabbits [20,21]. Notably, however, Meng et al. reported that TSA was not helpful with regards to term development and may have caused neonatal death of cloned rabbits [22].
The effects of HDACi agents other than TSA on rabbit cloning have apparently not been reported. For example, SCP, a novel histone deacetylase inhibitor, enhanced transcriptional activity and protein expression in mammalian cells [23]. In SCNT, SCP treatment is of low toxicity for embryo development and improved cloning efficiency by modifying histone acetylation patterns and alleviating aberrant gene expression, as demonstrated in mice, pigs and cattle [18,19,24]. However, there are apparently no reports regarding the use of SCP in culture of cloned rabbit embryos.
Oct-4 is the most important transcription factor required for pluripotent lineage formation and germ cell development in mice [25,26]. Inefficient reprogramming by SCNT was associated with abnormal Oct-4 reactivation [27]. Moreover, the acetylation level of H4K5 was an indicator of global gene activation. Previously, we documented Oct-4 expression patterns and level of H4K5ac in normally fertilized embryos [28].
The present study aimed to assess effects of a combination of TSA and SCP on in vitro developmental competence of cloned rabbit embryos. We first examined the effects of TSA or SCP and determined the optimal concentration of the individual HDACi. Development of cloned embryos treated with a single HDACi and the combined treatment were compared, and total cell numbers, levels of H4K5ac, and Oct-4 expression patterns of hatching blastocysts were compared to those in normal embryos.
2. Materials and methods
All chemicals were purchased from Sigma Chemical Co. (St. Louis, MO, USA) unless otherwise indicated. The media were prepared as described [29]. Briefly, Dulbecco’s phosphate-buffered saline (D-PBS; 15240-013, Gibco, Grand Island, NY, USA) containing 0.1% polyvinyl alcohol (PVA) was used for flushing oocytes from oviducts (PBS-PVA) and collecting cumulus-oocyte complexes (COCs) from ovarian follicles. Tissue and cell cultures were maintained in Dulbecco’s Minimum Eagle’s medium (DMEM; 11995-065, Gibco). Medium 199 (M199), which contains Earle’s salts, L-glutamine, 2.2 g/L sodium bicarbonate, and 25 mM HEPES (12340-014, Gibco), was supplemented with 10% fetal bovine serum (FBS; SH0070.03, Hyclone, Logan, UT, USA) to serve as the standard manipulation medium. Rabbit oocytes and embryos were cultured in B2 medium (Laboratories CCD, Paris, France) containing 2.5% FBS at 38.5 °C in 5% CO2 and humidified air.
2.1. Preparation of donor animals
Sexually mature (6- to 18-mo old) New Zealand White (NZW) or hybrid strain (NZW female mating with Rex male) female rabbits (oocyte donors) were superovulated as described [29]. Briefly, the present study applied two 3-mg, two 4-mg, and two 5-mg im injections of Folltropin-V (FSH; Bioniche Animal Health Canada, Belleville, ON, Canada) at 12-h intervals. An intravenous injection (200 IU) of human chorionic gonadotropin (hCG; Chorulon, animal use, Intervet Inc, Millsboro, DE, USA) was then used to induce ovulation.
The animal maintenance, care, and use procedures were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of National Taiwan University. The surgical procedures in this study were performed in accordance with the Guiding Principles for the Care and Use of Laboratory Animals [30].
2.2. Collection of follicular oocytes
The reproductive tracts including ovaries were excised from euthanized donor rabbits by midventral laparotomy for oocyte collection. Cumulus–oocyte complexes (COCs) were either collected from top of the ovulation site, the ovarian Graafian follicles, or flushed from the oviducts at 10–12 h post hCG injection (hpi). Rabbit COCs were treated with 0.5 mg/mL hyaluronidase in PBS-PVA for 1 min, and cumulus cells were completely removed from the oocytes by careful pipetting with a fine bore glass pipette. The mature oocytes with first polar body examined under a stereomicroscope were selected for nuclear recipients.
2.3. Preparation of donor cells from rabbit tissues
The skin notches were taken from the ear of an adult NZW rabbit. Cell culture was performed as described [29]. Briefly, a skin sample with a diameter of 0.5 cm was cut into four or five pieces, washed, placed in a Falcon cell culture dish (Falcon 3001, Becton Dickinson Labware, Oxnard, CA, USA) containing 10% FBS DMEM, and cultured at 37 °C in 5% CO2 humidified air. Fibroblast monolayers formed around the tissue explants in 5 to 7 d. Fibroblasts were cultured until confluence was reached. During passaging, cultured cells were thoroughly washed with Dulbecco’s PBS and then gently digested with 0.05% trypsin-EDTA for 3 min at 37 °C. Trypsinization was terminated by adding 10% FBS in DMEM and washing. The cell suspension was then centrifuged at 450 xg for 5 min, and cells were seeded into new dishes, where they were maintained for an additional 3 to 5 d. Fibroblasts at passages 8 to 10 were used for NT after being serum starved in 0.5% FBS DMEM for 7 to 9 d. Prior to NT, cells were disassociated by trypsinization, re-suspended in 0.5% FBS in DMEM, and allowed to recover for approximately 1 h at 37 °C.
2.4. Nuclear transfer, activation, and embryo culture
The micromanipulations were carried out using a standard procedure [29]. The steps of enucleation, donor cell transfer, and electrical membrane fusion were completed within 2 to 3 h. For enucleation, an incision was made in the zona pellucida using a sharp-tipped enucleation needle with the closed end, and the polar body with an appropriate amount of surrounding cytoplasm was extruded by pressing oocytes with the enucleation needle. Successful enucleation was confirmed by ultraviolet-light exposure of the presumed karyoplasts under fluorescent microscope. A donor cell with a diameter of 18 to 20 μm was selected and transferred into the perivitelline space of an enucleated rabbit oocyte. Donor cell-cytoplasm pairs were pre-incubated in 0.3 M mannitol supplemented with 0.1 mM CaCl2 and 0.1 mM MgCl2 for 2–3 min and then transferred into an electrical chamber that had wires at a 1 mm distance and contained the same fusion medium. Cell fusion was induced by applying three direct current (DC) pulses of 3.2 kV/cm with a 20 μs -interval using a BTX 2001 Electro Cell Manipulator (Biotechnologies & Experimental Research Inc., San Diego, CA, USA). Following the DC pulses, oocytes were incubated for 60–80 min at 38.5 °C.
The fused cloned embryos were activated by the same electrical DC setting for cell fusion and subsequently incubation in M199 containing 10% FBS, 2.0 mM 6-dimethylaminopurine (6-DMAP) and 5 μg/mL cycloheximide (CHX) for 1 h. For the control groups, the flushed oocytes were parthenogenetically activated by the same activation regime at 16–18 hpi without SCNT. In addition, superovulated rabbits were mated with fertile NZW males and served as embryo donors. At 18 hpi, the reproductive tracts of embryo donors were flushed to harvest zygotes for in vitro culture. For each batch of experiments, five NZW female rabbits used as oocyte donors, and two more rabbits served as embryo donors, according to the experimental design.
Cloned, parthenogenetically activated (PAR) and fertilized (IVC) embryos were cultured in the B2 medium with 2.5% FBS to investigate their in vitro developmental potential. Cleavage rates were recorded 18 h post culture, and 2- to 4-celled embryos were cultured in the same B2 medium (Laboratories CCD, Paris, France) for an additional 4 d to monitor blastocyst development at 38.5 °C in 5% CO2 humidified air.
2.5. Treatment of TSA and SCP
The TSA and SCP were dissolved in DMSO and prepared as stock solutions and stored at –20 °C. According to the experimental design, various concentrations of TSA (0, 25, 50 and 100 nM) or/and SCP (0, 50, 250 and 500 nM) in 2.5% FBS B2 medium were used to treat activated cloned rabbit embryos for 6 h at 38.5 °C in 5% CO2 and humidified air. The treated NT embryos were then washed thoroughly and cultured in 2.5% FBS B2 medium for subsequent development.
2.6. Immunocytochemistry and image processing of cloned embryos
Hatching blastocysts (hBLs) from each group were fixed with fresh 4% paraformaldehyde in DPBS for 10 min and stored in DPBS at 4 °C until ready for processing [28]. Briefly, after washing in DPBS for 10 min, embryos were permeabilized by 0.5% Triton-X 100 for approximately 30 min, and then washed in 0.25% DPBS/Tween 20 (PBST) for 30 min at room temperature. Then, DPBS supplemented with 2% bovine serum albumin for 1 h at room temperature was used to block nonspecific binding sites. Immunostaining was performed by incubation with the primary monoclonal antibody against Oct-4 (1:150; MAB4401; Millipore, Billerica, MA, USA) at 4 °C overnight. Samples were then washed in PBST three times for 15 min at room temperature and incubated with secondary antibody Alexa Fluor 488 (A11029, Invitrogen, Carlsbad, CA, USA, 1:500) for 1 h at 37 °C. For H4K5ac staining, the embryos were further incubated primary monoclonal antibody against H4K5ac (monoclone, ab51997, Abcam, Cambridge Science Park, Cambridge, UK, 1:250) for 1.5 h at room temperature and then treated with secondary antibody Alexa Fluor 594 (A21207, Invitrogen; 1:500) for 1 h at 37 °C. After washing in PBST for 30 min at room temperature, embryos were subsequently incubated with DAPI (100 ng/mL) for 10 min and mounted in DPBS plus 50 % glycerol, and observed with a laser scanning confocal microscopy (Olympus IX71 with UltraVIEW Confocal Software, PerkinElmer Inc, Covina, CA, USA). Volocity (Version 5.3.1, Improvision) and ImageJ (version 1.45b, National Institutes of Health) were performed for intensity analysis of these images. For H4K5ac intensity of the whole embryo, image planes of each individual embryo were merged by Volocity without any modification on fluorescent intensity. Images were first converted to 8-bit gray scale and the background value was eliminated by the background subtract function. The nuclear H4K5ac intensities of integrated fluorescent were measured by manually outlining all nuclei of embryos at hBL stage using ImageJ and then quantized by the ratios of H4K5ac to DAPI (DNA) signals as reported [31].
2.7. Categorization of rabbit blastocysts and its Oct-4 expression patterns
Based on the observation of ICM morphology and the spatial expression pattern of Oct-4 signals in rabbit blastocysts, hBLs were categorized into four groups (Table 1). Class I embryos were presumably of best quality, i.e. with obvious ICM structures, intensive signals in the ICM cells, and diminished signals in the TE cells. Class II embryos were of second best quality, with obvious ICM structures and intensive Oct-4 signals in the ICM cells; however, these embryos had high Oct-4 signals in the TE cells, indicating aberrant Oct-4 expression in the TE cells. Class III embryos displayed only indistinct ICM structure accompanied with a scattered Oct-4 signal, indicating aberrant Oct-4 expressions in both ICM and TE cells. Class IV embryos were considered of worst quality, with unobservable ICM structure and barely detectable Oct-4 signals throughout the embryos, indicating failure of correct linage formation and reactivation of Oct-4. Of note, we did not observe other classes (i.e. other combination of ICM structure, Oct-4 in ICM, and Oct-4 in TE) of embryos in the present study; therefore we did not include such other possible categories in Table 1.
Table 1.
Categorization of rabbit blastocysts based on their ICM structure and Oct-4 expression patterns in ICM and TE cells.
| Category | ICM structure | Oct-4 signal in ICM cells | Oct-4 signal in TE cells |
|---|---|---|---|
| Class I | Obvious | High | Low |
| Class II | Obvious | High | High |
| Class III | Indistinct | Scattered | Scattered |
| Class IV | Not observed | Barely detectable Oct-4 signals throughout the embryo | |
2.8. Experimental designs
Experiment 1: Comparison of preimplantation development of cloned rabbit embryos treated with various TSA concentrations
In Experiment 1, cloned embryos were treated with TSA at 0, 25, 50, or 100 nM for 6 h and subsequently cultured for 5 d to assess cleavage (18 h), morulae (72 h) and blastocyst (120 h) development. Developmental rates of cloned embryos were compared with that of PAR embryos obtained by the same activation treatment of matured oocytes.
Experiment 2: Comparison of the preimplantation development of cloned rabbit embryos treated with various SCP concentrations
In Experiment 2, cloned embryos were treated with SCP at 0, 50, 250, or 500 nM for 6 h and subsequently cultured for 5 d to examine cleavage, morulae and blastocyst development. Developmental rates of cloned embryos were compared with that of PAR embryos obtained by the same activation treatment of matured oocytes.
Experiment 3: Comparison of the preimplantation development of cloned rabbit embryos treated with TSA and SCP
Cloned embryos were treated with 50 nM TSA (NTTSA), 250 nM SCP (NTSCP) or the combination of 50 nM TSA and 250 nM SCP (NTTSASCP) for 6 h. Rates of cleavage, morulae and blastocyst development were compared to those of IVC, or PAR embryos obtained by the same culture condition, or cloned without HDACi treatment (NT). On Day 5 of culture (5 dpc), the hBL stage embryos were collected from various groups for evaluating the total cell number, acetylation levels of H4K5 and Oct-4 expression pattern by immunostaining.
2.9. Statistical analysis
Rates of in vitro embryo development were analyzed by General Linear Model in SPSS (Version 11.0, IBM Corporation, Chicago, IL, USA). Differences of H4K5ac intensity in hBLs by different HDACi treatments were subjected to one-way ANOVA in SPSS. The data of Oct-4 characterization were pooled from at least three replicates and analyzed by a Chi square test in Prism (Version 5.0C, GraphPad Software, Inc., La Jolla, CA, USA). For all analyses, P < 0.05 was considered significant.
3. Results
3.1. In vitro development of cloned embryos treated with various TSA concentrations
Various concentrations of TSA (0, 25, 50 and 100 nM) were used to treat cloned embryos. Rates of cleavage (75.0 – 86.1%) and morula formation (26.8 – 46.3%) were not different between TSA treatments (Table 2). However, there were higher rates of cleavage (89.3%), morula (89.3%) and blastocyst (85.7%) formation in the PAR group compared to those in SCNT groups, regardless of the TSA concentrations (P < 0.05). Blastocyst development of cloned embryos was highest with the supplementation of 50 nM TSA (31.7%), which was higher than the untreated group (0 nM TSA, 12.2%; P < 0.05).
Table 2.
The effects of various concentrations of TSA on the in vitro development of cloned rabbit embryos.
| Treatments | No. embryos cultured (replicates) | No. (%) cleavage | No. (%)morula | No. (%)Blastocyst |
|---|---|---|---|---|
| NT | 41 (3) | 32 (78.0)a | 11 (26.8)a | 5 (12.2)a |
| NTTSA25 | 36 (3) | 31 (86.1)a | 11 (30.6)a | 7 (19.4)ab |
| NTTSA50 | 41 (3) | 32 (78.0)a | 19 (46.3)a | 13 (31.7)b |
| NTTSA100 | 36 (3) | 27 (75.0)a | 13 (36.1)a | 9 (25.0)ab |
| PAR | 56 (3) | 50 (89.3)b | 50 (89.3)b | 48 (85.7)c |
Within a column, values without a common superscript differed (P<0.05).
Each datum was derived from three independent replicates. Oocytes were collected from superovulated ova donors at 10–12 h post human choriogonadotropin (hCG) injection (hpi) and used for SCNT. Cultured fibroblasts were used as nuclear donors. Fused NT embryos were activated by electric pulses and a chemical combination of 6-DMAP and CHX for 1 h. Cloned embryos were then treated without HDACi agents (NT) or treated with different concentrations of TSA (i.e. 25, 50, or 100 nM) for 6 h and arranged for development in vitro. The rates of cleavage, morula and blastocyst were evaluated and compared with that of parthenogenetically activated (PAR) embryos. The percentage of embryonic development was calculated based upon the number of cultured embryos.
3.2. In vitro development of cloned embryos treated with various SCP concentrations
Cloned embryos were treated with various concentrations of SCP (0, 50, 250 and 500 nM) and examined for their developmental potential (Table 3). Cleavage rates were not different between SCP treatments (68.0 – 87.5%). There were higher rates (P < 0.05). of cleavage (92.4%), morula (87.9%) and blastocyst (81.8%) formation in the PAR group than those in SCNT groups treated with 0, 50 or 500 nM of SCP. Notably, morula (67.4%) and blastocyst (40.7%) development appeared highest when 250 nM SCP was applied, and the blastocyst rate was higher than that in the untreated group (0 nM SCP, 14.0%; P < 0.05).
Table 3.
The effects of various concentrations of SCP on in vitro development of cloned rabbit embryos.
| Treatments | No. embryos cultured (replicates) | No. (%)cleavage | No. (%) morula | No. (%) Blastocyst |
|---|---|---|---|---|
| NT | 50 (3) | 34 (68.0)a | 13 (26.0)a | 7 (14.0)a |
| NTSCP50 | 32 (3) | 27 (84.4)a | 10 (31.3)a | 3 (9.4)a |
| NTSCP250 | 86 (3) | 70 (81.4)a | 58 (67.4)bc | 35 (40.7)b |
| NTSCP500 | 32 (3) | 28 (87.5)a | 13 (40.6)ab | 7 (21.9)ab |
| PAR | 66 (3) | 61 (92.4)b | 58 (87.9)c | 54 (81.8)c |
Within a column, values without a common superscript differed (P<0.05).
Each datum was derived from three independent replicates. Oocytes were collected from superovulated ova donors at 10–12 h post human choriogonadotropin (hCG) injection (hpi) and used for SCNT. Cultured fibroblasts were used as nuclear donors. Fused NT embryos were activated by electric pulses and a chemical combination of 6-DMAP and CHX for 1 h. Cloned embryos were then treated without HDACi agents (NT) or treated with different concentrations of SCP (i.e. 50, 250, or 500 nM) for 6 h and arranged for development in vitro. The rates of cleavage, morula and blastocyst were evaluated and compared with that of parthenogenetically activated (PAR) embryos. The percentage of embryonic development was calculated based upon the number of cultured embryos.
3.3. In vitro development of cloned embryos treated with TSA and SCP
To examine whether the combination of histone deacetylation inhibitors could further improve cloning efficiency, cloned embryos were treated with various HDACi treatments (i.e. NT, NTTSA, NTSCP, and NTTSASCP treatments), and compared to PAR and IVC embryos.
The morula (28.4%) and blastocyst (14.9%) rates were lowest in the NT group (Table 4). In addition, embryos in the NT group had small blastocoels (Fig. 1, A) and reduced blastocyst cell numbers (147.7±7.3). Treatment of TSA (50 nM) or SCP (250 nM) alone increased rates of morula (44.1.4–50.9%) and blastocyst (33.3–33.8%) formation compared to the NT group (P < 0.05), consistent with results in Experiments 1 and 2. It was noteworthy that single HDACi treatment had no obvious beneficial effects on total cell number (Fig. 1, B and C; Table 4). In the NTTSASCP group, the morula (58.6%) and blastocyst (49.4%) rates were further improved. Moreover, embryos in this group had a high number of total cells (203.7±14.4; Fig 1, D) in the hBLs, which was similar to that of PAR or IVC embryos (P>0.05), but much higher than the NT group (P<0.05; Table 4).
Table 4.
In vitro development of rabbit cloned embryos treated with HDACi agents
| Treatments | No. embryos cultured (replicates) | No. (%) cleavage | No. (%) morula | No. (%) blastocyst | Mean no.± SEM of cells in blastocysts |
|---|---|---|---|---|---|
| NT | 67 (3) | 49 (73.1)a | 19 (28.4)a | 10 (14.9)a | 147.7±7.3 |
| NTTSA | 57 (3) | 44 (77.2)a | 29 (50.9)b | 19 (33.3)b | 162.1±8.2 |
| NTSCP | 68 (3) | 48 (70.6)a | 27 (44.1)a | 23 (33.8)b | 158.9±9.0 |
| NTTSASCP | 87 (3) | 67 (77.0)a | 51 (58.6)b | 43 (49.4)b | 203.7±14.4 |
| PAR | 69 (3) | 62 (89.9)b | 60 (87.0)c | 55 (79.7)c | 206.3±12.9 |
| IVC | 84 (3) | 82 (97.6)b | 81 (96.4)c | 78 (92.9)c | 214.5±18.2 |
Within a column, values without a common superscript differed (P<0.05).
Each datum was derived from three independent replicates. Oocytes were collected from superovulated ova donors at 10–12 h post human choriogonadotropin (hCG) injection (hpi) and used for SCNT. Cultured fibroblasts were used as nuclear donors. Fused NT embryos were activated by electric pulses and a chemical combination of 6-DMAP and CHX for 1 h. Cloned embryos were then treated without HDACi agents (NT) or treated with 50 nM TSA (NTTSA), 250 nM SCP (NTSCP) or a combination of 50 nM TSA and 250 nM SCP (NTTSASCP) for 6 h and arranged for development in vitro. The rates of cleavage, morula and blastocyst were evaluated and compared with that of parthenogenetically activated (PAR) and fertilized (IVC) embryos. The percentage of embryonic development was calculated based upon the number of cultured embryos.
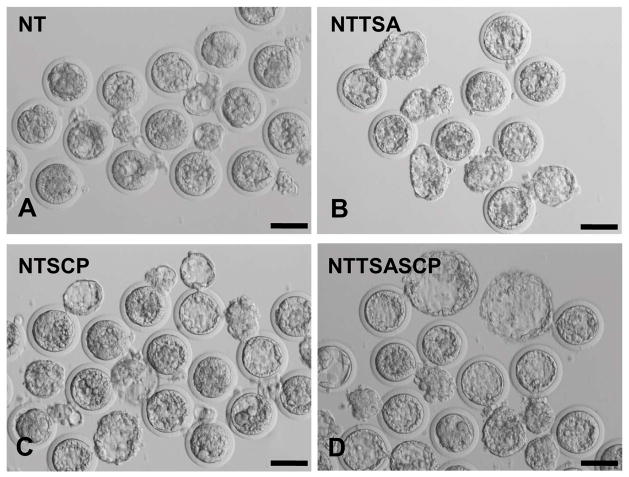
Fig 1. Blastocyst development of rabbit cloned embryos treated with different HDACi agents.
Reconstructed embryos treated without HDACi agents or with 50 nM TSA, 250 nM SCP, or a combination of 50 nM TSA and 250 nM SCP were cultured in vitro. Cloned embryos reached the hatching blastocyst stage on 5 dpc. The blastocyst morphology of each group was shown. There were smaller blastocysts in the NT (A), NTTSA (B) and NTSCP (C) groups, but more expanded hatching blastocysts in the NTTSASCP (D) group. Scale bar = 100 μm.
3.4. The effects of HDACi treatments on acetylation level of H4K5
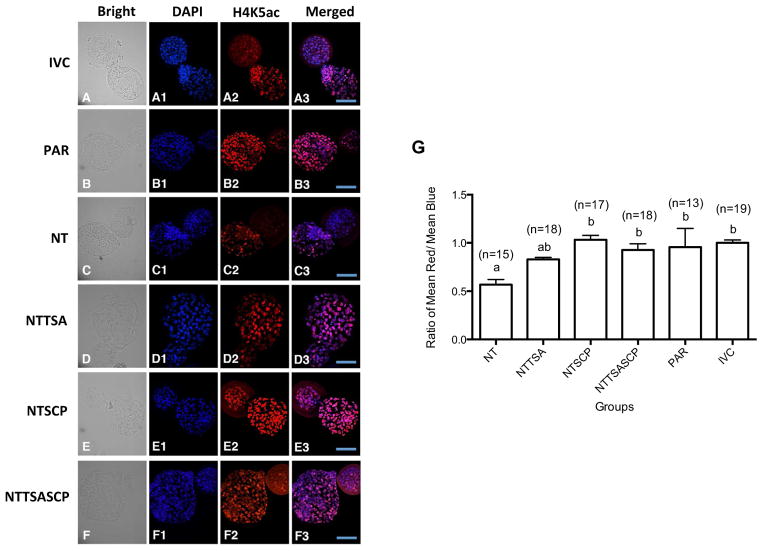
Alevels of histone H4K5 in rabbit D5 hBLs were examined by immunostaining. The intensity of H4K5ac (Fig. 2) for each hBL was scored as a mean ratio of H4K5ac (red stain) to DAPI counterstain of chromatin (blue stain). The H4K5ac score of untreated cloned embryos appeared lower than that in the PAR or IVC groups (P < 0.05). Treatment of TSA appeared to only slightly enhance acetylation level of H4K5 in cloned embryos (P > 0.05). However, in the NTSCP or NTTSASCP groups, the H4K5ac score was higher than that in untreated or TSA groups (P < 0.05) and similar to that in the PAR or IVC groups.
Fig 2. The effects of HDACi treatments on histone H4K5 acetylation at hatching blastocyst stage.
By immunostaining, hatching blastocysts derived from NT (A), NTTSA (B), NTSCP (C), or NTTSASCP (D) group were collected for examination of H4K5ac intensity and compared to PAR (E) or IVC (F) embryos. Blue and red signals represent DAPI counterstain and acetylation of H4K5 stain, respectively. ImageJ software was used for analysis and the H4K5ac intensity scores were determined by the ratios of H4K5ac to DAPI DNA signals (G). Scale bar=100 μm. a,bValues without a common superscript differed (P < 0.05).
3.5. Lineage formation and Oct-4 expression patterns in cloned hBLs
We examined Oct-4 expression in various cell lineages (i.e. ICM and TE) of the hBLs of different HDACi treatment groups (i.e. NT, NTTSA, NTSCP, and NTTSASCP treatments), and compared with that in the PAR or IVC embryos. The embryos were categorized as Class I (Fig. 3, A), Class II (Fig. 3, B), Class III (Fig. 3, C) and Class IV (Fig. 3, D).
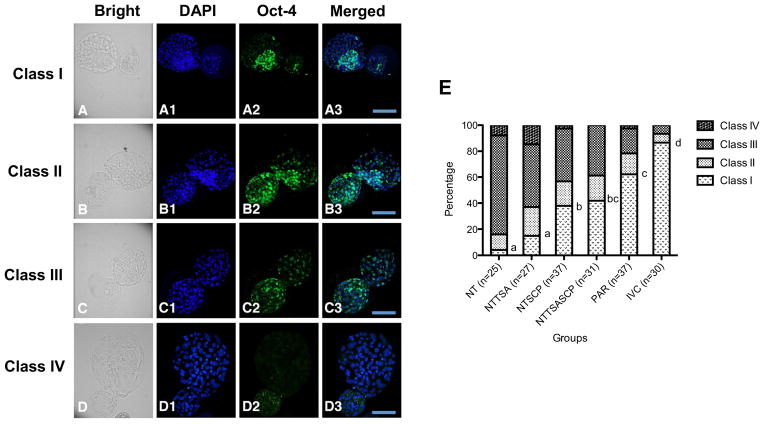
Fig 3. Oct-4 expression patterns in rabbit embryos treated with HDACi agents.
Oct-4 expression patterns in the cloned embryos were categorized as (A) Class I: evident ICM cells observed in the bright field or DAPI staining were accompanied with a more intensive Oct-4 signal as compared to that in the TE cells; (B) Class II: evident ICM cells were observed in the blastocysts but with similar Oct-4 intensity between ICM and TE cells; (C) Class III: ICM structure was indistinct and scattered Oct-4 signals were observed in the blastocysts; (D) Class IV: ICM structure was not identified and Oct-4 signal was weak or absent in the blastocysts. (G) The ratios of Oct-4 expression patterns in cloned embryos treated with different HDACi treatments were compared to those of PAR or IVC embryos. In the NT group, one out of 25 hBLs (4%) was classified as Class I, 12% (3/25) were classified as Class II, 76% (19/25) were classified as Class III, and 8% (2/25) were Class IV. In the NTTSA group, 14.8% (4/27) of hBLs were classified as Class I, 22.2% (6/27) were classified as Class II, 48.1% (13/27) were classified as Class III, and 14.8% (4/27) were classified as Class IV. In the NTSCP group, 37.8% (14/37) of hBLs were classified as Class I, 18.9% (7/37) were classified as Class II, 40.5% (15/37) were classified as Class III, and only one embryo was classified as Class IV (2.7%). In the NTTSASCP group, 41.9% (13/31) of hBLs were classified as Class I, 19.4% (6/31) were classified as Class II, and 38.7% (12/31) were classified as Class III. In the PAR group, 62.2% (23/37) of hBLs were classified as Class I, 16.2% (6/37) were classified as Class II, 18.9% (7/37) were classified as Class III, and one embryo was classified as Class IV (2.7%). In the IVC group, 86.7% (26/30) of hBLs were classified as Class I, 6.7% (2/30) were classified as Class II, and 6.7% (2/30) were classified as Class III. Scale bar=100 μm. a–dValues without a common letter differed (P < 0.05).
Previously, we documented the expression pattern of Oct-4 in hBLs of normally IVC embryos. Consistent with the previous observation in IVC embryos, all of the hBLs expressed Oct-4, and 86.7% (26 of 30 IVC embryos) were categorized as Class I (highest quality). Similarly, Oct-4 was expressed in all PAR hBLs, but the ratio of Class I was decreased to 62.2% (23/37), as shown (Fig. 3, E).
In contrast to IVC and PAR embryos, only 4% of hBLs (1/25) were categorized as Class I in the NT group. In most blastocysts, ICM structure was indistinct or not observed (84%, 21/25). When cloned embryos were treated with TSA alone, the percentage of Class I hBLs increased to 14.8% (4/27). Importantly, increased percentage of Class I was observed in the NTSCP treatment group (37.8%, 14/37) (P < 0.05). The combinational treatment of HDACi further increased the percentage of Class I embryos. In the NTTSASCP treatment group, we detected the Oct-4 signal in all cloned embryos and the percentage of Class I was as high as 41.9% (13/31).
4. Discussion
Successful animal cloning requires resetting of epigenetic modifications, such as erasure of epigenetic marks of a differentiated somatic cell, alteration of chromatin configuration, and activation of specific gene expression program to direct embryo development. The histone acetylation level of the somatic chromatin has been shown to rapidly decrease upon introduction into a metaphase recipient oocyte and then re-acetylate during formation of the pseudo-pronuclei. Although these dynamic changes of histone acetylation are similar to that of a fertilized embryo, the global level of acetylation in cloned embryos is generally lower than in normal embryos [32]. Incomplete reprogramming, e.g. abnormal histone acetylation levels, may result in the persistent expression of somatic genes derived from the donor cell, and failure to activate embryonic genes in SCNT embryos [2–4].
Previous reports demonstrated that TSA treatment increases histone acetylation in cloned embryos, which consequently results in normalized patterns of chromatin remodeling, histone modification and DNA replication, and improves transcriptional activity, gene expression and developmental competence in various species [14,16,33–37]. In the present study, TSA treatment appeared to increase in vitro developmental potential of cloned rabbit embryos reconstructed with fibroblast cells. Rates of morula and blastocyst formation were significantly increased by treatment of 50 nM TSA for 6 h, similar to the previous resultsin rabbits [20]. In that work, Shi et al. reported that comparing to the untreated control group embryos, the histone acetylation patterns in TSA-treated SCNT embryos were more similar to those of normally fertilized embryos [20]. In contrast, Meng et al. reported that TSA treatment did not improve cleavage and blastocyst rates, nor total cell numbers in cloned rabbit embryos [22]. Notably, Meng’s group used a different donor cell type, i.e. cumulus cells, and a different TSA treatment regime, i.e. 5 nM for 10 h, whereas our group and Shi’s group both used fibroblasts as donor cells and treated embryos with TSA at 50 nM for 6 h. Such discrepancies may have contributed to the different outcomes between studies. However, since we did not conduct in vivo experiments in the present study, we cannot completely exclude the possibility that TSA treatment may have negative effects on embryo development and may increase postnatal deaths [22,38]. Previous studies have indicated that HDACs do not have equal sensitivities to different HDACi agents [39], and the effects of HDACi can be cell-type dependent [40]. Therefore, different donor cell types, recipient oocytes, animal strains or species may have diverse responses to different HDACi agents, and may require different HDACi agent(s) to achieve optimal reprogramming efficiency. We hypothesize that the use of HDACi agents other than TSA may also improve cloning efficiency in rabbits. In the present study, we examined the effects of a novel HDACi, namely SCP. Similar to TSA, SCP is one of the hydroxamic acids and able to inhibit many HDACs [15]. Van Thuan et al. reported that SCP treatment enhances de novo synthesis of nascent mRNA in mouse SCNT embryos [18]. Even at extremely high concentrations (~2000 nM) or prolonged treatment periods (~48 h), SCP has very low toxicity to embryo development. In cloned cattle and pig embryos, SCP treatment can improve the histone acetylation patterns to a level comparable to those of fertilized embryos [24,38], and reduce the number of highly methylated nuclei [19]. It has been shown to greatly enhance the in vitro and in vivo developmental potential of reconstructed embryo in mice, pigs and cattle [17,18,24,38,41]. The present study revealed that rabbit clones treated with 250 nM SCP, the same concentration used in mice [18], led to the best blastocyst rates.
Oct-4 is an essential transcription factor required for preimplantation development [42]. In mice, Oct-4 expression begins at the 4- to 8-cell stage and becomes restricted to pluripotent lineage beyond blastocyst stage [43]. Deletion of Oct-4 in mice causes lethality, because the mutated embryos are unable to form ICM cells. We previously reported that rabbit zygotic Oct-4 starts to express at 8– to 16–cell stages, and increases afterwards until the compact morula stage [28]. Expression pattern of Oct-4 is a potential marker for cloned embryo quality. Aberrant Oct-4 expression patterns are often present in cloned mouse and bovine blastocysts [27,44–46]. In many cloned mouse embryos, Oct-4 expression initiated at the correct stage, but displayed an aberrant spatial distribution at the blastocyst stage. Specifically, Oct-4 transcripts form mosaic or ectopic patterns and fail to restrict into the ICM in cloned blastocysts. Furthermore, Oct-4 levels in cloned embryos apparently correlate well with the efficiency of outgrowth formation and embryonic stem cell derivation, suggesting that the failure of Oct-4 expression in mouse clones may result in an incorrect lineage determination and impair embryos development beyond implantation [27]. It was also reported that inefficient demethylation of the Oct-4 promoter in cloned embryos was associated with developmental retardation at early cleavage stages [47], implying that demethylation of the Oct-4 gene is one of the critical events during nuclear reprogramming and that the abnormal Oct-4 expression patterns indicate a general failure of cloned embryos to reset the epigenetic program after SCNT. Therefore, detection of Oct-4 expression in cloned embryos can be used to evaluate reprogramming efficiency, embryo quality and developmental potential of cloned embryos.
This was apparently the first study to investigate Oct-4 expression in cloned rabbit embryos. In the present study, the majority of cloned rabbit embryos had aberrant ICM structure accompanied with abnormal spatial distribution of Oct-4 signals. Without HDACi treatment, only a small percentage of cloned rabbit embryos (4%) were categorized as “Class I”, which had similar ICM structure and Oct-4 patterns with IVC embryos.
Both TSA and SCP increased the percentage of Class I embryos in the present study. Strikingly, SCP significantly improved the Oct-4 expression pattern in rabbit cloned hBLs, in which 56.8% (21/37) of embryos had a distinct ICM structure (Class I and Class II) and 37.8% (14/37) of embryos had a normal Oct-4 expression pattern (Class I) compared to 16% (4/25) and 4% (1/25) in the untreated group. This result was consistent with previous studies in pigs and cattle where SCP treatment corrected gene expression and increased Oct-4 transcripts in SCNT blastocysts [19,24].
The present study was apparently the first to treat cloned embryos with combined HDACi treatments (i.e. TSA and SCP) and demonstrated that co-treatment of TSA (50 nM) and SCP (250 nM) on rabbit cloned embryos for 6 h greatly increased acetylation score of H4K5 at the hBL stage. The blastocyst rate was higher than that in the NT, NTTSA or NTSCP groups. Moreover, similar to previous reports in pigs and rabbits [22,48], TSA or SCP treatment had no effect to increase total cell number of blastocysts. In contrast, the present study revealed that simultaneous treatment with TSA and SCP was competent to improve the blastocyst quality with significantly increased cell number. In addition, the highest percentage (41.9%, 13/31) of cloned embryos that were considered to normally express Oct-4 was observed. Therefore, the present study demonstrated that TSA and SCP treatments (alone or in combination) can not only improve blastocyst rates, but also embryo quality, as evidenced by an increased cell number and a proper Oct-4 distribution pattern at the blastocyst stage. Embryo transfer experiments are necessary in future studies to confirm if such in vitro improvements, especially those achieved by the combinational TSA and SCP treatment, will ultimately lead to better pregnancy rates and term rates of rabbit cloning.
In conclusion, the present study evaluated the combined effects of TSA and SCP on the in vitro development of cloned rabbit embryos. The concentration of the individual HDACi (i.e. TSA and SCP) was optimized for rabbit cloning. We compared developmental potential of cloned embryos exposed to various HDACi treatments (none, TSA, SCP, and TSA & SCP). Synergistic effects of TSA and SCP on nuclear reprogramming of cloned rabbit embryos occurred, manifested by the development and quality of treated embryos.
Acknowledgments
The authors gratefully acknowledge the confocal resources supplied from Dr. Song-Kun Shyue at Institute of Biomedical Sciences, Academia Sinica, Taipei, Taiwan. This study was supported by the National Science Council, Taiwan, R.O.C. (Grant Number 100-2313-B-002-045-MY3 and 100-2313-B-002-051-MY2 to Dr. Li-Ying Sung), and in part by a grant from the National Heart, Lung, and Blood Institute of the National Institutes of Health (Grant Number 5 R44 HL091605-03 to Dr. Fuliang Du).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Egli D, Birkhoff G, Eggan K. Mediators of reprogramming: transcription factors and transitions through mitosis. Nat Rev Molec Cell Biol. 2008;9:505–16. doi: 10.1038/nrm2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng RK, Gurdon JB. Maintenance of epigenetic memory in cloned embryos. Cell Cycle. 2005;4:760–3. doi: 10.4161/cc.4.6.1743. [DOI] [PubMed] [Google Scholar]
- 3.Ng RK, Gurdon JB. Epigenetic memory of active gene transcription is inherited through somatic cell nuclear transfer. PNAS. 2005;102:1957–62. doi: 10.1073/pnas.0409813102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ng RK, Gurdon JB. Epigenetic memory of an active gene state depends on histone H3. 3 incorporation into chromatin in the absence of transcription. Nat Cell Biol. 2008;10:102–9. doi: 10.1038/ncb1674. [DOI] [PubMed] [Google Scholar]
- 5.Cezar GG. Epigenetic reprogramming of cloned animals. Cloning Stem Cells. 2003;5:165–80. doi: 10.1089/153623003769645839. [DOI] [PubMed] [Google Scholar]
- 6.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33 (Suppl):245–54. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 7.Okada Y, Yamagata K, Hong K, Wakayama T, Zhang Y. A role for the elongator complex in zygotic paternal genome demethylation. Nature. 2010;463:554–8. doi: 10.1038/nature08732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science. 2010;329:78–82. doi: 10.1126/science.1187945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–10. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 10.Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, et al. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- 11.Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington's canal. Nat Rev Mol Cell Biol. 2009;10:526–37. doi: 10.1038/nrm2727. [DOI] [PubMed] [Google Scholar]
- 12.Chan MM, Smith ZD, Egli D, Regev A, Meissner A. Mouse ooplasm confers context-specific reprogramming capacity. Nat Genet. 2012;44:978–80. doi: 10.1038/ng.2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogura A, Inoue K, Wakayama T. Recent advancements in cloning by somatic cell nuclear transfer. Philos Trans R Soc Lond B Biol Sci. 2013;368:20110329. doi: 10.1098/rstb.2011.0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maalouf WE, Liu Z, Brochard V, Renard JP, Debey P, Beaujean N, et al. Trichostatin A treatment of cloned mouse embryos improves constitutive heterochromatin remodeling as well as developmental potential to term. BMC Dev Biol. 2009;9:11. doi: 10.1186/1471-213X-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ono T, Li C, Mizutani E, Terashita Y, Yamagata K, Wakayama T. Inhibition of class IIb histone deacetylase significantly improves cloning efficiency in mice. Biol Reprod. 2010;83:929–37. doi: 10.1095/biolreprod.110.085282. [DOI] [PubMed] [Google Scholar]
- 16.Bui HT, Wakayama S, Kishigami S, Park KK, Kim JH, Thuan NV, et al. Effect of trichostatin A on chromatin remodeling, histone modifications, DNA replication, and transcriptional activity in cloned mouse embryos. Biol Reprod. 2010;83:454–63. doi: 10.1095/biolreprod.109.083337. [DOI] [PubMed] [Google Scholar]
- 17.Kishigami S, Mizutani E, Ohta H, Hikichi T, Thuan NV, Wakayama S, et al. Significant improvement of mouse cloning technique by treatment with trichostatin A after somatic nuclear transfer. Biochem Biophys Res Commun. 2006;340:183–9. doi: 10.1016/j.bbrc.2005.11.164. [DOI] [PubMed] [Google Scholar]
- 18.Van Thuan N, Bui HT, Kim JH, Hikichi T, Wakayama S, Kishigami S, et al. The histone deacetylase inhibitor scriptaid enhances nascent mRNA production and rescues full-term development in cloned inbred mice. Reproduction. 2009;138:309–17. doi: 10.1530/REP-08-0299. [DOI] [PubMed] [Google Scholar]
- 19.Whitworth KM, Zhao J, Spate LD, Li R, Prather RS. Scriptaid corrects gene expression of a few aberrantly reprogrammed transcripts in nuclear transfer pig blastocyst stage embryos. Cell Reprogram. 2011;13:191–204. doi: 10.1089/cell.2010.0087. [DOI] [PubMed] [Google Scholar]
- 20.Shi LH, Ai JS, Ouyang YC, Huang JC, Lei ZL, Wang Q, et al. Trichostatin A and nuclear reprogramming of cloned rabbit embryos. J Anim Sci. 2008;86:1106–13. doi: 10.2527/jas.2007-0718. [DOI] [PubMed] [Google Scholar]
- 21.Shi LH, Miao YL, Ouyang YC, Huang JC, Lei ZL, Yang JW, et al. Trichostatin A (TSA) improves the development of rabbit-rabbit intraspecies cloned embryos, but not rabbit-human interspecies cloned embryos. Dev Dyn. 2008;237:640–8. doi: 10.1002/dvdy.21450. [DOI] [PubMed] [Google Scholar]
- 22.Meng Q, Polgar Z, Liu J, Dinnyes A. Live birth of somatic cell-cloned rabbits following trichostatin A treatment and cotransfer of parthenogenetic embryos. Cloning Stem Cells. 2009;11:203–8. doi: 10.1089/clo.2008.0072. [DOI] [PubMed] [Google Scholar]
- 23.Su GH, Sohn TA, Ryu B, Kern SE. A novel histone deacetylase inhibitor identified by high-throughput transcriptional screening of a compound library. Cancer Research. 2000;60:3137–42. [PubMed] [Google Scholar]
- 24.Wang LJ, Zhang H, Wang YS, Xu WB, Xiong XR, Li YY, et al. Scriptaid improves in vitro development and nuclear reprogramming of somatic cell nuclear transfer bovine embryos. Cell Reprogram. 2011;13:431–9. doi: 10.1089/cell.2011.0024. [DOI] [PubMed] [Google Scholar]
- 25.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–91. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 26.Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–62. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 27.Boiani M, Eckardt S, Scholer HR, McLaughlin KJ. Oct4 distribution and level in mouse clones: consequences for pluripotency. Genes Dev. 2002;16:1209–19. doi: 10.1101/gad.966002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen CH, Chang WF, Liu CC, Su HY, Shyue SK, Cheng WT, et al. Spatial and temporal distribution of Oct-4 and acetylated H4K5 in rabbit embryos. Reprod Biomed Online. 2012;24:433–42. doi: 10.1016/j.rbmo.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sung LY, Chen CH, Xu J, Lin TA, Su HY, Chang WF, et al. Follicular oocytes better support development in rabbit cloning than oviductal oocytes. Cell Reprogram. 2011;13:503–12. doi: 10.1089/cell.2011.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Institute of Laboratory Animal Research, Commission on Life Sciences, National Research Council. Guide for the Care and Use of Laboratory Animals. Washington, DC: The National Academics Press; 1996. [Google Scholar]
- 31.Wee G, Koo DB, Song BS, Kim JS, Kang MJ, Moon SJ, et al. Inheritable histone H4 acetylation of somatic chromatins in cloned embryos. J Biol Chem. 2006;281:6048–57. doi: 10.1074/jbc.M511340200. [DOI] [PubMed] [Google Scholar]
- 32.Wang F, Kou Z, Zhang Y, Gao S. Dynamic reprogramming of histone acetylation and methylation in the first cell cycle of cloned mouse embryos. Biol Reprod. 2007;77:1007–16. doi: 10.1095/biolreprod.107.063149. [DOI] [PubMed] [Google Scholar]
- 33.Fan Y, Jiang Y, Chen X, Ou Z, Yin Y, Huang S, et al. Derivation of cloned human blastocysts by histone deacetylase inhibitor treatment after somatic cell nuclear transfer with beta-thalassemia fibroblasts. Stem Cells and Dev. 2011;20:1951–9. doi: 10.1089/scd.2010.0451. [DOI] [PubMed] [Google Scholar]
- 34.Cui XS, Xu YN, Shen XH, Zhang LQ, Zhang JB, Kim NH. Trichostatin A modulates apoptotic-related gene expression and improves embryo viability in cloned bovine embryos. Cell Reprogram. 2011;13:179–89. doi: 10.1089/cell.2010.0060. [DOI] [PubMed] [Google Scholar]
- 35.Li X, Kato Y, Tsuji Y, Tsunoda Y. The effects of trichostatin A on mRNA expression of chromatin structure-, DNA methylation, and development-related genes in cloned mouse blastocysts. Cloning Stem Cells. 2008;10:133–42. doi: 10.1089/clo.2007.0066. [DOI] [PubMed] [Google Scholar]
- 36.Li J, Svarcova O, Villemoes K, Kragh PM, Schmidt M, Bogh IB, et al. High in vitro development after somatic cell nuclear transfer and trichostatin A treatment of reconstructed porcine embryos. Theriogenology. 2008;70:800–8. doi: 10.1016/j.theriogenology.2008.05.046. [DOI] [PubMed] [Google Scholar]
- 37.Kohda T, Kishigami S, Kaneko-Ishino T, Wakayama T, Ishino F. Gene expression profile normalization in cloned mice by trichostatin A treatment. Cell Reprogram. 2012;14:45–55. doi: 10.1089/cell.2011.0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao J, Hao Y, Ross JW, Spate LD, Walters EM, Samuel MS, et al. Histone deacetylase inhibitors improve in vitro and in vivo developmental competence of somatic cell nuclear transfer porcine embryos. Cell Reprogram. 2010;12:75–83. doi: 10.1089/cell.2009.0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drummond DC, Noble CO, Kirpotin DB, Guo Z, Scott GK, Benz CC. Clinical development of histone deacetylase inhibitors as anticancer agents. Annu Rev Pharmacol Toxicol. 2005;45:495–528. doi: 10.1146/annurev.pharmtox.45.120403.095825. [DOI] [PubMed] [Google Scholar]
- 40.Taddei A, Maison C, Roche D, Almouzni G. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat Cell Biol. 2001;3:114–20. doi: 10.1038/35055010. [DOI] [PubMed] [Google Scholar]
- 41.Panda SK, George A, Saha A, Sharma R, Singh AK, Manik RS, et al. Effect of scriptaid, a histone deacetylase inhibitor, on the developmental competence of Handmade cloned buffalo (Bubalus bubalis) embryos. Theriogenology. 2012;77:195–200. doi: 10.1016/j.theriogenology.2011.07.033. [DOI] [PubMed] [Google Scholar]
- 42.Ovitt CE, Scholer HR. The molecular biology of Oct-4 in the early mouse embryo. Mol Hum Reprod. 1998;4:1021–31. doi: 10.1093/molehr/4.11.1021. [DOI] [PubMed] [Google Scholar]
- 43.Palmieri SL, Peter W, Hess H, Scholer HR. Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev Biol. 1994;166:259–67. doi: 10.1006/dbio.1994.1312. [DOI] [PubMed] [Google Scholar]
- 44.Daniels R, Hall V, Trounson AO. Analysis of gene transcription in bovine nuclear transfer embryos reconstructed with granulosa cell nuclei. Biol Reprod. 2000;63:1034–40. doi: 10.1095/biolreprod63.4.1034. [DOI] [PubMed] [Google Scholar]
- 45.Daniels R, Hall VJ, French AJ, Korfiatis NA, Trounson AO. Comparison of gene transcription in cloned bovine embryos produced by different nuclear transfer techniques. Mol Reprod Dev. 2001;60:281–8. doi: 10.1002/mrd.1089. [DOI] [PubMed] [Google Scholar]
- 46.Kishigami S, Hikichi T, Van Thuan N, Ohta H, Wakayama S, Bui HT, et al. Normal specification of the extraembryonic lineage after somatic nuclear transfer. FEBS Lett. 2006;580:1801–6. doi: 10.1016/j.febslet.2006.02.036. [DOI] [PubMed] [Google Scholar]
- 47.Yamazaki Y, Fujita TC, Low EW, Alarcon VB, Yanagimachi R, Marikawa Y. Gradual DNA demethylation of the Oct4 promoter in cloned mouse embryos. Mol Reprod Dev. 2006;73:180–8. doi: 10.1002/mrd.20411. [DOI] [PubMed] [Google Scholar]
- 48.Zhao J, Whyte J, Prather RS. Effect of epigenetic regulation during swine embryogenesis and on cloning by nuclear transfer. Cell Tissue Res. 2010;341:13–21. doi: 10.1007/s00441-010-1000-x. [DOI] [PubMed] [Google Scholar]